Abstract
Escherichia coli strain DIER was constructed for estrogen detection by inserting an estrogen-sensitive intein (VMAER intein) into the specific site of the constitutively expressed chromosomal lacZ gene. This VMAER intein was generated by replacing the endonuclease region of the Saccharomyces cerevisiae VMA intein with the estrogen binding region of the human estrogen receptor α (hERα). When there were estrogens or analogs, the splicing of the VMAER intein was induced to produce the mature LacZ protein, which was detected through a β-galactosidase colorimetric assay. Eight typical chemicals (17-β-estradiol, bisphenol A, chrysene, 6-OH-chrysene, benz[a]anthracene, pyrene, progesterone, and testosterone) were detected using this DIER strain, and the whole detection procedure was accomplished in 2 h. Their 50% effective concentrations (EC50), relative estrogenic activities, and estradiol equivalency factors were calculated and were quite consistent with those detected with the yeast estrogen screening (YES) system. Furthermore, the estrogenic activities of the synthetic musk samples extracted from the wastewater and waste sludge of a sewage treatment plant of Shanghai (China) were detected, and their results were comparable to those obtained from the YES system and gas chromatography-mass spectrometry (GC-MS). In conclusion, the DIER bioassay could fill a niche for the efficient, rapid, high-throughput screening of estrogenic compounds and has potential for the remote, near-real-time monitoring of environmental estrogens.
Environmental deposition of natural, pharmaceutical, and synthetic chemicals with estrogenic activities is associated with numerous human and wildlife physiological disorders, prompting the development of various assays to screen for estrogenic potencies (12, 18, 30).
Many chemical methods and bioassays have been developed for estrogen detection. Conventional methods, such as liquid chromatography-mass spectrometry (LC-MS), gas chromatography-mass spectrometry (GC-MS), and high-performance liquid chromatography (HPLC), are accurate and have been widely used. Since chemicals with estrogenic activities have different chemical structures and properties, chemical methods are not often suitable for fast detection (26). Bioassays have been developed based on the estrogenic receptor (ER)-activated expression of a reporter gene through estrogen-responsive sequences (ERE), including in vivo mammalian assays and in vitro assays (6, 21, 28, 42). The in vitro assays often include the following categories: competitive ligand binding assays, cell proliferation assays, postconfluent cell accumulation, induction of protein expression/enzyme activities, and the recombinant receptor/reporter gene assays (42). Among them, the recombinant receptor/reporter gene assays are designed to detect the induction or repression of a biological process via specific endocrine receptors. They usually have high responsiveness and sensitivity and can be used to assess the relative potencies of alleged receptor-mediated agonists and antagonists. A widely used receptor/reporter assay is the yeast estrogen screening (YES) system (29), which has successfully detected many estrogenic chemicals, such as polychlorinated biphenyls (PCB), hydroxylated derivatives, polynuclear aromatic hydrocarbons (PAH), and others (14, 32-34). It has also been used to detect environmental estrogenic compounds, such as those in samples from wastewater treatment systems and from dairy manure (14, 27). The main drawback of these biosensors is that their detection procedures are somewhat complex and the detection time is relatively long. It usually takes several days to incubate the reporter cells with the samples in order to accumulate enough reporter protein and produce a measurable signal, which is not really suitable for large-scale sample screening.
Inteins have been called “protein introns”; an intein is a protein element found as an in-frame insertion within the sequence of a particular host gene (20, 23). It possesses the ability to excise itself posttranslationally from its protein host and ligate the flanking peptide sequences with a native peptide bond via a very efficient self-catalyzed reaction, called protein splicing (20, 23). In general, inteins' insertion into the host protein can inactivate the host protein, and only after protein splicing can the host protein's activity be restored (5, 39). Up to the present, many inteins have been found in different prokaryotic and eukaryotic organisms, and their structures and properties have many similarities (24). For example, most inteins have two regions, the splicing region and the endonuclease region, and the latter one is not essential for protein splicing (4). Several inteins have been shown to retain their activities after being transferred into nonnative genes or cell hosts, such as the widely used Saccharomyces cerevisiae VMA intein (20, 31). And the requirement for efficient protein splicing of the host protein is that the first amino acid of the C-terminal extein should be a cysteine (Cys), serine (Ser), or threonine (Thr) residue (24). All these characteristics make it possible for inteins to be used as molecular “switches” to control the activities of the arbitrary target proteins. In recent years, attempts have been made to generate tunable inteins, such as the temperature-sensitive intein alleles or the chimeric intein triggered by 4-hydroxytamoxifen (4-HT) or by human thyroid hormone (2, 35, 43), to control protein activity more flexibly. Because of their efficient splicing ability and controllable flexibility, inteins have been widely used in the protein engineering fields for protein expression and activity control, protein ligation, protein cyclization, protein modification, drug discovery, and other functions (3, 20, 25). However, up to now, there has been no report of an intein being used in estrogen detection.
In this work, an Escherichia coli strain, DIER (detection strain with intein VMAER), was constructed for estrogen detection based on an artificial estrogen-sensitive VMAER intein and β-galactosidase colorimetric assay. This VMAER intein contained the splicing region of the S. cerevisiae VMA intein and the estrogen-binding domain of hERα. When there were estrogenic hormones or synthetic analogs present, the VMAER intein could splice itself out from the LacZ protein efficiently, so that this DIER strain detected the chemicals' estrogenic activities by β-galactosidase colorimetric assay. Eight typical chemicals and the environmental samples extracted from wastewater and waste sludge were detected using this strain, and the results were compared with those obtained from the YES system and GC-MS.
MATERIALS AND METHODS
Strains and reagents.
