Abstract
Propolis, a natural product of plant resins, is used by the bees to seal holes in their honeycombs and protect the hive entrance. However, propolis has also been used in folk medicine for centuries. Here, we apply the power of Saccharomyces cerevisiae as a model organism for studies of genetics, cell biology, and genomics to determine how propolis affects fungi at the cellular level. Propolis is able to induce an apoptosis cell death response. However, increased exposure to propolis provides a corresponding increase in the necrosis response. We showed that cytochrome c but not endonuclease G (Nuc1p) is involved in propolis-mediated cell death in S. cerevisiae. We also observed that the metacaspase YCA1 gene is important for propolis-mediated cell death. To elucidate the gene functions that may be required for propolis sensitivity in eukaryotes, the full collection of about 4,800 haploid S. cerevisiae deletion strains was screened for propolis sensitivity. We were able to identify 138 deletion strains that have different degrees of propolis sensitivity compared to the corresponding wild-type strains. Systems biology revealed enrichment for genes involved in the mitochondrial electron transport chain, vacuolar acidification, negative regulation of transcription from RNA polymerase II promoter, regulation of macroautophagy associated with protein targeting to vacuoles, and cellular response to starvation. Validation studies indicated that propolis sensitivity is dependent on the mitochondrial function and that vacuolar acidification and autophagy are important for yeast cell death caused by propolis.
INTRODUCTION
Propolis is a natural product of plant resins collected by honeybees (Apis mellifera) from various plant sources. It is used by the bees to seal holes in their honeycombs and protect the hive entrance (22, 24, 82). Propolis has been used in folk medicine for centuries. Its chemical composition is quite complex since more than 300 compounds, such as polyphenols, phenolic aldehydes, sequiterpene quinines, coumarins, amino acids, steroids, and inorganic compounds, have been identified in propolis samples. Propolis has cytotoxic (53), anti-herpes virus (83), antitumor (34), radical scavenging (4), antimicrobial (38, 60), antiprotozoan (55), and anti-HIV (30) activity and suppressive effects of dioxin toxicity (60). More recently, evidence has shown that propolis can be used to treat Candida fungal infections (1, 17, 59, 68, 76).
There is an urgent need to discover novel antifungal drugs. There are currently five different classes of antifungal drugs classified based on their mechanisms of action (37). Most of these drugs affect the cell membrane or cell wall integrity through inhibition of either ergosterol synthesis or β-glucan/chitin synthesis, respectively. A further complication to this limited spectrum of drugs is the acquisition of resistance by the pathogenic fungi observed in clinical settings (26, 31, 57). While amphotericin B has fungicidal activity, the azoles are generally fungistatic agents for Candida spp. and fungicidal agents for Aspergillus spp. (52). The echinocandins (caspofungin, micafungin, and anidulafungin) exhibit fungicidal activity against Candida spp. and fungistatic activity against Aspergillus spp. (25). There are very few studies investigating the relationship between antifungal drugs and cell death (for a review, see reference 63). Recently, Saccharomyces cerevisiae has become an excellent model system to characterize cell death (for a review, see reference 14). The apoptotic core machinery is quite conserved in this organism, and multiple yeast orthologues of mammalian apoptotic proteins have been identified (14). In addition, several cell death pathways have been characterized, and assays for apoptotic and/or necrotic cell death are being currently used (14).
The identity and the mechanism of action of the chemical compounds present in the propolis are not known. To identify the cellular targets of any bioactive compounds is a very difficult task. In the last years, however, all the genome resources that were developed for S. cerevisiae (www.yeastgenome.org) provided an excellent genomic platform for drug development and drug target identification (7, 28, 50, 85). In addition, all these genetic tools have made it possible to study and construct genetic interaction networks for identification of specific drug targets (9, 16). Here, we apply the power of S. cerevisiae as a model organism for studies of genetics, cell biology, and genomics in order to understand how propolis affects fungi at the cellular level. First, we evaluate how propolis can affect S. cerevisiae cell survival by assessing several genetic determinants involved in apoptosis and/or necrosis in this organism. As a complementary step, we have used the yeast nonessential gene deletion library to investigate possible cell targets for propolis.
MATERIALS AND METHODS
Strains, media, and culture methods.
