Abstract
The Saccharomyces cerevisiae SNF1 protein kinase, a member of the SNF1/AMP-activated protein kinase (AMPK) family, is activated by three kinases, Sak1, Tos3, and Elm1, which phosphorylate the Snf1 catalytic subunit on Thr-210 in response to glucose limitation and other stresses. Sak1 is the primary Snf1-activating kinase and is associated with Snf1 in a complex. Here we examine the interaction of Sak1 with SNF1. We report that Sak1 coimmunopurifies with the Snf1 catalytic subunit from extracts of both glucose-replete and glucose-limited cultures and that interaction occurs independently of the phosphorylation state of Snf1 Thr-210, Snf1 catalytic activity, and other SNF1 subunits. Sak1 interacts with the Snf1 kinase domain, and nonconserved sequences C terminal to the Sak1 kinase domain mediate interaction with Snf1 and augment the phosphorylation and activation of Snf1. The Sak1 C terminus is modified in response to glucose depletion, dependent on SNF1 activity. Replacement of the C terminus of Elm1 (or Tos3) with that of Sak1 enhanced the ability of the Elm1 kinase domain to interact with and phosphorylate Snf1. These findings indicate that the C terminus of Sak1 confers its function as the primary Snf1-activating kinase and suggest that the physical association of Sak1 with SNF1 facilitates responses to environmental change.
INTRODUCTION
The SNF1/AMP-activated protein kinase (AMPK) family is important in metabolic control. In mammals, AMPK coordinates energy homeostasis, regulating lipid and glucose metabolism, and is activated by increases in AMP during metabolic stress, by hormones, and by drugs used in the treatment of type 2 diabetes (for a review, see references 11 and 21). In the yeast Saccharomyces cerevisiae, the SNF1 protein kinase is activated in response to glucose depletion and other environmental stresses (for a review, see reference 5). SNF1 regulates the transcription of many genes and the activity of metabolic enzymes and is involved in the utilization of alternate carbon sources, meiosis and sporulation, aging, and invasive growth.
SNF1 is heterotrimeric, as is AMPK. The kinase comprises the Snf1 (α) catalytic subunit, one of three β subunit isoforms (Gal83, Sip1, or Sip2), and the Snf4 (γ) subunit. Activation requires phosphorylation of conserved Thr-210 on the activation loop of Snf1 by any of three Snf1-activating kinases, Sak1, Tos3, and Elm1 (8, 16, 22). These kinases exhibit similarity within their catalytic domains, but their C-terminal regions are not conserved, and deletion of these regions affects Snf1-dependent growth phenotypes and invertase expression (17). Biochemical studies of these kinases purified from cells grown under different conditions did not reveal differences in their activities (18). SNF1 is negatively regulated by the Reg1-Glc7 protein phosphatase 1 (PP1), which is targeted to Snf1 by the Reg1 subunit and affects phosphorylation of Thr-210 (9, 12, 14, 23). It has been proposed that access of PP1 to Thr-210 is regulated (18). In addition, mutations that alter the Snf4 and β subunits of SNF1 relieve glucose inhibition of Thr-210 phosphorylation (15).
The three Snf1-activating kinases have overlapping functions in regulating SNF1, and all three cognate genes must be deleted to confer a substantial growth defect on nonpreferred carbon sources (8, 22). However, under all growth and environmental stress conditions tested, Sak1 was the major kinase responsible for phosphorylation and activation of Snf1, as judged by analysis of kinase-deficient mutants (6–8, 13). In addition, Sak1, but not the other two kinases, controls the nuclear enrichment of SNF1 containing the Gal83 β subunit in response to carbon stress, which requires the phosphorylation of Thr-210 (6). In a previous study, of the three kinases, only Sak1 copurified with Snf1 in a stable complex; most of the Sak1 protein was associated with Snf1, but only a fraction of the total Snf1 protein was in a complex with Sak1 (3). In that study, Elbing and colleagues obtained comparable levels of copurification from both glucose- and sucrose-grown cultures (3); although using different procedures, Nath et al. found increased association under glucose-limiting conditions (16).
These findings suggest that the physical association of Sak1 with SNF1 is responsible for its key functional role. Here we have examined their interaction under different conditions, determined the regions of each kinase that interact, and assessed the requirements for Snf1 Thr-210, Snf1 catalytic activity, and the other SNF1 subunits. We have further explored the role of sequences C terminal to the Sak1 kinase domain by replacing the sequence distal to the Elm1 and Tos3 kinase domains with the Sak1 sequence. Our findings support the view that the C terminus of Sak1 is responsible for its function as the primary Snf1-activating kinase. We also report SNF1-dependent modification of Sak1 and weak interaction of Sak1 with PP1.
MATERIALS AND METHODS
Strains and plasmids.