E. coli strains and the plasmids used in this work are listed in Table 1. All E. coli strains were cultured in Luria-Bertani (LB) medium at 32°C, and ampicillin was used at 100 μg/ml if needed. Sucrose was present at 10%. The recombinant yeast strain BJ3505 (7) was used for yeast estrogen screening (Table 1). The yeast medium contained yeast nitrogen base without amino acids (6.7 g/liter), plus dextrose (20 g/liter), leucine (60 mg/liter), and histidine (20 mg/liter). 17-β-Estradiol (98% purity), progesterone (99% purity), and testosterone (98% purity) were purchased from Sigma-Aldrich Chemical Company (St. Louis, MO). Bisphenol A (BPA) (99% purity), chrysene (98.7% purity), 6-OH-chrysene (100% purity), benz[a]anthracene (99.5% purity), and pyrene (98.6% purity) were purchased from AccuStandard, Inc. These eight chemicals were prepared as 10-mg/ml stock solutions in ethanol or acetone. All other chemicals were analytical grade.
TABLE 1.
Strains and plasmids used in this work
| Strain/plasmid | Description | Source or reference |
|---|---|---|
| E. coli DY329 | W3110ΔlacU169 nadA::Tn10 gal490 lcI857Δ(cro-bioA) | 41 |
| S. cerevisiae BJ3505 | MATαpep4::HIS3 prb1-Δ1.6R his3-Δ200 lys2-801 trp1-Δ101 ura3-52 (can1) | 7 |
| E. coli DYSA | DY329 (sacB-bla)::lacZ | This work |
| E. coli DYGL | DYSA (PgapA-lacZ-6×His):: (sacB-bla) | This work |
| E. coli DIER | DY329 (PgapA-lacZ-VMAER-6×His)::lacZ | This work |
| pUC-PgapA | pUC18 derivative with E. coli gapA promoter, Apr | 15 |
| pEX18Ap | Plasmid containing sacB and bla gene, Apr | 8 |
| pKT:ΔI-SMER | Plasmid containing hERαbr-interrupted M. tuberculosis recA intein, Apr | 35 |
| pUC-inteinER | pUC18 derivative with hERαbr interrupted VMA intein, Apr | This work |
Standard DNA manipulation.
Oligonucleotides were synthesized at Invitrogen Ltd. (Shanghai, China) and are listed in Table S1 in the supplemental material. Restriction enzymes and DNA-modifying enzymes were purchased from TaKaRa Biocompany (Dalian, China). PCRs were carried out in 50-μl reaction solutions, containing 1× PCR buffer, 200 μM deoxynucleoside triphosphates (dNTPs), 0.5 μM (each) primer, 2.5 U LA-Taq DNA polymerase (TaKaRa, Dalian, China), and certain amounts of templates (plasmid DNA, 10 ng; genomic DNA, 50 ng) at 94°C for 5 min, cycled 30 times at 94°C for 30 s, 55°C for 1 min, and 72°C for 1 to 5 min (depending on the length of the amplified DNAs), with a final extension step of 72°C for 7 min. The PCR products were separated by agarose gel electrophoresis and recovered with the QIAquick gel extraction kit (Qiagen, Shanghai, China). Plasmid DNAs were isolated using the QIAprep Mini-spin kit (Qiagen, Shanghai, China), and genomic DNA was obtained using the QIAamp DNA minikit (Qiagen, Shanghai, China). Other general techniques for restriction enzyme manipulation, molecular cloning, and agarose gel electrophoresis were carried out with standard protocols (11). Homologous recombination was carried out with the E. coli DY329 strain as described before (41). All the constructs were confirmed by DNA sequencing (Invitrogen, Shanghai, China).
Construction of strain DIER.
Homologous recombination was used to insert hERαbr (the estrogen binding region of hERα, residues Ser301 to Thr553) into the VMA intein (15). The fragment hERαbr, amplified from the plasmid pKT:ΔI-SMER (35), and the fragment IntS, amplified from the plasmid pUC-intein (15), were coelectrotransformed into DY329 (Table 1) competent cells, and the positive plasmid contained an hERαbr-interrupted VMA intein. This chimeric intein was named the VMAER intein, and this plasmid was called pUC-inteinER (see Fig. S1a in the supplemental material).
To generate a constitutively expressed lacZ gene, first the fragment SAL, amplified from the plasmid pEX18Ap (8), was electrotransformed into DY329 competent cells to generate strain DYSA (see Fig. S1b in the supplemental material). The lacZ fragment amplified from the plasmid pME6522 (15) was inserted into the plasmid pUC-PgapA (15) to generate the plasmid pUC-PgapA-lacZ (Table 1; see also Fig. S1b). Then, the fragment lacZR, amplified from the plasmid pUC-PgapA-lacZ, was electrotransformed into DYSA competent cells to generate strain DYGL (Table 1; see also Fig. S1b). Next, fragments SA123 and SA329, amplified from the plasmid pEX18Ap, were separately electrotransformed into DYGL competent cells to generate strains DYGLSA123 and DYGLSA329 (Table 1; see also Fig. S1b). Finally, two fragments (VMAER123 and VMAER329) amplified from the plasmid pUC-inteinER were electrotransformed into the competent cells of strains DYGLSA123 and DYGLSA329, respectively. The positive recombinants were named strain DIER123 and strain DIER329, accordingly (Table 1; see also Fig. S1b).
Splicing efficiency analysis of the VMAER intein.
Cells of strains DIER123 and DIER329 were grown in 10 ml LB medium to an optical density at 600 nm (OD600) of ≈1.0. The in vivo analysis was performed by adding 1 mg/liter 17-β-estradiol to the culture, which was incubated for various time periods. Proteins were purified using a nickel-nitrilotriacetic acid (Ni-NTA) column, and 10-μl protein aliquots were detected by SDS-PAGE (16). Similarly, the in vitro analysis was achieved by purifying the proteins first and then adding 1 mg/liter 17-β-estradiol to the protein solution with treatment for different periods of time. After that, 10-μl protein aliquots were analyzed using SDS-PAGE.
