Abstract
Incompatibility group P1 (IncP-1) plasmid diversity was evaluated based on replication initiator protein (TrfA) phylogeny. A new and highly divergent clade was identified. Replication assays indicated that TrfA of recently discovered IncP-1 plasmids from Xylella fastidiosa and Verminephrobacter eiseniae initiated plasmid replication using cognate or heterologous origins of replication.
Incompatibility group P-1 (IncP-1) plasmids encode backbone modules for replication, stable inheritance, and conjugation plus DNA transfer, as well as accessory modules conferring environmental adaptations (5). Five IncP-1 subgroups (α, β, γ, δ, and ɛ) have been described previously (2, 8, 17, 20). Recently, an IncP-1 plasmid (pXF-RIV11; GenBank accession no. GU938457) from the plant-pathogenic bacterium Xylella fastidiosa was characterized (22) and shown to be related to pVEIS01 (GenBank accession no. CP000543) from the earthworm symbiont Verminephrobacter eiseniae (16). Neither has been assigned to a subgroup, as the gene complements and organizations of pXF-RIV11 and pVEIS01 are sufficiently different from those of other IncP-1 plasmids.
The IncP-1 replication module consists of trfA, encoding a replication initiator protein (TrfA), and the origin of replication (oriV) (1, 15, 21). As TrfA is the only plasmid-encoded protein required for replication, all IncP-1 plasmids bear trfA. Thus, divergence among TrfA homologues may be informative with respect to evolutionary history and subgroup classification (2, 8). Here, we examine TrfA phylogeny to determine the relationships of pXF-RIV11 and pVEIS01 with IncP-1 plasmids from a wide variety of bacteria.
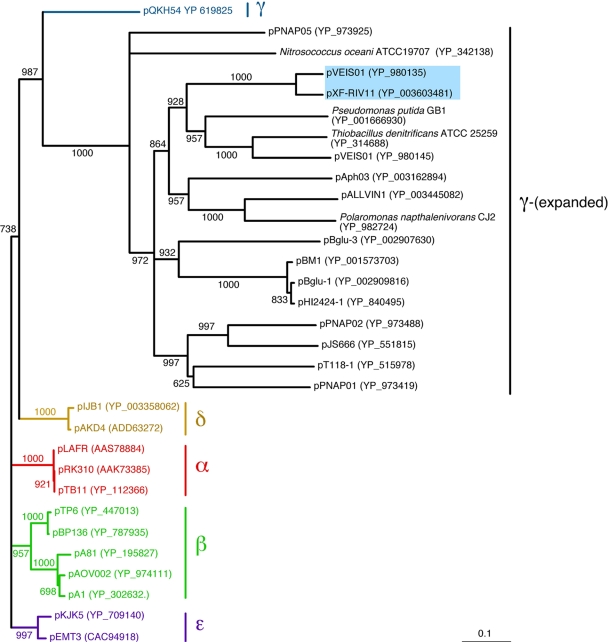
The pXF-RIV11 TrfA sequence was used as a query in BLAST searches of the GenBank protein database. Subjects returned (Table 1) included TrfA from established members of the five subgroups and numerous homologues not assigned to a subgroup. TrfA amino acid sequences were aligned, and neighbor-joining analysis was performed using Clustal X (10). Eighteen unclassified TrfA sequences (including pXF-RIV11 and pVEIS01) formed a clade sharing a most recent common ancestor with TrfA of pQKH54, the archetype and sole recognized member of subgroup γ (7), in what is referred to here as the γ-(expanded) subgroup (Fig. 1). Whereas the diversity of TrfA within subgroups α, β, δ, and ɛ was limited, genetic distances among homologues clustering with TrfA of pQKH54 were substantially greater, as indicated by branch lengths (Fig. 1).
TABLE 1.
