Abstract
Many eukaryotic proteins are posttranslationally modified by the esterification of cysteine thiols to long-chain fatty acids. This modification, protein palmitoylation, is catalyzed by a large family of palmitoyl acyltransferases that share an Asp-His-His-Cys Cys-rich domain but differ in their subcellular localizations and substrate specificities. In Trypanosoma brucei, the flagellated protozoan parasite that causes African sleeping sickness, protein palmitoylation has been observed for a few proteins, but the extent and consequences of this modification are largely unknown. We undertook the present study to investigate T. brucei protein palmitoylation at both the enzyme and substrate levels. Treatment of parasites with an inhibitor of total protein palmitoylation caused potent growth inhibition, yet there was no effect on growth by the separate, selective inhibition of each of the 12 individual T. brucei palmitoyl acyltransferases. This suggested either that T. brucei evolved functional redundancy for the palmitoylation of essential palmitoyl proteins or that palmitoylation of some proteins is catalyzed by a noncanonical transferase. To identify the palmitoylated proteins in T. brucei, we performed acyl biotin exchange chemistry on parasite lysates, followed by streptavidin chromatography, two-dimensional liquid chromatography-tandem mass spectrometry protein identification, and QSpec statistical analysis. A total of 124 palmitoylated proteins were identified, with an estimated false discovery rate of 1.0%. This palmitoyl proteome includes all of the known palmitoyl proteins in procyclic-stage T. brucei as well as several proteins whose homologues are palmitoylated in other organisms. Their sequences demonstrate the variety of substrate motifs that support palmitoylation, and their identities illustrate the range of cellular processes affected by palmitoylation in these important pathogens.
INTRODUCTION
The attachment of palmitic acid to a protein substrate can have dramatic effects on its function by targeting it to specific membrane domains, promoting association with lipid microdomains, regulating responsiveness to upstream signaling, and/or protecting it from degradation (22). However, the extent of protein palmitoylation in eukaryotes has long been underappreciated, due in part to the technical difficulty in detecting this modification. Metabolic labeling with [3H]palmitate requires immunoprecipitation and prolonged autoradiography exposure. Further complicating palmitoyl protein analysis is the inherent difficulty in candidate identification. While there are a few sequences that are highly predictive of protein palmitoylation (notably, a cysteine in close proximity to an N-terminal myristoylation site or a C-terminal CaaX box prenylation site), these are present in only a minority of palmitoyl proteins. Considerable advances have recently been made by instead applying an unbiased proteomic approach, first in Saccharomyces cerevisiae (34) and then in rat neurons (18). These approaches have yielded several important findings. First, detection of palmitoylation for a given protein provides a possible mechanism for the regulation of its activity, trafficking, or expression. Second, an expanded data set allows for pattern detection in substrate sequences that may predict enzyme-substrate relationships. For example, the detection of an FWC carboxy-terminal tripeptide on amino acid permeases in yeast led to the discovery of those proteins all being modified by the same enzyme, Pfa4 (34). Finally, given the many known regulatory roles for palmitoylation in eukaryotic biology, palmitoyl proteome identification provides insight into the specific cellular processes affected by this modification. The discovery of a critical role for palmitoylation in regulating dendritic spine morphogenesis was prompted by the detection of an unusual Cdc42 splice variant in the rat neuron palmitoyl proteome (18).
Just as the identification of palmitoylation substrates has been slowed by technical obstacles, so too has the identification of enzymatic mediators. Palmitoyl acyltransferase (PAT) activity is tightly associated with cellular membranes and is often unstable upon extraction, hindering purification and identification of these enzymes by conventional biochemical approaches. The observation that certain proteins could spontaneously undergo palmitoylation in vitro advanced the argument for enzyme-independent palmitoylation. Ultimately, however, genetic screens in yeast were successful in identifying the first bona fide PATs (23, 33). Both of these enzymes, Erf2/Erf4 and Akr1p, were found to share an Asp-His-His-Cys Cys-rich domain (DHHC-CRD) motif, which was subsequently shown to play a direct role in PAT activity. The DHHC-CRD gene family has since been shown to be responsible for most, if not all, protein palmitoylation in yeast (34). This gene family is highly conserved among eukaryotes, with 7 members in yeast, 22 in mice, 23 in humans, and 12 in T. brucei (10). A comprehensive analysis of DHHC-CRD protein localization in human embryonic kidney 293T cells demonstrated these enzymes to have distinct and restricted subcellular localizations (25), a property that might underlie the differential trafficking effects on individual protein substrates. We previously identified the T. brucei calflagin PAT, TbPAT7, and demonstrated its role in calflagin trafficking to the flagellar, rather than pellicular, membrane (9, 10). Here, we expand on these earlier investigations of T. brucei protein palmitoylation to assess both the enzymes that catalyze this modification and the substrates that comprise the entire palmitoyl proteome.
MATERIALS AND METHODS
Parasites.
All T. brucei strains described in this study are derivatives of the procyclic 29-13 strain or the bloodstream-form single-marker cell line, both of which have been engineered to coexpress the bacteriophage T7 polymerase and tet repressor (38). Parasite culture methods and generation of tetracycline-inducible PAT RNA interference (RNAi) cell lines have been previously described (10).
Growth in culture.
