Abstract
Listeria monocytogenes contains (i) epidemic clone (EC) strains, which have been linked to the majority of listeriosis outbreaks worldwide and are overrepresented among sporadic cases in the United States, and (ii) strains commonly isolated from ready-to-eat foods that carry a mutation leading to a premature stop codon (PMSC) in inlA, which encodes the key virulence factor internalin A (InlA). Internalin A binds certain isoforms of the cellular receptor E-cadherin to facilitate crossing the intestinal barrier during the initial stages of an L. monocytogenes infection. Juvenile guinea pigs, which express the human isoform of E-cadherin that binds InlA, were intragastrically challenged with a range of doses of (i) an EC strain associated with a listeriosis outbreak or (ii) a strain carrying a PMSC mutation in inlA. Recovery of L. monocytogenes from tissues (i.e., liver, spleen, mesenteric lymph nodes, and ileum) was used to develop strain-specific dose-response curves on the basis of individual and combined organ data. Modeling of individual and combined organ data revealed an approximate 1.2 to 1.3 log10 increase in the median infectious dose for the strain carrying a PMSC in inlA relative to that for the EC strain. Inclusion of the strain parameter significantly improved the goodness of fit for individual and combined organ models, indicating a significant shift in median infectious dose for guinea pigs challenged with an inlA PMSC strain compared to that for guinea pigs challenged with an EC strain. Results from this work provide evidence that the L. monocytogenes dose-response relationship is strain specific and will provide critical data for enhancement of current risk assessments and development of future risk assessments.
Listeria monocytogenes is a facultative intracellular pathogen that is the etiological agent of the human food-borne disease listeriosis. Invasive listeriosis may lead to life-threatening clinical manifestations of disease, such as septicemia, meningitis, encephalitis, and spontaneous abortions or stillbirths in pregnant women (41). The elderly, young children, pregnant women and their fetuses, and individuals with definite immune-compromising circumstances (e.g., cancer, organ transplant, and HIV-infected or AIDS patients) are the most susceptible to a systemic L. monocytogenes infection leading to invasive listeriosis (45). While listeriosis cases are relatively rare compared to the incidence of other food-borne illnesses, such as salmonellosis, listeriosis cases usually lead to hospitalization (85% to 90%) and often result in death (20 to 30%). In addition, listeriosis was projected to be responsible for nearly 30% of all deaths attributed to known pathogens in the United States per annum (23), supporting the suggestion that listeriosis clearly represents a significant public health concern in the United States.
Molecular subtyping studies (i.e., DNA band- and sequence-based typing studies) consistently showed that L. monocytogenes isolates cluster into four divergent genetic lineages, termed lineages I, II, III, and IV (2, 26, 32, 34, 48, 49). Previous molecular epidemiology studies suggest that L. monocytogenes genetic lineages and clonal groups within those lineages differ in their associations with human disease and isolation from foods (12, 15, 25, 26, 27, 39, 40, 47, 48, 49). For example, three highly clonal L. monocytogenes strains within lineage I (termed epidemic clones I, Ia, and II) that belong to serotype 4b have been linked to the majority of listeriosis epidemics worldwide (9, 17) and have frequently been isolated from sporadic listeriosis cases in the United States (12). In contrast, multiple strains representing lineage II are significantly overrepresented among isolates from ready-to-eat (RTE) foods but have rarely or never been linked to human disease (12). While no genetic markers have been mechanistically related to enhanced virulence of epidemic clone strains, at least 18 naturally occurring single nucleotide polymorphisms (SNPs) leading to a premature stop codon (PMSC) in the key L. monocytogenes virulence gene inlA have been identified worldwide to date (7, 13, 14, 16, 27, 30, 33, 35, 38, 43, 44, 46).
The virulence factor internalin A (InlA), encoded by inlA, facilitates uptake of L. monocytogenes by host cells expressing certain isoforms of E-cadherin, making the interaction between InlA and E-cadherin a critical first step for crossing the intestinal barrier during the initial stages of an L. monocytogenes infection (21). L. monocytogenes isolates carrying a PMSC mutation in inlA demonstrate reduced invasion of Caco-2 human intestinal epithelial cells in vivo (7, 27, 30, 33, 44, 46). A previous study by our group characterized a set of paired isogenic mutants with and without an inlA PMSC using an intragastric guinea pig infection model to demonstrate that inlA PMSC mutations appear to be causally associated with attenuated mammalian virulence (24). Another recent study by Roldgaard and coworkers (37) also described notably reduced L. monocytogenes counts in select internal organs from guinea pigs challenged with the laboratory strain LO28, which carries a PMSC mutation in inlA, compared to the counts in guinea pigs challenged with two other strains encoding a full-length InlA. Combined, DNA sequence, virulence phenotype, and epidemiology studies showed that a significant proportion (between 30 and 45%) of L. monocytogenes isolates from RTE foods and RTE food production environments carry a virulence-attenuating PMSC mutation in the key virulence gene inlA (11, 14, 24, 27, 43, 44, 46), suggesting that humans are commonly exposed to virulence-attenuated L. monocytogenes strains through consumption of contaminated RTE foods.
Although the infectious dose for L. monocytogenes in humans is not precisely known, it has been suggested to be high (e.g., >6 log10 CFU), on the basis of the minimum infectious dose administered to elicit a response in animal infection experiments (45). However, a leading hypothesis is that a range of doses as opposed to an infectious dose threshold can cause an infection with various probabilities. More specifically, as the dose of L. monocytogenes consumed by an individual increases, the likelihood that an infection will occur also increases (3, 8, 42). Existing L. monocytogenes risk assessments assume some heterogeneity in virulence (8, 42, 50); however, they do not mechanistically relate strain-specific genetic characteristics to virulence and, as a result, may overestimate the virulence of strains carrying a PMSC in inlA and underestimate the virulence of epidemic clone strains. A critical need thus exists to incorporate strain-specific infectious dose data that mechanistically relate virulence to defined strain-specific genetic characteristics in order to revise current and develop future L. monocytogenes risk assessments. Unlike mice and rats, guinea pigs carry the same isoform of E-cadherin as humans, which binds InlA (20), making the guinea pig a suitable animal model to probe the strain-specific virulence characteristics for L. monocytogenes carrying different inlA allelic types. Although a recent study by Williams and coworkers (51) modeled the dose-response relationship of L. monocytogenes after oral exposure in pregnant guinea pigs, the study relied on a single L. monocytogenes strain and used fetal stillbirths to define the dose-response curve. As a result, the objective of this study was to intragastrically infect juvenile male guinea pigs and determine the infection status of four tissues (i.e., liver, spleen, ileum, and mesenteric lymph nodes) to model strain-specific dose-response curves for a fully invasive serotype 4b outbreak-associated L. monocytogenes strain belonging to epidemic clone II and a serotype 1/2a L. monocytogenes strain carrying the most broadly distributed PMSC in inlA.