IncP-1 plasmid subgroupings based on TrfA phylogeny
| Subgroup and plasmid (size in bp) | Host species (strain) | TrfA protein accession no. |
|---|---|---|
| α | ||
| pRK2 derivative pRK310a (19,041) | Pseudomonas sp.b | AAK73385 |
| pTB11 (68,869) | Uncultured bacterium | YP_112366 |
| pRK2 derivative pLAFRa (20,352) | Pseudomonas sp.b | AAS78884 |
| β | ||
| pTP6 (54,344) | Uncultured bacterium | YP_447013 |
| pA1 (46,557) | Sphingomonas sp. (A1) | YP_302632 |
| pA81 (98,192) | Achromobacter xylosoxidans (A8) | YP_195827 |
| pBP136 (41,268) | Bordetella pertussis (clinical isolate) | YP_787935 |
| pAOV002 (63,609) | Acidovorax sp. (JS42) | YP_974111 |
| γ | ||
| pQKH54 (69,966) | Epilithic bacterium | YP_619825 |
| δ | ||
| pIJB1 (99,448) | Burkholderia cepacia (2a) | YP_003358062 |
| pAKD4 (56,803) | Uncultured bacterium | ADD63272 |
| ɛ | ||
| pKJK5 (54,383) | Uncultured bacterium | YP_709140 |
| pEMT3 (unknown) | Uncultured bacterium | CAC9491 |
| γ-(expanded) | ||
| pXF-RIV11 (25,105) | Xylella fastidiosa (Riv11) | YP_003603481 |
| pVEIS01 (31,194) | Verminephrobacter eiseniae (EF01-2) | YP_980135 |
| pVEIS01 (31,194) | Verminephrobacter eiseniae (EF01-2) | YP_980145 |
| NAc | Thiobacillus denitrificans (ATCC 25259) | YP_314688 |
| pALLVIN1 (102,242) | Allochromatium vinosum (DSM180; ATCC 17899) | YP_003445082 |
| NA | Polaromonas naphthalenivorans (CJ2) | YP_982724 |
| pPNAP02 (190,172) | Polaromonas naphthalenivorans (CJ2) | YP_973488 |
| pPNAP01 (353,291) | Polaromonas naphthalenivorans (CJ2) | YP_973419 |
| pPNAP05 (58,808) | Polaromonas naphthalenivorans (CJ2) | YP_973925 |
| NA | Pseudomonas putida (GB1) | YP_001666930 |
| pAph03 (37,695) | “Candidatus Accumulibacter phosphatis” (clade IIA, UW-1) | YP_003162894 |
| pBglu-3 (141,067) | Burkholderia glumae(BGR1) | YP_002907630 |
| pJS666 (360,405) | Polaromonas sp. (JS666) | YP_551815 |
| pT118-1 (257,447) | Rhodoferax ferrireducens (T118) | YP_515978 |
| pBM1 (167,422) | Burkholderia multivorans (ATCC 17616) | YP_001573703 |
| pBglu-1 (133,591) | Burkholderia glumae (BGR1) | YP_002909816 |
| pHI2424-1 (164,857) | Burkholderia cenocepacia (HI2424) | YP_840495 |
| NA | Nitrosococcus oceani (ATCC 19707) | YP_342138 |
Cloning vector with trfA derived from pRK2, originally isolated from Pseudomonas sp.
Host of pRK2 parent plasmid.
NA, not applicable; integrated in host chromosome.
FIG. 1.
Phylogeny of IncP-1 TrfA homologues. Presented is a neighbor joining tree (1,000 bootstrap iterations) based on alignment of TrfA amino acid sequences. Taxa are indicated at branch tips by the IncP-1 plasmid name (or bacterial species name if trfA was integrated into the host chromosome) followed by the GenBank protein accession number in parentheses. Branches and taxa previously assigned to IncP-1 subgroups α, β, γ, δ, and ɛ are color coded. Eighteen taxa included in subgroup γ-(expanded) are designated on the right. Numbers along branches indicate bootstrap support of distal node; nodes with >60% bootstrap support were collapsed to polytomies; the bar at lower right corresponds to a genetic distance of 0.1. The colored box denotes taxa used to construct minimal replicons bearing cognate or heterologous IncP-1 replication module elements.
Bahl et al. (2) suggested that the known diversity of IncP-1 plasmids could be skewed by discovery methods, especially those based on accessory module phenotype (11, 24). Metagenomic analyses and mating trapping strategies have yielded IncP-1 sequences from environmental samples (2, 3, 9, 18, 19, 23). However, trapping strategies may be biased, as assays requiring plasmid mobilization limit discovery to those able to propagate in experimental hosts. Metagenomic methods lack phenotypic biases described above, as exemplified by PCR amplification of trfA sequences representing all five subgroups from total community DNA isolated from wastewater (2). Nonetheless, PCR-based methods are biased due to primer design (26). To illustrate this last point, all three primer sets employed by Bahl et al. (2) shared limited sequence in common with pXF-RIV11 trfA such that sequences of γ-(expanded) subgroup plasmids would not have been amplified.