For 2-bromopalmitate inhibition, T. brucei was seeded at a density of 106/ml for procyclic forms or 105/ml for blood forms and cultured for 18 to 24 h in the presence of indicated concentrations of the chemical inhibitor 2-bromopalmitate (Sigma-Aldrich, St. Louis, MO). Cells were enumerated by visual inspection of culture suspension loaded on a Neubauer hemacytometer. Growth was calculated relative to vehicle control. Fifty percent inhibitory concentration (IC50) calculations were performed using the program GraphPad Prism (GraphPad Software, La Jolla, CA). The same range of concentrations was also incubated for 12 to 16 h with log-phase Escherichia coli, and growth was ascertained by subtracting the optical density at 600 nm (OD600) of cultures with the drug plus LB alone from the OD600 of growing cultures. For RNAi experiments, cells were seeded at an initial density of 5 × 104/ml for procyclic forms or 5 × 103/ml for bloodstream forms and cultured in the presence or absence of 1 μg/ml tetracycline (Sigma). At the indicated time points, cell density was measured with a Z1 series cell counter (Beckman-Coulter, Fullerton, CA), and cells were diluted as necessary to maintain logarithmic-phase growth. The cumulative cell number was calculated as the density at a given time point multiplied by the cumulative dilution factor.
RNA analysis.
Total RNA was isolated from 1 × 107 parasites by extraction with 1 ml TRIzol reagent (Invitrogen, Carlsbad, CA) and treated with amplification-grade DNase I (Invitrogen) for 15 min at room temperature. Poly(T) primers were used to generate cDNA from 0.5 to 1.0 μg total RNA with the SuperScript first-strand synthesis for reverse transcription-PCR (RT-PCR) system (Invitrogen). Gene-specific primers (sequences are available upon request) were then used to amplify a 250- to 350-bp region outside the RNAi target sequence in order to avoid amplification from the induced double-stranded RNA (dsRNA). For each set of PCR primers, a pilot experiment was conducted to determine an optimal number of cycles that enabled detection in the linear range of the assay.
Acyl biotin exchange.
Total cellular palmitoylated proteins were purified as previously described (10) with the following modifications: a total of 3.2 × 109 cells per replicate were harvested and lysed in 10 ml lysis buffer (50 mM Tris-Cl [pH 7.4], 150 mM NaCl, 5 mM EDTA) with 1.7% Triton X-100. Following clarification by centrifugation at 1,000 × g for 10 min at 4°C, three aliquots of 3 ml each were processed in parallel. Immediately prior to the exchange reaction, each pellet was resuspended in 400 μl solubilization buffer, pooled, and split into 600-μl samples for parallel labeling under experimental and control conditions. Following removal of residual label, pellets were resuspended in 300 μl 2% SDS buffer (50 mM Tris-HCl [pH 7.4], 5 mM EDTA, 2% SDS) and then mixed with 5.7 ml lysis buffer and added batch-wise in 1.5-ml aliquots to four tubes containing 40 μl prewashed streptavidin-agarose beads. For the final wash step, all beads were collected into a single tube. Samples were eluted by incubating beads in 110 μl elution buffer (lysis buffer containing 0.2% Triton X-100, 0.1% SDS, 1% β-mercaptoethanol) at 37°C for 15 min with vortexing every 1 to 2 min to keep beads in suspension. Samples were finally centrifuged for 1 min at 20,000 × g, and 100 μl of supernatant was transferred to a new tube that was stored at −80°C until further analysis.
LC-MS/MS analysis.
Streptavidin eluates were precipitated with trichloroacetic acid, and the pellets were dissolved in 20 ml 0.4 M NH4HCO3 containing 8 M urea. Samples were reduced with 5 mM dithiothreitol for 15 min at 50°C, alkylated with 10 mM iodoacetamide for 30 min at room temperature, and diluted with high-performance liquid chromatography (LC)-grade water to obtain a 1 M final concentration of urea. Samples were digested for 16 h with 2 μg sequencing-grade trypsin (Sigma-Aldrich). The reaction was stopped with 1 μl formic acid. Samples were desalted with POROS R2 ziptips (17) and dried with a vacuum centrifuge. Samples were then redissolved in 50 μl 0.1% formic acid (FA) and 20 μl was subjected to two-dimensional LC-tandem mass spectrometry (2D LC-MS/MS) analysis, using an Eksigent 1D-plus nano-LC apparatus coupled to a Thermo Fisher LTQ XL/ETD-MS system, equipped at the front end with a Triversa/Advion electrospray ionization nanosource. For the nano-LC separation, the sample was loaded into a strong-cation exchange (SCX) trap column (5 μl; Optimized Technologies), and the elution was carried out by injecting increasing concentrations of NaCl (0, 25, 50, 100, 200, and 500 mM NaCl in 5% acetonitrile [ACN]–0.5% FA) with the autosampler and washing for 130 min with 5% ACN–0.5% formic acid at 1.5 μl/min (E. S. Nakayasu, A. F. Marques, and I. C. Almeida, unpublished data). The peptides eluted from the SCX column were trapped and washed in an in-house packed C18 reverse-phase capillary column (1 cm by 75 μm; Phenomenex Luna C18, 5 μm). The separation was performed in an in-house packed capillary C18 reverse-phase column (20 cm by 75 μm; Phenomenex Luna C18, 5 μm) in a linear gradient (solvent A, 5% ACN–0.1% FA; solvent B, 80% ACN–0.1% FA; with 5 to 40% solvent B for 200 min, 50 to 90% B for 1 min, 5 min in B, 90 to 5% B for 1 min, and 20 min in 5% B). The MS system was set to perform one full scan (400 to 2,000 m/z range) followed by MS/MS of the 10 most abundant parent ions (isolation width, 3.0 m/z; 35% normalized collision energy). The dynamic exclusion was set to collect each parent ion twice and then excluding them for 30 s.
Bioinformatic analysis.