MATERIALS AND METHODS
Bacterial isolates for guinea pig infection experiments.
Male juvenile guinea pigs were previously challenged with the standard laboratory control strain 10403S (1) at a dose of 10 log10 CFU in the studies by Nightingale et al. (24) and Oliver et al. (31) or a dose of 10.2 log10 CFU in the study by Garner et al. (10). Data from these previous studies are summarized in Table 1 to describe the infectivity of challenge by a laboratory control strain at a dose of 10 log10 CFU or higher. In the current study, two L. monocytogenes strains were selected for intragastric guinea pig infections to represent the range of strain-specific virulence within this pathogen on the basis of available molecular subtyping, virulence phenotype, and epidemiological data. Specifically, we selected a serotype 1/2a L. monocytogenes strain (CSUFSL N1-040) isolated from an RTE food sample carrying a virulence-attenuating inlA PMSC mutation. Strain CSUFSL N1-040 belongs to ribotype DUP-1062A, which carries the most common inlA PMSC mutation found among L. monocytogenes isolates from RTE foods samples in the United States (12, 27). We also selected a serotype 4b L. monocytogenes strain (CSUFSL N1-054) that was isolated from a patient associated with the 1998 to 1999 listeriosis outbreak and that belonged to ribotype DUP-1044A (epidemic clone II) (9, 17). Strain CSUFSL N1-054 was characterized by in vitro invasion assays using the Caco-2 human intestinal epithelial and HepG2 human hepatic cell lines and was found to represent a consistent highly invasive strain across both cell lines under different bacterial growth conditions (36).
TABLE 1.
Percent organs infected for both the fully virulent outbreak-associated and natural virulence-attenuated strains at each dose at which animals were infected
| Straina | Dose | No. of animals positive/no. tested (%) |
% total organs infected | % animals infected | Reference or source | |||
|---|---|---|---|---|---|---|---|---|
| Liver | Spleen | Mesenteric lymph node | Ileum | |||||
| 10403S | 1 × 1010 | 9/9 (100) | 9/9 (100) | 9/9 (100) | 9/9 (100) | 100 | 100 | 10, 24, 31 |
| CSU N1-054 | 1 × 108 | 5/5 (100) | 5/5 (100) | 5/5 (100) | 3/5 (60) | 90 | 100 | This study |
| 1 × 107 | 4/9 (44.4) | 1/9 (11.1) | 3/9 (33.3) | 3/9 (33.3) | 30 | 44.4 | This study | |
| 1 × 106 | 6/11 (54.5) | 2/11 (18.2) | 3/11 (27.3) | 0/11 (0) | 25 | 63.6 | This study | |
| 1 × 105 | 0/15 (0) | 0/15 (0) | 0/15 (0) | 0/15 (0) | 0 | 0 | This study | |
| CSU N1-040 | 1 × 1010 | 3/3 (100) | 3/3 (100) | 3/3 (100) | 3/3 (100) | 100 | 100 | 24 |
| 1 × 108 | 4/10 (40) | 2/10 (20) | 2/10 (20) | 1/10 (10) | 22.5 | 60 | This study | |
| 1 × 107 | 4/10 (40) | 0/10 (0) | 3/10 (30) | 0/10 (0) | 17.5 | 50 | This study | |
| 1 × 106 | 0/5 (0) | 0/5 (0) | 0/5 (0) | 0/5 (0) | 0 | 0 | This study | |
10403S is a standard laboratory control strain (1); CSUFSL N1-054 is an outbreak-associated epidemic clone lineage II strain associated with the 1998-1999 listeriosis outbreak linked to SaraLee (9, 17). CSUFSL N1-040 is a strain that carries the most broadly distributed mutation leading to a premature stop codon in inlA (12, 27). 10403S is a standard laboratory control strain that was used in a previous study (10, 24, 31). Male juvenile guinea pigs were challenged with 10403S at a dose of 10 log10 CFU in the studies by Nightingale et al. (24) and Oliver et al. (31) or a dose of 10.2 log10 CFU in the study of Garner et al. (10).
Bacterial growth conditions.
Bacterial cultures used for guinea pig infection experiments were grown to stationary phase (optical density at 600 nm = 1 plus 3 h) at 37°C with aeration (shaking at 250 rpm). Aliquots (100 ml) of stationary-phase bacterial cultures were concentrated by centrifugation (7,500 × g for 10 min) and resuspended in 20 ml of phosphate-buffered saline (PBS) containing 20% glycerol. Aliquots of the resuspended cultures (1 ml) were then frozen at −80°C until use in animal infection experiments as previously described (10). Bacterial viability and numbers were determined by plating appropriate serial dilutions of three individual tubes from each culture preparation on brain heart infusion agar in duplicate, prior to guinea pig challenge experiments.
Intragastric guinea pig challenge experiments.
An intragastric guinea pig infection model was used to generate empirical data to model the infectious dose response of a serotype 1/2a L. monocytogenes strain carrying an inlA PMSC (CSUFSL N1-040) and a serotype 4b outbreak-associated L. monocytogenes strain (CSUFSL N1-054). Animal infection experiments were conducted according to a protocol approved by Colorado State University's Animal Care and Use Committee (protocol 06-266A-02) and were performed as detailed previously (10, 24, 31). Briefly, male juvenile (weight, 300 g) pathogen-free Hartley guinea pigs (Charles River Laboratories, North Wilmington, MA) were housed in individual cages and acclimated for at least 5 days at Colorado State University's Laboratory Animal Resource Center (LAR; Fort Collins, CO). Animals were fasted for 12 h prior to being anesthetized with isoflurane and challenged with either the inlA PMSC or epidemic clone L. monocytogenes strain at the intended dose. More specifically, anesthetized guinea pigs were gavaged with a rubber catheter (Viagon, Norristown, PA), which was used to deliver 1.5 ml of a calcium carbonate solution (83 g/liter) to buffer stomach pH, and 1 ml of inoculum (containing a target log10 CFU dose of the intended L. monocytogenes strain carried in PBS), followed by 1 ml of PBS. Guinea pigs were challenged with a range of doses, from 5 log10 to 10 log10 CFU, where the number of animals used for each challenge dose was determined to reduce uncertainty, with particular emphasis placed on low doses. At least five animals were analyzed for each challenge dose, and additional animals were infected as necessary to minimize uncertainty. Animals were weighed daily after infection and were euthanized by CO2 asphyxiation at 72 h postchallenge.