It is remarkable that 17 of 18 TrfA homologues belonging to subgroup γ-(expanded) were discovered by genome sequencing projects (4, 13, 16, 25). The 18th (pXF-RIV11) was discovered by direct extraction from cultured bacteria without selection for phenotype (22). As discovery by genome sequencing projects is not based on the specific genotype/phenotype of resident plasmids associated with a bacterial genome, this newly recognized diversity of TrfA suggests that mating trapping strategies and metagenomic surveys were biased against discovery of subgroup γ-(expanded) plasmids.
Genes for four TrfA homologues are integrated into chromosomes of their respective hosts (Table 1). This observation, coupled with the lack of functional analyses for most plasmid-borne TrfA homologues of subgroup γ-(expanded), raises the question as to whether these divergent homologues initiate plasmid replication. To partially address this question, plasmids containing the minimal IncP-1 replication module of pXF-RIV11 or pVEIS01 inserted into the Escherichia coli cloning vector pCR2.1 were constructed (12). Replication in E. coli was driven by the pCR2.1 ori. Replication in X. fastidiosa strain Temecula1 was driven by inserted IncP-1 replication modules; both trfA and oriV were required.
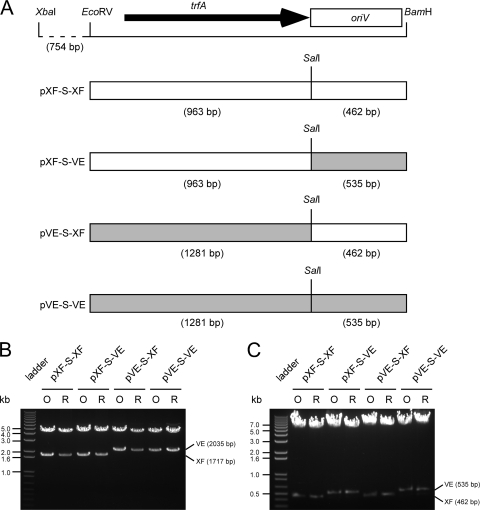
As TrfA homologues encoded by pXF-RIV11 (YP_003603482) and pVEIS01 (YP_980135) share 87% amino acid identity, the two replication initiator proteins may be functionally interchangeable. To test this hypothesis, plasmids containing cognate or heterologous combinations of trfA and oriV were constructed and tested for replication in X. fastidiosa (Fig. 2). Constructs bearing cognate replication elements of pXF-RIV11 (pXF-S-XF) or pVEIS01 (pVE-S-VE) contained an inserted SalI site (to facilitate replication element exchange) between the trfA stop codon and cognate oriV. Constructs bearing heterologous replication elements contained trfA from pXF-RIV11 and oriV from pVEIS01 (pXF-S-VE) or trfA from pVEIS01 and oriV from pXF-RIV11 (pVE-S-XF). The pemI/pemK addiction system of pXF-RIV11 (12) was present on all constructs to confer stable inheritance in X. fastidiosa.
FIG. 2.
Construction and replication of plasmids bearing cognate or heterologous IncP-1 replication elements. (A) Locations and sources of IncP-1 replication module elements (trfA and oriV). Plasmid names are indicated on the left. Regions derived from pXF-RIV11 are indicated as white boxes; regions derived from pVEIS01 are indicated as gray boxes. Locations of relevant endonuclease restriction sites are indicated; distances in base pairs separating endonuclease restriction sites are indicated parenthetically. (B and C) Restriction endonuclease profiles of plasmids bearing cognate (pXF-S-XF and pVE-S-VE) or heterologous (pXF-S-VE and pVE-S-XF) IncP-1 replication module elements and used to transform Xylella fastidiosa (O) or rescued from X. fastidiosa by transformation of Escherichia coli (R). Note that length polymorphism of fragments bearing trfA (B) or oriV (C) verifies the source (XF = pXF-RIV11; VE = pVEIS01) of the respective IncP-1 replication module elements.