The following S. cerevisiae strains were used: BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) (10), BY4741A and BY4742 rho−/0 (both MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [rho−/0]) (this study), YJL208C/NUC1 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 GFP::nuc1) (Invitrogen), YJR048W/CYC1 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 GFP::cyc1) (Invitrogen), and ΔYCA1 (MATα his3Δ1 leuΔ0 met15Δ0 ura3Δ0 yca1::kanMX4) (49). The following media were used: the complete medium YPD agar (2% [wt/vol] glucose, 1% [wt/vol] yeast extract, 2% [wt/vol] peptone, and 2% [wt/vol] agar), YPGal agar (2% [wt/vol] galactose, 1% [wt/vol] yeast extract, 2% [wt/vol] peptone, and 2% [wt/vol] agar), minimal medium SC agar (0.7% [wt/vol] yeast nitrogen base [DIFCO], 2% [wt/vol] glucose, 0.1g/liter l-leucine, 0.1g/liter l-lysine, 0.1g/liter l-tryptophan, 0.05g/liter l-histidine, 2% [wt/vol] agar), and YPD, YPGal, and SC liquid media with the same composition as above but without agar. Rosella strains were constructed by the transformation of c-Rosella plasmid (67) into the BY4742 strain. Transformants were selected in SC agar minimal medium.
Preparation of the propolis standardized extract.
Three batches of propolis standardized extract (PSE), 14004/10, 14401/10, and 010/08, were produced by Apis Flora Company (EPP-AF; Ribeirão Preto, SP, Brazil). The extracts were standardized using a propolis blend composed of raw material obtained from several sites of Brazil (A. A. Beretta-Silva, A. C. Meda, and M. E. T. Ferreira, 1 February 2005, Brazilian patent application PI 0405483), such as the states of Minas Gerais, São Paulo, Paraná, Santa Catarina, and Rio Grande do Sul. Propolis (blend of raw material) was kept in a freezer for 12 h and ground to a fine powder in a blender, and its particle size was standardized using a 42-mesh sieve. It was then extracted using hydroalcoholic solution (7:3), with dynamic maceration followed by a percolation process and finally by filtration. The PSE obtained represents 11% (wt/vol) of dry residue and chemical composition standardized qualitatively and quantitatively by reverse-phase high-pressure liquid chromatography (RP-HPLC) by analyzing the following compounds: caffeic, p-coumaric, and cinnamic acids; isosakuranetin; and artepillin C (ARC) (see Table S1 in the supplemental material).
Screening of the yeast deletion library.
For the screening of the yeast deletion library (84; http://www-sequence.stanford.edu/group/yeast_deletion_project/) a set of approximately 4,800 haploid deletions in nonessential genes of S. cerevisiae were screened for propolis sensitivity. Mutants were inoculated from stock cultures in 96-well master plates at −80°C and were grown at 30°C in YPD medium containing 200 μg/ml of Geneticin (G418; Sigma, St. Louis, MO) and then stored at 4°C. To determine which mutants were sensitive to propolis, the cells were spotted on YPD plates plus 0.68% ethanol or alcoholic propolis extract (Apis Flora, Ribeirão Preto, SP, Brazil) at 0.25% and 0.50% (vol/vol). Plates were incubated at 30°C for 5 days.
RNA isolation and real-time PCR.
Exponential (9 h) growth phase BY4742 cells were harvested and suspended (107 cells ml−1) in YPD liquid medium containing 0.125% (vol/vol) alcoholic propolis extract. The treatment was carried out for 5 and 10 min at 30°C with mechanical shaking (200 rpm). Then, yeast cells were washed with distilled water, and RNA was isolated. For total RNA isolation, the yeast cells were disrupted by vortexing with glass beads, and total RNA was extracted with Trizol reagent (Invitrogen). Ten micrograms of RNA from each treatment was then fractionated in 2.2 M formaldehyde–1.2% (wt/vol) agarose gel, stained with ethidium bromide, and then visualized with UV light. The presence of intact 25S and 18S rRNA bands was used as a criterion to assess the integrity of the RNA. RNase-free DNase I treatment for the real-time PCR experiments was carried out as previously described by Semighini et al. (72).
All the PCRs were performed using an ABI 7500 Fast Real-Time PCR System (Applied Biosystems) and TaqMan Universal PCR Master Mix kit (Applied Biosystems). The reactions and calculations were performed according to Semighini et al. (72). The primers and Lux fluorescent probes (Invitrogen) used in this work were the following: ATG8_263RL (5′-CGGCTATATGGCAGACATCAACGC-FAM-G-3′) and ATG8_263RL/235FU (5′-CCCTGAGAAGGCCATCTTCATTT-3′) for YBL078C/ATG8; ATG14_892FL (5′-CGGACAACAAGATGAAGTGTAGGTC-FAM-G-3′) and ATG14_892FL/910RUa (5′-AACCATCGAGTACAACATCCCAAA-3′) for YBR128C/ATG14; and ACT1_366FL (CGGCTGGTTCTGGTATGTGTAAAGC-FAM-G) and ACT1_366FL/385RU (ATGGGAAGACAGCACGAGGAG) for YFL039C/ACT1.
Optical and confocal microscopy.