The S. cerevisiae strains used in this work are listed in Table 1. Plasmids are listed in Table 2. Standard genetic methods were used.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype |
|---|---|
| W303-1A | MATaade2 can1 his3 leu2 trp1 ura3 |
| W303-1B | MATα ade2 can1 his3 leu2 trp1 ura3 |
| MCY4908 | W303-1A snf1Δ10 |
| YL1045 | W303-1B REG1-8×myc::TRP1 |
| YL1112 | W303-1B SNF1-8×myc::TRP1 reg1Δ::HIS3 sak1Δ::KanMX4 |
| YL1140 | W303-1B sak1Δ::KanMX4 SNF1-8×myc::TRP1 |
| YL1157 | W303-1A SNF1-3×HA::URA3 |
| YL1161 | W303-1B sak1Δ::KanMX4 REG1-8×myc::TRP1 |
| YL1163 | W303-1B sak1Δ::KanMX4 REG1-8×myc::TRP1 snf1Δ::LEU2 |
| YL1167 | W303-1B SNF1-8×myc::URA3 gal83Δ::TRP1 sip1Δ::KanMX6 sipΔ2::KanMX4 snf4Δ::hphMX4 |
| YL1176 | W303-1A snf1Δ10 reg1(1-400)-myc::TRP1 |
| YL1179 | W303-1A snf1Δ10 reg1(1-740)-myc::TRP1 |
| YL1180 | W303-1A snf1Δ10 REG1-myc::TRP1 |
| YL1181 | W303-1A SNF1-3×HA |
| YL1185 | W303-1B SNF1-8×myc::TRP1 sak1Δ::KanMX4 tos3Δ::KanMX4 elm1Δ::ADE2 |
| YL1213 | W303-1B sak1Δ::KanMX4 tos3Δ::KanMX4 elm1Δ::URA3 snf1Δ::LEU2 |
Table 2.
Plasmids used in this study
| Plasmid | Vector | Promoter | Expressed protein | Reference or source |
|---|---|---|---|---|
| pWS93 | ADH1 | 3×HA | 20 | |
| pSB16 | pWS93 | ADH1 | 3×HA-Reg1 | 19 |
| pRH104 | pWS93 | ADH1 | 3×HA-Sak1 | 6 |
| pRJ65 | pEG202 | ADH1 | LexA-Reg1 | 23 |
| pRS316-Sak1-5V5 | pRS316 | SAK1 | Sak1-5×V5 | 17 |
| pRS314-Sak1-5V5 | pRS314 | SAK1 | Sak1-5×V5 | M. Schmidt |
| pYL199 | pRS313 | SNF1 | Snf1-8×myc | This study |
| pYL201 | pRS313 | SNF1 | Snf1-3×HA | This study |
| pYL228 | pRS313 | SNF1 | Snf1T210A-3×HA | This study |
| pYL229 | pRS313 | SNF1 | Snf1K84R-3×HA | This study |
| pYL241 | pRS315 | ADH1 | 3×HA | This study |
| pYL306 | pRS315 | ADH1 | 3×HA-Sak1(1-519), HA-Sak1KD | This study |
| pYL307 | pRS315 | ADH1 | 3×HA-Sak1(501-1142) | This study |
| pYL308 | pRS315 | ADH1 | 3×HA-Sak1 | This study |
| pYL322 | pRS316 | SAK1 | Sak1Δ(501-740)-5×V5 | This study |
| pYL348 | pRS316 | SAK1 | Sak1(1-1130)-5×V5 | This study |
| pYL352 | pRS313 | SNF1 | Snf1(1-356)-8×myc | This study |
| pYL359 | pRS313 | SNF1 | Snf1(301-633)-8×myc | This study |
| pYL360 | pRS313 | SNF1 | Snf1(488-633)-8×myc | This study |
| pYL362 | pRS315 | ADH1 | 3×HA-Sak1(741-1130) | This study |
| pYL386 | pRS316 | ELM1 | Elm1-5×V5 | This study |
| pYL387 | pRS316 | ELM1 | Elm1(1-420)-5×V5, Elm1KD-V5 | This study |
| pYL388 | pRS316 | TOS3 | Tos3-5×V5 | This study |
| pYL389 | pRS316 | TOS3 | Tos3(1-350)-5×V5, Tos3KD-V5 | This study |
| pYL391 | pRS316 | ELM1 | Elm1(1-420)-Sak1(519-1142)-5×V5 | This study |
| pYL393 | pRS316 | TOS3 | Tos3(1-350)-Sak1(519-1142)-5×V5 | This study |
| pYL408 | pRS316 | ELM1 | Elm1(1-420)-Sak1(741-1142)-5×V5 | This study |
| pYL410 | pRS316 | SAK1 | Sak1(1-519)-5×V5, Sak1KD-V5 | This study |
| pYL411 | pRS313 | SNF1 | Snf1(1-309)-8×myc | This study |
Preparation of cell extracts and coimmunoprecipitation assays.