Plasmids purified from E. coli JM109 were used to transform X. fastidiosa strain Temecula1 by electroporation (14). Plasmid pUCLAa (6) was used as a positive control; no DNA was used as a negative control. X. fastidiosa transformants were selected on PD3 medium containing 5 μg/ml kanamycin (22). Transformants (four per construct) were picked and grown for 10 to 14 days in liquid PD3 medium containing 5 μg/ml kanamycin. Plasmid DNA extracted from subcultured X. fastidiosa transformants (22) was used to transform E. coli.
Heterologous combinations of replication elements derived from pXF-RIV11 and pVEIS01 were competent for replication in X. fastidiosa (Fig. 2). Samples of the original plasmids used to transform X. fastidiosa and samples of plasmids rescued from X. fastidiosa by transformation of E. coli were digested with restriction enzymes to generate fragments that differed in size, depending upon whether IncP-1 replication elements were derived from pXF-RIV11 or from pVEIS01. These results indicated that specificity determinants of TrfA for oriV recognition were conserved and that consensus sequence differences (22) in oriV-iterated elements of pXF-RIV11 (TTACCGTCGCAGCATCCT) and pVEIS01 (TTACCGTCGTAGCATCCGC) did not prevent recognition by the heterologous TrfA. Modified plasmids bearing cognate combinations of trfA and oriV (pXF-S-XF and pVE-S-VE) replicated in X. fastidiosa (Fig. 2), demonstrating that alteration of spacing (SalI site insertion) between replication elements was tolerated. Although trfA and oriV are adjacent to one another in both pXF-RIV11 and pVEIS01, some IncP-1 plasmids have accessory modules inserted at this locus (5). These observations suggest that this same locus in pXF-RIV11, pVEIS01, and shuttle vector derivatives may be used to insert foreign sequences.
Acknowledgments
We thank Keira Neumann, Stephanie Underwood, and Kunbo Zhang for technical assistance.
Mention of proprietary or brand names is necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by USDA implies no approval to the exclusion of others that also may be suitable.
Footnotes
Published ahead of print on 4 February 2011.
REFERENCES
- 1.Ayres, E. K., V. J. Thomson, G. Merino, D. Balderes, and D. H. Figurski. 1993. Precise deletions in large bacterial genomes by vector-mediated excision (VEX). The trfA gene of promiscuous plasmid RK2 is essential for replication in several gram-negative hosts. J. Mol. Biol. 230:174-185. [DOI] [PubMed] [Google Scholar]
- 2.Bahl, M. I., M. Burmølle, A. Meisner, L. H. Hansen, and S. J. Sørensen. 2009. All incP-1 plasmid subgroups, including the novel ɛ subgroup, are prevalent in the influent of a Danish wastewater treatment plant. Plasmid 62:134-139. [DOI] [PubMed] [Google Scholar]
- 3.Bahl, M. I., L. H. Hansen, A. Goesmann, and S. J. Sørensen. 2007. The multiple antibiotic resistance incP-1 plasmid pKJK5 isolated from a soil environment is phylogenetically divergent from members of previously established α, β, and δ sub-groups. Plasmid 58:31-43. [DOI] [PubMed] [Google Scholar]
- 4.Beller, H. R., et al. 2006. The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans. J. Bacteriol. 188:1473-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dennis, J. J. 2005. The evolution of IncP catabolic plasmids. Curr. Opin. Biotechnol. 16:291-298. [DOI] [PubMed] [Google Scholar]
- 6.Guilhabert, M. R., V. J. Stewart, and B. C. Kirkpatrick. 2006. Characterization of putative rolling-circle plasmids from the Gram-negative bacterium Xylella fastidiosa and their use as shuttle vectors. Plasmid 55:70-80. [DOI] [PubMed] [Google Scholar]
- 7.Haines, A. S., et al. 2006. Plasmids from freshwater environments capable of incQ retrotransfer are diverse and include pQKH54, a new IncP-1 subgroup archetype. Microbiology 152:2689-2701. [DOI] [PubMed] [Google Scholar]
- 8.Harada, K. M., Y. Aso, W. Hashimoto, B. Mikami, and K. Murata. 2006. Sequence and analysis of the 46.6-kb plasmid pA1 from Sphingomonas sp. A1 that corresponds to the typical Inc-P1β plasmid backbone without any accessory gene. Plasmid 56:11-23. [DOI] [PubMed] [Google Scholar]