After incubation under the appropriate conditions for each experiment, slides were mounted and visualized in a fluorescence microscope. For YJL208C/NUC1, YJR048W/CYC1, and Rosella strain microscopy, stationary (16 h)-growth-phase cells were harvested and suspended (107 cells ml−1) in SC liquid medium for YJL208C and YJR048W and in YPD liquid medium for Rosella containing 0.125% (vol/vol) alcoholic propolis extract; the treatments were carried out for 15, 10, and 90 min, respectively, at 30°C with mechanical shaking (200 rpm). A positive control for Rosella staining was performed by growing cells for 16 h at 30°C in YPD medium and transferring them to starvation medium (0.17% [wt/vol] yeast extract, 0.1% [wt/vol] KH2PO4, 0.01% [wt/vol] CaCl2, 0.0005% [wt/vol] FeCl3, 0.07% [wt/vol] MgCl2, 0.05% [wt/vol] NaCl, 2% [wt/vol] glucose) to induce autophagy. After 4 h of incubation at 30°C, protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP; at 10 μM) was added in pH 7.5 buffer for 10 min. Incubation of starved cells before imaging in pH 7.5 buffer containing the protonophore CCCP resulted in an increase in green fluorescence emission in the vacuole (67). For positive control to apoptosis in the NUC1 microscopy, we used 0.4 mM H2O2 for 20 h (13). Cells were washed with phosphate-buffered saline (PBS) and incubated for 5 min in a solution supplemented with 100 ng/ml of 4′,6′-diamidino-2-phenylindole (DAPI; Sigma Chemical, St. Louis, MO). After incubation with the dye, cells were washed with PBS buffer for 10 min at room temperature, and then the slides were mounted. Slides were viewed with a Carl Zeiss (Jena, Germany) microscope using a 100× magnification oil immersion objective lens (EC Plan-Neofluar; numerical aperture, 1.3). In order to verify the accumulation of reactive oxygen species (ROS), cells were stained with 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA). Yeast cells (107 cells ml−1) were grown in 50-ml tubes in 10 ml of YPD medium for 9 or 16 h at 30°C and exposed to 0.125% propolis for 5 or 10 min. Cells were washed with YPD medium and incubated with prewarmed PBS buffer containing 10 μM H2DCFDA (Invitrogen, Molecular Probes) for 30 min in the dark at 37°C. Slides were mounted, and images were analyzed by fluorescent microscopy at room temperature. Slides were viewed with a Carl Zeiss (Jena, Germany) microscope using a 100× oil immersion objective lens (filter set 46 HE YFP; excitation wavelength, 500/20 nm; emission wavelength, 535/30 nm). Phase-contrast for the bright-field images and fluorescent images were captured with an AxioCam camera (Carl Zeiss), processed using the AxioVision software, version 3.1, and saved as TIFF files. Further processing was performed using Adobe Photoshop, version 7.0 (Adobe Systems Incorporated, CA).
In terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays, slides were visualized in a confocal microscope. Images were analyzed using a Leica TCS SP5 spectral laser scanning confocal microscope (Leica Microsystems, Heidelberg, Germany) at the Laboratory of Confocal Microscopy, Faculty of Medicine of Ribeirão Preto (FMRP), University of São Paulo, São Paulo, Brazil, using a 63× water immersion objective lens, with laser line 488 nm for green fluorescent protein (GFP) and 405 nm for DAPI. Images were captured by direct acquisition, with the Z step ranging from 0.5 to 2 μm with the Leica LAS AF software (Leica Microsystems), and additional processing was carried out using Adobe Photoshop, version 7.0.
Induction of cell death.
YPD liquid medium was inoculated with BY4742 cells for 16 h at 30°C. Grown cells were centrifuged and inoculated in 50 ml of fresh YPD liquid medium in 250-ml flasks to a final concentration of 5 × 105 cells ml−1. Cells were then grown for 9 h (exponential; 2.0 ×107 cells ml−1) and 16 h (stationary; 3.0 × 108 cells ml−1). The cells in exponential and stationary phases were harvested and inoculated again in 50 ml of fresh YPD liquid medium in 250-ml flasks to a final concentration of 107 cells ml−1 and exposed to either 0.68% ethanol or 0.125% (vol/vol) alcoholic propolis extract. The treatment was carried out for 5, 10, and 20 min at 30°C with mechanical shaking (200 rpm). Then, the cells were washed with phosphate-buffered saline (PBS) to remove propolis, and the procedures were initiated for cell death detection. Cell viability was determined by plating appropriate cell concentrations and counting the number of grown colonies.
TUNEL assay.
