Abstract
Marker removal strategies were developed for Thermoanaerobacterium saccharolyticum to select against the pyrF gene and the pta and ack genes. The pta- and ack-based haloacetate selective strategy was subsequently used to create strain M0355, a markerless Δldh Δpta Δack strain that produces ethanol at a high yield.
Low-cost biological conversion of lignocellulosic biomass would accelerate the emergence of a cellulosic biofuel industry (3, 6). Improvements to hydrolyzing enzymes and fermenting organisms are particularly attractive process investments, as they can result in improved yield and productivity without increasing capital or operating costs (7, 13, 14).
Currently, microbial catalysts are being developed to directly convert cellulose and xylan to biofuels (7). Thermoanaerobacter and Thermoanaerobacterium species are well suited for such development, owing to their ability to hydrolyze and ferment xylan and soluble cellodextrins (1, 2, 8, 12). Genetic tools, including plasmids (9), antibiotic markers (11), and a natural competence-based transformation system (10), have been described to manipulate these organisms; however, a system for genetic marker removal is essential for enactment of further modifications and creation of strains for industrial applications.
Strains and vectors.
The strains, plasmids, and PCR products used for transformation are listed in Table S1 in the supplemental material. The plasmids were constructed using standard techniques with Escherichia coli TOP10 (Invitrogen, Madison, WI), with the primers listed in Table S2. The linear DNA PCR products used to transform Thermoanaerobacterium saccharolyticum were constructed through an initial round of amplification followed by a second round of PCR-based fusion through homology regions designed into the first-round primer 5′ ends (5).
Transformation of T. saccharolyticum.
Transformation was performed via a natural competence protocol as described previously (10).
Selective conditions and media.
T. saccharolyticum was grown at 55°C, and manipulations were performed in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI). T. saccharolyticum was grown in modified DSMZ medium 122 (10), with selection-specific modifications. For kanamycin resistance selection, 200 μg/ml kanamycin sulfate was added and the pH was adjusted to 6.7. For 5-fluoroorotic acid (5-FOA) resistance selection, 5.7 mM 5-FOA (Zymo Research, Orange, CA) was added and the pH was adjusted to 5.0. For uracil autotrophy selection, an M122-defined medium with the yeast extract replaced with 2× RPMI 1640 vitamins (Sigma R7256) and 1× minimal essential medium (MEM) amino acids (Sigma M5550) was used, and the pH was adjusted to 6.7. For haloacetate resistance selection, the M122-defined medium was used with the addition of sodium chloroacetate (CA) at 0.2 mM or sodium fluoroacetate (FA) at 25 mM, and the pH was adjusted to 5.0. For selections with toxic analogues, no more than 2 × 106 cells per ml of solid media were added during the selection. All biochemicals were from Sigma-Aldrich (St. Louis, MO), and yeast extract was from BD Difco (Franklin Lakes, NJ).
Fermentation conditions and media.
For batch cultivation in 1-liter-working-volume bioreactors (Sartorius-Stedim, Bohemia, NY), TSC1 medium was used. This medium contained, per liter, the following: 50 g cellobiose, 2.0 g sodium citrate tribasic dihydrate, 1.9 g (NH4)2SO4, 0.1 g FeSO4·7H2O, 1.0 g MgSO4, 1.0 g KH2PO4, 0.1 gCaCl2·2H2O, and 8.5 g yeast extract. The pH was maintained at 5.8 with the addition of 2.5 M NH4OH, temperature was maintained at 55°C, agitation was performed at 150 rpm, and an initial N2 purge was performed to remove air from the bioreactor. Fermentation metabolite concentrations were determined via high-performance liquid chromatography (HPLC) using an Aminex HPX-87H column (Bio-Rad Laboratories, Hercules, CA).
To create a removable marker, selection conditions that would link growth to the removal of a gene via the T. saccharolyticum homologous recombination machinery were developed.
Deletion of pyrF.