Microbiological analysis of guinea pig organs.
Guinea pig infection status was evaluated by microbiologically analyzing select organs (i.e., liver, spleen, mesenteric lymph nodes, and ileum) to detect the presence of L. monocytogenes as previously described (10, 24, 31). Ileum tissues were washed with PBS three times and treated with Dulbecco's modified Eagle medium (Gibco, Invitrogen, Carlsbad, CA) supplemented with gentamicin (150 μg/ml) for 90 min, prior to homogenization and L. monocytogenes enumeration, in order to eliminate extracellular microflora. All organs were homogenized in PBS using sterile blending units (Semimicro; Eberback, Ann Arbor, MI). Organ homogenates (10 ml) were selectively enriched in 90 ml of Listeria enrichment broth (LEB; Difco, Sparks, MD) at 30°C for 48 h, aliquots of selective enrichments (50 μl) were streaked onto Oxford plates (Difco; Oxoid, Hampshire, United Kingdom), and plates were incubated at 30°C for 48 h. Tissues were considered to be presumptively positive for L. monocytogenes if colonies with typical Listeria morphology were observed on Oxford plates. Up to five presumptive positive colonies on an Oxford plate representing each presumptive positive organ were confirmed by screening for the presence of an hly fragment that is unique to L. monocytogenes (29). A tissue sample was considered positive for L. monocytogenes if both typical Listeria morphology on Oxford plates and the hly fragment that is unique to L. monocytogenes were detected.
Infectious dose-response curve modeling.
Dose-response curves were modeled on the basis of the presence or absence L. monocytogenes in four tissues (i.e., liver, spleen, mesenteric lymph nodes, and ileum) for animals challenged with a range of doses for each strain. Dose-response curves were constructed with either a log-logistic (LL), beta Poisson (BP), or exponential (EX) model to fit the raw individual and combined organ data using the maximum-likelihood method implemented in the Splus (version 6.2) program. The median infectious dose (the dose required to infect 50% of tissue samples [ID50]) was calculated for each respective tissue using the LL and BP models; however, the ID50 was determined for the spleen using the LL and EX models, because the BP model failed to converge for the spleen data. For the LL model, the infectivity rate, P(d), was given by the formula
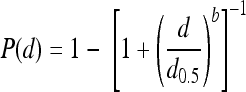 |
where d is dose and d0.5 is the ID50 for the L. monocytogenes strain. The BP model has the form 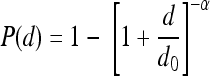 where α and d0 are BP model parameters. Using the BP model, the ID50 is calculated with the equation
where α and d0 are BP model parameters. Using the BP model, the ID50 is calculated with the equation 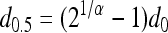 . A limiting model of the BP model is the EX model, which has the form
. A limiting model of the BP model is the EX model, which has the form 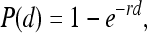 where r is the EX model parameter, and where the ID50 is calculated as follows:
where r is the EX model parameter, and where the ID50 is calculated as follows: 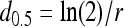 . The BP model for individual organs is given by the following equation:
. The BP model for individual organs is given by the following equation: 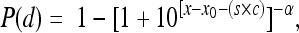 where x is log(d), x0 is log(d0), and s is a term used to combine the mathematical expressions of the BP model for two different types of strains; s is equal to 1 for the outbreak-associated strain (CSUFSL N1-054) and is equal to 0 for the virulence-attenuated strain carrying a PMSC in inlA. The parameter c estimates the shift in the model between strains. A negative value for the parameter c indicates that a decreased inoculum level is required to establish an organ infection for the outbreak-associated strain (CSUFSL N1-054) relative to the inoculum of the virulence-attenuated strain (CSUFSL N1-040) at corresponding dose levels. Using the same notation, the EX model was parameterized as follows when the spleen data were fitted:
where x is log(d), x0 is log(d0), and s is a term used to combine the mathematical expressions of the BP model for two different types of strains; s is equal to 1 for the outbreak-associated strain (CSUFSL N1-054) and is equal to 0 for the virulence-attenuated strain carrying a PMSC in inlA. The parameter c estimates the shift in the model between strains. A negative value for the parameter c indicates that a decreased inoculum level is required to establish an organ infection for the outbreak-associated strain (CSUFSL N1-054) relative to the inoculum of the virulence-attenuated strain (CSUFSL N1-040) at corresponding dose levels. Using the same notation, the EX model was parameterized as follows when the spleen data were fitted: 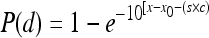 . The BP model for combined models is as follows:
. The BP model for combined models is as follows: 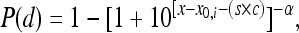 where the parameter x0,i, which is equal to log(d0,i) (where i is the organ type), differs across organs. Thus, for the combined model there is a common shift between strains for each organ and a common shape parameter across organs. The individual LL model uses the notation
where the parameter x0,i, which is equal to log(d0,i) (where i is the organ type), differs across organs. Thus, for the combined model there is a common shift between strains for each organ and a common shape parameter across organs. The individual LL model uses the notation 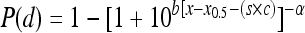 and the combined LL uses the notation
and the combined LL uses the notation 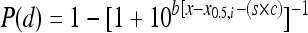 . Solutions to the maximized nonlinear likelihood equations provided parameter estimates for the three models on the basis of the raw individual and combined organ data (Tables 2 and 3). The level of agreement between the model-predicted values and the observed values was quantified by applying the deviance function for quantal dose-response models. P values were calculated for each model (including and excluding the strain) to indicate which model fit the raw individual and combined organ data most appropriately.
. Solutions to the maximized nonlinear likelihood equations provided parameter estimates for the three models on the basis of the raw individual and combined organ data (Tables 2 and 3). The level of agreement between the model-predicted values and the observed values was quantified by applying the deviance function for quantal dose-response models. P values were calculated for each model (including and excluding the strain) to indicate which model fit the raw individual and combined organ data most appropriately.
TABLE 2.