Cells were grown to mid-log phase in selective synthetic complete (SC) media containing 2% glucose, and an aliquot of the culture was collected by rapid filtration to maintain the phosphorylation state of Thr-210. The remainder of the culture was collected by rapid filtration, resuspended in SC media plus 0.05% glucose for 10 min, and collected by filtration. For immunoblot analysis, whole-cell extracts were prepared as described previously (6). For coimmunoprecipitation assays, the extraction buffer used was 50 mM HEPES (pH 7.5), 150 mM KCl, 0.5% Triton X-100, 1 mM dithiothreitol (DTT), 5 mM sodium pyrophosphate, 50 mM NaF, 2 mM phenylmethylsulfonyl fluoride, and Complete protease inhibitor cocktail (Roche). About 0.25 μg anti-hemagglutinin (HA) (12CA5; Roche), anti-myc (9E10; Santa Cruz), or anti-V5 (Invitrogen) antibody was used in each immunoprecipitation reaction. Antibodies were incubated with 0.2 mg (for anti-HA) or 0.8 to 1 mg (for anti-myc and anti-V5) of cell extract at 4°C for 1 to 2 h and incubated with 10 μl protein G Sepharose 4 Fast Flow (GE Healthcare) at 4°C for 1 h. After the beads were washed with 1 ml extraction buffer 3 times, proteins were eluted by boiling in 40 μl 2× SDS-PAGE loading dye for immunoblot analysis.
Immunoblot analysis.
Proteins (5 to 10 μg) from whole-cell extracts or immunopurified samples were separated by SDS-PAGE and analyzed by immunoblotting using anti-V5, anti-myc, anti-myc-horseradish peroxidase (HRP), anti-HA, anti-Snf1 (2), and anti-Thr(P)-172-AMPK (Cell Signaling Technologies) antibodies. Proteins were detected by using antibody to the epitope tag unless otherwise noted. ECL Plus (GE Healthcare) was used for visualization. Before the membrane was reprobed, it was incubated in 0.2 M glycine, pH 2, for 10 min.
RESULTS
SNF1 coimmunopurifies with Sak1 independently of glucose availability and phosphorylation state of Snf1 Thr-210.
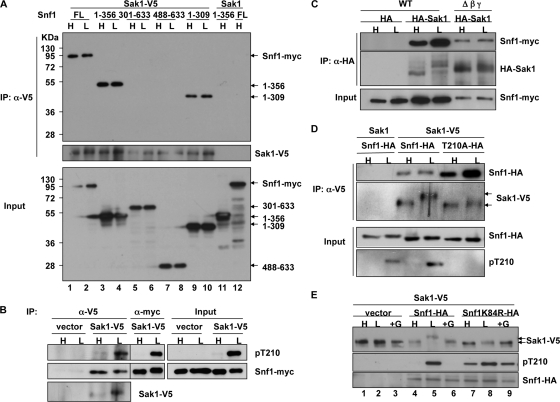
We examined the association of Sak1 with SNF1 under conditions of different levels of glucose availability by using rapid filtration, which preserves the phosphorylation state of Snf1 Thr-210, to collect cells. C-terminally tagged Sak1-V5 and Snf1-myc were expressed from their native promoters in snf1Δ sak1Δ cells. Cultures were grown to exponential phase in 2% (high) glucose and shifted to 0.05% (low) glucose for 10 min. We immunoprecipitated Sak1-V5 from cell extracts with anti-V5 antibody, resolved the copurifying proteins by SDS-PAGE, and assayed for Snf1-myc by immunoblot analysis. The two proteins copurified from extracts of both glucose-replete and glucose-depleted cultures (Fig. 1A, lanes 1 and 2), indicating that Sak1 binds to Snf1 constitutively, not in response to a low-glucose signal.
Fig. 1.
Interaction of Sak1 and Snf1. Cells expressing the indicated proteins were grown in SC media plus 2% (high [H]) glucose, and an aliquot was shifted to 0.05% (low [L]) glucose for 10 min. For panel E, an aliquot of glucose-depleted cells was replenished with glucose to 2% (+G) for 5 min. Cells were collected by rapid filtration, and extracts were prepared. (A) Sak1-V5 and either full-length (FL) Snf1-myc or myc-tagged partial Snf1 polypeptides containing the indicated residues were expressed from their native promoters on centromeric plasmids in snf1Δ sak1Δ cells. Sak1-V5 was immunoprecipitated (IP) from extracts, and copurifying proteins were resolved by 10% SDS-PAGE and analyzed by immunoblotting. α, anti. (B) Sak1-V5 was expressed in sak1Δ tos3Δ elm1Δ SNF1-myc cells, which expressed Snf1-myc from the genomic locus. Control cells expressed HA from empty vector. Sak1-V5 was immunoprecipitated, and copurifying proteins were resolved on 6% SDS-PAGE and analyzed by immunoblotting. Phosphorylated Thr-210 (pT210) of Snf1 was detected with anti-phospho-Thr-172-AMPK; the antibody is specific for Thr-210 (see panel D). Snf1-myc was also immunopurified with anti-myc, and 10% of the sample was loaded. All lanes are from the same blot. (C) HA-Sak1 was overexpressed in gal83Δ sip1Δ sip2Δ snf4Δ SNF1-myc (Δβγ) cells or SNF1-myc (wild-type [WT]) cells. HA-Sak1 was immunopurified, and proteins were resolved by 6% SDS-PAGE and analyzed by immunoblotting. (D) Sak1-V5 and Snf1-HA or Snf1T210A-HA were expressed from their native promoters on centromeric plasmids in snf1Δ sak1Δ cells. Analysis was done as described for panel A, except using 6% SDS-PAGE. Control cells expressed Sak1 and Snf1-HA from genomic loci. (E) Proteins were expressed in snf1Δ sak1Δ tos3Δ elm1Δ cells. Proteins were analyzed by 6% SDS-PAGE and immunoblotted.