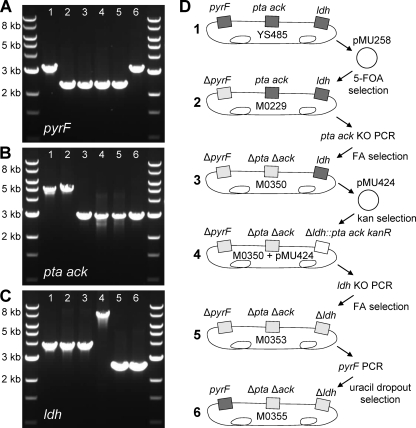
Orotidine-5′-phosphate decarboxylase (pyrF), a key step in pyrimidine biosynthesis, has been used for marker cycling systems for many microbial hosts. The toxic analogue 5-fluoroorotic acid (5-FOA) was tested with T. saccharolyticum for selective resistance after transformation with pMU258, a plasmid containing fused flanking regions of pyrF. Many 5-FOA-resistant colonies were screened by colony PCR, and a few were found to have DNA bands at the predicted size of a ΔpyrF locus. One was further isolated, confirmed by sequencing, and designated M0229 (Fig. 1).
FIG. 1.
Colony PCR analysis of the pyrF (A), pta ack (B), and ldh (C) loci during construction of the markerless ethanologen strain. (D) Diagram showing genomic modifications, transforming vectors, and selection conditions. Note that the pyrF deletion is not essential for creation of the markerless ethanologen strain M0355. PCR was performed with primers external to areas of homologous recombination. For each panel: lane 1, wild-type YS485; lane 2, M0229 (ΔpyrF); lane 3, M0350 (ΔpyrF Δpta Δack); lane 4, M0350(pMU424) (ΔpyrF Δpta Δack Δldh::pta ack Kanr); lane 5, M0353 (ΔpyrF Δpta Δack Δldh); lane 6, M0355 (Δpta Δack Δldh). The DNA size marker is a 1-kb New England BioLabs DNA ladder.
Deletion of pta and ack.
Deletion of the phosphotransacetylase (pta) and acetate kinase (ack) genes was also tested as a marker removal strategy. Strain ALK2 (11), with a Δpta Δack::Kan locus, was tested in parallel with wild-type T. saccharolyticum for Δpta Δack-dependent resistance to haloacetate compounds. Initial screening in a rich medium yielded a high rate of haloacetate resistance not specific to the absence of pta and ack. Subsequently, M122-defined medium at pH 5.0 was used as a background for haloacetate selection; under these conditions, fluoroacetate (FA) at 25 to 50 mM and chloroacetate (CA) at 0.1 to 0.2 mM created an environment where growth depended on the absence of the pta and ack genes, and spontaneous haloacetate-resistant cells occurred at a frequency of less than 10−7 per CFU from the wild-type population. We preferred the use of CA to that of FA, as CA yielded comparable results with T. saccharolyticum and is less toxic to human health than FA (4). The ΔpyrF strain M0229 was arbitrarily chosen for subsequent generation of a markerless Δpta Δack locus via transformation with 3 μg of the pta ack knockout (KO) PCR product and selection for CA resistance. One ΔpyrF Δpta Δack colony was reisolated on regular medium, confirmed again by colony PCR, and designated strain M0350.
Marker removal system and construction of a marker-free ethanologen.
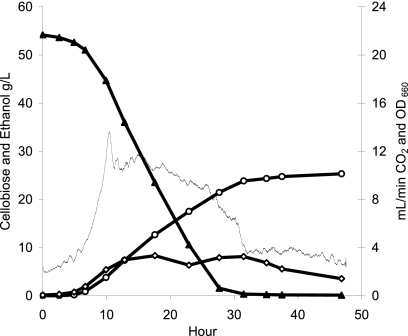
Strain M0350 was transformed with pMU424, which has the pta, ack, and kanamycin resistance genes flanked by DNA fragments homologous to the T. saccharolyticum ldh gene. Next, the ldh KO PCR product was transformed with CA selection to create strain M0353 (ΔpyrF Δpta Δack Δldh), and finally, strain M0353 was transformed with the pyrF PCR product and selected on M122 defined medium to create strain M0355 (Δpta Δack Δldh). The generation of deletions and insertions was tracked by PCR with primers external to the sites of homologous recombination, as shown in Fig. 1, and the presence of organic acid pathway genes could also be followed by end product formation, as shown in Table 1. Strain M0355 ferments 50 g/liter of cellobiose (Fig. 2), with a yield of 0.44 ± 0.00 g ethanol per g glucose equivalent substrate and a maximum volumetric productivity of 1.13 ± 0.12 g ethanol liter−1 h−1, as determined from duplicate runs. The ability to make markerless deletions will allow for more-ambitious genetic engineering projects with this organism, and strain M0355 should be a useful starting point for the creation and study of biocatalysts for the conversion of lignocellulosic biomass.