The resulting MS/MS spectra (800 to 3,500 Da, minimum of 10 counts) were converted to DTA files and submitted to a database search using Sequest (Bioworks 3.3.1; Thermo Fisher Scientific). The database was comprised of T. brucei, human keratin, and porcine trypsin sequences (all in correct and reverse orientations) downloaded from GenBank on 1 July 2008. The parameters for the database search were as follows: (i) 2.0 and 1.0 Da for peptide and fragment mass tolerance; (ii) trypsin cleavage at both termini and one missed cleavage allowed; (iii) methionine oxidation, cysteine carbamidomethylation, and N-ethylmaleimide derivatization as possible modifications. The Sequest results were filtered in Bioworks with DCn (delta correlation) values of ≥0.05, peptide probability of ≤0.05, and Xcorr of ≥1.5, 2.0, or 2.5 for singly, doubly, and triply charged peptides, respectively. After filtering, peptide sequences were assembled into protein sequences by using an in-house-written Perl script, searching the peptides using the BLAST algorithm and considering only those with 100% identity. After assembling, the results were filtered using the sum of the peptide Xcorr of 3.5, which provided a false-discovery rate (FDR) for protein identification of 1.6%. The relative protein quantification was done by spectral count, and statistical analysis was carried out using QSpec (3). Sequences were analyzed for predicted N-terminal myristoylation, palmitoylation sites, and transmembrane domains using Myristoylator (2), CSS-Palm 2.0 (30), and TMHMM 2.0 (19), respectively.
Data set and protein sequences.
The raw LC-MS/MS data files and protein sequence database are available at the Proteome Commons Tranche network (https://proteomecommons.org/dataset.jsp?i=cnyVALIAf%2FzGw1EFUwyh PJRyHqOjKqxyoVBtIvy%2BkCof8r6xd93Wd9zzAKZzSO1FBfj3Bbv%2FJclFE5cbNNwJmf3snjQAAAAAAAACZg%3D%3D). The identified proteins were deposited in TriTrypDB (tritrypdb.org).
RESULTS
Growth inhibition of T. brucei by 2-bromopalmitate.
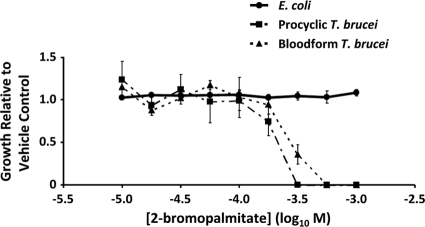
We first employed a pharmacologic approach to inhibit palmitoylation in T. brucei. Procyclic and bloodstream-form cells were incubated for 48 h in the presence of a range of concentrations of the nonmetabolizable palmitate analogue 2-bromopalmitate. Growth was analyzed relative to the vehicle control (Fig. 1). Both life cycle stages of the parasite were sensitive to 2-bromopalmitate treatment, with a calculated IC50 of 197 μM (95% confidence interval, 127 to 304 μM) for the procyclic life cycle stage and 226 μM (95% confidence interval, 147 to 348 μM) for the bloodstream life cycle stage. Growth was completely inhibited at concentrations consistent with the dose required in mammalian cells to inhibit palmitoylation (31). In contrast, 2-bromopalmitate had no effect on the growth of E. coli, in which protein palmitoylation is not known to occur.
Fig. 1.
Growth inhibition of procyclic and bloodstream life cycle stages of T. brucei by 2-bromopalmitate. T. brucei cultures were initiated at a density of 1 × 106/ml for strain 29-13 procyclic parasites or 1 × 105/ml for single-marker strain bloodstream-form parasites. After 24 h in the presence of various concentrations of the palmitoylation inhibitor 2-bromopalmitate or vehicle control, parasite density was determined by Neubauer hemocytometry. Cell density was normalized to the vehicle control, and data are presented as the means ± standard errors of the means of triplicate experiments.
Inhibition of specific PATs does not affect in vitro growth cell morphology, or motility.
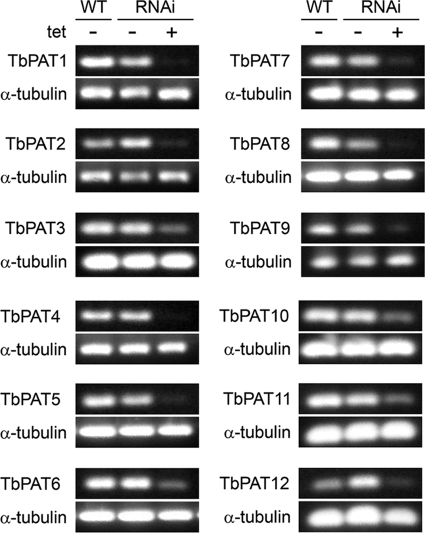
To investigate the requirement of individual palmitoyl acyltransferases for the viability and growth of T. brucei, we analyzed a series of cell lines containing an integrated, drug-inducible construct for RNAi inhibition of each PAT. The target genes were selected on the basis of their containing a DHHC-CRD motif (Pfam ID 01529). Target sequences were chosen to avoid off-target inhibition (29). These cell lines were previously used to identify the PAT that palmitoylates the calflagins, a necessary step in their trafficking to the flagellar membrane (10). We measured the transcript levels of each PAT in the parental cell line and in the RNAi line by semiquantitative RT-PCR (Fig. 2). Primers were selected to amplify transcripts in regions outside that targeted by RNAi to avoid detection of dsRNA. For each cell line, RNAi inhibition was successful in reducing transcript levels. While there were low levels of transcription following RNAi in a few of the lines, our experience with TbPAT7 indicates that these residual levels are not sufficient to maintain palmitoylation (10).
Fig. 2.
Depletion of individual PAT mRNAs by RNA interference. Semiquantitative RT-PCR was conducted on RNA samples isolated from either parental 29-13 (wild-type [WT]) cells, or individual PAT RNAi cell lines cultured with or without tetracycline for 2 days. Labels along the vertical axes indicate the primer set used to monitor mRNA levels, with α-tubulin serving as a control.