We next addressed the possibility that phosphorylation of Thr-210 is coupled to dissociation of Sak1 from SNF1, which is not excluded by the previous findings because the fraction of Snf1 that is phosphorylated in response to glucose depletion is unknown. Although the association of Sak1 with Snf1 is sufficiently stable for copurification, studies of subcellular localization suggest that their interaction is dynamic (6, 24). Both SNF1 and Sak1 are localized in the cytosol in glucose-grown cells. In response to glucose depletion, SNF1 containing the major β subunit, Gal83, becomes enriched in the nucleus. This nuclear enrichment requires Sak1 and phosphorylation of Thr-210, but Sak1 remains cytosolic, raising the possibility that Sak1 phosphorylates SNF1 and dissociates before nuclear entry. Biochemical studies showed that most of the Sak1 protein is associated with Snf1 but that only a fraction of the total Snf1 protein is in complex with Sak1 (3); hence, the dissociated Sak1 protein could bind to another cytosolic, unphosphorylated Snf1 protein. If only a fraction of Snf1 molecules were phosphorylated in response to glucose depletion, redistribution of Sak1 to unphosphorylated Snf1 would yield the observed constitutive association of Sak1 with Snf1 and cytosolic localization of Sak1.
To determine whether Sak1 binds to phosphorylated Snf1, we immunoprecipitated Sak1-V5 from extracts of glucose-replete and glucose-depleted sak1Δ tos3Δ elm1Δ SNF1-myc cell cultures and subjected the copurifying proteins to immunoblot analysis. Phosphorylated Snf1 Thr-210 was detected using antibody to phosphorylated Thr-172 of AMPK. Phosphorylated Snf1-myc copurified with Sak1-V5 from glucose-depleted cell extracts (Fig. 1B). In a control experiment, anti-myc was added to directly immunoprecipitate Snf1-myc. These results show that Sak1 binds to phosphorylated Snf1 and suggest that another mechanism, unrelated to the phosphorylation state of Thr-210, is responsible for the retention of Sak1 in the cytosol. In this experiment, proteins were analyzed by SDS-PAGE in 6% polyacrylamide, and we noted that the mobility of Sak1 was decreased when wild-type cells were depleted of glucose.
Sak1 binds to the Snf1 kinase domain.
To determine the roles of the kinase domain and C-terminal region of Snf1 in binding to Sak1, we expressed partial Snf1 polypeptides with a C-terminal myc tag. Two polypeptides containing the N-terminal Snf1 kinase domain (residues 1 to 309 and 1 to 356) coimmunopurified efficiently with Sak1-V5 (Fig. 1A, lanes 3, 4, 9, and 10). In contrast, polypeptides containing C-terminal Snf1 sequences that interact with the β subunit (residues 488 to 633) or both the β and γ subunits (residues 301 to 633) (1, 10) did not copurify (Fig. 1A, lanes 5 to 8). Similar results were obtained when cells were grown in high glucose or shifted to low glucose.
A previous study detected the binding of the recombinant Snf1 kinase domain to Sak1 in vitro but did not detect the copurification of Snf1 with Sak1-TAP from cell extracts lacking the β and γ subunits of SNF1, despite the expression of Sak1-TAP from a multicopy (2μm) plasmid (4). Because levels of Snf1 are reduced when the SNF1 holoenzyme is not intact, we revisited this issue. We were able to detect the coimmunoprecipitation of Snf1-myc with HA-Sak1, overexpressed from the ADH1 promoter on a centromeric plasmid, from extracts of gal83Δ sip1Δ sip2Δ snf4Δ SNF1-myc (Δβγ) cells (Fig. 1C). Samples were rerun with serial 3-fold dilutions of the wild-type sample to confirm that the absence of the β and γ subunits had no major effect on coprecipitation of Snf1-myc with HA-Sak1 (data not shown). We note that Snf1 has little or no catalytic activity in a snf4Δ mutant (25). These findings indicate that the Snf1 kinase domain mediates the stable association of Sak1 with the SNF1 holoenzyme in vivo.
To test whether the binding of Sak1 to Snf1 requires the phosphorylation site Thr-210, we expressed a kinase-dead mutant protein with Ala substituted for Thr-210, Snf1T210A-HA, from the native promoter in snf1Δ sak1Δ cells. Snf1T210A-HA coimmunopurified with Sak1-V5, and in this experiment, recovery exceeded that of wild-type Snf1-HA (Fig. 1D). Analysis of Thr-210 phosphorylation confirmed the identity of the mutant protein, as well as the specificity of the antibody. The kinase-dead mutant Snf1K84R-HA, with Arg substituted for Lys in the ATP-binding site, also copurified (data not shown). These results show that stable binding of Sak1 to Snf1 does not require the phosphorylation site or SNF1 activity.
Glucose-regulated modification of Sak1 depends on SNF1 activity.
