Abstract
The G1/S transition is a critical control point for cell proliferation and involves essential transcription complexes termed SBF and MBF in Saccharomyces cerevisiae or MBF in Schizosaccharomyces pombe. In the fungal pathogen Candida albicans, G1/S regulation is not clear. To gain more insight into the G1/S circuitry, we characterized Swi6p, Swi4p and Mbp1p, the closest orthologues of SBF (Swi6p and Swi4p) and MBF (Swi6p and Mbp1p) components in S. cerevisiae. The mbp1Δ/Δ cells showed minor growth defects, whereas swi4Δ/Δ and swi6Δ/Δ yeast cells dramatically increased in size, suggesting a G1 phase delay. Gene set enrichment analysis (GSEA) of transcription profiles revealed that genes associated with G1/S phase were significantly enriched in cells lacking Swi4p and Swi6p. These expression patterns suggested that Swi4p and Swi6p have repressing as well as activating activity. Intriguingly, swi4Δ/Δ swi6Δ/Δ and swi4Δ/Δ mbp1Δ/Δ strains were viable, in contrast to the situation in S. cerevisiae, and showed pleiotropic phenotypes that included multibudded yeast, pseudohyphae, and intriguingly, true hyphae. Consistently, GSEA identified strong enrichment of genes that are normally modulated during C. albicans-host cell interactions. Since Swi4p and Swi6p influence G1 phase progression and SBF binding sites are lacking in the C. albicans genome, these factors may contribute to MBF activity. Overall, the data suggest that the putative G1/S regulatory machinery of C. albicans contains novel features and underscore the existence of a relationship between G1 phase and morphogenetic switching, including hyphal development, in the pathogen.
INTRODUCTION
The G1 phase of the cell cycle is the stage when cells commit to mitosis and proliferation or embark on developmental pathways in response to environmental and internal cues (54, 81). A crucial feature of the G1/S transition involves transcriptional regulation (64). In mammals, cyclin-dependent kinase (CDK) activity involving CDK4/cyclin D acts upstream of the G1/S circuit and functions to inactivate retinoblastoma protein (pRb), a repressor of the E2F G1/S transcription factor. Activation of E2F triggers a transcriptional cascade that is required for cell proliferation (38). In Saccharomyces cerevisiae, the CDK Cdc28p associates with the G1 cyclin Cln3p and activates the SBF/MBF G1/S transcription factor complexes, which are composed of ankyrin repeat proteins (9). Specifically, SBF is composed of the transcriptional activator Swi6p in association with the DNA binding factor Swi4p and mediates G1/S early events, such as cyclin expression, budding, and cell wall deposition. SBF is activated through Cdc28p/Cln3p-mediated inactivation of Whi5p, a repressor of SBF and, thus, the functional equivalent of pRB (24, 29). MBF is composed of Swi6p in combination with the DNA binding element Mbp1p (77). MBF represses its targets, many of which are associated with DNA replication, outside of G1 phase with the assistance of the corepressor Nrm1p (28). SBF and MBF bind SCB (Swi4/6 cell cycle binding box) and MCB (MluI cell cycle box) elements, respectively (17), although cross-binding can occur (12). Their numerous targets include other transcription factors, TOS4, HCM1, and FKH2, for example, and the G1 cyclins CLN1 and CLN2 (41). SBF/MBF activity is also regulated by Bck2p and Stb1p (25, 31). In Schizosaccharomyces pombe, a single MBF complex mediates G1/S transcription and is composed of the activating factor Cdc10p and at least two DNA binding elements, Res1p and Res2p (9, 19, 55, 84). SBF and/or MBF complexes are crucial for G1/S control in fungi, since their absence results in a G1 phase arrest and/or inviable cells (3, 11, 44, 55, 60).
The G1/S circuitry is also linked to development. CDK inhibitors or downregulation of CDK activators can induce a G1 phase arrest, which precedes cell differentiation (54). For example, in S. cerevisiae, Cln3p is downregulated and G1 phase is blocked or delayed in response to external signals that trigger meiosis, conjugation tube formation, and pseudohyphal growth (35, 81). In higher organisms, G1/S regulators, including pRB, E2F, and CDK inhibitors, can also regulate differentiation independent of their cell cycle functions (30, 54). Thus, G1 phase is tightly coordinated with development in most systems, and G1/S regulatory factors can play independent roles in controlling developmental events.
Candida albicans is one of the most prevalent fungal pathogens in humans. Its ability to differentiate into a variety of cell types, including white phase yeast, mating-competent opaque phase yeast, pseudohyphae, hyphae, or chlamydospores (79), is a crucial virulence-determining trait (49, 66). Thus, it is important to identify the mechanisms underlying basic cell proliferation and differentiation in this organism. However, a detailed picture of the G1/S circuit based on functional analyses is lacking. Cote et al. (26) reported cell cycle-dependent transcription patterns in opaque yeast cells of C. albicans, which were most similar to those in S. cerevisiae. However, some unique features in putative G1/S circuitry were noted, including the potential involvement of fungal-specific genes (26). Moreover, sequence orthologues of key players, including BCK2, WHI5, and NRM1, are lacking, the G1 cyclin Cln3p is essential for G1/S progression (7, 22), unlike in S. cerevisiae, and the C. albicans genome lacks predicted SCB elements, which raises the possibility that an MBF complex mediates G1/S transcription (26). Thus, a framework of the G1/S circuit in C. albicans is emerging, but identification of additional players and crucial functional studies are required. It is also not known whether this circuitry is the same in white and opaque cells; some aspects of cell cycle regulation differ between C. albicans cell types (7, 67).
It is less clear how the G1/S circuit may be integrated with development in C. albicans. Hyphal growth is stimulated by environmental conditions and mediated by several signaling pathways (79). However, maintenance of hyphal growth does not correlate with one cell cycle stage (37) and there are conflicting data on whether hyphal initiation is restricted to G1 phase (37, 70). Orthologues of G1/S-associated factors in other systems can influence hyphal growth in C. albicans, including the cyclins Ccn1p and Hgc1p, the CDK Cdc28p, and the F-box protein Cdc4p (2, 50, 76, 83). With the exception of Ccn1p, however, none of these factors were shown to influence G1/S phase itself. Depletion of the G1 cyclin Cln3p, on the other hand, resulted in a G1 phase arrest in yeast cells and subsequent development of hyphae and pseudohyphae (7, 22), supporting a link between G1 phase and hyphal development. Depletion of Cln3p in S. cerevisiae, on the other hand, did not activate development (27, 59), and blocking/delaying other cell cycle stages in C. albicans did not result in true hyphae (1, 5, 6, 10, 13, 14, 18, 47, 68, 75, 80, 83).
In order to gain new insight into the G1/S circuitry and potential mediators of Cln3p in C. albicans, we characterized orthologues of Swi6p, Swi4p, and Mbp1p. Our results suggest that Swi4p and Swi6p are important for cell proliferation, but in contrast to the situation in other organisms, their combined function is not essential for G1/S progression. We also demonstrate that Swi4p and Swi6p influence morphogenesis, including hyphal differentiation. Together, our results reveal unique variations in the cell cycle circuitry of C. albicans and support the existence of a relationship between G1 phase and hyphal development.
MATERIALS AND METHODS
Media and growth conditions.
Candida albicans strains were grown at 30°C on solid or in liquid glucose minimal medium (0.67% yeast nitrogen base without amino acids, 2% glucose) supplemented with all amino acids except during selection for URA3, HIS1, or ARG4 phototrophs. For MET3 conditional strains, cells were grown in inducing (−MC) or repressing (+MC) minimal medium with or without 2.5 mM methionine and 0.5 mM cysteine, respectively (20). For analysis of cell phenotype, cells were grown overnight in minimal medium, diluted the following day to an optical density at 600 nm (OD600) of 0.1 in fresh medium, and incubated at 30°C.
Strain construction.
Strains, oligonucleotides, and plasmids are listed in Tables 1, 2, and 3, respectively. In order to construct a strain lacking SWI4, alleles were replaced with URA3 and HIS1 markers in strain BWP17, using 2-step PCR fusion constructs (62, 82). Fragments approximately 750 bp in length corresponding to the 5′ and 3′ flanks of SWI4 were amplified with oligonucleotides BH10F and BH10R and BH14F and BH14R, respectively. A HIS1 fragment from plasmid pBS-CaHIS1 was amplified with oligonucleotides BH13F and BH13R. The products were amplified with oligonucleotides BH10F and BH14R to produce a 2,916-bp product that was transformed into strain BWP17, resulting in strain BH180 (swi4Δ::HIS1/SWI4). To delete the second copy of SWI4, a PCR fusion construct containing the same 5′ and 3′ flanks as described above and a 1,765-bp URA3 fragment, amplified from plasmid pBS-CaURA3 with oligonucleotides BH13F and BH13R, was utilized. The final 2,926-bp construct was transformed into strain BH180, resulting in strain BH185 (swi4Δ::HIS/swi4Δ::URA3). A prototrophic control strain (BH440) was created by sequentially transforming strain BWP17 with plasmids pBS-CaHIS and pBS-CaURA3.
Table 1.