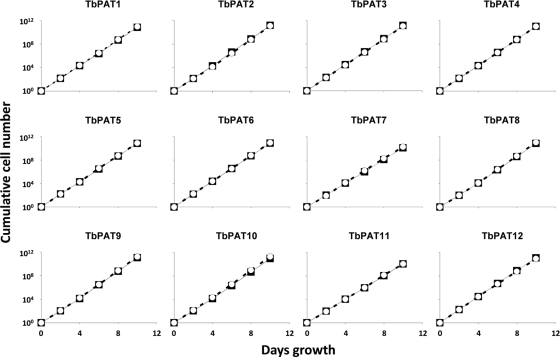
We performed phenotypic analysis on the 12 PAT RNAi lines. We first tested for an effect on parasite viability and/or proliferation by analyzing growth curves. Both procyclic-stage and bloodstream-stage parasites were seeded at a density that supports logarithmic growth. RNAi was induced by the addition of tetracycline to the culture medium. Cell counts were determined for induced and uninduced cultures over a 10-day period, with cultures diluted as necessary to prevent a shift into stationary phase. In contrast to the potent growth inhibition observed with 2-bromopalmitate treatment, none of the 12 individual RNAi lines, in either life cycle stage, exhibited a growth defect in culture (Fig. 3). Gross observation of parasite morphology and motility likewise did not uncover any defect in these mutants. Thus, while T. brucei palmitoylation on a global level appears essential, these cells are not exquisitely sensitive to inhibition of any single enzyme. From a therapeutic standpoint, therefore, it appears more promising to target critical palmitoylation substrates rather than enzymatic regulators. With this in mind, we next sought to identify all of the proteins modified by these enzymes.
Fig. 3.
T. brucei growth is not affected by RNAi depletion of any single PAT mRNA. Tetracycline-inducible PAT RNAi cell lines were cultured at an initial density of 5 × 103 cells/ml and propagated for 10 days in the presence (closed squares) or absence (open circles) of 1 μg/ml tetracycline. Every 48 h, cell density was quantified with an automated cell counter and cells were diluted as necessary to prevent entry into stationary phase.
Purification and identification of total cellular palmitoyl proteins.
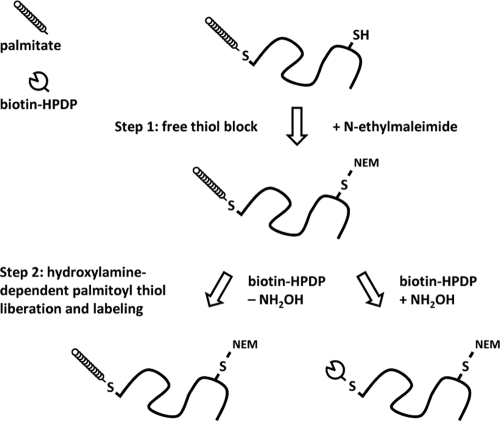
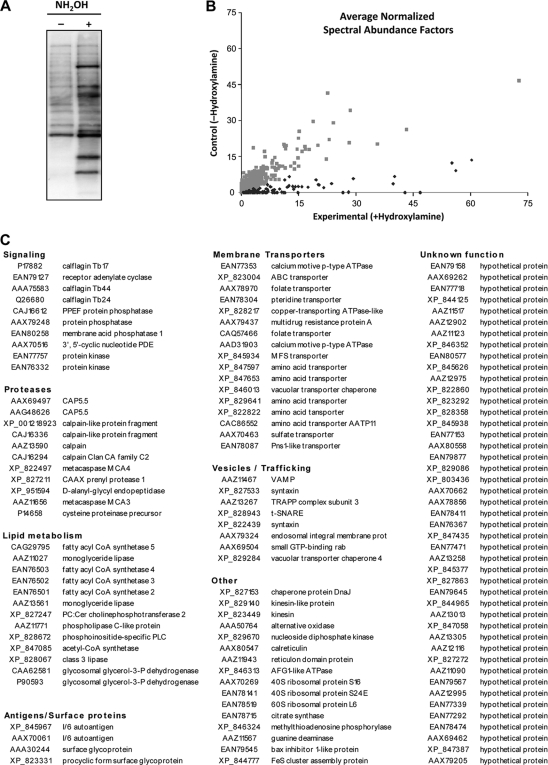
To purify T. brucei palmitoylated proteins, we first performed an acyl biotin exchange reaction on procyclic-stage T. brucei whole-cell lysates (Fig. 4) (36). In this reaction, the nucleophilic reagent hydroxylamine (NH2OH) is used to cleave protein-palmitoyl bonds, liberating a reactive thiol that is labeled by coincubation with N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (biotin-HPDP). To prevent nonspecific labeling, a preincubation step treats lysates with N-ethylmaleimide, which reacts with free thiols to generate an NH2OH-insensitive bond. As a control for nonspecific labeling, half of the sample is treated with the labeling reaction mixture minus NH2OH. The resulting sample can then be either analyzed directly by probing with streptavidin-horseradish peroxidase (HRP) (Fig. 5A) or purified by streptavidin-agarose chromatography. Elution of biotinylated proteins from streptavidin was performed by incubation with β-mercaptoethanol, which reduces the protein-HPDP linkage. Immunoblotting with a panel of antibodies against control proteins previously demonstrated the sensitivity and specificity of this approach in T. brucei (10).
Fig. 4.