Chemical detection with strain DIER and the YES system.
Strains DIER123, DIER329, and DYSL were cultured in 10 ml LB medium to an OD600 of ≈0.6 and were supplied with 10 μl chemical solutions (17-β-estradiol, BPA, pyrene, chrysene, 6-OH-chrysene, benz[a]anthracene, progesterone, and testosterone). A series of 1:10 dilutions ranging from 10−10 g/liter to 10−1 g/liter were used. After a 2-h incubation, cultures were collected to perform the β-galactosidase assay, and β-galactosidase activity was calculated and expressed in Miller units (19). The β-galactosidase comparative activity (the β-galactosidase activity relative to the background level) was calculated by the equation (Miller units of sample − Miller units of medium)/Miller units of medium.
Yeast strain BJ3505 was used to perform the YES system detection as described by K. W. Gaido (7), and a modified procedure was used as mentioned previously (17). The eight chemicals were detected. The β-galactosidase activity was expressed as Vmax (ΔOD420/min) divided by cell density (OD590), and the β-galactosidase comparative activity was calculated by the equation (β-galactosidase activity of sample − β-galactosidase activity of medium)/β-galactosidase activity of medium.
The dose-response profiles of these chemicals were plotted. The x axis was the log of the chemical's concentration, and the y axis was the β-galactosidase comparative activity. If the β-galactosidase comparative activity was lower than 0.1, the data point was not shown. The 50% effective concentration (EC50) was calculated from seven individual experiments; the mean and standard deviation were calculated.
Permeability analysis of strain DIER.
DIER cells were cultured in 10 ml LB medium to an OD600 of ≈0.6, and 1 mg/liter chemicals (17-β-estradiol, BPA, pyrene, chrysene, 6-OH-chrysene, benz[a]anthracene, progesterone, and testosterone) were supplied. After a 2-h incubation, the cultures were collected and washed five times with Millipore water. The cells were disrupted by sonication on ice, and the cell lysates were collected by centrifugation at 4°C, 12,000 rpm, for 30 min. Then the lysates were detected using the YES system and GC-MS. Similar detection was also performed with the yeast BJ3505 lysates.
Environmental sample analysis.
Environmental samples were collected and extracted from the wastewater and waste sludge of a sewage treatment plant of Shanghai (China) as described previously (40). The samples were dissolved in hexane and were analyzed using the DIER strain, the BJ3505 yeast strain, and GC-MS. One hundred microliters of extracted samples were used, and the same amounts of hexane and medium were used as controls. The detection procedures with DIER detection and the yeast BJ3505 strain were the same as those described above. GC-MS analysis was performed as described before (40).
RESULTS
E. coli DIER bioassay.
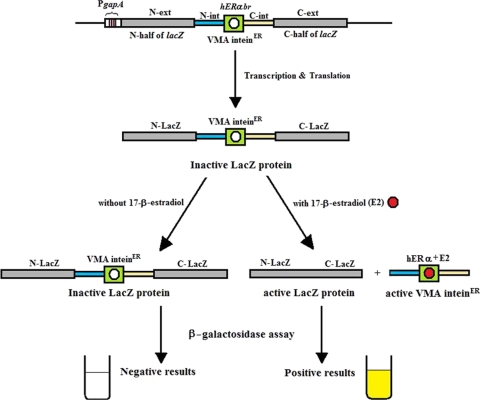
In this work, an E. coli strain, DIER, was constructed for estrogen detection based on an estrogen-sensitive intein (VMAER intein) inserted into the constitutively expressed lacZ gene. When estrogen or analogs are added, they bind to the hERα receptor region of the VMAER intein to induce the splicing of VMAER intein and produce the mature LacZ protein. Otherwise, no splicing happens and no active LacZ protein is generated. The estrogenic activities of chemicals were detected and quantified by β-galactosidase assay (Fig. 1).
FIG. 1.
Strategy of the DIER bioassay. The E. coli gapA promoter was used to control the expression of LacZ interrupted with the VMAER intein (containing the intein splicing region and the binding region of the hERα gene). In the presence of estrogen or analogs, the splicing of VMAER intein is induced and the active LacZ protein is produced to generate positive results in the β-galactosidase assay. Otherwise, no β-galactosidase activity can be detected from the chimeric protein.
As we described before, the splicing efficiency of the VMA intein was very high, whether in its original host or in the nonnative host background (15). This is the reason that the VMA intein was selected to generate this estrogen-sensitive intein. Previous research has confirmed that inserting the ligand binding region of the hERα protein into the M. tuberculosis recA intein can generate estrogen-sensitive intein (35). Therefore, we used this coding region (from Ser301 to Thr553) to replace the endonuclease region of the VMA intein (from K206 to G387), which was not involved in the splicing process (1). This chimeric intein was denoted the VMAER intein (Fig. 1; see also Fig. S1 in the supplemental material).
To generate estrogen-sensitive LacZ for detection, the VMAER intein was inserted into the specific site of the constitutively expressed full-length lacZ gene, not only to abolish the activity of the LacZ protein completely but also to control the maturation of the LacZ protein by ligand binding and VMAER intein splicing. First, strain DYGL containing the constitutively expressed full-length lacZ gene was generated by integrating the promoter of the gapA gene, the full-length lacZ gene, and the 6×His tag into strain DYSA (Table 1; see also Fig. S1 in the supplemental material). Then, the insertion site of the VMAER intein was selected. Although the splicing process for the intein was entirely autonomous, the host protein residue immediately adjacent to the C terminus of the intein was critical for splicing (23). To meet the requirement for efficient splicing, two insertion sites were selected. One site was between Gly122 and Cys123; the other was between Ala328 and Cys329. There were two reasons for this selection. First, the residue after the C terminus of the VMA intein was cysteine in the original protein context, and its splicing efficiency was high when the host residue proximate to the N terminus of intein was alanine or glycine (23). Another reason was that these two sites are located in the essential region of the LacZ protein (9), and the insertion of intein at these sites may block protein activity completely. The insertion of the VMAER intein into the two sites of the lacZ gene in strain DYGL produced strains DIER123 and DIER329 (Table 1; see also Fig. S1).