Parameter values and model goodness of fit for models based on individual organ data
| Organ | Model | Parameter valuesa |
Model goodness of fit |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Including strain |
Excluding strain |
||||||||
| ID50 | c | a | b | Deviance (dofb) | P value | Deviance (dof) | P value | ||
| Liver | Beta Poisson | 7.68 | −1.22 | 0.33 | 1 | 11.2 (5) | 0.05 | 17.4 (6) | 0.01 |
| Log-logistic | 7.83 | −1.25 | 1 | 0.64 | 11.8 (5) | 0.04 | 18.6 (6) | 0.004 | |
| Spleen | Exponential | 8.54 | −1.26 | 6.2 (6) | 0.40 | 22.0 (7) | 0.003 | ||
| Log-logistic | 8.59 | −1.33 | 1 | 1.04 | 7.6 (5) | 0.18 | 18.4 (6) | 0.005 | |
| Mesenteric lymph | Beta Poisson | 8.10 | −1.19 | 0.40 | 1 | 11 (5) | 0.05 | 16.2 (6) | 0.01 |
| node | Log-logistic | 8.25 | −1.24 | 1 | 0.69 | 9.6 (5) | 0.09 | 16.2 (6) | 0.01 |
| Ileum | Beta Poisson | 8.91 | −1.36 | 0.96 | 1 | 2.4 (5) | 0.79 | 12.4 (6) | 0.05 |
| Log-logistic | 8.87 | −1.34 | 1 | 1.06 | 2.6 (5) | 0.76 | 11.8 (6) | 0.07 | |
ID50, median infectious dose for the strain carrying an inlA PMSC mutation (CSUFSL N1-040); c, shift in ID50 for the outbreak-associated epidemic clone strain (CSUFSL N1-054) relative to CSUFSL N1-040; a, intercept of the dose-response curve; b, slope of the dose-response curve.
dof, degrees of freedom.
TABLE 3.
Parameter values and model goodness of fit for models based on combined organ data
| Model | Parameter values |
Model goodness of fit including strain |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ID50a |
c | a | b | Deviance (dofb) | P value | ||||
| Liver | Spleen | Mesenteric lymph node | Ileum | ||||||
| Beta Poisson | 7.716 | 8.692 | 8.169 | 8.973 | −1.32 | 0.451 | 1 | 36 (26) | 0.09 |
| Log-logistic | 7.828 | 8.736 | 8.27 | 9.031 | −1.33 | 1 | 0.772 | 34.8 (26) | 0.12 |
ID50, median infectious dose for the strain carrying an inlA PMSC mutation (CSUFSL N1-040); c, the shift in ID50 for the outbreak-associated epidemic clone strain (CSUFSL N1-054) relative to CSUFSL N1-040; a, the intercept of the dose-response curve; b, slope of the dose-response curve.
dof, degrees of freedom.
RESULTS AND DISCUSSION
Previous molecular epidemiology studies suggested that L. monocytogenes contains at least two distinct subpopulations that differ in virulence, including (i) epidemic clone strains, which have been associated with the majority of listeriosis outbreaks worldwide and which are overrepresented among sporadic cases in the United States, and (ii) strains carrying virulence-attenuating PMSC mutations in inlA, which are common in RTE foods but which are associated with human disease only on very rare occasions (12, 14, 25, 26, 27, 35, 43, 44, 46, 47, 48). Current L. monocytogenes risk assessments (8, 42, 50), which assume that an L. monocytogenes population with nonuniform variation in virulence is present in RTE foods, therefore likely underestimate the virulence of epidemic clone strains and overestimate the virulence of strains carrying a PMSC in inlA. A critical need thus exists to incorporate dose-response data for strain-specific genetic characteristics as they mechanistically relate to virulence (e.g., encoding a full-length or truncated InlA) to revise current and develop future L. monocytogenes risk assessments. In the current study, guinea pigs were challenged with a range of log10 doses of either an outbreak-associated epidemic clone L. monocytogenes strain or a strain carrying a mutation leading to a PMSC in inlA. Recovery of L. monocytogenes from four internal organs (i.e., liver, spleen, mesenteric lymph nodes, and ileum) was used to develop strain-specific dose-response curves. Application of different models to develop strain-specific dose-response curves for individual and combined organ data sets showed notable differences in the median infectious dose for an outbreak-associated epidemic clone L. monocytogenes strain and that for a strain carrying a PMSC mutation in inlA.
L. monocytogenes strains show notable differences in median infectious doses required to cause a systemic infection.
Male juvenile guinea pigs were intragastrically infected with either an epidemic clone strain or a strain carrying a PMSC in inlA at doses ranging from 5 log10 CFU to 10 log10 CFU. Recovery of L. monocytogenes from four tissues (liver, spleen, mesenteric lymph nodes, and ileum) was used to determine the infection status of each animal (Table 1). Consistent with existing L. monocytogenes risk assessments based on nonpregnant animal challenge studies, recovery of L. monocytogenes in the spleen and/or liver was used to indicate a systemic L. monocytogenes infection in the current study (8, 42). Overall, we observed the greatest difference in the infection status of spleens from animals challenged with the same dose of either the epidemic clone strain or the strain carrying a PMSC in inlA. For example, L. monocytogenes was recovered from 100% of spleens collected from animals challenged with a dose of 8 log10 CFU of the epidemic clone strain, while only 20% of spleens from animals challenged with 8 log10 CFU of a strain carrying a PMSC in inlA tested positive for L. monocytogenes. Additionally, L. monocytogenes was not detected in spleen tissues from animals challenged with a dose of ≤7 log10 CFU of the inlA PMSC strain, while lowering the challenge dose of the epidemic clone strain to 5 log10 CFU was required to observe 0% infected spleen tissues (Table 1). A recent study using the pregnant guinea pig model showed a similar trend in the spleen infection status of dams challenged with a serotype 1/2a strain encoding a full-length InlA (determined in the current study) compared to the spleen infection status of male juvenile guinea pigs challenged in the current study with the epidemic clone strain, which also encodes a full-length InlA. Specifically, L. monocytogenes was recovered from 75% of spleen tissues from pregnant guinea pigs challenged with 8 log10 CFU of the 1/2a strain, while 0% of spleen tissues from dams challenged at a dose of 5 log10 CFU contained L. monocytogenes at 21 days postchallenge (51).
In the current study, other tissues (i.e., liver, mesenteric lymph node, and ileum) showed similar trends with respect to strain-specific differences in tissue infection status at corresponding challenge doses; however, the minimum dose resulting in infection of these other tissues differed by approximately 1 log10 CFU between the epidemic clone strain and the strain carrying a PMSC in inlA (Table 1). For the administered dose of 7 log10 CFU, L. monocytogenes was recovered at similar proportions for liver (approximately 40%) and mesenteric lymph node (approximately 30%) tissues from animals challenged with either the outbreak strain or the strain carrying a PMSC in inlA (Table 1). Animals appeared to begin to clear L. monocytogenes from the ileum by the time that they were euthanized at 72 h postchallenge, as the fewest number of samples representing this tissue were infected following challenge by either L. monocytogenes strain compared to the infection status of samples of the other tissues at 72 h postchallenge. More specifically, animals challenged with 6 log10 CFU of the epidemic clone strain (CSUFSL N1-054) had infected livers, spleens, and mesenteric lymph nodes, but they did not have infected ileum tissues. A similar trend was observed for animals challenged with the strain carrying an inlA PMSC mutation (CSUFSL N1-040); when animals were challenged with this strain at 7 log10 CFU, the livers and mesenteric lymph nodes were infected but the spleens and ileums were not. These observations support the important role of InlA in crossing the intestinal barrier during the initial stages of an infection in order to later infect deeper tissues (e.g., liver and spleen) to establish a systemic infection (19).