Candida albicans strains used in this study
| Strain | Genotype | Parent/source |
|---|---|---|
| BWP17 | ura3Δ::imM434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG arg4Δ::hisG/arg4Δ::hisG | 33 |
| BH101 | swi6Δ::HIS1/SWI6 | BWP17 |
| BH104 | swi4Δ::hisG-URA3-hisG/SWI4 | BWP17 |
| BH113 | swi4Δ::hisG/SWI4 | BWP17 |
| BH115 | swi4Δ::hisG/SWI4 | BWP17 |
| BH120 | swi6Δ::HIS1/swi6Δ::URA3 | BWP17 |
| BH137 | mbp1Δ::HIS1/MBP1 | BWP17 |
| BH140 | swi4Δ::hisG/SWI4 swi6Δ::HIS1/SWI6 | BWP17 |
| BH150 | swi4Δ::hisG/MET3::SWI4-ARG4 | BWP17 |
| BH160 | swi4Δ::hisG/MET3::SWI4-ARG4 swi6Δ::HIS1/SWI6 | BWP17 |
| BH180 | swi4Δ::HIS1/SWI4 | BWP17 |
| BH185 | swi4Δ::URA3/swi4Δ::HIS1 | BWP17 |
| BH190 | swi4Δ::hisG/MET3::SWI4-ARG4 swi6::HIS1/swi6::URA3 | BWP17 |
| BH261 | mbp1Δ::HIS1/mbp1Δ::URA3 | BWP17 |
| BH270 | swi4Δ::hisG/MET3::SWI4-ARG4 mbp1Δ::HIS1/MBP1 | BWP17 |
| BH277 | swi4Δ::hisG/MET3::SWI4-ARG4 mbp1Δ::HIS1/mbp1Δ::URA3 | BWP17 |
| BH339 | swi4Δ::hisG/swi4Δ::URA3 | BWP17 |
| BH342 | swi4Δ::hisG/swi4Δ::URA3 mbp1Δ::HIS1/MBP1 | BWP17 |
| HH62 | swi4Δ::hisG/swi4Δ::URA3 mbp1Δ::HIS1/mbp1Δ::ARG4 | BWP17 |
| BH420 | BWP17 (pRM100 pBS-CaARG4) | BWP17 |
| BH440 | BWP17 (pBS-CaHIS1 pBS-CaURA3) | BWP17 |
| CB540 | swi4Δ::URA3/SWI4 | BWP17 |
| CB547 | swi4Δ::URA3/MET3::SWI4-ARG4 | BWP17 |
| CB557 | swi6Δ::URA3/MET3::SWI6-HIS1 | BWP17 |
| CB600 | SWI6/MET3::SWI6-HIS1 | BWP17 |
| AG160 | swi4Δ::hisG/swi4Δ::URA3 swi6Δ::HIS1/SWI6 | BWP17 |
| AG168 | swi4Δ::hisG/swi4Δ::URA3 swi6Δ::HIS1/swi6Δ::ARG4 | BWP17 |
| KMCa4a | mbp1Δ::URA3/MET3::MBP1-ARG4 | BWP17 |
Table 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| BH2F | GAAGATTCATTGATATGTGTGGTAAGGCAC |
| BH2R | CCAGCGTTTATAATGATAACGTTCAGCTTC |
| BH3F | GAAGCTGAACGTTATCATTATAAACGCTGGTATAGGGCGAATTGGAGCTC |
| BH3R | CACGGGGAATTAGAAGTATACATGTGTTCGGACGGTATCGATAAGCTTGA |
| BH4F | CGAACACATGTATACTTCTAATTCCCCGTG |
| BH4R | TTCCACATCCATACTAAATCTTACTACAGC |
| BH7F | GTAACAATACCTTTATCAGAGGATTCACCC |
| BH7R | GATGTGATGGGTTGATAAATGAAATGAGCG |
| BH8F | CGCTCATTTCATTTATCAACCCATCACATCTATAGGGCGAATTGGAGCTC |
| BH8R | GGATGCTGGTAAAGTAGTAGAGTATGATAAGACGGTATCGATAAGCTTGA |
| BH9F | TTATCATACTCTACTACTTTACCAGCATCC |
| BH9R | GTTCCTATTATCTGTTGCTTGTGTTGCCCA |
| BH10F | AGCAGTATCTACATGAGAATTAATCAGATG |
| BH10R | TTGGTATAACATACATTTGAGTAGGTAGCT |
| BH11F | AGCTACCTACTCAAATGTATGTTATACCAAGGATCCCCCCTTTAGTAAGA |
| BH11R | ACACCTTCTCATAATTGAGGACTCGTTCATCATGTTTTCTGGGGAGGGTA |
| BH12F | ATGAACGAGTCCTCAATTATGAGAAGGTGT |
| BH12R | CGATCAATGTATTTCCATTGTCAATCGGTT |
| BH13F | AGCTACCTACTCAAATGTATGTTATACCAATATAGGGCGAATTGGAGCTC |
| BH13R | TTGTTCCGATTTAATTTCCCCCATCTATCGGACGGTATCGATAAGCTTGA |
| BH14F | CGATAGATGGGGGAAATTAAATCGGAACAA |
| BH14R | AATATTTGTGTTGGCCACATTTGAGTCTGA |
| CB115F | CCAAATGGGATATATATGAAGATTCATTGATATGTGTGGTAAGGCACAACTTACACTCTAGCATACCCAAATGGTTTCGGTATAGGGCGAATTGGAGCTC |
| CB115R | CAAACTCCTGAAGCTAGAGTAGATAGATATTTCCAGTTGTTGGGCAAAGACAAGAATACCGACAAATTAGATTTGAATGAGACGGTATCGATAAGCTTGA |
| CB119F | ACGCGTCGACTCTAATTGACATGGATACGA |
| CB119R | ACGCTAGAGCTCTCTGGATTAGTCACATCTTC |
| CB120F | ACGCTAGGATCCTCGTACTGGCAATGTATAACT |
| CB120R | ACGCTATAGATCTCATCGGTCTAGATTGTAATAT |
| CB122F | CTACTACATAATGTCTGAACCTCCCCAAGTATTTCGAGCTACCTACTCAAATGTATGTTATACCAATGTTTCTTAACTTCTATAGGGCGAATTGGAGCTC |
| CB122R | CAATGCACTAAAGTTCTAGATCCTCGCAGATAATAAAGTTGTTTGATACATGAACATCGAGTGCCTTTTCTAATATTGGAGACGGTATCGATAAGCTTGA |
| CB127F | CCAAATGGGATATATATGAAGATTCATTGATATGTGTGGTAAGGCACAACTACACTCTAGCATACCCAAATGGTTTCGGGGATCCTGGAGGATGAGGG |
| CB127R | ATTGTTGTTATTTATATGGGTCTCGAATAATTTTTGTTGAATTGACTGTGTTGTCAAGTCACCAATGTGTATAGGAGAATCCATGTTTTCTGGGGAGGGT |
| CB129F | CGTTCAGCTTCCTTTCAATGAAATAAGTAT |
| CB129R | TCAGCAGACACAACAAGATACTGATACTTG |
| CB130F | GAACACATGTATACTTCTAATTCCCCGTGT |
| CB130R | ATTTGAGGCAGCTTCGACAGGCCACGTATT |
| CB131F | ATACTTATTTCATTGAAAGGAAGCTGAACGTATAGGGCGAATTGGAGCTC |
| CB131R | ACACGGGGAATTAGAAGTATACATGTGTTCGACGGTATCGATAAGCTTGA |
Table 3.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pBS-CaURA3 | pBluescript CaURA3 | A. J. P. Brown |
| pBS-CaHIS1 | pBluescript CaHIS1 | C. Bachewich |
| pBS-CaARG4 | pBluescript CaARG4 | C. Bachewich |
| pFA-MET3p-CaURA3 | pFA-MET3p-CaURA3 | 36 |
| pFA-MET3p-CaHIS1 | pFA-MET3p-CaHIS1 | 36 |
| pFA-MET3p-CaARG4 | pFA-MET3p-CaARG4 | 36 |
| p5921 | pUC18-hisG-URA3-hisG | W. A. Fonzi |
| pRM100 | pUC19 URA3HIS1 | J. Pla |
| pCB180 | pUC18-SWI4 | This study |
| pCB181 | pUC18-hisG-URA3-hisG with SWI4 flanks | This study |
In order to confirm that the phenotype of strain BH185 was due to the deletion of SWI4, a conditional strain carrying a single copy of SWI4 under the control of the MET3 promoter was created. A 3-kb fragment containing the SWI4 open reading frame and approximately 1 kb of 3′ and 5′ flanking sequences was amplified from genomic DNA (gDNA) with oligonucleotides CB119F and CB119R and cloned into SalI/SacI sites of pUC18, producing plasmid pCB180. Primers CB120F and CB120R were then used to amplify the flanking and vector sequences from pCB180, into which the BamHI/BglII hisG-URA3-hisG cassette (p5921) (33) was cloned, resulting in plasmid pCB181. The SWI4 deletion construct was liberated using SalI and SacI restriction enzymes and transformed into strain BWP17. The resulting strain, BH104, was grown overnight in yeast extract-peptone-dextrose (YPD) medium and then plated onto 5-fluoroorotic acid (5-FOA) to select for URA3− auxotrophs. PCR screening confirmed strain BH113 (swi4Δ::hisG/SWI4). To place the second copy of SWI4 under the control of the MET3 promoter (20), fragments corresponding to the 5′ and 3′ flanks of SWI4 were amplified using oligonucleotides BH10F and BH10R and BH12F and BH12R, respectively. The MET3 promoter from plasmid pFA-CaARG4-MET3 (36) was amplified with oligonucleotides BH11F and BH11R. The final 4,895-bp construct was amplified from the three PCR products using oligonucleotides BH10F and BH12R and transformed into strain BH115, producing strain BH150 (swi4Δ::hisG/MET3::SWI4-ARG4). Alternatively, oligonucleotides CB122F and CB122R amplified a URA3-containing deletion construct from pBS-CaURA3, which was transformed into strain BWP17, resulting in strain CB540 (swi4Δ::URA3/SWI4). The second copy was placed under the control of the MET3 promoter as described above, resulting in strain CB547 (swi4Δ::URA3/MET3::SWI4-ARG4).
In order to delete one copy of MBP1, a similar PCR fusion strategy was utilized. PCR fragments corresponding to the 5′ and 3′ flanks of MBP1 were amplified using oligonucleotides BH7F and BH7R and BH9F and BH9R, respectively. A HIS1 fragment was amplified from plasmid pBS-CaHIS1 using oligonucleotides BH8F and BH8R. The final 3,006-bp fusion construct was amplified with oligonucleotides BH7F and BH9R and transformed into strain BWP17, resulting in strain BH137 (mbp1Δ::HIS1/MBP1). The second copy of MBP1 was replaced with a similar PCR fusion construct containing a URA3 fragment that was amplified from plasmid pBS-CaURA3 with oligonucleotides BH8F and BH8R. The final 3,016-bp fusion product was transformed into strain BH137, resulting in strain BH261 (mbp1Δ::HIS1/mbp1Δ::URA3).