DNA strand breaks were demonstrated by TUNEL with an In Situ Cell Death Detection Kit, Fluorescein, from Roche. This procedure was according to Madeo et al. and Ribeiro et al. (47, 65) with some modifications. Yeast cells were fixed with 3.7% (vol/vol) formaldehyde for 30 min at room temperature and washed three times with PBS, and cell walls were digested with 15 U/ml lyticase, from Arthrobacter luteus (crude 100,000 U; 362 units/mg; Sigma) at 37°C for 60 min. Ten microliters of the cell suspensions was applied to a microscope slide and allowed to dry for 30 min at 37°C. The slides were rinsed with PBS, incubated in permeabilization solution (0.1% [vol/vol] Triton X-100 and 0.1% [wt/wt] sodium citrate) for 2 min on ice, and rinsed twice with PBS. For a positive control the endonuclease DNase I (Sigma) was applied to the cells on microscope slides. The slides were placed in a humidified box for 1 h at 37°C and then washed twice in PBS. Slides were subsequently incubated with 10 μl of TUNEL reaction mixture, containing terminal deoxynucleotidyltransferase and fluorescein isothiocyanate (FITC)-dUTP, for 60 min at 37°C. They were rinsed three times with PBS and incubated for 5 min in a solution supplemented with 100 ng/ml of DAPI (Sigma Chemical, St. Louis, MO). After incubation with the dye, they were washed with PBS buffer for 10 min at room temperature and then rinsed in distilled water, mounted, and visualized in a confocal microscope.
Annexin and PI assays.
Phosphatidylserine exposure was detected by an annexin-V-Fluos staining kit (Roche), as described by Madeo et al. (48) with some modifications. Cells were harvested and washed with sorbitol buffer (1.2 M sorbitol, 0.5 mM MgCl2, 35 mM K2HPO4, pH 6.8). Cell walls were digested with 15 U of lyticase (Sigma) in sorbitol buffer for about 30 min at 37°C. Cells were then washed twice with binding buffer (10 mM HEPES/NaOH, pH 7.4, 140 mM NaCl, 2.5 mM CaCl2) containing 1.2 M sorbitol (binding-sorbitol buffer). To 38-μl cell suspensions in binding-sorbitol buffer, 2 μl of annexin V (Roche) and 2 μl of a propidium iodide (PI) working solution (50 μg/ml) were added, and the mixture was incubated for 15 min at room temperature. The cells were then washed and resuspended in binding-sorbitol buffer. Finally, the slides were mounted with the cell suspensions. Microscope settings and image acquisitions were as described above using epifluorescence microscopy. For quantitative assessment of annexin V-PI staining, at least 100 yeast cells were counted per sample. For the apoptosis positive control, the cells were treated with 80 mM acetic acid, pH 3.0, for 200 min (44), and for the necrosis positive control, the cells were fixed with fixative solution (3.7% formaldehyde, 50 mm sodium phosphate buffer, pH 7.0, 0.2% Triton X-100) for 30 min at room temperature.
Mitochondrial fractionation studies.
For the extraction of mitochondrial proteins, mitochondria were isolated from S. cerevisiae cells based on the a previously described procedure (23, 64). Cells grown for 16 h in YPGal liquid (at 30°C), either untreated or exposed to 0.125% propolis for 10 min at 30°C, were harvested at 3,500 × g for 5 min and were washed once with 1.2 M sorbitol. Then, washed cells were suspended in digestion buffer at a concentration of 10 g of cells (wet weight) in 50 ml of buffer (1.2 M sorbitol, 0.06 M phosphate, pH 7.5, 1 mM EDTA, 15 mM β-mercaptoethanol, and 50 mg Zymolyase 20T [MP Biomedicals]) and incubated at 37°C for 2 h to be converted to spheroplasts. Spheroplasts were washed with 1.2 M sorbitol and then gently homogenized in a resuspension buffer (0.6 M sorbitol, 20 mM HEPES, pH 7.4, and 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) using an Elvehjem-Potter homogenizer. The homogenate was centrifuged at 2,500 × g for 5 min at 4°C. The supernatant was centrifuged again at 2,500 × g for 5 min at 4°C two additional times, and then it was centrifuged at 12,000 × g for 10 min to sediment mitochondria. The supernatant (cytoplasmic portion) was collected, and the mitochondrial pellet was resuspended gently with resuspension buffer and washed four additional times at 2,000 × g for 5 min at 4°C. The final mitochondrial pellet was resuspended gently in 0.6 M sorbitol–20 mM HEPES. Protein concentration was determined by Biuret assay (43) using bovine serum albumin (BSA) as a standard.
Samples were then analyzed by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Samples were prepared for SDS-PAGE by the addition of 1× sample buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, and 5% bromophenol blue) and heating at 100°C for 3 min. Twenty micrograms of total protein from each sample was loaded into each lane of a 10% SDS-PAGE gel. After separation of the proteins, the gel was blotted onto a pure nitrocellulose membrane (0.2-μm pore size; Bio-Rad), and after being blocked in 5% dried milk in TBS-T buffer (10 mM Tris-HCl, 150 mM NaCl, pH 8.0, and 0.05% Tween 20), the membrane was probed with the antibody rabbit anti-cytochrome c (CNAT against native cytochrome c from yeast; Sigma) at a 1:200 dilution in TBS-T buffer for 1 h at room temperature. The membrane was washed four times for 5 min each with TBS-T buffer and then incubated with a 1:5,000 dilution of goat anti-rabbit IgG peroxidase-labeled antibody (KPL) for 1 h. After being washed, the blot was developed by use of the SuperSignal Ultra chemiluminescence detection system (Pierce) and recorded by the use of Hyperfilm ECL (Amersham Biosciences).