In order to compare strain-specific tissue infectivity at high challenge doses, data from our previous studies were also considered. The standard laboratory control strain 10403S, which encodes a full-length InlA, and the strain carrying a PMSC in inlA (CSUFSL N1-040) were previously administered to male juvenile guinea pigs at a dose of 10 to 10.2 log10 CFU (10, 24, 31) using the same protocol employed in the current study. Both 10403S and the strain carrying a PMSC in inlA (CSUFSL N1-040) were recovered from 100% of organs (on the basis of the findings for all four organs) from all guinea pigs challenged with a dose of ≥10 log10 CFU. Another recently completed study showed that challenge of male juvenile guinea pigs with four wild-type strains representing each L. monocytogenes genetic lineage [I, II (10403S), IIIA, and IIIB] at a dose of 10 log10 CFU (using the same protocol described here) led to infection of all four tissues in all animals (31). Collectively, these studies show that intragastrically administering a dose of ≥10 log10 CFU of L. monocytogenes, regardless of inlA allelic type (e.g., encoding a full-length or truncated InlA), to male juvenile guinea pigs leads to the establishment of a systemic L. monocytogenes infection at 72 h postchallenge.
The percentage of total organs infected within each animal challenged by either L. monocytogenes strain declined as the dose administered was lowered (Table 1); however, the decline was notably more rapid in the group of animals challenged with the strain carrying a PMSC mutation in inlA, whereas the decline in the percentage of tissues infected for animals challenged with the epidemic clone strain was more gradual (Table 1). More specifically, the epidemic clone strain (CSUFSL N1-054) was recovered from 90% of organs from animals infected with 8 log10 CFU, 30% of organs from animals infected with 7 log10 CFU, 25% of organs from animals infected with 6 log10 CFU, and 0% of organs from animals infected at a dose of 5 log10 CFU. In contrast, the strain carrying a PMSC in inlA was recovered from 100% of organs from guinea pigs infected at ≥10 log10 CFU (9, 23, 30), 22.5% of organs from animals infected at 8 log10 CFU, 17.5% of organs from animals infected at 7 log10 CFU, and 0% of organs from animals infected at a dose of 6 log10 CFU (Table 1). Evaluating the animal as a whole, L. monocytogenes was recovered from at least one tissue of 100% of animals challenged with 8 log10 CFU of the epidemic clone strain (CSUFSL N1-054), while only 60% of animals challenged with the same dose of the inlA PMSC strain (CSUFSL N1-040) were characterized by at least one infected tissue. Additionally, we observed at least one infected tissue for 63.6% of animals challenged with CSUFSL N1-054 at a dose of 6 log10 CFU, while L. monocytogenes was not isolated from any tissues collected from animals challenged with the same dose of CSUFSL N1-040 (Table 1). These results support the suggestion that L. monocytogenes strains demonstrate notable differences in the challenge dose required to infect different tissues within an animal and to establish a systemic infection.
Dose-response modeling showed a shift in the median infectious dose between strains.
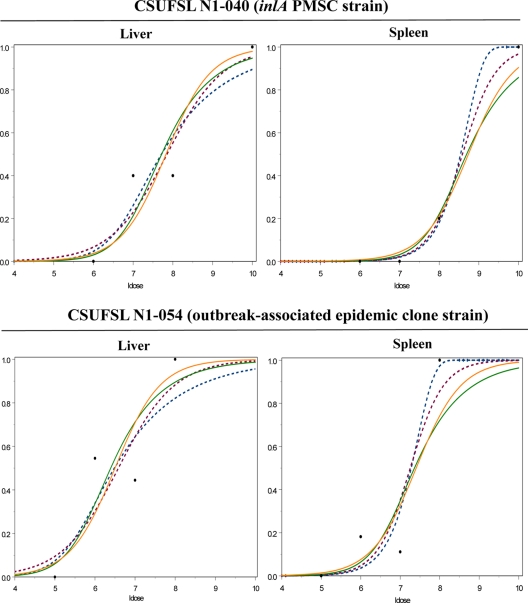
Dose-response curves fit from data generated in this study (Fig. 1) allowed us to solve for the model fit by the data using the maximum-likelihood approach to provide model parameter estimates. Model parameter estimates for models fit to individual organ (Table 2) and combined organ (Table 3) data sets include the ID50, the slope (b) and intercept (a) of each dose-response curve, and c, which indicates the shift in ID50 in relation to that for the strain carrying a PMSC in inlA (CSUFSL-N1-040). The ID50s for guinea pigs infected with the strain carrying an inlA PMSC mutation (CSUFSL N1-040) ranged from 7.68 log10 CFU (BP model, liver data) to 8.91 log10 CFU (BP model, ileum data) on the basis of individual organ models. In contrast, the median ID50s for animals challenged with the epidemic clone strain (CSUFSL N1-054) derived from the individual organ models ranged from 6.46 log10 CFU (BP model, liver data) to 7.55 log10 CFU (BP model, ileum data) (Table 2). These individual organ models estimated that the value of c ranged from 1.22 log10 CFU to 1.36 log10 CFU (Table 2). For example, modeling of the spleen data using the EX model estimated an ID50 of 8.54 log10 CFU for guinea pigs challenged with an L. monocytogenes strain carrying a PMSC mutation in inlA, while an ID50 of 7.28 log10 CFU was estimated for the epidemic clone strain (Table 2; Fig. 1). Consistent with the models based on the individual organ data, combined organ model data also estimated an approximate 1.3-log10-CFU shift between the two strains (Table 3; Fig. 1).
FIG. 1.
Dose-response of L. monocytogenes for infectivity of select tissues (i.e., liver and spleen). Dose-response curves on the top represent those for organs from animals infected with a virulence-attenuated isolate carrying a PMSC in inlA (CSUFSL N1-040), and dose-response curves on the bottom represent those for organs from animals infected with an outbreak-associated epidemic clone strain (CSUFSL N1-054). For all dose-response curves, the proportion of infected tissues is indicated on the y axis and the log10 CFU dose (ldose) is indicated on the x axis. The dose-response curves for tissue infectivity were constructed with either a log-logistic, beta Poisson, or exponential (for spleen data) model fit to the raw individual and combined organ data. Raw organ data are represented by the large black solid dots. Dose-response curves fit to individual organ data are represented by dashed lines. More specifically, the log-logistic model is represented by the purple dashed lines, and the beta Poisson model is represented by the blue dashed lines (in the case of the spleen, however, the blue dashed lines represent the exponential model). Dose-response curves fit to combined organ data are represented by solid lines. More specifically, the log-logistic model is represented by the solid orange line, and the beta Poisson model is represented by the solid green line.