In order to create a SWI6 deletion strain, a HIS1-containing construct containing 80 bp complementary to the 5′ and 3′ flanks of SWI6 was amplified with oligonucleotides CB115F and CB115R from pBS-CaHIS1 and transformed into strain BWP17, resulting in strain BH101 (swi6Δ:: HIS1/SWI6). The second copy of SWI6 was deleted using a PCR fusion construct. Oligonucleotides BH2F and BH2R and BH4F and BH4R amplified 5′ and 3′ flanking fragments of SWI6, respectively. The URA3 marker from pBS-CaURA3 was amplified with oligonucleotides BH3F and BH3R. The final 2,721-bp fusion construct was amplified with oligonucleotides BH2F and BH4R and transformed into strain BH101, resulting in strain BH120 (swi6Δ::HIS1/swi6Δ::URA3). A strain containing a single conditional copy of SWI6 was created by placing one allele of SWI6 under the control of the MET3 promoter, using a construct created with oligonucleotides CB127F and CB127R that contained 80 bp complementary to regions immediately up- and downstream of the SWI6 start codon, respectively, and 20 bp homologous to plasmid pFA-MET3-HIS1 (36). The final product was transformed into strain BWP17, producing strain CB600. The second allele of SWI6 was deleted using a PCR fusion construct. Oligonucleotides CB129F and CB129R and CB130F and CB130R amplified the 5′ and 3′ flank of SWI6, respectively. The URA3 marker from pBS-CaURA3 was amplified with oligonucleotides CB131F and CB131R. The final 3,000-bp fusion construct was amplified with oligonucleotides CB129F and CB130R and transformed into strain CB600, resulting in strain CB557 (swi6Δ::URA3/MET3::SWI6-HIS1).
In order to create a strain depleted of both Swi4p and Swi6p, the first copy of SWI6 was deleted from strain BH115 (swi4Δ::hisG/SWI4) using a PCR construct created with oligonucleotides CB115F and CB115R, as described above. The final 1,508-bp product was transformed into strain BH115, resulting in strain BH140 (swi4Δ::hisG/SWI4 swi6Δ::HIS1/SWI6). The second copy of SWI4 was then placed under the control of the MET3 promoter using a PCR fusion construct created with oligonucleotides BH10F and BH10R, BH11F and BH11R, and BH12F and BH12R, as described above. The final 4,895-bp product was transformed into strain BH140, resulting in strain BH160 (swi4Δ::hisG/MET::SWI4-ARG4 swi6Δ::HIS1/SWI6). The second copy of SWI6 was deleted using a PCR fusion construct created from oligonucleotides BH2F and BH2R, BH3F and BH3R, and BH4F and BH4R as described above, and the final 2,721-bp product was transformed into strain BH160, resulting in strain BH190 (swi4Δ::hisG/MET::SWI4-ARG4 swi6Δ::HIS1/swi6Δ::URA3). A prototrophic control strain was created by sequentially transforming plasmids pRM100 and pBS-CaARG4 into strain BWP17, resulting in strain BH420. To confirm the phenotype of strain BH190, a strain with both SWI4 and SWI6 deleted was created. The remaining SWI4 allele in strain BH113 was replaced with a URA3-containing construct created with oligonucleotides BH10F, BH10R, BH13F, BH13R, BH14F, and BH14R and pBS-CaURA3, resulting in strain BH339 (swi4Δ::hisG/swi4Δ::URA3). A SWI6 allele was then replaced with a HIS1-containing construct, created with oligonucleotides CB115F and CB115R and pBS-CaHIS1, resulting in strain AG160 (swi4Δ::hisG/swi4Δ::URA3 swi6Δ::HIS1/SWI6). The remaining SWI6 allele was replaced with an ARG4-containing construct created with oligonucleotides CB115F and CB115R and pBS-CaARG4. The final product was transformed into strain AG160, resulting in strain AG168 (swi4Δ::hisG/swi4Δ::URA3 swi6Δ::HIS1/swi6Δ::ARG4).
To create a strain that lacked SWI4 and MBP1, the first copy of MBP1 was deleted from strain BH150 (swi4Δ::hisG/MET::SWI4-ARG4) by transforming a HIS1-containing PCR fusion construct as described above into strain BH150, resulting in strain BH270 (swi4Δ::hisG/MET::SWI4-ARG4 mbp1Δ::HIS1/MBP1). The second copy of MBP1 was replaced with a URA3 marker as described above, resulting in strain BH277 (swi4Δ::hisG/MET::SWI4-ARG4 mbp1Δ::URA3/mbp1Δ::HIS1). To confirm the phenotype of strain BH277 under repressing conditions, a strain with both SWI4 and MBP1 deleted was created. A copy of MBP1 was deleted from strain BH339 using a HIS1 replacement construct as described above, resulting in strain BH342 (swi4Δ::hisG/swi4Δ::URA3 mbp1Δ::HIS1/MBP1). The second copy of MBP1 was replaced with a PCR fusion construct as described above but containing an ARG4 marker that was amplified with oligonucleotides BH8F and BH8R. The final 3,785-bp product was transformed into strain BH342, resulting in strain HH62 (swi4Δ::hisG/swi4Δ::URA3 mbp1Δ::HIS1/mbp1Δ::ARG4).
PCRs were performed with Expand long template polymerase (Roche). Cells were transformed using lithium acetate (23, 36) with modifications. Transformation mixtures were incubated overnight at 30°C and heat shocked at 43°C for 15 to 60 min prior to plating on selective medium. gDNA was extracted according to the method of Rose et al. (65a). All strains were screened by PCR and by Southern blot analysis using a DIG (digoxigenin) hybridization system kit (Roche) (data not shown).
Transcriptional profiling.
For transcription profiling of strains lacking Swi4p and Swi6p, overnight cultures of strains BH190 and BH420 were diluted to an OD600 of 0.2 in 10 ml of repressing medium and collected after 7 h of incubation at 30°C. Cell pellets were fast-frozen and stored at −80°C until use. Microarray hybridizations were conducted with four pairs of RNA preparations produced from independent cultures. RNA from cell pellets was extracted using a MasterPure yeast RNA purification kit (Epicentre Biotechnologies, InterScience, Markham, ON, Canada). Total RNA (40 μg) was used in direct labeling with dCTP linked to Cy3 or Cy5. Sample labeling and hybridizations to oligonucleotide microarrays were performed as described previously (57). Slides were scanned and quantified with an Axon GenePix pro 4.0 scanner (Axon Instruments, Inc., Sunnyvale, CA). Data normalization using LOWESS and statistical analyses were performed with GeneSpring, version 7.3 (Agilent Technologies, Santa Clara, CA). Genes with a significant change in transcript abundance were identified in a Volcano plot using a 1.7-fold-change cutoff along with a t test function using as the confidence level a P value of <0.05. To construct pie chart distributions of the total set of modulated genes, the data were manually sorted into select categories. The data were also sorted according to biological processes using the gene ontology (GO) Slim Mapper at Candida Genome Database (CGD) (http://www.candidagenome.org/cgi-bin/GO/goTermMapper). Significant enrichment of genes within a particular process was determined by comparing the number of modulated genes within Swi4p- and Swi6p-depleted cells that grouped to a particular process to the total number of genes within the genome that sorted to the same process, using the Fisher exact test. Alternatively, we used gene set enrichment analysis (GSEA) (56, 72) to compare a ranked list of genes modulated in the Swi4p- and Swi6p-depleted cells to 29 lists of 64 to 558 genes that exhibit cell cycle-dependent periodic expression in C. albicans opaque cells (26) or significant changes during polar morphogenesis, including yeast-to-hypha transitions and the production of highly elongated buds due to M or S phase arrest (5, 34, 43, 53, 58, 71). We used the “classic” Enrichment statistic setting and calculated the false discovery rate (FDR) by performing 10,000 permutations. Selected result graphs are shown, and the complete GSEA output folder is included as GSEA data file S1 in the supplemental material.
Cell imaging.
Nuclei and septa were visualized by fixing cells in 70% ethanol for 1 h, followed by incubation in 1 μg/ml 4′,6′diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma) for 20 min. After rinsing with double-distilled water, cells were incubated in 1 μg/ml calcofluor white (Sigma) for 10 min. Nomarski differential interference contrast (DIC) and fluorescent images were obtained with a Leica DM6000B microscope (Leica Microsystems Canada, Inc., Richmond Hill, ON, Canada) equipped with a Hamamatsu ORCA ER camera (Hamamatsu Photonics, Hamamatsu City, Japan) using 63× or 100× objectives and DAPI (460-nm) filter sets. Images were captured with Openlab software (Improvision, Inc., PerkinElmer).
RESULTS
Cells lacking SWI4 or SWI6 demonstrate dramatic changes in growth pattern, unlike cells lacking MBP1.
Candida albicans open reading frames (ORFs) 19.5855, 19.4545, and 19.4725 are annotated as MBP1, SWI4 and SWI6, respectively, in the Candida Genome Database (http://candidagenome.org) and are 29, 23, and 26% identical to orthologues in S. cerevisiae at the protein level. The factors contain ankyrin repeat domains, similar to SBF/MBF components in other fungi (9). However, leucine zippers found in Swi4p and Swi6p of S. cerevisiae (69) are lacking, and all three C. albicans factors contain a KilA-N DNA binding domain, which is absent in Swi6p of S. cerevisiae.
The functions of Mbp1p, Swi4p, and Swi6p in C. albicans were determined by sequentially replacing alleles with URA3 and HIS1 markers in strain BWP17, creating strains BH261, BH185, and BH120, respectively. An isogenic control strain, BH440, and conditional strains for MBP1 (KMCa4a), SWI4 (BH150), and SWI6 (CB557), respectively, were also constructed. To determine effects on cell growth, overnight cultures were diluted into fresh minimal medium and incubated for 7 h at 30°C. Most mbp1Δ/Δ cells (BH261) were normal in morphology, but 7.4% (n = 309) were elongated, compared to 0.3% (n = 343) in the control strain (Fig. 1). When the conditional MBP1 strain KMCa4a was incubated in inducing (−MC) or repressing (+MC) medium, most cells were in a normal yeast form (see Fig. S1 in the supplemental material). Our results agree with large-scale screens of C. albicans mutants which showed that mbp1Δ/Δ colonies grew normally (40, 61). In S. cerevisiae, the absence of MBP1 resulted in a 20% increase in cell volume and a 5% increase in the proportion of budded cells (12). We did not quantify cell volumes, but the length-to-width measurements of mbp1Δ/Δ cells (20.1 ± 1.2 μm2 [mean ± standard error of the mean], n = 100) were similar to those of control cells (18.5 ± 0.5 μm2, n = 101) (Fig. 2). Thus, Mbp1p is not essential for normal cell growth and has only a mild influence on morphology in C. albicans.