As noted above, Sak1 showed decreased mobility when wild-type cells were depleted of glucose. A change in mobility was also observed when cells were exposed to other environmental stresses that activate SNF1, including growth in glycerol, exposure to alkaline pH, and exposure to 1 M sodium chloride (data not shown). Sak1 did not display a similar change in mobility in snf1Δ cells expressing kinase-dead Snf1T210A (Fig. 1D), Snf1K84R (Fig. 1E, lanes 7 to 9), or no Snf1 protein (Fig. 1E, lanes 1 to 3). Subsequent studies localized this modification to the C terminus of Sak1 (Fig. 2C and 3).
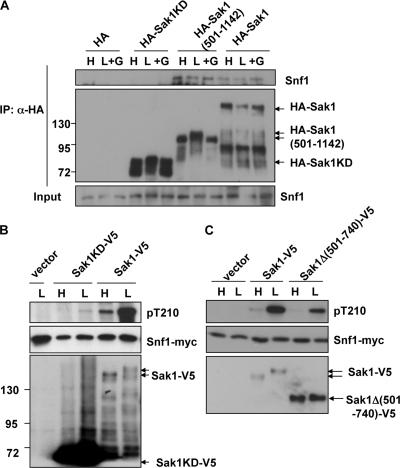
Fig. 2.
The Sak1 C-terminal region interacts with Snf1. Cells were grown as described in the legend to Fig. 1. (A) Proteins were overexpressed in WT cells. HA-Sak1 polypeptides were immunopurified from extracts, and proteins were resolved by 8% SDS-PAGE and subjected to immunoblot analysis. Untagged Snf1 protein was detected with anti-Snf1. Molecular mass markers are noted on the left in kilodaltons. (B, C) Proteins were expressed from the native promoter from centromeric plasmids in sak1Δ tos3Δ elm1Δ SNF1-myc cells. Proteins were resolved on 6% SDS-PAGE and immunoblotted. Phosphorylated Thr-210 (pT210) was detected with anti-phospho-Thr-172-AMPK.
To determine whether this Snf1-dependent modification of Sak1 affects the phosphorylation of Thr-210 by Sak1, we expressed Sak1-V5 and kinase-dead Snf1K84R-HA from their native promoters in snf1Δ sak1Δ tos3Δ elm1Δ cells. Cells were grown in high glucose, subjected to glucose depletion for 10 min, and replenished with 2% glucose for 5 min. Immunoblot analysis showed that glucose depletion did not result in a change of mobility for Sak1 comparable to that observed in the presence of wild-type Snf1-HA (Fig. 1E, compare lanes 5 and 8). Nonetheless, Thr-210 of Snf1K84R was phosphorylated to an extent similar to that of wild-type Snf1-HA upon glucose depletion (Fig. 1E, lanes 5 and 8). Thus, SNF1 activity is not required for phosphorylation of Thr-210 by Sak1, consistent with the function of Sak1 as the primary Snf1-activating kinase. Low-level phosphorylation of Snf1K84R in glucose-grown cells (Fig. 1E, lane 7) was also observed with cells expressing only Tos3 and Elm1, neither of which exhibiting a similar mobility shift (Fig. 3), and thus is unlikely to result from the absence of the modification of Sak1.
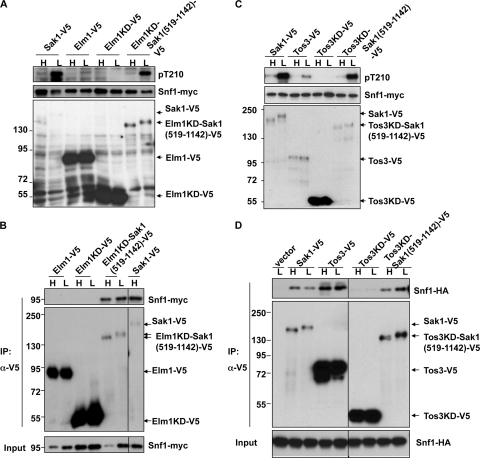
Fig. 3.
The Sak1 C terminus promotes interaction of Elm1KD and Tos3KD with Snf1. Proteins were expressed from their native promoters in sak1Δ tos3Δ elm1Δ SNF1-myc cells (A to C) or sak1Δ snf1Δ cells that also expressed Snf1-HA from its native promoter on a centromeric plasmid (D). (A, C) Proteins were analyzed by 6% SDS-PAGE and immunoblotting. (B, D) Sak1-V5 was immunopurified, and copurifying proteins were similarly analyzed. Molecular mass markers are noted on the left in kilodaltons.
The C-terminal region of Sak1 interacts with Snf1 and enhances phosphorylation.
To identify the region of Sak1 that is responsible for its stable interaction with Snf1, we overexpressed HA-tagged Sak1 polypeptides in wild-type cells and prepared extracts after growth under different conditions. The Sak1 polypeptide was immunoprecipitated, and copurifying proteins were analyzed by immunoblotting. Snf1 copurified with HA-Sak1 and with HA-Sak1(501-1142), containing the C-terminal region, but not with the Sak1 kinase domain (HA-Sak1KD, residues 1 to 519), although the last was more highly expressed (Fig. 2A). Experiments using sak1Δ tos3Δ elm1Δ SNF1-myc cells confirmed these results (data not shown). These findings indicate that the C-terminal region is necessary and sufficient for interaction with Snf1.