Schematic representation of the acyl biotin exchange reaction. Acyl biotin exchange relies on the sensitivity to hydroxylamine (NH2OH) treatment of the covalent thioester linkage between palmitic acid moieties and cysteine residues of protein substrates. In the first step, parasite lysates are treated with N-ethylmaleimide to block reactive free thiols. The sample is then split in half and incubated with the thiol-labeling reagent biotin-HPDP in the presence or absence of NH2OH. Liberation of thiols by NH2OH cleavage enables specific labeling, while the other half of the sample serves as a control for nonspecific labeling due to incomplete preblock, or later, nonspecific purification during streptavidin chromatography.
Fig. 5.
The T. brucei palmitoyl proteome. (A) Acyl biotin exchange samples prepared from procyclic-stage T. brucei were fractionated by SDS-PAGE, transferred to nitrocellulose membranes, and probed with streptavidin-HRP to detect total lysate labeling under each condition. (B) Individual proteins purified by streptavidin chromatography from acyl biotin exchange samples were identified by LC/MS-MS. Their relative abundances were quantified and normalized for experimental and control conditions. In this graphical representation, each point represents an individual protein, with its coordinates indicating its abundance under each condition. Thus, proteins clustered along the x axis show high NH2OH dependence for their purification and, by extension, palmitoylation. Gray dots represent those proteins deemed insignificant by statistical analysis, while black dots represent proteins that met the threshold for significance. (C) The T. brucei palmitoyl proteome (n = 124 proteins) is listed, with corresponding NCBI accession numbers and functional groupings.
To identify total proteins purified under experimental (with NH2OH) and control (without NH2OH) conditions, acyl biotin exchange samples were subjected to protein identification by LC-MS/MS. Peptides mapping to each protein under each condition from triplicate analyses were then converted to normalized spectral abundance factors (40) (Fig. 5B) and analyzed for significance by QSpec, which uses a hierarchical Bayes estimation of a generalized linear mixed-effects model to assess significance (3). A total of 1,672 proteins were identified, and most of them were present in similar proportions under control and experimental conditions (Fig. 5B; see also Tables S1 and S2 in the supplemental material). These likely represent proteins for which N-ethylmaleimide blockade of free thiols was incomplete or proteins that nonspecifically associated with streptavidin-agarose beads. A subset, however, was present in significantly greater abundance under experimental conditions. A total of 132 proteins met the threshold for significance for being palmitoylated, with a false-discovery rate cutoff of 1% (Fig. 5C; see alsoo Table S3 in the supplemental material). These included all of the known procyclic-stage T. brucei palmitoyl proteins—calflagins Tb17, Tb24, and Tb44 (10), CAP5.5 (15), and PPEF phosphatase (24), as well as three proteins, phosphoinositide-specific phospholipase C and two calpain-like protein fragments, which have homologues in the closely related Trypanosoma cruzi (12) and Leishmania major (35), respectively, that are palmitoylated at Cys residues conserved in the T. brucei proteins. To cull for false-positive identifications, we next removed eight noteworthy proteins for which an alternative hydroxylamine-sensitive thioester linkage has been reported. These included ubiquitination enzymes, which form a transient thioester link with ubiquitin (14, 27), and pyruvate dehydrogenase, whose lipoic acid prosthetic group accepts acetyl moieties via thioester linkage. We also detected two palmitoyl acyltransferases, TbPAT2 and TbPAT11, which are expected to form a transient DHHC-palmitate intermediate during catalysis. After removing these proteins, we were left with a list of 124 high-confidence palmitoyl proteins (Fig. 5C; see also Table S3).
Characteristics of the T. brucei palmitoyl proteome.
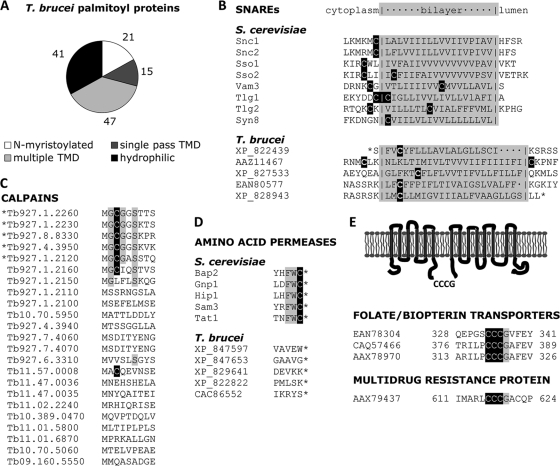
Among the 124 palmitoyl proteins identified in this study (Fig. 5C; Table S3), a variety of sequence contexts for palmitoylation were identified (Fig. 6A). A total of 21 palmitoyl proteins (17%) have sequences predictive of N-myristoylation. In each case, a cysteine residue could be found within five residues of the myristoylated glycine. These are highly likely to be dually acylated. Another 15 (12%) and 47 (38%) palmitoyl proteins in T. brucei have single or multiple predicted transmembrane domains. These are proteins for which palmitoylation is likely to serve a secondary role in modulating protein sorting or association with lipid microdomains. The remaining 41 (33%) proteins, on the other hand, are otherwise hydrophilic. Palmitoylation for this subset likely serves as the main anchor for membrane attachment.
Fig. 6.
Characteristics of T. brucei palmitoyl proteins. (A) Each palmitoyl protein was analyzed for alternative means of membrane association. The proportion belonging to each classification is displayed. (B) As in the yeast proteome, multiple SNAREs with single-pass transmembrane domains were detected. The sequence of the transmembrane domain for each SNARE is displayed with its predicted topology. (C) The N-terminal sequence of each calpain in the T. brucei genome is displayed. Only those calpains sharing a conserved MGCGxxS N terminus were identified in the palmitoyl proteome (indicated with an asterisk). (D) Although several amino acid permeases were identified in the T. brucei palmitoyl proteome, none of them contained the FWC C-terminal tripeptide common to palmitoylated yeast permeases. (E) Three different proteins of the folate/biopterin family were identified as palmitoylated, and each shares a CCCG tetrapeptide in a cytoplasmic loop. Likewise, the multidrug resistance protein A has multiple transmembrane domains and a cytoplasmic CCCG tetrapeptide.