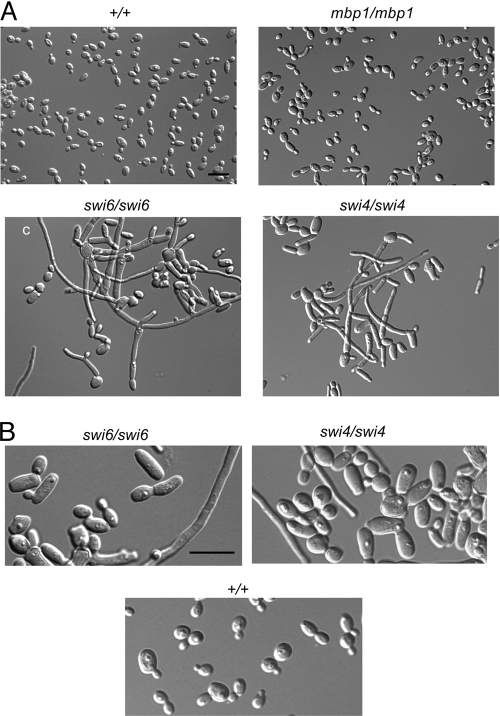
Fig. 1.
Deletion of SWI4 or SWI6 results in dramatic changes in growth pattern, including cell enlargement and induction of fila-ments, in contrast to deletion of MBP1. (A) Cells from strains BH440 (MBP1/MBP1 SWI6/SWI6 SWI4/SWI4 URA3+ HIS1+), BH261 (mbp1Δ::URA3/mbp1Δ::HIS1), BH120 (swi6Δ::URA3/swi6Δ::HIS1), and BH185 (swi4Δ::URA3/swi4Δ::HIS1) were incubated in minimal medium overnight and then diluted into fresh medium and incubated for 7 h at 30°C. (B) Strains BH120, BH185, and BH440 at higher magnification. Bar = 10 μm.
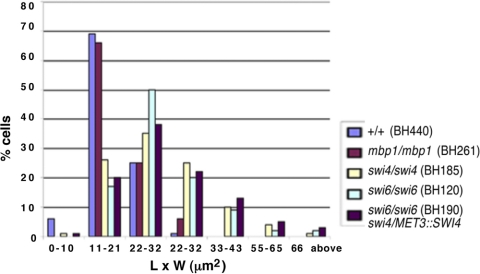
Fig. 2.
Distribution of yeast size, represented by length-to-width measurements (μm2), in cells lacking Swi4p, Swi6p, or Mbp1p. Cells from strains BH440 (MBP1/MBP1 SWI6/SWI6 SWI4/SWI4 URA3+ HIS1+), BH261 (mbp1Δ::URA3/mbp1Δ::HIS1), BH120 (swi6Δ::URA3/swi6Δ::HIS1), and BH185 (swi4Δ::URA3/swi4Δ::HIS1) were incubated in liquid glucose minimal medium overnight and then diluted into fresh medium and incubated for 7 h at 30°C. Strain BH190 (swi6Δ::URA3/swi6Δ::HIS1 swi4Δ::hisG/MET3::SWI4-ARG4) was incubated in repressing medium for 7 h.
In contrast, cells lacking SWI4 (BH185) or SWI6 (BH120) showed a dramatic and pleiotropic change in growth pattern, including enlarged budding yeast cells and a diversity of elongated and filamentous cells (Fig. 1). Length-to-width measurements of yeast cells in strains lacking SWI4 (30.6 ± 1.3 μm2, n = 102) and SWI6 (30.9 ± 1.2 μm2, n = 100) were greater than those of control cells (18.5 ± 0.5 μm2, n = 101) (Fig. 2). In addition, approximately 46% (n = 193) and 44% (n = 144) of cells from strains BH185 and BH120, respectively, were elongated or filamentous. Similarly, in large-scale phenotypic screens (40, 61), swi4Δ/Δ colonies and cells showed growth defects, although colony morphology on solid spider medium was not affected. We also found that swi4Δ/Δ and swi6Δ/Δ colonies did not show dramatic changes in morphology on solid minimal medium (data not shown), which suggests that environmental conditions, including liquid versus solid medium, can influence the phenotype. Consistent with the deletion phenotypes, conditional SWI4 (BH150) and SWI6 (CB557) strains grew predominantly in the yeast form under inducing conditions and showed growth and phenotype defects in repressing medium, although the effects were not as severe in strain CB557 (see Fig. S1 in the supplemental material). Thus, Swi6p and Swi4p influence yeast size and growth patterns in a similar manner, whereas Mbp1p has only a minor effect. The dramatic increase in yeast cell size raises the possibility of a delay in G1 phase and a role for Swi6p and Swi4p in G1/S regulation. In comparison, swi4Δ or swi6Δ cells in S. cerevisiae also demonstrated cell enlargement, bud defects, slow growth, and some elongation but did not produce filaments as seen in the C. albicans mutants (32, 51, 60).
Cells lacking both Swi6p and Swi4p or Swi4p and Mbp1p are viable and do not arrest in G1 phase.
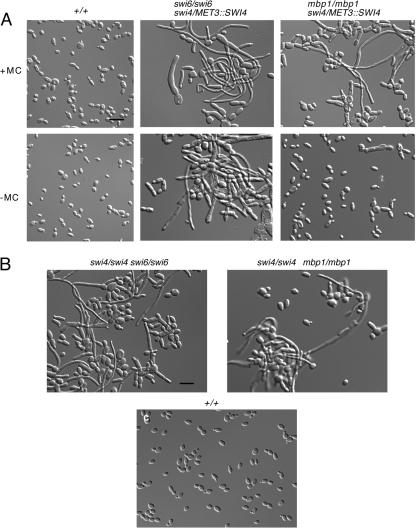
Factors comprising MBF in S. pombe or SBF/MBF in S. cerevisiae are crucial for cell cycle entry; cells lacking either Cdc6, Res1, and Res2, Swi4p and Swi6p, or Swi4p and Mbp1p are inviable and/or blocked in division (3, 11, 44, 55, 60). In order to obtain additional evidence that Swi4p and Swi6p play a role in G1/S regulation in C. albicans, we created a conditional strain lacking both copies of SWI6 and carrying a single copy of SWI4 under the control of the MET3 promoter (BH190), as well as an isogenic control strain (BH420). Overnight cultures of cells were incubated in fresh inducing or repressing medium for 7 h at 30°C. Under repressing conditions, strain BH190 was viable and showed yeast cell enlargement comparable to that in the single deletion strains, based on length-to-width measurements (33.1 ± 1.3 μm2, n = 100) (Fig. 2 and 3A). However, the number of elongated or filamentous cells was moderately higher in repressing medium than in inducing medium (66%, n = 150, versus 37%, n = 230, respectively). In contrast, control cells (BH420) were in a normal yeast form (Fig. 3A). In confirmation of the phenotype, subsequent deletion of both SWI6 and SWI4 (AG168) resulted in similar effects (Fig. 3B). Thus, in contrast to the situation in S. cerevisiae, the combined functions of Swi4p and Swi6p are important but not essential for cell proliferation in C. albicans.
Fig. 3.
Depletion of both Swi4p and Swi6p or Swi4p and Mbp1p results in viable cells with phenotypes similar to those of swi4Δ/Δ or swi6Δ/Δ cells. (A) Cells from strains BH420 (SWI6/SWI6 SWI4/SWI4 URA3+ HIS1+ ARG4+), BH190 (swi6Δ::URA3/swi6Δ::HIS1 swi4Δ::hisG/MET3::SWI4-ARG4), and BH277 (mbp1Δ::URA3/mbp1Δ::HIS1 swi4Δ::hisG/MET::SWI4-ARG4) were incubated in inducing (−MC) or repressing medium (+MC) for 7 h at 30°. (B) Strains AG168 (swi4Δ::hisG/swi4Δ::URA3 swi6Δ::ARG4/swi6Δ::HIS1), HH62 (swi4Δ::hisG/swi4Δ::URA3 mbp1Δ::ARG4/mbp1Δ::HIS1), and BH420 were cultured in minimal medium overnight and then diluted into fresh medium and incubated for 7 h at 30°C.
We next investigated whether the combined functions of Swi4p and Mbp1p were essential for growth. A conditional strain (BH277) lacking both copies of MBP1 and carrying a single copy of SWI4 under the control of the MET3 promoter was created. In inducing medium, few cells (6.9%, n = 153) showed elongation, comparable to mbp1Δ/Δ cells (Fig. 3A). In repressing medium, cells remained viable and appeared similar to swi4Δ/Δ cells (Fig. 3A), with 37% (n = 230) in an elongated or filamentous form. Since strain BH277 was viable, a strain with SWI4 and MBP1 deleted was constructed (HH62). The phenotype was similar to that of strain BH277 under repressing conditions, with 30.8% (n = 221) elongated or filamentous cells (Fig. 3B). The slight reduction in filamentation compared to that seen in other mutants could reflect a mild synergistic effect. Collectively, these data show that Mbp1p plays a minor role in growth, that Swi4p and Mbp1p are not highly redundant in function, and that cells lacking both factors can progress through the cell cycle, unlike the situation in S. cerevisiae (44).
The gene expression patterns in cells lacking Swi6p and Swi4p suggest that these factors influence G1/S progression.
Cote et al. (26) reported the transcription profiles of synchronized opaque phase cells as they pass through the cell cycle, which consisted of four waves of expression. Each wave was represented by a specific set of keynote genes, many of which resembled those expressed at similar cell cycle stages in S. cerevisiae (40). The four waves were also associated with putative core transcription factor regulators, including Fkh2p (S/G2), Mcm1p (G2/M), and Ace2p (M/G1). The wave of G1/S-associated gene expression was suggested to be mediated by a single ankyrin motif-containing MBF complex, as seen in S. pombe, since MCB elements were present in promoters of the G1/S cluster of genes and the C. albicans genome lacked SCB elements (26). Since Swi4p and Swi6p in C. albicans have pronounced and similar effects on cell size and growth pattern, our results raise the possibility that these factors may be critical components of MBF in C. albicans, with Mbp1p playing only a minor role. To obtain additional evidence for this model, we first investigated whether the MCB-containing G1 cyclins PCL2 and CCN1 (15, 26) were downregulated in cells lacking Swi4p and Swi6p. Overnight cultures of swi6Δ/swi6Δ swi4Δ/MET3::SWI4 (BH190) and SWI4/SWI4 SWI6/SWI6 (BH420) cells were incubated in fresh repressing medium for 7 h at 30°C, and RNA was extracted for Northern blot analyses. Both cyclins were repressed in strain BH190 (Fig. 4A), suggesting that Swi4p and Swi6p influence G1/S progression.