Oxygen consumption.
Mitochondrial and spheroplast oxygen consumption was monitored on a computer-interfaced Clark-type electrode at 25°C with 1 mM NADH as a substrate in the presence of mitochondria at 100 μg/ml protein concentration. Mitochondria were isolated from S. cerevisiae cells grown in YPGal medium. Propolis was added to a 0.125% final concentration. In order to block cytochrome c oxidase respiration, 1 mM KCN was added at the end.
PPPI network design and topological analysis.
The survival data gathered from yeast deletion strains submitted to propolis treatment was used to obtain information about how the deleted genes and their products interact in the context of physical protein-protein interaction (PPPI) networks in S. cerevisiae. To this end, data mining screening and network design of PPPI networks were performed using Cytoscape software, version 2.6.3 (74). For this purpose, we used the PPPI data of S. cerevisiae available in the STRING database, version 8.3 (http://string.embl.de), using the following parameters: active prediction methods all enabled except text mining; no more than 50 interactions; medium confidence score (0.400); and network depth equal to 1 with addition of new nodes until saturation of the network. The PPPI networks obtained from this first screening were then combined in a unique PPPI network by employing the union function of the Cytoscape core plug-in Merge Networks (Table 1). This major PPPI network was then analyzed with molecular complex detection (MCODE) software (3), a Cytoscape plug-in (at http://www.cytoscape.org/plugins2.php) in order to detect subnetworks or clusters of proteins that could represent distinct biologic processes. The parameters used for MCODE to generate the subnetworks were as follows: loops included; degree cutoff of 2; deletion of single connected nodes from cluster (haircut option enable); expansion of cluster by one neighbor shell allowed (fluff option enable); node density cutoff of 0.1; node score cutoff of 0.2; k-core of 2; and maximum depth of network of 100.
Table 1.
Specific gene ontology classes derived from PPPI network observed in cluster 6
| Biological process | GO identification no. | P valuea | Corrected P valueb | kc | fd |
|---|---|---|---|---|---|
| Cell division | 51301 | 2.95 × 10−14 | 2.56 × 10−11 | 36 | 323 |
| Cell budding | 7114 | 1.31 × 10−13 | 5.68 × 10−11 | 19 | 82 |
| Asexual reproduction | 19954 | 4.16 × 10−13 | 1.20 × 10−10 | 19 | 87 |
| Reproduction of a single-celled organism | 32505 | 6.45 × 10−13 | 1.40 × 10−10 | 19 | 89 |
| Establishment of cell polarity | 30468 | 1.23 × 10−12 | 1.67 × 10−10 | 20 | 104 |
| Mitochondrial electron transport, ubiquinol to cytochromec | 6122 | 2.79 × 10−11 | 2.11 × 10−9 | 8 | 11 |
| Cytokinesis | 16288 | 2.91 × 10−11 | 2.11 × 10−9 | 19 | 109 |
| Cell cycle | 7049 | 2.13 × 10−9 | 1.05 × 10−7 | 40 | 566 |
| Electron transport chain | 22900 | 8.55 × 10−7 | 1.61 × 10−5 | 8 | 31 |
| pH reduction | 45851 | 2.63 × 10−5 | 3.08 × 10−4 | 6 | 24 |
| Vacuolar acidification | 7035 | 2.63 × 10−5 | 3.08 × 10−4 | 6 | 24 |
| Negative regulation of transcription from RNA polymerase II promoter by glucose | 433 | 3.30 × 10−4 | 2.63 × 10−3 | 3 | 6 |
P values were calculated by the hypergeometric distribution of one ontology class visualized in the network.
Calculated values based on P values obtained after FDR was applied.
Total number of proteins found in the network which belong to a gene ontology.
Total number of proteins that belong to a specific gene ontology.
GO analysis.
Gene ontology (GO) clustering analysis was performed using Biological Network Gene Ontology (BiNGO) (50) software, a Cytoscape plug-in available at http://chianti.ucsd.edu/cyto_web/plugins/index.php. The degree of functional enrichment for a given cluster and category was quantitatively assessed (P value) by hypergeometric distribution (66), and a multiple test correction was applied using the false-discovery rate (FDR) (5) algorithm, fully implemented in BiNGO software. Overrepresented biological process categories were generated after FDR correction, with a significance level of 0.05.
RESULTS
Propolis induces cell death in S. cerevisiae.