Our previous study also predicted notable differences in the concentrations of L. monocytogenes epidemic clone subtypes and subtypes carrying a PMSC in inlA from a large survey of >30,000 RTE food samples collected from retail sources. Specifically, isolates carrying a PMSC in inlA were found at a concentration of up to 10,000-fold higher than that of isolates representing epidemic clone strains (5, 11). A companion study recently completed by our group input subtype-specific prevalence and concentration distributions from a food survey along with epidemiologic and consumption data into established exponential dose-response models (4). Quantifiable differences in virulence, as measured by log10 r values (probability of a single L. monocytogenes cell causing illness), were observed between L. monocytogenes subtypes encoding a full-length InlA and subtypes carrying a PMSC in inlA. Specifically, exponential models based on L. monocytogenes concentrations found in samples from retail sources generated mean log10 r values of −8.1 and −10.7 for (i) subtypes encoding a full-length InlA and (ii) subtypes carrying a PMSC in inlA, respectively. Inclusion of an additional parameter to estimate the increase in L. monocytogenes concentration between that in retail source samples and that from consumption resulted in mean log10 r values of −10.44 and −13.75 for subtypes encoding a full-length InlA and subtypes carrying a PMSC in inlA, respectively (4). Findings from the current study provide further quantitative evidence that L. monocytogenes subtypes vary in their ability to cause a systemic L. monocytogenes infection, which may be mechanistically attributed to a defined genetic marker (i.e., presence of a mutation leading to a PMSC in inlA).
To quantify the level of agreement between model-predicted values and observed values, the deviance function for quantal dose-response models was applied. The deviance was calculated for each model (including and excluding the strain effect to account for the presence or absence of a PMSC in inlA) to indicate which model fit the raw individual and combined organ data most appropriately (Tables 2 and 3). When the strain effect was included, goodness of fit (Tables 2 and 3) assumes that there is a difference in dose-response between strains; therefore, P values approaching 1.0 indicate that the observed values explain the model-predicted values and that the strain effect is needed for the model to explain the data. Values given when the strain effect is excluded (Tables 2 and 3) group all data together assuming that strains behave the same; therefore, low P values (<0.05) indicate that there is a significant difference between model-predicted values and observed raw data. The goodness-of-fit assessments indicate that models derived from the spleen and ileum data show the most concordance between the predicted models and the observed data (Tables 2).
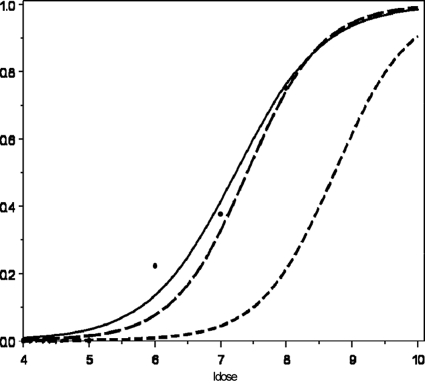
We also compared the strain-specific dose-response curves generated in the current study using the combined model, which uses all organs to estimate the b parameter in the LL model but which uses only the spleen data for ID50, to the dose-response data obtained from the study of Williams et al. (51) (Fig. 2). The study of Williams et al. (51) used an L. monocytogenes strain linked to a stillbirth in a rhesus monkey and obtained dose-response data using an oral pregnant guinea pig infection model. Although their study published an LL model to fit dose-response data on the basis of the dose resulting in fetal deaths, in the current study we used an LL model to fit the data from their study for infected maternal spleens (Fig. 2). Using the LL model for combined organ fits, we determined the ID50 for the outbreak-associated epidemic clone strain (CSUFSL N1-054) to be 7.41 log10 CFU of L. monocytogenes and the ID50 for the strain carrying a PMSC mutation in inlA (CSUFSL N1-040) to be 8.74 log10 CFU. The strain used by Williams et al. (51), which encodes a full-length InlA (as determined by inlA sequencing in this study), and the epidemic clone strain (CSUFSL N1-054) characterized here showed similar median infectious doses (Fig. 2). The slightly lower ID50 seen by Williams et al. (51) compared to the ID50 seen in this study may be explained by their use of a pregnant and therefore immune-suppressed animal infection model. The placenta is more susceptible to infection than other maternal organs, mostly due to the trophoblasts, which are involved in the vertical transmission of pathogens and which trap bacteria, which may thus provide a protective niche for bacterial survival (22). However, this analysis further extends the finding that L. monocytogenes strains carrying a PMSC in inlA demonstrate a notable increase in median infectious dose compared to that for strains encoding a full-length InlA.
FIG. 2.
Dose-response of L. monocytogenes for dam spleen infectivity (51) and combined models (which use all organs to estimate the b parameter in the log-logistic model but which use only the spleen data for ID50) for both the outbreak-associated (N1-054) and the virulence-attenuated (N1-040) strains used in the current study. The proportion of infected tissues is indicated on the y axis, and the log10 CFU dose (ldose) is indicated on the x axis. The dose-response curves for spleen infectivity were constructed with a log-logistic model. Raw spleen data from Williams et al. (51) are represented by the large black solid dots. The log-logistic model used to fit the dam spleen data from Williams et al. (51) is represented by the solid line; the log-logistic combined model for the outbreak-associated epidemic clone strain (CSUFSL N1-054) is represented by the line with longer dashes, and the log-logistic combined model for the strain carrying the most broadly distributed inlA PMSC (CSUFSL N1-040) is represented by the line with shorter dashes.
Conclusions.