Within these broader classifications of substrates, we identified more specific palmitoylation motifs. As was the case for the yeast palmitoyl proteome (34), we identified multiple SNARE proteins, each containing a single transmembrane domain with a Cys residue next to the cytoplasmic face of the lipid bilayer (Fig. 6B). Also consistent with the yeast palmitoyl proteome, the T. brucei palmitoyl proteome contained several amino acid permeases. These, however, lacked the characteristic C-terminal FWC tripeptide that serves as the acceptor site for the DHHC protein Pfa4 in yeast (Fig. 6D). Thus, while palmitoylation of amino acid permeases is shared between these organisms, the mechanism of this modification is divergent.
Previous investigators have noted the presence of an expanded calpain gene family in the T. brucei genome (11). These proteins lack the C-terminal calmodulin-like calcium-binding domain of mammalian calpains, but most retain a conserved protease motif. We detected five calpain or calpain-like sequences as palmitoylated. Notably, each of these has a sequence predictive of an N-terminal myristoylation/palmitoylation motif. Of the 17 calpain sequences in the genome lacking this motif, we detected none in the palmitoyl proteome (Fig. 6C). These findings both support the sensitivity and specificity of our results and confirm the previous suspicion that this subset of calpains undergoes palmitoylation.
A novel class of proteins with multiple individual hits in the T. brucei palmitoyl proteome consisted of transporters of the folate/biopterin family. These proteins have implications for drug development, since trypanosomes must take up from the environment either folate or biopterin, from which they synthesize folate de novo. Such scavenging renders Leishmania resistant to treatment with antifolates such as methotrexate (20). Despite considerable sequence divergence among them, each has a conserved CCCG tetrapeptide within a 75- to 100-amino-acid hydrophilic loop between transmembrane domains (Fig. 6E). None of the other Cys residues of these proteins is similarly conserved. In Leishmania, folate uptake is growth stage regulated (7) through an unknown posttranscriptional mechanism (4). For the main folate transporter in Leishmania infantum, FT1, the switch from logarithmic phase to stationary phase is accompanied by a redistribution of the protein from the plasma membrane to an internal compartment, where it then becomes degraded (32). The presence of these proteins in our palmitoyl proteome offers a possible explanation, in that palmitoylation might regulate this dynamic localization and degradation in a manner analogous to the stabilization and trafficking of several neuronal proteins by the mammalian PAT HIP14 (16, 39). Another multitransmembrane domain transporter in the T. brucei palmitoyl proteome is multidrug resistance protein A. Like the folate/pteridine transporters, this protein has a CCCG peptide in a region between two of its predicted transmembrane domains. Intriguingly, a homologue of this protein in Leishmania similarly appears to undergo growth stage-dependent redistribution to lysosomal compartments for degradation (6). We therefore propose that these proteins are common substrates for a single PAT whose activity confers growth stage-regulated responses.
DISCUSSION
The discovery of enzymatic regulators of protein palmitoylation, coupled with the development of biochemical methods for purifying palmitoylated proteins, has led to a rapid increase in our understanding of this important protein modification. Palmitoylation increases the lipophilicity of a protein, but many palmitoyl proteins are already associated with membranes via other lipid attachments or transmembrane domains. Thus, the secondary effects of palmitoylation in regulating sorting to different membrane domains, association with lipid rafts, binding partner interactions, and protein degradation are perhaps more significant consequences of this modification.
In T. brucei, treatment with 2-bromopalmitate suggested that palmitoylation serves an essential function in these cells. Although the toxicity of 2-bromopalmitate to mammalian cells limits its therapeutic potential, the considerable sequence divergence among PATs indicates the possibility for targeting individual enzymes. With this in mind, we tested individual PAT RNAi lines in an attempt to identify the enzyme(s) whose inhibition by 2-bromopalmitate was responsible for the effect of this compound on T. brucei growth. None of the 12 individual PAT mutants, however, exhibited a growth deficit in vitro. If palmitoylation is truly essential for T. brucei growth, as suggested by the effect of 2-bromopalmitate, there are three possible explanations for this finding. First, it may be that incomplete inhibition by our RNAi constructs preserved a small functional pool of enzyme that was sufficient to maintain the viable phenotype. While this same approach for TbPAT7 completely blocked calflagin palmitoylation and flagellar trafficking, there are surely different levels of inhibition achieved by each construct, so that incomplete inhibition of an essential PAT might preserve viability. Second, the survival of each individual PAT mutant could be a consequence of enzymatic redundancy. T. brucei may have evolved functional redundancy for catalysis of protein palmitoylation for those substrates essential for parasite viability and growth. Indeed, in yeast there are several proteins for which inhibition of multiple PATs is required to prevent palmitoylation (34), and none of the DHHC-CRD genes is individually essential (37). Finally, there might exist in T. brucei noncanonical PAT activity for essential palmitoyl proteins. While DHHC-CRD proteins are required for the great majority of palmitoylation in S. cerevisiae, at least one substrate (Bet3 in yeast, homologous to Tb927.8.5030 here) undergoes DHHC-independent palmitoylation. Simultaneous inhibition of all 12 PATs genetically could theoretically help distinguish among these possibilities, but the individual enzymes are too divergent at the sequence level to allow for targeting with a single RNAi construct.