- 9.Hill, K. E., A. J. Weightman, and J. C. Fry. 1992. Isolation and screening of plasmids from the epilithon which mobilize recombinant plasmid pD10. Appl. Environ. Microbiol. 58:1292-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 11.Jencova, V., et al. 2004. Chlorocatechol catabolic enzymes from Achromobacter xylosoxidans A8. Int. Biodeterior. Biodegradation 54:175-181. [Google Scholar]
- 12.Lee, M. W., E. E. Rogers, and D. C. Stenger. 2010. Functional characterization of replication and stability factors of an incompatibility group P-1 plasmid from Xylella fastidiosa. Appl. Environ. Microbiol. 76:7734-7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim, J. Y., et al. 2009. Complete genome sequence of Burkholderia glumae BGR1. J. Bacteriol. 191:3758-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumoto, A., G. M. Young, and M. M. Igo. 2009. Chromosome-based genetic complementation system for Xylella fastidiosa. Appl. Environ. Microbiol. 75:1679-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mei, J., S. Nenashski, and W. Firshein. 1995. Interactions of the origin of replication (oriV) and initiation proteins (TrfA) of plasmid RK2 with submembrane domains of Escherichia coli. J. Bacteriol. 177:6766-6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinel, N., S. K. Davidson, and D. A. Stahl. 2008. Verminephrobacter eiseniae gen. nov., sp. nov., a nephridial symbiont of the earthworm Eisenia foetida (Savigny). Int. J. Syst. Evol. Microbiol. 58:2147-2157. [DOI] [PubMed] [Google Scholar]
- 17.Schlüter, A., R. Szczepanowski, A. Pühler, and E. M. Top. 2007. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 31:449-477. [DOI] [PubMed] [Google Scholar]
- 18.Sen, D., et al. 2010. Comparative genomics of pAKD4, the prototype IncP-1δ plasmid with a complete backbone. Plasmid 63:98-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smalla, K., et al. 2006. Increased abundance of incP-1β plasmids and mercury resistance genes in mercury-polluted river sediments: first discovery of incP-1β plasmids with a complex mer transposon as the sole accessory element. Appl. Environ. Microbiol. 72:7253-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, C. A., and C. M. Thomas. 1987. Comparison of the organization of the genomes of phenotypically diverse plasmids of incompatibility group P: members of the IncPβ subgroup are closely related. Mol. Gen. Genet. 206:419-427. [DOI] [PubMed] [Google Scholar]
- 21.Stalker, D. M., C. M. Thomas, and D. R. Helinski. 1981. Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol. Gen. Genet. 181:8-12. [DOI] [PubMed] [Google Scholar]
- 22.Stenger, D. C., M. W. Lee, E. E. Rogers, and J. Chen. 2010. Plasmids of Xylella fastidiosa mulberry-infecting strains share extensive sequence identity and gene complement with pVEIS01 from the earthworm symbiont Verminephrobacter eiseniae. Physiol. Mol. Plant Pathol. 74:238-245. [Google Scholar]
- 23.Tennstedt, T., R. Szczepanowski, I. Krahn, A. Pühler, and A. Schlüter. 2005. Sequence of the 68,869 bp incP-1α plasmid pTB11 from a waste-water treatment plant reveals a highly conserved backbone, a Tn402-like integron and other transposable elements. Plasmid 53:218-238. [DOI] [PubMed] [Google Scholar]
- 24.Xia, X.-S., S. Aathithan, K. Oswiecimska, A. R. W. Smith, and I. J. Bruce. 1998. A novel plasmid pIJB1 possessing a putative 2,4-dichlorophenoxyacetate degradative transposon Tn5530 in Burkholderia cepacia strain 2a. Plasmid 39:154-159. [DOI] [PubMed] [Google Scholar]
- 25.Yagi, J. M., D. Sims, T. Brettin, D. Bruce, and E. L. Madsen. 2009. The genome of Polaromonas naphthalenivorans strain CJ2, isolated from coal tar-contaminated sediment, reveals physiological and metabolic versatility and evolution through extensive horizontal gene transfer. Environ. Microbiol. 11:2253-2270. [DOI] [PubMed] [Google Scholar]
- 26.Zheng, L., et al. 2008. Accumulating variation at conserved sites in potyvirus genomes is driven by species discovery and affects degenerate primer design. PLoS One 3:e1586. [DOI] [PMC free article] [PubMed] [Google Scholar]