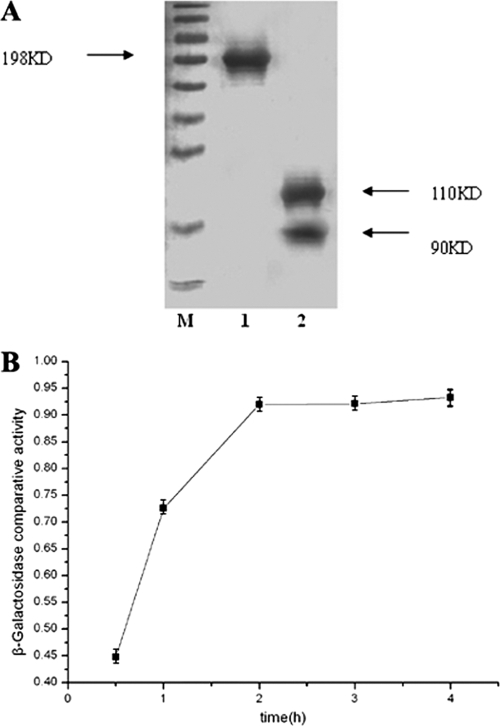
To study whether the VMAER intein was estrogen sensitive and whether the activity of the chimeric LacZ protein was controllable, the in vivo and in vitro expression of LacZ and the β-galactosidase activities were determined. Cells were induced with 1 mg/liter 17-β-estradiol for different times (1 h, 2 h, 3 h, 4 h, and 12 h). For in vitro analysis, a 198-kDa protein was purified from two strains, corresponding to the mass of the LacZ protein interrupted with the VMAER intein. After 2 h of induction of 17-β-estradiol, SDS-PAGE results showed that this 198-kDa band totally disappeared and two other bands (110 kDa and 90 kDa) were produced, equal to the mass of the full-length LacZ protein and the VMAER intein (Fig. 2 A). No β-galactosidase activity was detected in the purified 198-kDa protein solution before the induction. After a 2-h incubation with 17-β-estradiol, its β-galactosidase activity was similar to that of the 110-kDa protein solution purified from DYGL (data not shown). In vivo analysis showed that only the 110-kDa protein could be purified after the 17-β-estradiol induction (data not shown). In addition, results indicated that the splicing of the VMAER intein could be induced very quickly. Two hours of incubation with 17-β-estradiol was enough to induce efficient splicing (Fig. 2B). These results demonstrated that the VMAER intein was quite sensitive to estrogens and its efficient splicing could be induced very quickly. Then, the β-galactosidase activities of the two strains were analyzed, and strain DYGL was used as a positive control. When there was no estrogen present, the β-galactosidase activity could be detected only in DYGL cells. After the 17-β-estradiol induction, the β-galactosidase activities of DIER123 and DIER329 cells were almost equal to that of DYGL cells (data not shown). This further confirmed that the insertion of the VMAER intein could block the activity of the LacZ protein completely and efficient splicing of the VMAER intein induced by estrogens could produce mature and active LacZ, whose levels were similar to that of the wild type. Furthermore, the position effect of intein insertion was studied. Without an inducer, no β-galactosidase activity was detected in two strains. After induction with 17-β-estradiol at different concentrations, the β-galactosidase activity of strain DIER123 was almost equal to that of strain DYGL and a little higher than that of strain DIER329 (data not shown). This demonstrated that the position between Gly122 and Cys123 was more suitable for VMAER intein insertion. Therefore, strain DIER123 was designated strain DIER, which was quite sensitive to estrogen and could be used for fast estrogens detection.
FIG. 2.
Efficiency analysis of DIER bioassay. (A) In vitro protein purification results, analyzed by SDS-PAGE. M is the protein marker; lane 1 contains protein aliquots purified from the noninduced DIER strain; lane 2 contains the protein aliquots of lane 1 treated with 10 μl 1-g/liter 17-β-estradiol for 2 h. (B) Influence of inducing time on β-galactosidase comparative activity. The x axis shows the concentration of 17-β-estradiol, and the y axis shows β-galactosidase comparative activity. Values are means ± SD (n = 7).
Dose-response profiles.
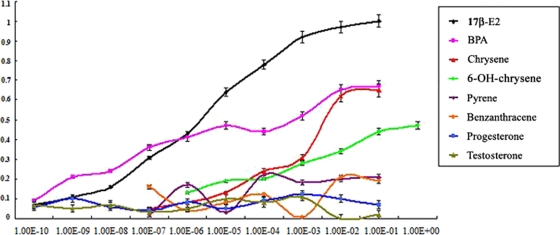
Eight chemicals were detected, and their dose-response curves were plotted. No β-galactosidase activity was induced by progesterone and testosterone, and the comparative β-galactosidase activities of the other six estrogenic chemicals were correlated with their concentrations in a direct ratio except in the case of benz[a]anthracene and pyrene (Fig. 3). The dose-response profiles of benz[a]anthracene and pyrene were not related to their concentrations but fluctuated erratically (Fig. 3). In addition, the lower limits of detection and the upper limits of detection for each chemical were not same and were related to their relative estrogenic activities (Table 2 ). These results were consistent with a previous report that BPA had strong estrogenic activity (22), chrysene and 6-OH-chrysene had moderate estrogenic activities (10, 44), the estrogenic activities of benz[a]anthracene and pyrene were weak (12), and progesterone and testosterone had no estrogenic activity (29). To verify the efficiency of the DIER bioassay, we also analyzed these eight chemicals using the widely used recombinant yeast strain BJ3505 (7) and compared the results of two assays (Table 2). We found that the results for each chemical analyzed with two bioassays were quite similar, and almost all were in the same order of magnitude, meaning that the DIER strain was sensitive to estrogenic chemicals and its β-galactosidase response could reflect the estrogenic activity difference (Table 2). Therefore, the DIER bioassay could be used to detect different estrogens or analogs, and the detection efficiency was related to the chemical's estrogenic activity.