A number of proteins in kinetoplastids had been previously reported to undergo thioacylation, including SMP-1 (35) and hydrophilic acylated surface proteins (5) of Leishmania major, protein phosphatases with EF hand domains of L. major and T. brucei (24), phosphoinositide-specific phospholipase C (PLC) (12, 26) and FCaBP of T. cruzi (13), and glycophosphatidylinositol-PLC (28), calflagins (8, 10), and CAP5.5 (15) of T. brucei. Our previous finding that calflagin palmitoylation was mediated by a single DHHC-CRD protein, TbPAT7, established the conservation in T. brucei of PAT activity by DHHC-CRD proteins (10). Here we identified a total of 124 palmitoyl proteins, of which very few are likely to represent statistical anomalies. Our confidence in these data is strengthened by the identification of each of the known positive controls expressed in the procyclic stage, as well as many proteins very likely to be palmitoylated on the basis of their similarity to known palmitoyl proteins.
It is important to note, however, that the NH2OH-dependent labeling and purification of proteins in this study would not distinguish between covalent attachment of palmitate or a different long-chain fatty acid. Although palmitate has been shown to be the predominant such modification in mammalian cells, thioesterification of stearate, palmitoleate, and oleate have also been observed (21a). This consideration is especially salient given the unique features of fatty acid synthesis in T. brucei (21) and the observation in this organism of protein S-myristoylation (1). Therefore, this palmitoyl proteome set might more aptly be termed a “thioacylome.” For the sake of consistency with previously published reports, we nevertheless use palmitoylation as a more general term referring to Cys thioester-linked fatty acids.
These findings are likely to apply to several other pathogenic kinetoplastid parasites. The PAT family of enzymes is well represented in each of T. brucei, T. cruzi, L. major, L. infantum, and Leishmania brasiliensis. Most PATs retain conservation in gene synteny among these species, but lineage-specific variation is also evident, including gene loss and tandem duplication. For a given PAT, sequence variation is highly conserved at the DHHC-CRD catalytic motif and transmembrane domains but divergent for hydrophilic loops of the protein. This pattern likely reflects the underlying function of each region, that while selective pressure on the DHHC-CRD was necessary for retention of catalytic function, the other hydrophilic loops of PATs may serve as selectivity filters for each enzyme. Thus, their divergence probably correlates with divergence of their substrates, possibly with the concordant acquisition and loss of substrates.
This work also lays the groundwork for a comprehensive mapping of individual protein substrates to their cognate PAT enzymes. As was shown for TbPAT7 and the calflagins, purification of each substrate by acyl biotin exchange/streptavidin should depend on the presence of its PAT. The wild-type palmitoyl proteome, therefore, should provide a baseline state from which we can determine the dependence of each palmitoyl protein on individual PAT expression, by acyl biotin exchange proteomics on each of the 12 PAT RNAi lines. Finally, the utility of this data set should only increase as more is learned about the function of individual proteins in T. brucei. Given the evolutionary distance of trypanosomes from better-studied eukaryotes, it is not surprising that such a large proportion of palmitoylated proteins are annotated as hypothetical proteins. As more is uncovered about the function of these proteins, palmitoylation will undoubtedly become implicated in an ever broader array of cellular processes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Amy Roth for advice on the palmitoyl proteome protocol and the Biomolecule Analysis Core Facility at the Border Biomedical Research Center/Biology/UTEP (funded by NIH/NCRR grant 5G12RR008124-16A1) for use of the LC-MS instrument.
This work was supported in part by NIH grants R01 AI46781 (D.M.E.), 5G12RR008124-A16 and 5G12RR008124-16A1S1, and R01 CA126239 (A.I.N.). B.T.E. was supported by predoctoral fellowships from the American Heart Association and the NIH (F30 HL094026). E.S.N. was supported in part by a George A. Krutilek Memorial Scholarship from the Graduate School at UTEP. T.J.P.S. was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo-FAPESP, Brazil.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 30 December 2010.
REFERENCES
- 1. Armah D. A., Mensa-Wilmot K. 1999. S-myristoylation of a glycosylphosphatidylinositol-specific phospholipase C in Trypanosoma brucei. J. Biol. Chem. 274:5931–5938 [DOI] [PubMed] [Google Scholar]
- 2. Bologna G., Yvon C., Duvaud S., Veuthey A. L. 2004. N-terminal myristoylation predictions by ensembles of neural networks. Proteomics 4:1626–1632 [DOI] [PubMed] [Google Scholar]
- 3. Choi H., Fermin D., Nesvizhskii A. I. 2008. Significance analysis of spectral count data in label-free shotgun proteomics. Mol. Cell Proteomics 7:2373–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cunningham M. L., Beverley S. M. 2001. Pteridine salvage throughout the Leishmania infectious cycle: implications for antifolate chemotherapy. Mol. Biochem. Parasitol. 113:199–213 [DOI] [PubMed] [Google Scholar]
- 5. Denny P. W., Gokool S., Russell D. G., Field M. C., Smith D. F. 2000. Acylation-dependent protein export in Leishmania. J. Biol. Chem. 275:11017–11025 [DOI] [PubMed] [Google Scholar]
- 6. Dodge M. A., et al. 2004. Localization and activity of multidrug resistance protein 1 in the secretory pathway of Leishmania parasites. Mol. Microbiol. 51:1563–1575 [DOI] [PubMed] [Google Scholar]
- 7. Ellenberger T. E., Beverley S. M. 1987. Biochemistry and regulation of folate and methotrexate transport in Leishmania major. J. Biol. Chem. 262:10053–10058 [PubMed] [Google Scholar]