Propolis is a complex product derived from plant resins and bee saliva. There are several chemical compounds present in this natural product that could potentially be responsible for its antibiotic properties. As a first step to understand whether individual components of propolis could play such role, we fractionated propolis extracts by HPLC and purified some of the most abundant chemical compounds (see Fig. S1 and Table S1 in the supplemental material). We have obtained similar fingerprints for three different batches of propolis (see Fig. S2). We tested S. cerevisiae growth in the presence of four of these chemical compounds (caffeic, p-coumaric, and cinnamic acids and isosakuranetin). We applied 1, 2, 5, 10, 25, 50, and 100 μg/ml of each compound and a mix of 100 μg/ml of each in microtiter plate liquid cultures. None of these treatments was able to inhibit the growth of S. cerevisiae BY4742, Candida albicans CAI4, or Candida parapsilosis ATCC 2201 (data not shown). Thus, taking into consideration the fact that the cell death effects of propolis could be due to a combination of chemical compounds and concentrations, we decided to investigate the cell death effects of propolis by concentrating our experiments on alcoholic extracts of propolis.
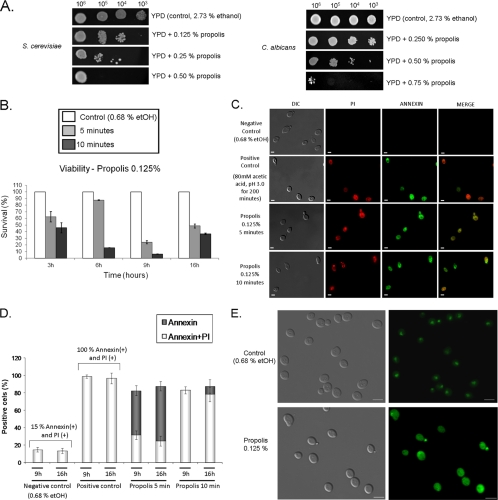
The first series of experiments was related to verifying a working propolis concentration and determining how yeast viability was affected. Since propolis is dissolved in ethanol (60% alcoholic extract), the control treatment was grown on 2.73% ethanol because 0.5% propolis has a final concentration of about 2.73% ethanol (Fig. 1A). Tenfold dilutions of S. cerevisiae cells suggested that 0.125% propolis could be an adequate choice as a subinhibitory concentration (Fig. 1A). All the controls for further experiments using 0.125% propolis as a treatment have 0.68% ethanol (what corresponds to 60% alcoholic extract for 0.125% propolis). We were not able to perform the determination of MICs by using microdilutions because the color of the propolis is dark, which interferes in the spectrophotometer determinations (data not shown). The inhibitory effects of propolis on S. cerevisiae are not affected by growth on lower oxygen concentrations, such as 1% O2 (data not shown). We also observed that C. albicans was more susceptible to concentrations of propolis between 0.50 and 0.75% (Fig. 1A). However, C. parapsilosis and Aspergillus fumigatus growth was not affected by the same concentrations of propolis (data not shown). Yeast cells in the initial mid-exponential (6 h; 1.2 × 106 cells ml−1) and stationary (16 h; 3.0 × 108 cells ml−1) phases are more resistant to 0.125% propolis extracts while cells in the exponential phase (9 h; 2.0 × 107 cells ml−1) are much more sensitive (Fig. 1B). In the next step, we investigated what was the main mechanism of cell death in the cells in the exponential and stationary phases, i.e., 9 and 16 h of growth, respectively, by observing the number of cells which are propidium iodide (PI) positive (PI+) or annexin and TUNEL positive (preliminary indications of necrosis or apoptosis, respectively).
Fig. 1.
Propolis induces apoptosis and late necrosis in S. cerevisiae. (A) Spot dilution assays for S. cerevisiae and C. albicans in YPD medium supplemented with different concentrations of propolis. The plates were incubated at 30°C for 72 h. (B) Viability of yeast cells in different growth cycle phases exposed to 0.125% propolis for 5 or 10 min. Cell viability was determined by plating appropriate cell concentrations and counting the number of colonies. The numbers are the average ± standard deviation of three independent experiments. (C) Annexin and PI assays for 16-h yeast cells exposed to 0.125% propolis. (D) Total numbers for annexin- and PI-positive cells for yeast cells exposed to 0.125% propolis. Bar, 5 μm. The numbers are the average ± standard deviation of three independent experiments. In each experiment, 100 cells were assessed for annexin and PI staining. Positive controls for annexin and PI were exposing the cells to acetic acid, pH 3.0, for 200 min and fixing them with fixation solution as described in the Materials and Methods section. (E) Nucleo-cytosolic translocation of Nhp6Ap-GFP in untreated S. cerevisiae cells or cells exposed to 0.125% propolis for 10 min. Bar, 5 μm.