Collectively, previous challenge studies using a murine model did not reveal consistent virulence phenotypes that might explain the predominance of certain L. monocytogenes strains that cluster within genetic lineage I (e.g., serotype 4b, epidemic clone strains) among listeriosis outbreaks and sporadic cases (6, 18, 28). Mutations leading to a PMSC in inlA are overrepresented among serotype 1/2a and 1/2c L. monocytogenes isolates belonging to genetic lineage II and are found among serotype 4b isolates only on very rare occasions (43, 46). Genetic markers associated with enhanced virulence of epidemic clone strains have not been described to date, and mutations leading to a PMSC in inlA appear to represent the only genetic markers that have been mechanistically related to virulence differences among L. monocytogenes subtypes. Results from the current study provide empirical dose-response data for L. monocytogenes subtypes with defined virulence characteristics, including a strain carrying a PMSC in inlA that represents a significant proportion of L. monocytogenes isolates recovered from RTE foods in the United States (12, 27, 43, 44, 46) and an epidemic clone strain that has been implicated in two recent multistate listeriosis outbreaks in the United States (9, 17). To our knowledge, this is the first study to report intragastric guinea pig challenge data that are based on well-characterized strains that carry genetic markers that are mechanistically related to virulence. In the current study, modeling of individual or combined organ data showed an approximate 1.3-log10 increase in the ID50 for the strain carrying a PMSC in inlA relative to that for the outbreak-associated epidemic clone strain. When subtype-specific prevalence and concentration distributions from food surveys, as well as epidemiologic and consumption data, were input into exponential models, r values shifted by 2.6 to 3.1 log10 (based on models without or with a parameter to estimate a concentration increase between samples from retail sources and consumption, respectively) for subtypes encoding a full-length InlA compared to the values for subtypes carrying a PMSC in inlA (4). Collectively, results from the current study and previous studies support a significant difference in the risk of sustaining a systemic L. monocytogenes infection from exposure to strains with defined virulence characteristics in RTE foods and that from exposure to strains without such virulence characteristics (i.e., with or without a PMSC in inlA), and data from this study are critical to revise current and develop future risk assessments that mechanistically relate strain-specific genetic characteristics and virulence to predict the risk of human disease.
Acknowledgments
The project was supported by the National Research Initiative of the USDA-Cooperative State Research, Education, and Extension Service-National Research Initiative grant number 2005-35201-16266 and USDA-Cooperative State Research, Education, and Extension Service Special Research Grant 2008-56341-8789.
We are grateful to M. A. Smith from the University of Georgia for providing the L. monocytogenes strain from the study of Williams et al. (51), for inlA sequencing to determine the inlA allelic type of this strain, and for generously sharing the dam spleen infection data from pregnant guinea pig challenge experiments. We thank LAR for fostering an environment of care and respect for animals, as well as maintaining the facilities and equipment to meet the needs both of the animals and of the research project. In addition, we are indebted to all members of the Colorado State University Food Safety Laboratory for assistance with this project.
Footnotes
Published ahead of print on 4 February 2011.
REFERENCES
- 1.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes: the influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 2.Brosch, R., J. Chen, and J. B. Luchansky. 1994. Identification of genomic divisions for Listeria monocytogenes and their correlation with serovar by pulsed-field electrophoresis. Appl. Environ. Microbiol. 60:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchanan, R. L., W. G. Damert, R. C. Whiting, and M. van Schothorst. 1997. Use of epidemiologic and food survey data to estimate a purposefully conservative dose-response relationship for Listeria monocytogenes levels and incidence of listeriosis. J. Food Prot. 60:918-922. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Y., et al. 2011. Variation in Listeria monocytogenes dose responses in relation to subtypes encoding a full-length or truncated internalin A. Appl. Environ. Microbiol. 77:1171-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y., et al. 2006. Attributing risk to Listeria monocytogenes subgroups: dose-response in relation to genetic lineages. J. Food Prot. 69:335-344. [DOI] [PubMed] [Google Scholar]
- 6.del Corral, F., R. L. Buchanan, M. M. Bencivengo, and P. H. Cooke. 1990. Quantitative comparison of selected virulence associated characteristics in food and clinical isolates of Listeria. J. Food Prot. 53:1003-1009. [DOI] [PubMed] [Google Scholar]
- 7.Felicio, M. T., T. Hogg, P. Gibbs, P. Teixeira, and M. Wiedmann. 2007. Recurrent and sporadic Listeria monocytogenes contamination in alheiras represents considerable diversity, including virulence-attenuated isolates. Appl. Environ. Microbiol. 73:3887-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Food and Agriculture Organization of the United Nations and World Health Organization. 2004. Risk assessment of Listeria monocytogenes in ready-to-eat foods. Technical report. Microbiological Risk Assessment Series 5. Food and Agriculture Organization of the United Nations, Rome, Italy, and World Health Organization, Geneva, Switzerland. http://www.fao.org/docrep/010/y5394e/y5394e00.htm.
- 9.Fugett, E., E. Fortes, C. Nnoka, and M. Wiedmann. 2006. International Life Sciences Institute North American Listeria monocytogenes strain collection: development of standard Listeria monocytogenes strain sets for research and validation studies. J. Food Prot. 69:2929-2938. [DOI] [PubMed] [Google Scholar]
- 10.Garner, M. R., K. E. James, M. C. Callahan, M. Wiedmann, and K. J. Boor. 2006. Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect. Immun. 74:876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gombas, D. E., Y. Chen, R. S. Clavero, and V. N. Scott. 2003. Survey of Listeria monocytogenes in ready-to-eat foods. J. Food Prot. 66:559-569. [DOI] [PubMed] [Google Scholar]
- 12.Gray, M. J., et al. 2004. Food and human isolates of Listeria monocytogenes form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handa-Miya, S., et al. 2007. Nonsense-mutated inlA and prfA not widely distributed in Listeria monocytogenes isolates from ready-to-eat seafood products in Japan. Int. J. Food Microbiol. 117:312-318. [DOI] [PubMed] [Google Scholar]
- 14.Jacquet, C., et al. 2004. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189:2094-2100. [DOI] [PubMed] [Google Scholar]
- 15.Jeffers, G. T., et al. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 16.Jonquieres, R., H. Bierne, J. Mengaud, and P. Cossart. 1998. The inlA gene of Listeria monocytogenes L028 harbors a nonsense mutation resulting in release of internalin. Infect. Immun. 66:3420-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 11:1811-1829. [DOI] [PubMed] [Google Scholar]
- 18.Lammerding, A. M., K. A. Glass, A. Gendron-Fitzpatrick, and M. P. Doyle. 1992. Determination of virulence of different strains of Listeria monocytogenes and Listeria innocua by oral inoculation of pregnant mice. Appl. Environ. Microbiol. 58:3991-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lecuit, M., et al. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722-1724. [DOI] [PubMed] [Google Scholar]