FIG. 3.
Dose-response curves of eight chemicals detected with DIER bioassay. DIER cells were cultured with eight chemicals (17-β-estradiol, BPA, chrysene, 6-OH-chrysene, pyrene, benz[a]anthracene, progesterone, and testosterone) at different concentrations for 2 h, and their β-galactosidase activities were determined. The concentrations ranged from 1 × 10−10 to 1 g/liter. The abscissa shows each chemical's concentration, and the ordinate shows the β-galactosidase comparative activity.
TABLE 2.
Detection results for eight chemicals with DIER and YES bioassaysa
| Strain | Chemical | Limit of detection (g/liter) | Linear scope (g/liter) | Relative estrogenic activity | Estradiol equivalency factor | Estrogenic activity |
|---|---|---|---|---|---|---|
| DIER | 17-β-Estradiol | 10−10 | 10−10-10−1 | 1.0 | 1.0 | Strong |
| BPA | 10−10 | 10−10-10−2 | 10−4 | 1.2 × 10−4 | Strong | |
| Chrysene | 10−7 | 10−6-10−1 | 10−4 | 0.98 × 10−4 | Moderate | |
| 6-OH-chrysene | 10−6 | 10−6-10−1 | 10−5 | 1.4 × 10−5 | Moderate | |
| Benz[a]anthracene | NDb | ND | ND | ND | Weak | |
| Pyrene | ND | ND | ND | ND | Weak | |
| Progesterone | ND | ND | ND | ND | No | |
| Testosterone | ND | ND | ND | ND | No | |
| BJ3505 | 17-β-Estradiol | 10−10 | 10−10-10−1 | 1.0 | 1.0 | Strong |
| BPA | 10−10 | 10−10-10−1 | 10−4 | 1.7 × 10−4 | Strong | |
| Chrysene | 10−6 | 10−5-10−1 | 10−4 | 1.13 × 10−4 | Moderate | |
| 6-OH-chrysene | 10−6 | 10−6-10−2 | 10−5 | 1.1 × 10−5 | Moderate | |
| Benz[a]anthracene | ND | ND | ND | ND | Weak | |
| Pyrene | ND | ND | ND | ND | Weak | |
| Progesterone | ND | ND | ND | ND | No | |
| Testosterone | ND | ND | ND | ND | No |
Values represent data from seven separate experiments.
ND, not detected (when relative β-galactosidase activity is lower than 0.1).
Permeability of strain DIER.
Since strain DIER was a living system, the permeability of the E. coli cell wall and the degradation of the E. coli transport or enzyme system might have influenced the detection results. To be sure of whether the chemicals' molecules entered the bacterial cell and whether their structures were changed, we determined the chemicals' concentrations in the lysates of DIER cells after 2 h of incubation by using the YES system and GC-MS. Results showed that over 95% of the chemicals' molecules were detected in the bacterial lysates, comparable to those from the yeast lysates. GC-MS results indicated that no degradation or structure change happened to these chemicals (data not shown). These results demonstrated that this DIER strain was permeable to the estrogenic chemicals and had no alteration of the chemicals, meaning it was fit for accurate and fast estrogen detection.
Environmental sample analysis.
To analyze the feasibility of the DIER assay for analysis of the environmental samples, we extracted the synthetic musk from the wastewater and sludge samples during various treatment steps of a sewage treatment plant and detected the estrogenic activities of these samples using three methods, the DIER strain, yeast strain, and GC-MS methods. The DIER results indicated that the β-galactosidase comparative activities ascended with the concentration of the synthetic musk and were correlated to the results of GC-MS. Similar results were also obtained using the yeast strain B3505 (Table 3 ).
TABLE 3.
Concentrations and β-galactosidase comparative activities of synthetic musk extracted from wastewater and sludge samples of sewage treatment steps
| Sample source | Concn of synthetic musk (g/liter) (GC-MS)a | β-Galactosidase comparative activity |
|
|---|---|---|---|
| DIER | YES | ||
| Sludge of anaerobic filter | 1.2 × 10−4 | 3.0 | 3.4 |
| Sludge of oxygen-poor filter | 9.5 × 10−5 | 2.7 | 2.6 |
| Returned sludge | 9.0 × 10−5 | 2.4 | 2.1 |
| Water inflow | 1.3 × 10−6 | 0.1 | 0.09 |
| Water of primary settling tank | 8 × 10−7 | 0.02 | 0.04 |
See reference 28.