- 8. Emmer B. T., Daniels M. D., Taylor J. M., Epting C. L., Engman D. M. 2010. Calflagin inhibition prolongs host survival and suppresses parasitemia in Trypanosoma brucei infection. Eukaryot. Cell 9:934–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emmer B. T., Maric D., Engman D. M. 2010. Molecular mechanisms of protein and lipid targeting to ciliary membranes. J. Cell Sci. 123:529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Emmer B. T., et al. 2009. Identification of a palmitoyl acyltransferase required for protein sorting to the flagellar membrane. J. Cell Sci. 122:867–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ersfeld K., Barraclough H., Gull K. 2005. Evolutionary relationships and protein domain architecture in an expanded calpain superfamily in kinetoplastid parasites. J. Mol. Evol. 61:742–757 [DOI] [PubMed] [Google Scholar]
- 12. Furuya T., Kashuba C., Docampo R., Moreno S. N. 2000. A novel phosphatidylinositol-phospholipase C of Trypanosoma cruzi that is lipid modified and activated during trypomastigote to amastigote differentiation. J. Biol. Chem. 275:6428–6438 [DOI] [PubMed] [Google Scholar]
- 13. Godsel L. M., Engman D. M. 1999. Flagellar protein localization mediated by a calcium-myristoyl/palmitoyl switch mechanism. EMBO J. 18:2057–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hershko A., Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479 [DOI] [PubMed] [Google Scholar]
- 15. Hertz-Fowler C., Ersfeld K., Gull K. 2001. CAP5.5, a life-cycle-regulated, cytoskeleton-associated protein is a member of a novel family of calpain-related proteins in Trypanosoma brucei. Mol. Biochem. Parasitol. 116:25–34 [DOI] [PubMed] [Google Scholar]
- 16. Huang K., et al. 2004. Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron 44:977–986 [DOI] [PubMed] [Google Scholar]
- 17. Jurado J. D., et al. 2007. Complement inactivating proteins and intraspecies venom variation in Crotalus oreganus helleri. Toxicon 49:339–350 [DOI] [PubMed] [Google Scholar]
- 18. Kang R., et al. 2008. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456:904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 20. Kundig C., Haimeur A., Legare D., Papadopoulou B., Ouellette M. 1999. Increased transport of pteridines compensates for mutations in the high affinity folate transporter and contributes to methotrexate resistance in the protozoan parasite Leishmania tarentolae. EMBO J. 18:2342–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee S. H., Stephens J. L., Paul K. S., Englund P. T. 2006. Fatty acid synthesis by elongases in trypanosomes. Cell 126:691–699 [DOI] [PubMed] [Google Scholar]
- 21a. Liang X., Lu Y., Wilkes M., Neubert T. A., Resh M. D. 2004. The N-terminal SH4 region of the Src family kinase Fyn is modified by methylation and heterogeneous fatty acylation: role in membrane targeting, cell adhesion, and spreading. J. Biol. Chem. 279:8133–8139 [DOI] [PubMed] [Google Scholar]
- 22. Linder M. E., Deschenes R. J. 2007. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8:74–84 [DOI] [PubMed] [Google Scholar]
- 23. Lobo S., Greentree W. K., Linder M. E., Deschenes R. J. 2002. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277:41268–41273 [DOI] [PubMed] [Google Scholar]
- 24. Mills E., Price H. P., Johner A., Emerson J. E., Smith D. F. 2007. Kinetoplastid PPEF phosphatases: dual acylated proteins expressed in the endomembrane system of Leishmania. Mol. Biochem. Parasitol. 152:22–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohno Y., Kihara A., Sano T., Igarashi Y. 2006. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta 1761:474–483 [DOI] [PubMed] [Google Scholar]
- 26. Okura M., et al. 2005. A lipid-modified phosphoinositide-specific phospholipase C (TcPI-PLC) is involved in differentiation of trypomastigotes to amastigotes of Trypanosoma cruzi. J. Biol. Chem. 280:16235–16243 [DOI] [PubMed] [Google Scholar]
- 27. Passmore L. A., Barford D. 2004. Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem. J. 379:513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paturiaux-Hanocq F., et al. 2000. A role for the dynamic acylation of a cluster of cysteine residues in regulating the activity of the glycosylphosphatidylinositol-specific phospholipase C of Trypanosoma brucei. J. Biol. Chem. 275:12147–12155 [DOI] [PubMed] [Google Scholar]
- 29. Redmond S., Vadivelu J., Field M. C. 2003. RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol. 128:115–118 [DOI] [PubMed] [Google Scholar]
- 30. Ren J., et al. 2008. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 21:639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Resh M. D. 2006. Use of analogs and inhibitors to study the functional significance of protein palmitoylation. Methods 40:191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Richard D., Leprohon P., Drummelsmith J., Ouellette M. 2004. Growth phase regulation of the main folate transporter of Leishmania infantum and its role in methotrexate resistance. J. Biol. Chem. 279:54494–54501 [DOI] [PubMed] [Google Scholar]
- 33. Roth A. F., Feng Y., Chen L., Davis N. G. 2002. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159:23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roth A. F., et al. 2006. Global analysis of protein palmitoylation in yeast. Cell 125:1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tull D., et al. 2004. SMP-1, a member of a new family of small myristoylated proteins in kinetoplastid parasites, is targeted to the flagellum membrane in Leishmania. Mol. Biol. Cell 15:4775–4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wan J., Roth A. F., Bailey A. O., Davis N. G. 2007. Palmitoylated proteins: purification and identification. Nat. Protoc. 2:1573–1584 [DOI] [PubMed] [Google Scholar]
- 37. Winzeler E. A., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906 [DOI] [PubMed] [Google Scholar]
- 38. Wirtz E., Leal S., Ochatt C., Cross G. A. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 39. Yanai A., et al. 2006. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat. Neurosci. 9:824–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zybailov B., et al. 2006. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5:2339–2347 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.