Early apoptosis is characterized by an increased number of annexin V-positive (A+) cells and PI negative (PI−) cells while in late apoptosis (leading to secondary necrosis), there is an increase in the number of A+ and PI+ cells. The primary necrosis is characterized by an increased number of annexin V-negative (A−) cells and PI+ cells. We have observed about 15% A+ and PI+ cells in the negative control (0.68% ethanol [EtOH]) while 100% A+ and PI+ cells in the positive control (acetic acid, pH 3.0, for 200 min). Upon 5 min of exposure to 0.125% propolis, cells grown for 9 and 16 h have an increased number of A+ cells (about 30% both A+ and PI+ and 80% A+), suggesting early apoptosis (Fig. 1B to D). Upon 10 min of exposure to 0.125% propolis, cells grown for 9 and 16 h have an increased number of A+ and PI+ (about 80% for both classes), suggesting late apoptosis, leading to secondary necrosis (Fig. 1B and C). However, the TUNEL-positive staining remained constant for both 5 and 10 min of exposure to 0.125% propolis (about 80%) (see Fig. S3 in the supplemental material).
Nuclear release of mammalian high-mobility group box-1 protein (HMGB1) is a defining feature of necrosis in mammalian cells (69). Since apoptotic cells can become leaky during further cultivation, resulting in PI positives, we determined the nucleo-cytosolic translocation of the yeast HMGB1 homologue Nhp6Ap in cells grown for 16 h exposed to 0.125% propolis for 5 and 10 min. Fluorescence microscopy showed the nucleo-cytosolic translocation of the GFP-tagged Nhp6Ap (Nhp6Ap-GFP) in 20 and 30% of the cells after exposure for 5 and 10 min to 0.125% propolis, respectively (Fig. 1E). Taken together, these results indicate that propolis is able to induce an apoptosis cell death response but that increased exposure to propolis provides a corresponding increase in the necrosis response.
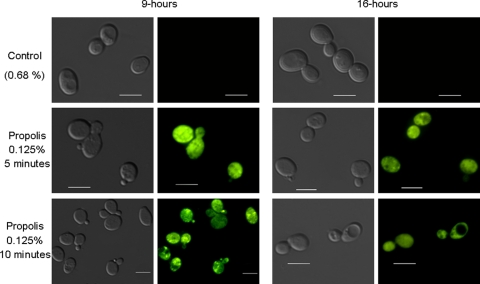
There is an accumulation of mitochondrion-produced reactive oxygen species (ROS) during yeast apoptosis (for reviews, see references 19, 58, and 62). To investigate the production of ROS during propolis-induced cell death in S. cerevisiae, we used 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA), a cell-permeable ROS indicator that is nonfluorescent until acetate groups are removed (H2DCF) by intracellular esterases and oxidation occurs within the cell. Although H2DCF can be oxidized by different ROS, it is mainly used to detect superoxides. Yeast cells grown for 9 or 16 h were either untreated or exposed to 0.125% propolis for 5 and 10 min, incubated with H2DCFDA, and examined by fluorescence microscopy (Fig. 2). The DCFDA-dependent green fluorescence was detected in about 10% of the 9- or 16 h-grown cells not exposed to propolis. Treatment of 9-h-grown cells with 0.125% propolis for 5 and 10 min yielded 70 to 80% fluorescence while treatment of 16-h-grown cells treated for 5 and 10 min produced 40 to 60% fluorescence (Fig. 2). These results indicate that there is a great accumulation of ROS during cell death induced by propolis.
Fig. 2.
Propolis induces the production of ROS. Yeast cells were grown for 9 or 16 h at 30°C in YPD medium and stained with H2DCFDA for 30 min at 37°C before fluorescent microscopy analysis. Bar, 5 μm.
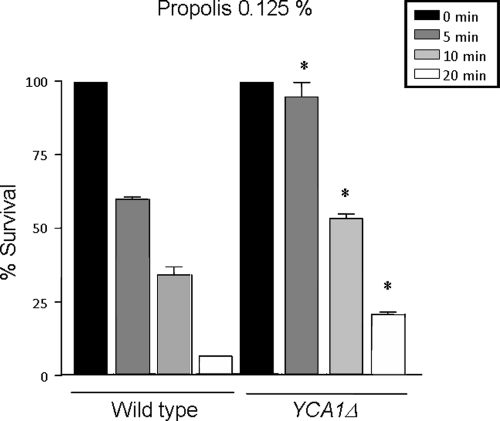
Caspases are members of a family known as cysteine proteases that are actively involved in cell death in eukaryotes (for a review, see reference 41). Although S. cerevisiae does not have caspases in its genome, a caspase-like protein named YCA1, fitting into the type I category of metacaspases, was identified and characterized previously (49, 54, 81). It has been shown that YCA1 is involved in several cell death stimuli (for a review, see reference 54). When yeast cells grown for 16 h were exposed to 0.125% propolis for 5, 10, and 20 min, there was a reduction in the cell viability of 40, 70, and 95% in the wild-type strain while in the YCA1Δ strain, this reduction was about 10, 50, and 80% (Fig. 3). These results strongly indicate that the YCA1 gene is important for propolis-mediated cell death.