To determine whether the C terminus of Sak1 is required for phosphorylation of Snf1, we expressed Sak1KD-V5 and Sak1-V5 from the native promoter in sak1Δ tos3Δ elm1Δ SNF1-myc cells and assayed phosphorylation of Snf1 Thr-210 by immunoblotting. Phosphorylation by Sak1KD-V5 in response to glucose depletion was barely detectable, despite the much higher protein levels of Sak1KD-V5 than of full-length Sak1-V5 (Fig. 2B). SNF1 catalytic activity was similarly low in glucose-depleted cells carrying Sak1KD-V5 (0.8 and 8.7 nmol phosphate incorporated into peptide/min/mg of protein for Sak1KD-V5 and Sak1-V5, respectively, as assayed by phosphorylation of a synthetic peptide [15]). Sak1Δ(501-740) phosphorylated Thr-210 at reduced levels relative to those of wild-type Sak1, although it was expressed at higher levels (Fig. 2C, note the lack of mobility shift when cells were depleted of glucose). These findings indicate that the C-terminal region of Sak1 is important for the phosphorylation and activation of Snf1.
The Sak1 C terminus promotes interaction of Elm1KD or Tos3KD with Snf1.
Analysis of deletion mutants lacking different Snf1-activating kinases showed that both Elm1 and Tos3 phosphorylate and activate Snf1 in response to glucose depletion but do so less effectively than Sak1 (6, 8). Elm1 and Tos3 exhibit sequence similarity to Sak1 within the kinase domain but have divergent C termini. Evidence that the C-terminal region of Sak1 interacts with Snf1 suggests that this region is responsible for its function as the primary Snf1-activating kinase. To test this idea, we examined whether replacement of the C terminus of Elm1 with the Sak1 C-terminal sequence would enhance the ability of the Elm1 kinase domain (Elm1KD) to interact with and phosphorylate Snf1.
Elm1 and Elm1KD (residues 1 to 420), tagged with V5, were expressed from the native promoter. Neither protein phosphorylated Snf1 Thr-210 effectively compared to Sak1-V5 (Fig. 3A), and Snf1-myc did not coimmunoprecipitate (Fig. 3B), consistent with evidence that Elm1 and Snf1 are not in a stable complex (3). Elm1KD-V5 was functional as assayed by suppression of the elongated morphology phenotype caused by elm1Δ (data not shown). To test whether the C-terminal region of Sak1 could enhance the function of Elm1KD toward Snf1, we fused Elm1KD to Sak1 residues 519 to 1142. Although levels of Elm1KD-Sak1(519-1142)-V5 were much lower than those of Elm1KD-V5, the fusion protein phosphorylated Snf1 Thr-210 in glucose-depleted sak1Δ tos3Δ elm1Δ cells at levels comparable to Sak1-V5 (Fig. 3A) and coimmunopurified with Snf1-myc (Fig. 3B).
Using coimmunoprecipitation, we detected interaction of Tos3-V5 with Snf1-HA when both were expressed from native promoters (Fig. 3D). These results differ from those of Elbing et al., who did not detect interaction of this pair (3), but this is not a significant discrepancy because only a small fraction of Tos3-V5 was associated with Snf1. Tos3KD-V5 (residues 1 to 350) did not phosphorylate Thr-210, and little or no Snf1-HA coimmunopurified (Fig. 3C and D). Fusion of the Sak1 C terminus to Tos3KD enhanced both phosphorylation of Thr-210 in glucose-depleted cells and copurification of Snf1-HA, despite much-reduced protein levels for Tos3KD-Sak1(519-1142)-V5 relative to those of Tos3KD-V5 (Fig. 3C and D).
These findings indicate that the C-terminal sequence is largely responsible for the function of Sak1 as the major Snf1-activating kinase. We also note that fusion proteins carrying Sak1 residues 519 to 1142 exhibited a mobility shift similar to that observed for Sak1 when cells were depleted of glucose.
Sak1 interacts with Reg1 independently of SNF1.