Results showed the concentrations of synthetic musk extracted from waste sludge samples were almost 100-fold higher than those from the wastewater, consistent with the high lipid solubility of synthetic musk. Also, the comparative estrogenic activity of synthetic musk was calculated according to the curve of 17-β-estradiol (Table 3). For example, the β-galactosidase comparative activity of the synthetic musk from water inflow was equivalent to that of 10−9 g/liter 17-β-estradiol. Since the concentration of synthetic musk detected by GC-MS was 1.3 μg/liter and the synthetic musk occupied about half of the extracted material, the estrogenic activity of the synthetic musk might be 10−4, almost equal to those of BPA and chrysene. In comparison with the results for other chemicals detected with the DIER bioassay, the detection limit of the synthetic musk was a little higher. The reason might be that current extraction methods not only could extract the synthetic musk but also could extract other chemicals with estrogenic activity. Also, since some calculated β-galactosidase comparative activities were out of the range of the 17-β-estradiol curve, dilution or reconcentration might be made before detection. These results confirmed that the DIER bioassay can be used for efficient primary screening and quantification of estrogenic environmental samples.
DISCUSSION
Environmental deposition of chemicals with estrogenic activities has prompted researchers to develop various methods for estrogenic potency screening (12). In this work, we constructed a DIER strain containing VMAER intein whose splicing activity was regulated by the presence of estrogen hormone or analogs. Designed according to other engineered allosteric enzyme prototypes, an artificial intein chimera was created by fusing an estrogen-hormone-binding domain with the splicing region of the VMA intein. The insertion of the binding domain abolished the splicing activity of the intein but allowed it to be restored later by addition of estrogen hormone or synthetic analogs. The resulting allosteric intein was then used to conditionally activate the LacZ protein in a dose-dependent manner, which could be detected with a β-galactosidase assay. Eight representative chemicals and some environmental samples were tested. Results indicated the DIER bioassay could detect estrogen in a very short time, and its sensitivity and accuracy were similar to those of the widely used YES bioassay.
In this hormone-induced intein, the hormone-binding domain was known to undergo conformational change and stability enhancement upon ligand binding (37). The fusion of the foreign domain with the splicing domain was designed to cause a structural perturbation and most likely affected its stability as well. These effects would abolish the splicing activity of the chimeric intein but allow it to be restored upon hormone binding. It is the first time that the posttranslational functional regulation of protein through ligand-controlled intein splicing has been used for estrogen detection. Compared with other bioassays, the DIER bioassay using E. coli as the detection strain has many advantages, such as fast growth speed, low culturing cost, a clear genetic background, and an easy operating procedure. These characteristics could simplify the detection process, reduce uncertain disturbances due to unclear background, and improve detection accuracy.
A previous report has confirmed that the estrogen-sensitive M. tuberculosis recA intein spliced efficiently when the estrogenic chemicals were added (36). In this work, we used the Sce VMA intein with high splicing efficiency to construct an estrogen-sensitive strain for universal use. In comparison with the estrogen response element (ERE)-lacZ reporter fusion in YES strains, this design reached the goal of the activity of the LacZ protein being regulated at the posttranscription level, which could diminish the uncertain influence of the inducible promoter on the expression of the reporter gene. Experimental results confirmed that no active LacZ protein was detected without estrogen. Only after ligand-induced splicing of the VMAER intein happened in the presence of hormone was LacZ protein activity detected, and it was similar to that of the wild type, implying that VMAER intein splicing was efficient and restrictive. This was the foundation of the DIER bioassay.
Another advantage of the DIER assay compared with other assays was speed. YES strains are widely used in estrogen detection for sensitivity and accuracy. But the problem with that assay is a long detection time. Regardless of the culture procedure, its incubation and detection process required at least 24 h. However, when the DIER strain was exposed to estrogens, protein splicing was triggered very quickly and the ligated extein product could be detected in <30 min (data not shown). Quantifiable β-galactosidase activity was observed in 60 min, and a 2-h induction could make it reach the maximum. This could speed the screening procedure greatly. Combined with a 96-cell plate and a fast screening machine, this assay is quite fit for large-scale detection of estrogens.
Using the DIER strain, six typical estrogenic chemicals (17-β-estradiol, BPA, chrysene, 6-OH-chrysene, benz[a]anthracene, and pyrene) were assayed, and their limits of detection results were in agreement with a previous report (12). And a lack of detection of progesterone and testosterone confirmed the sensitivity and specificity of the DIER bioassay for estrogen hormone or its analogs. These chemicals were widely present in the environment, with different estrogenic activities and carcinogenicities (38). For example, these chemicals can stimulate the growth of human breast cancer cells (36) or induce the expression of vitellogenin in fish (13). Since PAHs were easily degraded by mammalian cells or higher-animal cells, the principal detection methods for these compounds were chemical methods, such as HPLC and GC-MS (12), and there were few reports of the use of biological detection methods. The construction of this DIER bioassay might shed new light on the detection of PAHs. At the same time, the high estrogenic activities of the PAHs detected by this DIER bioassay should remind researchers to pay more attention to the risks of the estrogen-like activities of these chemicals not only to their carcinogenicity. In addition, we detected the estrogenic activity of synthetic musk extracted from the environmental samples of the wastewater and the waste sludge, using the DIER bioassay, the YES assay, and GC-MS. The results were in agreement with each other. From these results, we found that the estrogenic activity of the synthetic musk was quite high, almost similar to that of BPA or chrysene. And the concentration of the synthetic musk in waste sludge was much higher than that in the wastewater because of its high lipid solubility. Therefore, researchers should pay more attention to the environmental risks of waste sludge and try to improve sewage treatment technology.