- 20.Lecuit, M., et al. 1999. A single amino acid in E-cadherin is responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18:3956-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lecuit, M., H. Ohayon, L. Braun, J. Mengaud, and P. Cossart. 1997. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect. Immun. 65:5309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeMonnier, A., et al. 2007. ActA is required for crossing of the fetoplacental barrier by Listeria monocytogenes. Infect. Immun. 75:950-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mead, P. S., et al. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nightingale, K. K., et al. 2008. inlA premature stop codons commonly found in Listeria monocytogenes isolated from food are responsible for virulence attenuation and confer protective immunity against infection by fully virulent L. monocytogenes. Appl. Environ. Microbiol. 74:6570-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nightingale, K. K., et al. 2007. Novel method to identify source-associated phylogenetic clustering shows that Listeria monocytogenes includes niche-adapted clonal groups with distinct ecological preferences. J. Clin. Microbiol. 44:3742-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nightingale, K. K., K. Windhan, and M. Wiedmann. 2005. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 187:5537-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nightingale, K. K., K. Windham, K. E. Martin, M. Yeung, and M. Wiedmann. 2005. Selected Listeria monocytogenes subtypes commonly found in food show reduced invasion in human intestinal cells due to distinct nonsense mutations in inlA leading to expression of truncated and secreted internalin A. Appl. Environ. Microbiol. 71:8764-8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishibori, T., et al. 1995. Correlation between the presence of virulence-associated genes as determined by PCR and actual virulence to mice in various strains of Listeria spp. Microbiol. Immunol. 39:343-349. [DOI] [PubMed] [Google Scholar]
- 29.Norton, D. M. 2002. Polymerase chain reaction-based methods for detection of Listeria monocytogenes: toward real-time screening for food and environmental samples. J. AOAC Int. 85:505-515. [PubMed] [Google Scholar]
- 30.Olier, M., F. Pierre, J. P. Lemaitre, C. Divies, A. Rousset, and J. Guzzo. 2002. Assessment of the pathogenic potential of two Listeria monocytogenes human faecal carriage isolates. Microbiology 148:1855-1862. [DOI] [PubMed] [Google Scholar]
- 31.Oliver, H. F., R. H. Orsi, M. Wiedmann, and K. J. Boor. 2010. Listeria monocytogenes σB has a small core regulon and a conserved role in virulence but makes differential contributions to stress tolerance across a diverse collection of strains. Appl. Environ. Microbiol. 76:4216-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orsi, R., et al. 2008. Lineage specific recombination and positive selection in coding and intragenic regions contributed to evolution of the main Listeria monocytogenes virulence gene cluster. Infect. Genet. Evol. 8:566-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orsi, R. H., D. Ripoll, K. K. Nightingale, and M. Wiedmann. 2007. Recombination and positive selection contribute to evolution of Listeria monocytogenes inlA. Microbiology 153:2666-2678. [DOI] [PubMed] [Google Scholar]
- 34.Piffaretti, J. C., H. Kressebuch, M. Aeschbacher, J. Bille, E. Bannerman, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. U. S. A. 10:3818-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ragon, M., et al. 2008. A new perspective on Listeria monocytogenes evolution. PloS Pathog. 4:e1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts, A. J., S. K. Williams, M. Wiedmann, and K. K. Nightingale. 2009. Some Listeria monocytogenes outbreak strains demonstrate significantly lower invasion, inlA transcript levels, and swarming motility in vitro. Appl. Environ. Microbiol. 75:5647-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roldgaard, B. B., J. B. Andersen, T. B. Hansen, B. B. Christensen, and T. R. Licht. 2009. Comparison of three Listeria monocytogenes strain in a guinea-pig model simulating food-borne exposure. FEMS Microbiol. Lett. 291:88-94. [DOI] [PubMed] [Google Scholar]
- 38.Rousseaux, S., M. Olier, J. P. Lamaitre, P. Piveteau, and J. Guzzo. 2004. Use of PCR-restriction fragment polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl. Environ. Microbiol. 70:2180-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sauders, B. D., et al. 2009. Prevalence and molecular diversity of Listeria monocytogenes in retail establishments. J. Food Prot. 72:2337-2349. [DOI] [PubMed] [Google Scholar]
- 40.Sauders, B. D., et al. 2004. Distribution of Listeria monocytogenes molecular subtypes among human and food isolates from New York State shows persistence of human disease-associated Listeria monocytogenes strains in retail environments. J. Food Prot. 67:1417-1428. [DOI] [PubMed] [Google Scholar]
- 41.Schlech, W. F. 2000. Foodborne listeriosis. Clin. Infect. Dis. 31:770-775. [DOI] [PubMed] [Google Scholar]
- 42.United States Food and Drug Administration, United States Department of Agriculture Food Safety and Inspection Service, and Centers for Disease Control and Prevention. 2003. Quantitative assessment of the relative risk to public health from foodborne Listeria monocytogenes among selected categories of ready-to-eat foods. United States Food and Drug Administration, United States Department of Agriculture Food Safety and Inspection Service, and Centers for Disease Control and Prevention, Washington, DC. http://www.fda.gov/downloads/food/scienceresearch/researchareas/riskassessmentsafetyassessment/ucm197330.pdf.
- 43.Van Stelten, A., J. M. Simpson, T. J. Ward, and K. K. Nightingale. 2010. Revelation by single-nucleotide polymorphism genotyping that mutations leading to a premature stop codon in inlA are common among Listeria monocytogenes isolates from ready-to-eat foods but not human listeriosis cases. Appl. Environ. Microbiol. 76:2783-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Stelten, A., and K. K. Nightingale. 2008. Development and implementation of a multiplex single-nucleotide polymorphism genotyping assay for detection of virulence-attenuating mutations in the Listeria monocytogenes virulence-associated gene inlA. Appl. Environ. Microbiol. 74:7365-7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Charkraborty, G. Dominguez-Bernal, W. Goebel, B. Zorn-Gonzalez, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward, T. J., et al. 2010. Molecular and phenotypic characterization of Listeria monocytogenes from U.S. Department of Agriculture Food Safety and Inspection Service surveillance of ready-to-eat foods and processing facilities. J. Food Prot. 73:861-869. [DOI] [PubMed] [Google Scholar]
- 47.Ward, T. J., T. F. Ducey, T. Usgaard, K. A. Dunn, and J. P. Bielawski. 2008. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl. Environ. Microbiol. 74:7629-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward, T. J., et al. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 15:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiedmann, M., et al. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams, D., J. Castleman, C.-C. Lee, B. Mote, and M. A. Smith. 2009. Risk of fetal mortality after exposure to Listeria monocytogenes based on dose-response data from pregnant guinea pigs and primates. Risk Anal. 29:1495-1505. [DOI] [PubMed] [Google Scholar]
- 51.Williams, D., E. A. Irvin, R. A. Chmielewski, J. F. Frank, and M. A. Smith. 2007. Dose-response of Listeria monocytogenes after oral exposure in pregnant guinea pigs. J. Food Prot. 70:1122-1128. [DOI] [PubMed] [Google Scholar]