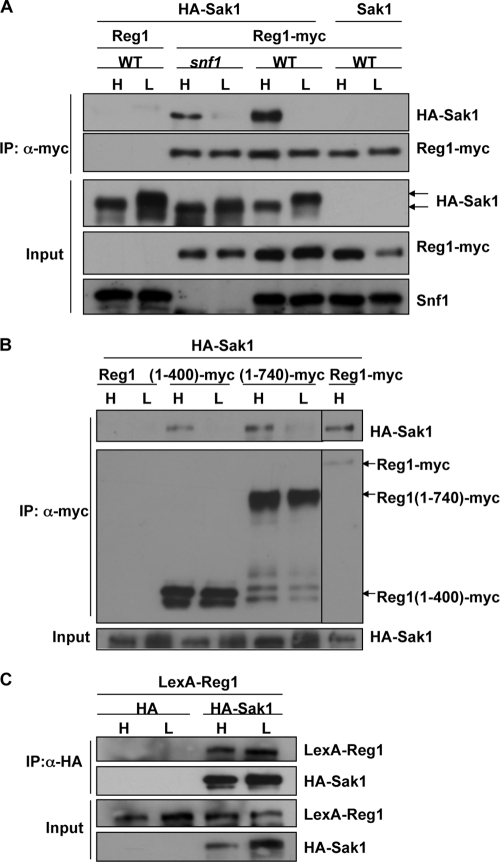
Like Sak1, the Reg1 subunit of Reg1-Glc7 PP1 interacts physically with Snf1 and is phosphorylated in a Snf1-dependent manner (12, 19), but previous experiments have not addressed the interdependence of the interactions among Reg1-Glc7, Sak1, and SNF1. We did not detect coimmunoprecipitation of Sak1 and Reg1 from extracts of cells expressing these proteins at native levels. However, when HA-Sak1 was overexpressed and Reg1-myc was expressed from the genomic locus, the two coimmunoprecipitated from extracts of glucose-grown wild-type and snf1Δ cells, but not from glucose-depleted cells (Fig. 4A). HA-Sak1 coimmunopurified with truncated Reg1(1-400)-myc and Reg1(1-740)-myc, expressed from the genomic locus in snf1Δ cells, and did so more efficiently from extracts of glucose-grown cells (Fig. 4B). Overexpressed LexA-Reg1 copurified with HA-Sak1, in the absence of all SNF1 subunits, from extracts of snf1Δ gal83Δ sip1Δ sip2Δ snf4Δ cells; copurification was observed using both glucose-replete and glucose-depleted cultures, which may reflect the overexpression of both proteins (Fig. 4C). Consistent with this idea, HA-Sak1 also copurified with Reg1(1-197), which is more highly expressed than the other truncated polypeptides, and in this case, the growth condition had no effect (data not shown). These findings indicate that Sak1 can interact with Reg1 independently of SNF1 and suggest that the interaction is regulated in response to glucose signals.
Fig. 4.
Interaction of Sak1 with Reg1. (A) HA-Sak1 was overexpressed in snf1Δ REG1-myc or SNF1 REG1-myc (WT) cells expressing Reg1-myc from the genomic locus. Proteins were immunopurified from extracts with anti-myc, resolved by 6% SDS-PAGE, and immunoblotted. Snf1 was detected with anti-Snf1. Control cells expressed untagged Reg1 or Sak1 from genomic loci. (B) HA-Sak1 was overexpressed in snf1Δ REG1, snf1Δ reg1(1-400)-myc, snf1Δ reg1(1-740)-myc, and snf1Δ REG1-myc cells, which expressed the indicated Reg1 polypeptide from the genomic locus. Full-length Reg1 has 1,014 residues. Analysis was done as described for panel A, except with 8% SDS-PAGE. (C) HA-Sak1 and LexA-Reg1 were overexpressed in snf1Δ gal83Δ sip1Δ sip2Δ snf4Δ cells, immunopurified with anti-HA, and immunoblotted as described for panel A.
DISCUSSION
We have examined the interaction of Sak1, the primary Snf1-activating kinase, with SNF1. Sak1 and the Snf1 catalytic subunit coimmunopurify from extracts of cells grown under conditions of both high and low glucose availability, and their interaction occurs independently of Thr-210 or its phosphorylation state, Snf1 catalytic activity, and other SNF1 subunits. Sak1 interacts with the Snf1 kinase domain, and the region C terminal to the Sak1 kinase domain is necessary and sufficient for association with Snf1. Deletion of the Sak1 C terminus impaired phosphorylation and activation of Snf1 by Sak1, in accord with evidence that its deletion affected Snf1-dependent growth phenotypes and invertase expression (17). We further showed that replacement of the C terminus of Elm1 with the Sak1 sequence enhanced the ability of the Elm1 kinase domain to interact physically with Snf1 and to phosphorylate Snf1 on Thr-210. Analogous replacement studies with Tos3 yielded similar results. These findings support the idea that the nonconserved C-terminal region of Sak1 is responsible for its function as the principal Snf1-activating kinase. Association of the activating kinase with SNF1 most likely facilitates the response to changing environmental conditions, as the activating kinase is poised to phosphorylate Thr-210.
Previous studies showed that SNF1 containing Gal83 becomes enriched in the nucleus in response to glucose depletion, dependent on phosphorylation of Thr-210, while Sak1 remains cytosolic (6). We addressed the possibility that phosphorylation is coupled to dissociation of Sak1 from SNF1, which could account for the retention of Sak1 in the cytosol; however, we found that Sak1 binds phosphorylated Snf1, suggesting that another mechanism is responsible. Our data do not exclude the possibility that the affinity of Sak1 for Snf1 is altered by the phosphorylation of Thr-210.
We also report that Sak1 exhibits Snf1-dependent modification, presumably phosphorylation, in response to glucose depletion. The C terminus of Sak1 is the major site of modification, as judged by the mobility of deletion and fusion constructs. The significance of this modification, if any, remains unclear. It may affect the functional interaction of Sak1 and SNF1 in some manner not yet identified. Another possibility is that it represents a mechanism by which SNF1 activity affects another, unknown function of Sak1. It is also possible that a modification without functional consequence results from the close association of Sak1 with the Snf1 catalytic domain.