Although the rapidity, accuracy, and efficiency of this DIER bioassay were quite good, other potential limitations still existed, like the lack of detection of benz[a]anthracene and pyrene. We thought that the E. coli cell wall and transport system might selectively decrease a particular chemical's potency or remain fully impermeable to it. Therefore, we constructed several mutation strains (lptD mutation, lpxC mutation, and tolC mutation) in which the cell permeability and sensitivity to antibiotics and chemicals should be increased. But little improvement was found in the detection of the two chemicals (data not shown). Cell permeability analysis confirmed that E. coli cells did not have any obvious selectivity for certain chemicals, and the cell wall or transportation system of strain DIER had almost no influence on permeability or detection. A lack of structure modification further confirmed that the detection results really reflected the chemicals' true estrogenic activity, which may be related to their structural differences and binding diversity. Therefore, E. coli strain DIER could provide a universal and accurate assay for estrogen detection. As to the nondetection of benz[a]anthracene and pyrene, it should be noted that the results of the DIER and YES bioassays agreed in this regard. Therefore, some unknown reasons may limit detection, and other work is needed.
Another potential improvement is use of the purified His-tagged VMAER intein-LacZ protein in estrogen detection. Using a protein solution directly may be much preferable in the analytical/diagnostic world. Bacterial strain bioassay versus proteins in a field assay have their own advantages and disadvantages. Proteins in a field assay have a cleaner background and easier validation and may be more compatible with existing diagnostic automation. However, the His-tagged VMAER intein-LacZ protein is a large protein molecule; it is very difficult to purify and get a large amount of protein for large-scale detection. And the difficulty in protein storage and the instability of protein activity may give uncertainty in the detection results. One of the strengths of the bacterial strain assay is that it makes the E. coli cell a portable diagnostic machine and there is no need to purify protein. It is also much easier for bacteria to be stored, and it can reflect the true chemical situation in a living system. Although we have confirmed that the bacterial cells have little influence on the chemicals' properties, some unknown factors inside the cell may have disturbed detection. Anyway, there are very few reports of engineered bacteria as analytical or diagnostic tools for estrogen detection, and this work goes a long way. Also, there is still room to optimize this work for an even more important usage.
In a word, the DIER bioassay is a fast and accurate means of initial analysis. It is unrealistic to use only a single assay to screen and distinguish thousands of chemicals. Rather, a combination of several assays might be more effective.
Conclusions.
Environmental estrogens were associated with human and wildlife physiological disorders, and researchers were asked to develop new assays for screening of various chemicals with estrogenic potencies. The estrogen-sensitive DIER bioassay was effective for the in vitro determination of chemicals with estrogenic activity in a short time and was especially fit for the large-scale primary screening of environmental samples.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (no. 30900010 and 30870512), a grant from the Ph.D. Programs Foundation of the Ministry of Education of China (grant no. 20090073120066), and the Major State Basic Research Development Program of China (973 Program; grant no. 2009CB118906).
Footnotes
Published ahead of print on 11 February 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Brenzel, S., T. Kurpiers, and H. D. Mootz. 2006. Engineering artificially split inteins for applications in protein chemistry: biochemical characterization of the split Ssp DnaB intein and comparison to the split Sce VMA intein. Biochemistry 45:1571-1578. [DOI] [PubMed] [Google Scholar]
- 2.Buskirk, A. R., Y. C. Ong, Z. J. Gartner, and D. R. Liu. 2004. Directed evolution of ligand dependence: small-molecule-activated protein splicing. Proc. Natl. Acad. Sci. U. S. A. 101:10505-10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheriyan, M., and F. B. Perler. 2009. Protein splicing: a versatile tool for drug discovery. Adv. Drug Deliv. Rev. 61:899-907. [DOI] [PubMed] [Google Scholar]
- 4.Chong, S., and M. Q. Xu. 1997. Protein splicing of the Saccharomyces cerevisiae VMA intein without the endonuclease motifs. J. Biol. Chem. 272:15587-15590. [DOI] [PubMed] [Google Scholar]
- 5.Daugelat, S., and W. R. Jacobs, Jr. 1999. The Mycobacterium tuberculosis recA intein can be used in an ORFTRAP to select for open reading frames. Protein Sci. 8:644-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang, H., et al. 2000. Quantitative comparisons of in vitro assays for estrogenic activities. Environ. Health Perspect. 108:723-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaido, K. W., et al. 1997. Evaluation of chemicals with endocrine modulating activity in a yeast-based steroid hormone receptor gene transcription assay. Toxicol. Appl. Pharmacol. 143:205-212. [DOI] [PubMed] [Google Scholar]
- 8.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson, R. H., X. J. Zhang, R. F. DuBose, and B. W. Matthews. 1994. Three-dimensional structure of beta-galactosidase from E. coli. Nature 369:761-766. [DOI] [PubMed] [Google Scholar]
- 10.Jonsson, G., I. C. Taban, K. B. Jørgensen, and R. C. Sundt. 2004. Quantitative determination of de-conjugated chrysene metabolites in fish bile by HPLC-fluorescence and GC-MS. Chemosphere 54:1085-1097. [DOI] [PubMed] [Google Scholar]