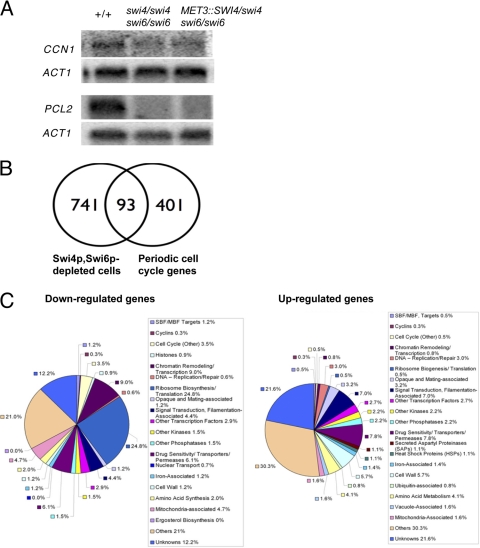
Fig. 4.
Gene expression patterns in cells depleted of Swi6p and Swi4p. (A) Northern analysis demonstrating downregulation of G1 cyclins CCN1 and PCL2 in cells depleted of Swi6p and Swi4p. A total of 20 μg of RNA from strains BH420 (SWI4/SWI4 SWI6/SWI6 URA3+ HIS1+ ARG4+), AG168 (swi4Δ::hisG/swi4Δ::URA3 swi6Δ::ARG4/swi6Δ::HIS1), and BH190 (swi6Δ::URA3/swi6Δ::HIS1 swi4Δ::hisG/MET3::SWI4-ARG4) grown under repressing conditions was loaded on the gel. ACT1 was used a loading control. (B) Venn diagrams comparing significantly modulated genes in strain BH190 with the total number of genes in C. albicans that show cell cycle-dependent periodic expression patterns (26). Strains BH190 and BH420 were incubated in repressing medium for 7 h and processed for microarray analysis. Data obtained from four microarray chips representing four separate samples were normalized with Lowess using GeneSpring software. Significantly modulated genes were determined based on a 1.7-fold cutoff and t test function with a P value of <0.05. (C) Pie categorization of significantly modulated genes in cells depleted of Swi4p and Swi6p. Gene names and functions were identified through GeneSpring analysis and updated using the Candida Genome Database (CGD) at http://candidagenome.org/. Different colors represent different categories as indicated. Genes were manually categorized according to a single function, although several have multiple functions.
In order to gain more evidence that Swi4p and Swi6p contribute to G1/S regulation, we used oligonucleotide microarrays to measure the transcription profiles of Swi4p- and Swi6p-depleted cells, despite the fact that asynchronous growth and a pleiotropic phenotype could dilute relevant cell cycle stage-specific expression patterns. Indeed, when a similar approach was used with swi4Δ cells of S. cerevisiae, very few SBF targets were identified, which was attributed in part to the lack of cell synchrony (42). However, it is difficult to obtain synchronous cell populations and subsequent time course-based transcription profiles in C. albicans (26), and some expression patterns may be strong enough to overcome the barriers imposed by asynchrony and pleiotropic phenotypes. Overnight cultures of swi6Δ/swi6Δ swi4Δ/MET3::SWI4 and SWI4/SWI4 SWI6/SWI6 cells were thus incubated in repressing medium for 7 h at 30°C and processed for microarray analysis. Significantly modulated genes were initially obtained using a 1.7-fold cutoff and a t test function with a P value of <0.05 (see Table S1 in the supplemental material for a complete list of genes). Based on this method, a small proportion (11.0%) of genes was found to overlap with the total set of periodically expressed cell cycle-regulated genes of Cote et al. (26) (Fig. 4B). Of these, 31.0% corresponded to the G1/S cluster, while 25.0, 22.5, and 21.5% corresponded to S/G2, G2/M, and M/G1 clusters, respectively (see Table S2 in the supplemental material).
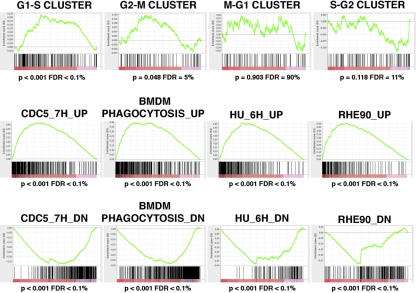
To confirm whether G1/S cluster genes were significantly enriched in Swi4p- and Swi6p-depleted cells, we used gene set enrichment analysis (GSEA) (56, 72), a computational method that determines whether defined sets of genes exhibit a statistically significant bias in their distribution within a ranked gene list. Since GSEA is a rank-based method that is not limited to an arbitrarily defined set of significantly modulated genes, it allows us to detect more subtle changes in transcript profiles. In addition to calculating a P value for any observed enrichment, this method also calculates a false discovery rate (FDR) by performing a permutation analysis on 10,000 randomly distributed datasets. The Broad Institute maintains an extensive web page on this tool, including a detailed guide on interpreting GSEA results (http://www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html). We thus compared a ranked list of the 5,675 genes whose transcript profiles were measured in cells lacking Swi4p and Swi6p to the distribution of the four cell cycle gene clusters defined by Cote et al. (26), as well as 25 gene sets representing morphogenetic switching, including the yeast-to-hypha transition, in C. albicans (5, 34, 43, 53, 58, 71). As seen in Fig. 5 and in Table S3 in the supplemental material, genes of the G1/S cluster showed a highly significant bias toward upregulation (normalized enrichment score [NES] = 3.41, P < 0.0001, FDR < 0.01%) in cells lacking Swi4p and Swi6p. Fifty-three percent of the G1/S cluster genes were present in the leading edge, which is defined as genes that were ranked at an earlier position than the maximal enrichment score. We also observed an enrichment of the upregulated G2/M cluster genes, although it was not as strong as the G1/S cluster. There was no significant enrichment of the S/G2 and M/G1 cluster genes. These results suggest that cultures lacking Swi4p and Swi6p have a greater proportion of cells in the G1/S phase than a culture of control cells, supporting a role for Swi4p and Swi6p in G1/S regulation. Impaired progression through G2/M is also possible but not as clear.
Fig. 5.
Gene set enrichment analysis (GSEA) of the transcriptional profile of Swi4p- and Swi6p-depleted cells. A ranked list of genes modulated in Swi4p- and Swi6p-depleted cells was compared to 29 gene sets of 64 to 558 genes that exhibit cell cycle-dependent periodic expression in C. albicans opaque cells (26) or significant changes during polar morphogenesis, including yeast-to-hypha transitions and the production of highly elongated buds due to M or S phase arrest (5, 34, 43, 53, 58, 71). Genes in the ranked list are organized along the x axis with upregulated genes to the left and downregulated genes to the right. The positions of the genes in each gene set are illustrated by the vertical black bars, while the green curve represents the cumulative value of the enrichment score (y axis). Graphs of selected results are shown, while the complete GSEA output folder is included as GSEA data file S1 in the supplemental material. BMDM, bone marrow-derived monocytes; RHE90, reconstituted human epithelial cells; HU, hydroxyurea.
Of the genes that were strongly modulated and in common with the periodic cell cycle set of Cote et al. (26), the MCB-containing G1 cyclin PCL2 was the most strongly downregulated gene (see Tables S1 and S2 in the supplemental material). CCN1 was not included in the list, in contrast to the Northern blot results. However, its exclusion was due to the fact that it has a very low expression level and was only detected on two of the microarray experiments, where it was strongly downregulated. Additional downregulated MCB-containing factors include the G1/S keynote genes HCM1 and GIN4 (26), the chromatin cohesin factor MCD1, and genes of unknown function. More G1/S-associated genes were upregulated, in agreement with the GSEA results, including the keynote gene YOX1 (26), a target of SBF in S. cerevisiae, and factors associated with DNA synthesis and DNA repair (RNR1, DUT1, PMS1, MLH1, and TERT) and other functions (see Table S2 in the supplemental material). These expression patterns raise the possibility that Swi4p and Swi6p have repressing and inducing activities. In support of this, MBF is a transcriptional repressor of genes predominantly associated with DNA replication and repair in S. cerevisiae; the absence of MBP1 or SWI4 and MBP1 resulted in elevated levels of DNA repair genes (12, 28). Moreover, the absence of Res1 or Res2 in S. pombe resulted in the repression or induction, respectively, of MBF target genes (11). Few other MCB-containing genes which were not identified by Cote et al. (26) were modulated in cells lacking Swi4p and Swi6p. These included uncharacterized genes (orf19.6048, orf19.4664, and orf19.413), a putative ion exchanger (orf19.2397), and a transcriptional repressor of hyphal formation that is responsive to DNA damage (RFX2). Of the S/G2 cluster genes, the putative central G2/M regulator FKH2 (26) was repressed, as well as histones (HHO1 and HTA3), gamma tubulin (TUB4), and other factors, most of which had unknown functions. The strongly modulated G2/M-associated genes did not include cell cycle regulatory factors, with the exception of a spindle midzone-associated protein (ASE1) (see Table S2 in the supplemental material). M/G1 factors notably included the keynote gene CDC6, encoding a component of the prereplicative complex (see Table S2). Intriguingly, the putative central regulator of the M/G1 cluster genes, MCM1, was also repressed. Thus, the expression profiles revealed modulation of some key MCB-containing genes, as well as several core putative regulators of subsequent cell cycle phases. Modulation of genes associated with other cell cycle stages could reflect additional functions for Swi4p and Swi6p, particularly in G2/M phase, based on GSEA results. Alternatively, this may represent indirect effects of the initial impairment in G1/S progression; cell cycle-dependent transcription patterns are regulated by a hierarchy of sequentially expressed networks of transcription factors (64), and in S. cerevisiae, SBF/MBF indirectly act on genes in subsequent cell cycle stages through their transcription factor targets and subsequent effectors (41).
Cells lacking Swi4p and Swi6p demonstrated strong modulation of additional genes associated with many other cellular processes (Fig. 4C; also see Table S1 in the supplemental material). Although the majority of these genes lack MCB elements and the responses likely reflect indirect effects, some of the expression patterns further support an influence on G1 phase. For example, ribosome biogenesis and RNA metabolism genes were downregulated (see Table S1) and identified by gene ontology (GO) term analysis as being the most significantly enriched in Swi4p- and Swi6p-depleted cells (Table 4). In addition, orthologues of TOR (target of rapamycin) pathway-dependent regulators of cell size, proliferation, and ribosome biogenesis, including SFP1, SCH9 (48), and TBF1 (39), were repressed in Swi4p- and Swi6p-depleted cells (see Table S1). Regulation of ribosome biogenesis is not clear in C. albicans, but it is intriguing that ribosome biogenesis is sensed and initiated at start in S. cerevisiae (16). PES1, a pescadillo orthologue important for yeast growth in C. albicans (67) but not identified in the periodic data set of Cote et al. (26), was also downregulated (see Table S1). Paradoxically, the G1 cyclin CLN3 was induced in cells lacking Swi4p and Swi6p (see Table S1). While Yox1p negatively regulates CLN3 expression in S. cerevisiae (41), both of these factors were induced in Swi4p- and Swi6p-depleted cells of C. albicans, demonstrating another example of subtle rewiring within the otherwise generally similar framework of cell cycle expression patterns in the two organisms (26). Collectively, these results demonstrate that Swi4p and Swi6p influence G1/S phase progression and, thus, may contribute to MBF activity.