Finally, we examined the interaction of Sak1 with the Reg1 subunit of Reg1-Glc7 PP1, which has roles in glucose regulation and in dephosphorylation of Snf1. Sak1 interacted with Reg1 independently of SNF1, but this interaction did not appear to be strong, as detection required overexpression of one of the partners, and may reflect a catalytic relationship. It is possible that Sak1 phosphorylates Reg1 or that Reg1-Glc7 dephosphorylates Sak1 or both. Growth in high glucose promoted the interaction of several constructs, although uncertainty regarding the effects of overexpression precludes any firm conclusion. Further studies will be required to determine whether the interaction of Reg1 and Sak1 affects regulation of SNF1 activity or affects other functions of Reg1-Glc7 or Sak1 that are unrelated to the SNF1 pathway.
ACKNOWLEDGMENTS
We thank Martin Schmidt for providing Sak1-V5 plasmids and Amparo Ruiz, Milica Momcilovic, and Zheng Hu for valuable discussion.
This work was supported by NIH grant GM34095 to M.C.
Footnotes
Published ahead of print on 7 January 2011.
REFERENCES
- 1. Amodeo G. A., Rudolph M. J., Tong L. 2007. Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1. Nature 449:492–495 [DOI] [PubMed] [Google Scholar]
- 2. Celenza J. L., Carlson M. 1986. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science 233:1175–1180 [DOI] [PubMed] [Google Scholar]
- 3. Elbing K., McCartney R. R., Schmidt M. C. 2006. Purification and characterization of the three Snf1-activating kinases of Saccharomyces cerevisiae. Biochem. J. 393:797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elbing K., Rubenstein E. M., McCartney R. R., Schmidt M. C. 2006. Subunits of the Snf1 kinase heterotrimer show interdependence for association and activity. J. Biol. Chem. 281:26170–26180 [DOI] [PubMed] [Google Scholar]
- 5. Hedbacker K., Carlson M. 2008. SNF1/AMPK pathways in yeast. Front. Biosci. 13:2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hedbacker K., Hong S. P., Carlson M. 2004. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol. Cell. Biol. 24:8255–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hong S. P., Carlson M. 2007. Regulation of Snf1 protein kinase in response to environmental stress. J. Biol. Chem. 282:16838–16845 [DOI] [PubMed] [Google Scholar]
- 8. Hong S. P., Leiper F. C., Woods A., Carling D., Carlson M. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. U. S. A. 100:8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hong S. P., Momcilovic M., Carlson M. 2005. Function of mammalian LKB1 and Ca2+/calmodulin-dependent protein kinase kinase α as Snf1-activating kinases in yeast. J. Biol. Chem. 280:21804–21809 [DOI] [PubMed] [Google Scholar]
- 10. Jiang R., Carlson M. 1997. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 17:2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kahn B. B., Alquier T., Carling D., Hardie D. G. 2005. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 1:15–25 [DOI] [PubMed] [Google Scholar]
- 12. Ludin K., Jiang R., Carlson M. 1998. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 95:6245–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCartney R. R., Rubenstein E. M., Schmidt M. C. 2005. Snf1 kinase complexes with different beta subunits display stress-dependent preferences for the three Snf1-activating kinases. Curr. Genet. 47:335–344 [DOI] [PubMed] [Google Scholar]
- 14. McCartney R. R., Schmidt M. C. 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276:36460–36466 [DOI] [PubMed] [Google Scholar]
- 15. Momcilovic M., Iram S. H., Liu Y., Carlson M. 2008. Roles of the glycogen-binding domain and Snf4 in glucose inhibition of SNF1 protein kinase. J. Biol. Chem. 283:19521–19529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nath N., McCartney R. R., Schmidt M. C. 2003. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 23:3909–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rubenstein E. M., McCartney R. R., Schmidt M. C. 2006. Regulatory domains of Snf1-activating kinases determine pathway specificity. Eukaryot. Cell 5:620–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rubenstein E. M., et al. 2008. Access denied: Snf1 activation loop phosphorylation is controlled by availability of the phosphorylated threonine 210 to the PP1 phosphatase. J. Biol. Chem. 283:222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanz P., Alms G. R., Haystead T. A., Carlson M. 2000. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 20:1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Song W., Carlson M. 1998. Srb/mediator proteins interact functionally and physically with transcriptional repressor Sfl1. EMBO J. 17:5757–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Steinberg G. R., Kemp B. E. 2009. AMPK in health and disease. Physiol. Rev. 89:1025–1078 [DOI] [PubMed] [Google Scholar]
- 22. Sutherland C. M., et al. 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13:1299–1305 [DOI] [PubMed] [Google Scholar]
- 23. Tu J., Carlson M. 1995. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 14:5939–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vincent O., Townley R., Kuchin S., Carlson M. 2001. Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism. Genes Dev. 15:1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woods A., et al. 1994. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J. Biol. Chem. 269:19509–19515 [PubMed] [Google Scholar]