- 11.Joseph, S., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Judson, R., et al. 2009. The toxicity data landscape for environmental chemicals. Environ. Health Perspect. 117:685-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwak, H. I., et al. 2001. Effects of nonylphenol, bisphenol A, and their mixture on the viviparous swordtail fish (Xiphophotus helleri). Environ. Toxicol. Chem. 20:787-795. [PubMed] [Google Scholar]
- 14.Layton, A. C., B. W. Gregory, J. R. Seward, T. W. Schultz, and G. S. Sayler. 2000. Mineralization of steroidal hormones by biosolids in wastewater treatment systems in Tennessee, USA. Environ. Sci. Technol. 34:3925-3931. [Google Scholar]
- 15.Liang, R. B., et al. 2007. A T7-expression system under temperature control could create temperature-sensitive phenotype of target gene in Escherichia coli. J. Microbiol. Methods 68:497-506. [DOI] [PubMed] [Google Scholar]
- 16.Liang, R. B., X. P. Liu, D. L. Pei, and J. H. Liu. 2007. Biochemical characterization and functional complementation of ribonuclease HII and ribonuclease HIII from Chlamydophila pneumoniae AR39. Microbiology 153:787-793. [DOI] [PubMed] [Google Scholar]
- 17.Lorenzen, A., et al. 2004. Survey of hormone activities in municipal biosolids and animal manures. Environ. Toxicol. 19:216-225. [DOI] [PubMed] [Google Scholar]
- 18.Markey, C. M., B. S. Rubin, A. M. Soto, and C. Sonnenschein. 2002. Endocrine disruptors: from Wingspread to environmental developmental biology. J. Steroid Biochem. Mol. Biol. 83:235-244. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Noren, C. J., J. Wang, and F. B. Perler. 2000. Dissecting the chemistry of protein splicing and its applications. Angew. Chem. Int. Ed. Engl. 39:450-466. [PubMed] [Google Scholar]
- 21.O'Connor, J. C., et al. 2002. Evaluation of tier I screening approaches for detecting endocrine-active compounds (EACs). Crit. Rev. Toxicol. 32:521-549. [DOI] [PubMed] [Google Scholar]
- 22.Palanza, P., L. Gioiosa, F. S. vom Saal, and S. Parmigiani. 2008. Effects of developmental exposure to bisphenol A on brain and behavior in mice. Environ. Res. 108:150-157. [DOI] [PubMed] [Google Scholar]
- 23.Paulus, H. 2000. Protein splicing and related forms of protein autoprocessing. Annu. Rev. Biochem. 69:447-496. [DOI] [PubMed] [Google Scholar]
- 24.Perler, F. B. 2002. InBase, the intein database. Nucleic Acids Res. 30:383-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perler, F. B. 2006. Protein splicing mechanisms and applications. IUBMB Life 58:63. [DOI] [PubMed] [Google Scholar]
- 26.Petrovic, M., E. Eljarrat, M. J. Lopez De Alda, and D. Barceló. 2004. Endocrine disrupting compounds and other emerging contaminants in the environment: a survey on new monitoring strategies and occurrence data. Anal. Bioanal. Chem. 378:549-562. [DOI] [PubMed] [Google Scholar]
- 27.Raman, D. R., et al. 2004. Estrogens content of dairy waste and swine wastes. Environ. Sci. Technol. 38:3567-3573. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Mozaz, S., M. P. Marco, M. J. Lopez de Alda, and D. Barceló. 2004. Biosensors for environmental monitoring of endocrine disruptors: a review article. Anal. Bioanal Chem. 378:588-598. [DOI] [PubMed] [Google Scholar]
- 29.Routledge, E. J., and J. P. Sumpter. 1996. Estrogenic activity of surfactants and some of their degradation products assessed using a recombinant yeast screen. Environ. Toxicol. Chem. 15:241-248. [Google Scholar]
- 30.Safe, S. H., et al. 2002. Problems for risk assessment of endocrine-active estrogenic compounds. Environ. Health Perspect. 110:925-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saleh, L., and F. B. Perler. 2006. Protein splicing in cis and in trans. Chem. Rec. 6:183-193. [DOI] [PubMed] [Google Scholar]
- 32.Schultz, T. W. 2002. Estrogenicity of biphenylols: activity in the yeast gene activation assay. Bull. Environ. Contam. Toxicol. 68:332-338. [DOI] [PubMed] [Google Scholar]
- 33.Schultz, T. W., and G. D. Sinks. 2002. Xenoestrogenic gene expression: structural feature of active polycyclic aromatic hydrocarbons. Environ. Toxicol. Chem. 21:783-786. [PubMed] [Google Scholar]
- 34.Schultz, T. W., G. D. Sinks, and M. T. D. Cronin. 2002. Structure-activity relationships for gene activation oestrogenicity: evaluation of a diverse set of aromatic compounds. Environ. Toxicol. 17:14-23. [DOI] [PubMed] [Google Scholar]
- 35.Skretas, G., and D. W. Wood. 2005. Regulation of protein activity with small-molecule-controlled inteins. Protein Sci. 14:523-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soto, A. M., H. Justicia, J. W. Wray, and C. Sonnenschein. 1991. Para-nonyl-phenol—an estrogenic xenobiotic released from modified polystyrene. Environ. Health Perspect. 92:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner, R. L., et al. 1995. A structural role for hormone in the thyroid hormone receptor. Nature 378:690-697. [DOI] [PubMed] [Google Scholar]
- 38.Wogan, G. N., S. S. Hecht, J. S. Felton, A. H. Conney, and L. A. Loeb. 2004. Environmental and chemical carcinogenesis. Semin. Cancer Biol. 14:473-586. [DOI] [PubMed] [Google Scholar]
- 39.Wood, D. W., W. Wu, G. Belfort, V. Derbyshire, and M. Belfort. 1999. A genetic system yields self-cleaving inteins for bioseparations. Nat. Biotechnol. 17:889-892. [DOI] [PubMed] [Google Scholar]
- 40.Yan, L., et al. 2008. The determination of trace personal care products (pcps) in water environment. Environ. Chem. 27:382-384. [Google Scholar]
- 41.Yu, D., et al. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zacharewski, T. 1997. In vitro bioassays for assessing estrogenic substances. Environ. Sci. Technol. 31:613-623. [Google Scholar]
- 43.Zeidler, M. P., et al. 2004. Temperature-sensitive control of protein activity by conditionally splicing inteins. Nat. Biotechnol. 22:871-876. [DOI] [PubMed] [Google Scholar]
- 44.Ziccardi, M. H., I. A. Gardner, and M. S. Denison. 2002. Application of the luciferase recombinant cell culture bioassay system for the analysis of polycyclic aromatic hydrocarbons. Environ. Toxicol. Chem. 21:2027-2033. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.