Table 4.
GO term analysis of significantly modulated genes in cells lacking Swi4p and Swi6pa
| GO categoryb | P valuec | GO genes in set (834 total)d | GO genes in genome (6,804 total)e |
|---|---|---|---|
| Ribosome biogenesis | 2.7E−13 | 88 | 746 |
| Biological process unknown | 8.3E−7 | 263 | 2,741 |
| RNA metabolic process | 1.7E−6 | 123 | 627 |
| Response to chemical stimuli | 4.5E−3 | 93 | 741 |
| Carbohydrate metabolism | 2.3E−3 | 47 | 787 |
Strains analyzed were BH190 (lacking Swi4p and Swi6p) and BH420 (wild type).
The GO type was biological process.
Significance of overrepresentation of GO categories represented in significantly modulated genes based on the Fisher exact test.
Number of genes within total significantly induced gene set of Swi6p- and Swi4p-depleted cells that are associated with the select GO category.
Total number of genes within the genome that associate with the selected GO category.
Swi4p and Swi6p influence morphogenesis, including differentiation of hyphae.
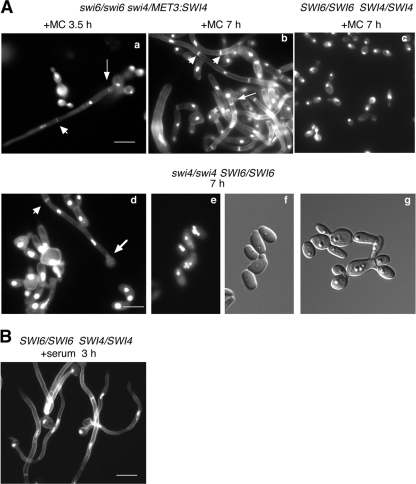
G1/S circuit components can regulate developmental processes independent of their cell cycle function. For example, E2F and pRb in mammals are linked to neuronal and adipocyte differentiation (54), whereas the SBF targets Tos4p and Yox1p in S. cerevisiae regulate genes associated with mating and pseudohyphal growth, respectively (41). However, the absence of SBF alone does not lead to changes in cell fate in the latter. To determine whether Swi4p and Swi6p also influenced developmental events in C. albicans, the phenotypes of cells lacking these factors were examined more closely. Strains lacking SWI4 (BH185) or SWI6 (BH120) or both (AG168 and BH190) demonstrated a pleiotropic phenotype. Of the yeast cells, many were multibudded (Fig. 1 and 6Ad and g), suggesting defects in cell separation similar to those of swi4Δ and swi6Δ cells of S. cerevisiae (60). DAPI staining demonstrated that most cells contained a single nucleus, but a proportion showed multinucleation (11.4%, n = 158, strain BH185; 9.4%, n = 180, strain BH190; and 3.8%, n = 160, strain BH420) (Fig. 6Ad and e), indicating deregulation of nuclear division in some cases. Intriguingly, some yeast cells were bean shaped, resembling opaque phase cells, and contained four nuclei (Fig. 6Ae and f). Most striking, however, was the presence of highly filamentous cells resembling elongated buds, pseudohyphae, true hyphae, or a combination of these cell types. The extent of filamentation increased when overnight cultures of cells were diluted into fresh medium and incubated for 7 h. The presence of true hyphae within the population was evident from filamentous cells containing unconstricted septa along their lengths (Fig. 6Ab and d, arrowheads). In addition, the first septa was positioned away from the bud neck in many of these cells (Fig. 6Aa and b, thin arrows), as seen in true hyphae that form in G1 phase (73, 74). Intriguingly, other filamentous cells demonstrated unconstricted septa in the subapical regions, but the first septa was located at the bud neck (Fig.6Ad, thick arrow), as described for pseudohyphae (73, 74). At earlier time points in fresh medium (3.5 h), the majority of cells were yeast or pseudohyphae. However, some true hyphae were also present at this time, based on the position and unconstricted appearance of the first septa (Fig.6Aa, thin arrow). Collectively, these results demonstrate that a variety of cell types and morphologies result from the absence of Swi4 and/or Swi6p, including true hyphal cells. The presence of pseudohyphal and hyphal features within single cells suggests that additional changes in fate may be taking place.
Fig. 6.
Yeast cells lacking Swi4p and/or Swi6p show pleiotropic changes in morphology and form true hyphae. (A) Cells of strain BH190 (swi6Δ::URA3/swi6Δ::HIS1 swi4Δ::hisG/MET3::SWI4-ARG4) (a, b), BH185 (swi4Δ::URA3/swi4Δ::HIS1) (d to g), and BH420 (SWI4/SWI4 SWI6/SWI6 URA3+ HIS1+ ARG4+) (c) were incubated for the indicated times in repressing medium at 30°C, fixed, and then stained with DAPI and calcofluor. Arrowheads indicate subapical, unconstricted septa, thin arrows show the first septa positioned away from the bud neck, and thick arrows demonstrate the first septa at the bud neck. (B) Strain BH420 was incubated for 3.0 h in the presence of 10% serum at 37°C for comparison of hyphal characteristics. Bars = 10 μm.
Consistent with the phenotypes of cells lacking Swi4p and Swi6p, transcription profiles based on a 1.7-fold cutoff and a P value of <0.05 revealed the induction of several hypha-associated and regulatory genes, including HWP1, RBR1, RBT1, PHR1, and IHD1, for example, at high levels (21, 43, 58) (see Table S4 in the supplemental material). This provides additional evidence that true hyphae can form upon the depletion of Swi4p and Swi6p. Some hyphal regulators were simultaneously repressed, including CZF1, TPK1, and PDE2 (see Table S4). When we used GSEA to compare the transcriptional profiles of the Swi4p- and Swi6p-depleted cells to the results of other microarray-based experiments based on morphogenetic switching, including yeast-to-hyphal cells, the strongest enrichments were seen with gene sets obtained from Cdc5p-depleted or hydroxyurea-treated cells, which form hypha-like elongated buds (5) (Fig. 5). Intriguingly, the analysis also revealed very strong enrichments of gene sets from the profiles of C. albicans interacting with host cells, such as primary mouse macrophages (53), reconstituted oral human epithelium (71), or polymorphonuclear leukocytes (34). In contrast, gene sets from laboratory-induced hyphae (43, 58) were not as enriched, mostly due to the fact that their upregulated genes tended to cluster at both the top and the bottom of the ranked gene list of Swi4p- and Swi6p-depleted cells (data not shown). These results suggest that Swi4p and Swi6p can influence the signaling associated with host cell-induced morphogenetic switching, including hyphal induction.
DISCUSSION
The G1/S transition serves as the gateway to cell proliferation but is not well characterized in C. albicans. Ankyrin-repeat motif proteins, including Swi4p, Swi6p, and Mbp1p in S. cerevisiae and Cdc10, Res1, and Res2 in S. pombe (9), comprise the major components of G1/S transcription factor complexes in fungi studied to date and are crucial for growth. Here, we show that the closest orthologues of Swi4p and Swi6p in C. albicans are important for cell proliferation and, thus, may contribute to MBF activity but that their combined function is, surprisingly, not essential for growth. We also show that these factors strongly influence morphogenesis, including hyphal differentiation, and expression patterns normally induced by interactions with host cells, supporting the existence of an important relationship between G1/S phase of the cell cycle and hyphal development in C. albicans.
Swi6p and Swi4p are important for yeast cell proliferation and may contribute to MBF activity.
Our results provide functional evidence that orthologues of Swi4p and Swi6p are important for yeast cell proliferation in C. albicans, influence G1/S progression and, thus, may contribute to MBF activity. First, key MCB-containing G1/S genes (26) were modulated in cells lacking Swi4p and Swi6p, including the G1 cyclins PCL2 and CCN1 (15), the transcription factor YOX1, and factors associated with DNA synthesis/repair, including RNR1. G1/S cluster genes were significantly enriched and represented the highest proportion of periodically expressed cell cycle genes modulated in the cells. The G2/M cluster genes were also enriched but to a lesser extent, which could imply a role for Swi4p and Swi6p in regulating additional cell cycle stages, given the rewiring in the cell cycle circuitry of the pathogen (15, 26). Alternatively, G2/M gene modulation may be an indirect response to a G1/S delay, since G1/S events influence transcription in subsequent cell cycle stages in S. cerevisiae (41), and the basic cell cycle expression program in C. albicans is very similar (26). The total number of MCB-containing genes modulated in cells lacking Swi4p and Swi6p was not high, which could imply that Swi4p and Swi6p do not function via interaction with MCB elements. Alternatively, asynchronous growth, pleiotropy in phenotype, and/or redundancy from other unknown factors could mask specific expression patterns, particularly those that are subtle. Indeed, transcription profiles of Δswi4 cells in S. cerevisiae did not reveal modulation of many SBF targets, which was attributed in part to asynchronous growth (42) and could also be due to compensation from Mbp1p (12). Our use of GSEA circumvented these problems in part, revealing significant enrichment of G1/S genes. Second, cells lacking Swi4p and/or Swi6p showed similar growth defects, including a dramatic increase in yeast cell size, suggesting a delay in G1 phase. Although an increase in size could occur by slowing G2/M phase, our results suggest that G1 phase is a major and possibly primary target of Swi4p and Swi6p. Third, additional expression patterns in cells lacking Swi6p and Swi4p, including repression of ribosome biogenesis genes, suggest a role for these factors in G1/S regulation. Ribosome biogenesis, controlled in part by the TOR pathway, is coordinated with cell cycle initiation in other organisms (46, 65) and feeds back on start in S. cerevisiae via Whi5p (16). Finally, the ability to form true hyphae upon depletion of Swi4p and Swi6p also supports a role for these factors in G1 phase progression. Previous work demonstrated that true hyphae with active cell cycles could form when G1 phase was blocked through depletion of the G1 cyclin CLN3 (7, 22). True hyphae also formed in the absence of the SCF ubiquitin ligase F-box protein Cdc4p, which has a G1/S-associated function in S. cerevisiae (2). In contrast, blocking or slowing other cell cycle phases in C. albicans results in elongated buds and/or pseudohyphal growth (15). Collectively, the results strongly suggest that Swi4p and Swi6p influence G1/S progression and, thus, may contribute to MBF activity. Future investigations involving chromatin immunoprecipitation-microarray (ChIP-chip) analysis will help to clarify the precise binding sequences and targets of these factors.