TABLE 1.
Twenty-four-hour fermentation profiles of strains leading to the markerless ethanologena
| Strain | Concn (g/liter) |
|||
|---|---|---|---|---|
| Cellobiose | Lactic acid | Acetic acid | Ethanol | |
| Wild type | 0.0 | 0.8 | 1.0 | 1.4 |
| M0350 | 2.5 | 0.8 | 0.1 | 0.6 |
| M0350(pMU424) | 0.0 | 0.0 | 0.9 | 1.7 |
| M0353 | 2.1 | 0.0 | 0.1 | 1.0 |
| M0355 | 2.0 | 0.0 | 0.0 | 0.8 |
The starting cellobiose concentration was 4.5 g/liter.
FIG. 2.
Representative 1-liter-batch fermentation with strain M0355. Cellobiose (▴), ethanol (○), cell mass (⋄), and CO2 (solid line).
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database under accession no. HM802207 (pyrF region), HM802208 (pta ack region), and HM802209 (ldh region). pMU258 and pMU424 sequences are included in Table S3 in the supplemental material.
Supplementary Material
Acknowledgments
This work was supported by the Mascoma Corporation and the U.S. Department of Energy under award no. DE-FC36-07G017057.
The growth medium TSC1 was developed by Haowen Xu.
This report was prepared as an account of work sponsored by an agency of the U.S. Government. Neither the U.S. Government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by trade name, trademark or manufacturer or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the U.S. Government or any agency thereof. The views and opinions of authors expressed herein do not necessarily state or reflect those of the U.S. Government or any agency thereof.
Footnotes
Published ahead of print on 11 February 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Breves, R., et al. 1997. Genes encoding two different β-glucosidases of Thermoanaerobacter brockii are clustered in a common operon. Appl. Environ. Microbiol. 63:3902-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demain, A. L., M. Newcomb, and J. H. Wu. 2005. Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69:124-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greene, N., et al. 2004. Growing energy: how biofuels can help end America's oil dependence. Report. Natural Resources Defense Council, New York, NY.
- 4.Hayes, F. D., R. D. Short, and J. E. Gibson. 1973. Differential toxicity of monochloroacetate, monofluoroacetate and monoiodoacetate in rats. Toxicol. Appl. Pharmacol. 26:93-102. [DOI] [PubMed] [Google Scholar]
- 5.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 6.Lynd, L. R., et al. 2008. How biotech can transform biofuels. Nat. Biotechnol. 26:169-172. [DOI] [PubMed] [Google Scholar]
- 7.Lynd, L. R., W. H. van Zyl, J. E. McBride, and M. Laser. 2005. Consolidated bioprocessing of cellulosic biomass: an update. Curr. Opin. Biotechnol. 16:577-583. [DOI] [PubMed] [Google Scholar]
- 8.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mai, V., and J. Wiegel. 2000. Advances in development of a genetic system for Thermoanaerobacterium spp.: expression of genes encoding hydrolytic enzymes, development of a second shuttle vector, and integration of genes into the chromosome. Appl. Environ. Microbiol. 66:4817-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw, A. J., D. A. Hogsett, and L. R. Lynd. 2010. Natural competence in Thermoanaerobacter and Thermoanaerobacterium species. Appl. Environ. Microbiol. 76:4713-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw, A. J., et al. 2008. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc. Natl. Acad. Sci. U. S. A. 105:13769-13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiegel, J., C. P. Mothershed, and J. Puls. 1985. Differences in xylan degradation by various noncellulolytic thermophilic anaerobes and Clostridium thermocellum. Appl. Environ. Microbiol. 49:656-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wooley, R., M. Ruth, D. Glassner, and J. Sheehan. 1999. Process design and costing of bioethanol technology: a tool for determining the status and direction of research and development. Biotechnol. Prog. 15:794-803. [DOI] [PubMed] [Google Scholar]
- 14.Wyman, C. E. 2007. What is (and is not) vital to advancing cellulosic ethanol. Trends Biotechnol. 25:153-157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.