Swi4p, Swi6p, and Mbp1p show unique features compared to orthologues in other systems.
Our results highlight key differences in the proposed functions of Swi4p and Swi6p compared to orthologues in other systems. A central finding is that the combined function of C. albicans Swi4p and Swi6p is not essential for cell proliferation. In contrast, the absence of both orthologues in S. cerevisiae results in nonviable cells; Mbp1p does not compensate (17, 44). Although we cannot rule out whether Mbp1p compensates for the lack of Swi4p and Swi6p in C. albicans, it plays a limited role in growth and shows low redundancy with Swi4p, based on the different phenotypes of swi4Δ/Δ and mbp1Δ/Δ cells and the lack of synergistic effects in the absence of both genes. This suggests that other factors may be involved in controlling cell proliferation. The viability of swi4Δ/Δ mbp1Δ/Δ cells of C. albicans is also novel and contradicts the lethal effects resulting from the absence of both orthologues in S. cerevisiae (44) or of both Res1p and Res2p in S. pombe (11, 55). It is possible that Swi6p in C. albicans can compensate under these conditions, since it contains a KilA-N DNA binding domain, unlike Swi6p in S. cerevisiae. However, the Swi6p orthologue Cdc10 in S. pombe also contains this domain but does not bind DNA directly or compensate for loss of Res1 and Res2 (78). Thus, additional elements may be involved in G1/S control in C. albicans. Candidate factors include the APSES domain-containing proteins Efg1p or Efh1p, which can bind MluI sites (63). However, there is currently no evidence supporting a role for either factor in regulating cell proliferation. Additional ankyrin-repeat domain proteins of unknown function and low homology to Swi4p, Swi6p, or Mbp1 exist in C. albicans, and we are currently exploring their potential contributions.
Another difference in the putative functions of Swi4p and Swi6p includes the possibility that they possess both activating and repressing activity, based on the repression and induction of G1/S cluster genes. More genes were induced, and these included factors associated with DNA synthesis and repair. Intriguingly, MBF in S. cerevisiae represses its targets, which include DNA synthesis and repair factors, in cell cycle phases other than G1/S with the assistance of the corepressor Nrm1p (27). In contrast, the SBF complex activates its targets. In S. pombe, the heteromeric MBF has two constitutive DNA binding elements, Res1 and Res2, but G1 transcripts increased or decreased in res1Δ versus res2Δ cells, respectively (11). Since the absence of Swi4p or Swi6p in C. albicans produced a similar phenotype, it is not likely that one is an activator while the other is a repressor. The putative dual function of Swi4p and Swi6p in regulating targets may be mediated by cofactors. In support of this, a candidate Nrm1p homologue exists in C. albicans that genetically interacts with and antagonizes putative Swi4p/Swi6p function (D. Kornitzer, personal communication).
If Swi4p, Swi6p, and other factors contribute to MBF activity, the resulting organization of this complex in C. albicans is not clear. The absence of Swi4p and Swi6p resulted in synergistic effects on filamentation, suggesting that these factors may act separately. However, this did not extend to cell cycle function, as the single and double mutants showed similar increases in yeast cell size. In addition, Swi4p and Swi6p physically interact in C. albicans (C. Bachewich, unpublished observations). Given that S. cerevisiae contains two G1/S transcription units as a potential consequence of undergoing a whole-genome duplication (26), it is unexpected that C. albicans, with its more simplified genome and absence of SCB elements, would also require several G1/S transcription complexes. The potential contribution of Mbp1p is also not clear, but it may be more important under different conditions or in different cell types, since Res2 of S. pombe contributes to mitotic cell proliferation but plays a stronger role during meiotic division (4, 55, 84). Overall, our results highlight potential variations in the putative G1/S regulatory circuit in C. albicans versus other fungi.
Swi4p and Swi6p play additional roles in morphogenesis and influence hyphal differentiation.
The absence of Swi4p and Swi6p resulted in pleiotropic morphologies, due in part to defects in budding. The pleiotropy could reflect variability in the levels of G1 cyclins and/or other regulatory factors which influence the timing of progression through G1/S phase and, perhaps, subsequent cell cycle stages, generating different cell shapes and types. The phenotype of cells lacking Swi4p and Swi6p was reminiscent of that of Grr1p mutants (47), with the exception that true hyphae could also form, based on strong modulation of several hyphal-associated genes and the presence of unconstricted septa. The relationship between hyphal development and G1 phase in C. albicans is complex, since there is conflicting information on whether hyphal initiation can occur at later cell cycle stages (15, 37, 70). A G1 phase-dependent bias for hyphal initiation could exist (15, 70), but strong environmental inducers, such as serum, may be able to override this relationship, allowing hyphal formation at other cell cycle stages (37). Consistent with this, hyphae and pseudohyphae form when yeast cells of C. albicans are arrested in G1 phase through depletion of the G1 cyclin Cln3p (7, 22). In contrast, arresting or slowing other cell cycle phases results in elongated buds or pseudohyphae, respectively (15). The presence of true hyphae within the filamentous population of cells lacking Swi6p and Swi4p suggests that these factors may mediate, in part, the effect of Cln3p on hyphal development. The transcription profiles of Swi4p- and Swi6p-depleted cells, which show strong enrichment of gene sets from profiles of C. albicans interacting with a diversity of host cells, further suggest that Swi4p and Swi6p function may contribute to host cell-induced morphogenetic switching, including hyphal induction. The factors may lie downstream of additional pathways, since Swi4p is a mitogen-activated protein kinase (MAPK) target (8, 45, 52). Alternatively, it is possible that hyphal growth is a general response to aspects associated with a G1 phase delay and that Swi4p and Swi6p play only an indirect role. The initial presence of pseudohyphal followed by hyphal characteristics in some cells supports this point. However, true hyphae could also form directly from yeast, and intriguingly, G1/S transcription regulators can independently influence developmental gene expression in other systems (41, 54). Many yeast cells lacking Swi4p and Swi6p also resembled opaque phase cells. Since a downstream target of SBF in S. cerevisiae functions as a repressor of mating, it is possible that Swi4p and Swi6p function and/or the G1/S circuit influence additional developmental pathways in C. albicans, which is currently under investigation.
In summary, we have demonstrated that Swi4p and Swi6p are important for cell proliferation and influence G1/S progression, suggesting that they may contribute to MBF-like activity in C. albicans. Our results also suggest that the emerging G1/S regulatory circuit in C. albicans has unique features compared to those in other ascomycetes and is linked to aspects of hyphal development, possibly through Swi4p and Swi6p function. Future work addressing the composition, regulation, and targets of MBF and the G1/S regulatory machinery in C. albicans will significantly advance our understanding of the thematic variations in how cells regulate basic proliferation and coordinate this process with developmental events, which are critical for virulence in an important fungal pathogen of humans.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Whiteway (Health Sector, Biotechnology Research Institute, NRC) and D. Kornitzer (Technion, ITT) for comments on the manuscript and P. Cote (Health Sector, Biotechnology Research Institute, NRC) for sharing bioinformatic data.
This work was supported by Canadian Institutes of Health Research Operating Bridge grant IGI-78908 (C.B.) and, in part, through Canadian Institutes of Health Research Team grant CTP79843 (C.B. and A.N.).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 21 January 2011.
REFERENCES
- 1. Andaluz E., Ciudad T., Gomez-Raja J., Calderone R., Larriba G. 2006. Rad52 depletion in Candida albicans triggers both the DNA-damage checkpoint and filamentation accompanied by but independent of expression of hypha-specific genes. Mol. Microbiol. 59:1452–1472 [DOI] [PubMed] [Google Scholar]
- 2. Atir-Lande A., Gildor T., Kornitzer D. 2005. Role for the SCFCDC4 ubiquitin ligase in Candida albicans morphogenesis. Mol. Biol. Cell 16:2772–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aves S. J., Durkacz B. W., Carr A., Nurse P. 1985. Cloning, sequencing and transcriptional control of the Schizosaccharomyces pombe cdc10 “start” gene. EMBO J. 4:457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ayte J., et al. 1995. The Schizosaccharomyces pombe MBF complex requires heterodimerization for entry into S phase. Mol. Cell. Biol. 15:2589–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bachewich C., Nantel A., Whiteway M. 2005. Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol. Microbiol. 57:942–959 [DOI] [PubMed] [Google Scholar]
- 6. Bachewich C., Thomas D. Y., Whiteway M. 2003. Depletion of a polo-like kinase in Candida albicans activates cyclase-dependent hyphal-like growth. Mol. Biol. Cell 14:2163–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bachewich C., Whiteway M. 2005. Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukarot. Cell 4:95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baetz K., Moffat J., Haynes J., Chang M., Andrews B. 2001. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol. Cell. Biol. 21:6515–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bahler J. 2005. Cell-cycle control of gene expression in budding and fission yeast. Annu. Rev. Genet. 39:69–94 [DOI] [PubMed] [Google Scholar]
- 10. Bai C., Ramanan N., Wang Y. M., Wang Y. 2002. Spindle assembly checkpoint component CaMad2p is indispensable for Candida albicans survival and virulence in mice. Mol. Microbiol. 45:31–44 [DOI] [PubMed] [Google Scholar]
- 11. Baum B., Wuarin J., Nurse P. 1997. Control of S-phase periodic transcription in the fission yeast mitotic cycle. EMBO J. 16:4676–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bean J. M., Siggia E. D., Cross F. R. 2005. High functional overlap between MluI cell-cycle box binding factor and Swi4/6 cell-cycle box binding factor in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics 171:49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bensen E. S., Clemente-Blanco A., Finley K. R., Correa-Bordes J., Berman J. 2005. The mitotic cyclins Clb2p and Clb4p affect morphogenesis in Candida albicans. Mol. Biol. Cell 16:3387–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bensen E. S., Filler S. G., Berman J. 2002. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot. Cell 1:787–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berman J. 2006. Morphogenesis and cell cycle progression in Candida albicans. Curr. Opin. Microbiol. 9:595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bernstein K. A., Bleichert F., Bean J. M., Cross F. R., Baserga S. J. 2007. Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Mol. Biol. Cell 18:953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Breeden L., Nasmyth K. 1987. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell 48:389–397 [DOI] [PubMed] [Google Scholar]
- 18. Butler D. K., et al. 2006. The GRR1 gene of Candida albicans is involved in the negative control of pseudohyphal morphogenesis. Fungal Genet. Biol. 43:573–582 [DOI] [PubMed] [Google Scholar]
- 19. Caligiuri M., Beach D. 1993. Sct1 functions in partnership with Cdc10 in a transcription complex that activates cell cycle START and inhibits differentiation. Cell 72:607–619 [DOI] [PubMed] [Google Scholar]
- 20. Care R. S., Trevethick J., Binley K. M., Sudbery P. E. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34:792–798 [DOI] [PubMed] [Google Scholar]
- 21. Carlisle P. L., et al. 2009. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc. Natl. Acad. Sci. U. S. A. 106:599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chapa y Lazo B., Bates S., Sudbery P. 2005. The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot. Cell 4:90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen D. C., Yang B. C., Kuo T. T. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83–84 [DOI] [PubMed] [Google Scholar]
- 24. Costanzo M., et al. 2004. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117:899–913 [DOI] [PubMed] [Google Scholar]
- 25. Costanzo M., Schub O., Andrews B. 2003. G1 transcription factors are differentially regulated in Saccharomyces cerevisiae by the Swi6-binding protein Stb1. Mol. Cell. Biol. 23:5064–5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cote P., Hogues H., Whiteway M. 2009. Transcriptional analysis of the Candida albicans cell cycle. Mol. Biol. Cell 20:3363–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cross F. R. 1988. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol. Cell. Biol. 8:4675–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Bruin R. A., et al. 2006. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol. Cell 23:483–496 [DOI] [PubMed] [Google Scholar]
- 29. de Bruin R. A., McDonald W. H., Kalashnikova T. I., Yates J., III, Wittenberg C. 2004. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117:887–898 [DOI] [PubMed] [Google Scholar]
- 30. De Falco G., Comes F., Simone C. 2006. pRb: master of differentiation. Coupling irreversible cell cycle withdrawal with induction of muscle-specific transcription. Oncogene 25:5244–5249 [DOI] [PubMed] [Google Scholar]
- 31. Ferrezuelo F., Aldea M., Futcher B. 2009. Bck2 is a phase-independent activator of cell cycle-regulated genes in yeast. Cell Cycle 8:239–252 [DOI] [PubMed] [Google Scholar]
- 32. Flick K., Wittenberg C. 2005. Multiple pathways for suppression of mutants affecting G1-specific transcription in Saccharomyces cerevisiae. Genetics 169:37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fonzi W. A., Irwin M. Y. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fradin C., et al. 2005. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56:397–415 [DOI] [PubMed] [Google Scholar]
- 35. Gari E., et al. 2001. Whi3 binds the mRNA of the G1 cyclin CLN3 to modulate cell fate in budding yeast. Genes Dev. 15:2803–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gola S., Martin R., Walther A., Dunkler A., Wendland J. 2003. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20:1339–1347 [DOI] [PubMed] [Google Scholar]
- 37. Hazan I., Sepulveda-Becerra M., Liu H. 2002. Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol. Biol. Cell 13:134–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ho A., Dowdy S. F. 2002. Regulation of G(1) cell-cycle progression by oncogenes and tumor suppressor genes. Curr. Opin. Gen. Dev. 12:47–52 [DOI] [PubMed] [Google Scholar]
- 39. Hogues H., et al. 2008. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol. Cell 29:552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Homann O. R., Dea J., Noble S. M., Johnson A. D. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 5:e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Horak C. E., et al. 2002. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 16:3017–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Iyer V. R., et al. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533–538 [DOI] [PubMed] [Google Scholar]
- 43. Kadosh D., Johnson A. D. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koch C., Moll T., Neuberg M., Ahorn H., Nasmyth K. 1993. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science 261:1551–1557 [DOI] [PubMed] [Google Scholar]
- 45. LaFayette S. L., et al. 2010. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 6:e1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lempiainen H., Shore D. 2009. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 21:855–863 [DOI] [PubMed] [Google Scholar]
- 47. Li W. J., et al. 2006. The F-box protein Grr1 regulates the stability of Ccn1, Cln3 and Hof1 and cell morphogenesis in Candida albicans. Mol. Microbiol. 62:212–226 [DOI] [PubMed] [Google Scholar]
- 48. Liu W., Zhao J., Li X., Li Y., Jiang L. 2010. The protein kinase CaSch9p is required for the cell growth, filamentation and virulence in the human fungal pathogen Candida albicans. FEMS Yeast Res. 10:462–470 [DOI] [PubMed] [Google Scholar]
- 49. Lo H. J., et al. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949 [DOI] [PubMed] [Google Scholar]
- 50. Loeb J. D., Sepulveda-Becerra M., Hazan I., Liu H. 1999. A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol. Cell. Biol. 19:4019–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lowndes N. F., Johnson A. L., Breeden L., Johnston L. H. 1992. SWI6 protein is required for transcription of the periodically expressed DNA synthesis genes in budding yeast. Nature 357:505–508 [DOI] [PubMed] [Google Scholar]
- 52. Madden K., Sheu Y. J., Baetz K., Andrews B., Snyder M. 1997. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275:1781–1784 [DOI] [PubMed] [Google Scholar]
- 53. Marcil A., et al. 2008. Analysis of PRA1 and its relationship to Candida albicans-macrophage interactions. Infect. Immun. 76:4345–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miller J. P., Yeh N., Vidal A., Koff A. 2007. Interweaving the cell cycle machinery with cell differentiation. Cell Cycle 6:2932–2938 [DOI] [PubMed] [Google Scholar]
- 55. Miyamoto M., Tanaka K., Okayama H. 1994. res2+, a new member of the cdc10+/SWI4 family, controls the “start” of mitotic and meiotic cycles in fission yeast. EMBO J. 13:1873–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mootha V. K., et al. 2003. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267–273 [DOI] [PubMed] [Google Scholar]
- 57. Nantel A. 2006. Microarrays for studying pathogenicity in Candida albicans, p. 181–209 In Kavanagh K. (ed.), Medical mycology: cellular and molecular techniques. Wiley Press, Hoboken, NJ [Google Scholar]
- 58. Nantel A., et al. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nash R., Tokiwa G., Anand S., Erickson K., Futcher A. B. 1988. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 7:4335–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nasmyth K., Dirick L. 1991. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell 66:995–1013 [DOI] [PubMed] [Google Scholar]
- 61. Noble S. M., French S., Kohn L. A., Chen V., Johnson A. D. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42:590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Noble S. M., Johnson A. D. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4:298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Noffz C. S., Liedschulte V., Lengeler K., Ernst J. F. 2008. Functional mapping of the Candida albicans Efg1 regulator. Eukaryot. Cell 7:881–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Orlando D. A., et al. 2008. Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature 453:944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pestov D. G., Strezoska Z., Lau L. F. 2001. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G1/S transition. Mol. Cell. Biol. 21:4246–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65a. Rose M. D., Winston F., Hieter P. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 66. Saville S. P., et al. 2006. Inhibition of filamentation can be used to treat disseminated candidiasis. Antimicrob. Agents Chemother. 50:3312–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shen J., Cowen L. E., Griffin A. M., Chan L., Kohler J. R. 2008. The Candida albicans pescadillo homolog is required for normal hypha-to-yeast morphogenesis and yeast proliferation. Proc. Natl. Acad. Sci. U. S. A. 105:20918–20923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shi Q. M., Wang Y. M., Zheng X. D., Lee R. T., Wang Y. 2007. Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol. Biol. Cell 18:815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sidorova J., Breeden L. 1993. Analysis of the SWI4/SWI6 protein complex, which directs G1/S-specific transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Soll D. R., Herman M. A., Staebell M. A. 1985. The involvement of cell wall expansion in the two modes of mycelium formation of Candida albicans. J. Gen. Microbiol. 131:2367–2375 [DOI] [PubMed] [Google Scholar]
- 71. Spiering M. J., et al. 2010. Comparative transcript profiling of Candida albicans and Candida dubliniensis identifies SFL2, a C. albicans gene required for virulence in a reconstituted epithelial infection model. Eukaryot. Cell 9:251–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Subramanian A., et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sudbery P., Gow N., Berman J. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317–324 [DOI] [PubMed] [Google Scholar]
- 74. Sudbery P. E. 2001. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 41:19–31 [DOI] [PubMed] [Google Scholar]
- 75. Trunk K., et al. 2009. Depletion of the cullin Cdc53p induces morphogenetic changes in Candida albicans. Eukaryot. Cell 8:756–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Umeyama T., Kaneko A., Niimi M., Uehara Y. 2006. Repression of CDC28 reduces the expression of the morphology-related transcription factors, Efg1p, Nrg1p, Rbf1p, Rim101p, Fkh2p and Tec1p and induces cell elongation in Candida albicans. Yeast 23:537–552 [DOI] [PubMed] [Google Scholar]
- 77. Verma R., Smiley J., Andrews B., Campbell J. L. 1992. Regulation of the yeast DNA replication genes through the MluI cell cycle box is dependent on SWI6. Proc. Natl. Acad. Sci. U. S. A. 89:9479–9483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Whitehall S., Stacey P., Dawson K., Jones N. 1999. Cell cycle-regulated transcription in fission yeast: Cdc10-Res protein interactions during the cell cycle and domains required for regulated transcription. Mol. Biol. Cell 10:3705–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Whiteway M., Bachewich C. 2007. Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 61:529–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wightman R., Bates S., Amornrrattanapan P., Sudbery P. 2004. In Candida albicans, the Nim1 kinases Gin4 and Hsl1 negatively regulate pseudohypha formation and Gin4 also controls septin organization. J. Cell Biol. 164:581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wittenberg C., La Valle R. 2003. Cell-cycle-regulatory elements and the control of cell differentiation in the budding yeast. Bioessays 25:856–867 [DOI] [PubMed] [Google Scholar]
- 82. Yang L., et al. 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3:1359–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zheng X., Wang Y., Wang Y. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23:1845–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhu Y., Takeda T., Whitehall S., Peat N., Jones N. 1997. Functional characterization of the fission yeast Start-specific transcription factor Res2. EMBO J. 16:1023–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.