Fig. 3.
Metacaspase YCA1 mutant is more tolerant to propolis than the corresponding wild-type strain. Viability of 16-h yeast wild-type and YCA1 mutant strains treated with 0.125% propolis (*, P < 0.05) was determined by plating appropriate cell concentrations and counting the number of colonies. The results are the means ± standard deviation of three independent experiments.
Cytochrome c is released during propolis-induced cell death.
We have also investigated if other components of the cell death machinery are being activated during propolis-induced cell death in S. cerevisiae. We concentrated our attention on two of these putative determinants: CYC1 and NUC1. In mammalian cells, at least two major apoptotic pathways have been described: (i) the intrinsic pathway that needs the involvement of the mitochondria, and (ii) the extrinsic pathway where mitochondria are bypassed and caspases are activated (for a review, see reference 21). One of the hallmarks of the intrinsic pathway is the release of apoptogenic factors such as cytochrome c to the cytosol and the consequent assembly organized by this protein of the high-molecular-weight complex, the mitochondrial apoptosome, that activates caspases (for a review, see reference 21). In S. cerevisiae cells undergoing an apoptotic process induced by acetic acid, translocation of cytochrome c to the cytosol was observed (45). We performed cell fractionation studies to determine if cytochrome c was translocated to the cytoplasm during exposure of yeast cells grown for 16-h (16-h yeast cells) to 0.125% propolis for 10 min (Fig. 4). When the yeast cells were not exposed to propolis, all the cytochrome c was located in the mitochondrial protein extracts while upon propolis addition, there was a significant presence of cytochrome c in the cytoplasm protein extracts (Fig. 4, right panel). There are corresponding protein amounts in each cell fraction extract, as shown by Coomassie staining (Fig. 4, left panel).
Fig. 4.
Exposure of yeast cells to propolis induces cytochrome c release into the cytoplasm. Yeast cells were grown for 16 h at 30°C and left untreated or exposed to 0.125% propolis for 10 min at 30°C. The cells were harvested and fractionated (mitochondria and cytoplasm) as described in the Materials and Methods section, and proteins were run on a polyacrylamide gel. At left is a Coomassie-stained gel with the same amount of proteins that were transferred to a nitrocellulose filter (right panel). This membrane was probed with the antibody rabbit anti-cytochrome c.
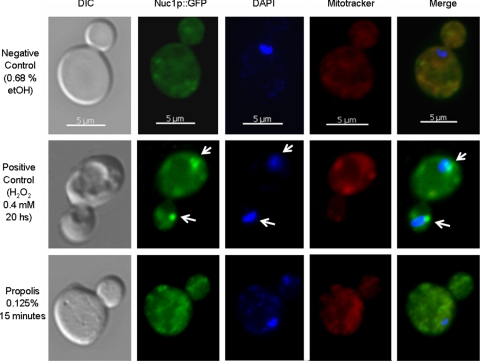
Endonuclease G (EndoG) has been described as a mitochondrial endonuclease that digests both DNA and RNA (12, 13, 61). Upon apoptosis induction, translocation of mammalian EndoG and its S. cerevisiae homologue NUC1 to the nucleus coincides with large-scale DNA fragmentation (12, 13, 41, 61). We have not observed increased sensitivity of NUC1Δ yeast cells to propolis (data not shown). Next, we examined if Nuc1p would translocate to the nucleus when yeast cells were exposed to propolis (Fig. 5). When 16-h yeast cells were not exposed to propolis, the Nuc1p-GFP showed a diffuse mitochondrion filament-like distribution, as confirmed by MitoTracker staining (Fig. 5, first row). In yeast cells exposed to hydrogen peroxide, part of the Nuc1p-GFP was translocated to the nucleus (DAPI staining), as previously shown by Büttner et al. (13), but it also accumulated in the mitochondria as indicated by MitoTracker staining (Fig. 5, second row). In contrast, when yeast cells were exposed to 0.125% propolis for 15 min, about 100% of the yeast cells showed a Nuc1p-GFP dot-like distribution, suggesting mitochondrial fragmentation, as again confirmed by MitoTracker staining (Fig. 5, third row). Moreover, the Nuc1p-GFP did not translocate to the nuclei, as shown by DAPI staining (Fig. 5, third row). These results strongly indicate that cytochrome c, but not Nuc1p, is involved in propolis-mediated cell death in S. cerevisiae.
Fig. 5.
Nuc1p is not involved in propolis sensitivity. Yeast Nuc1-GFP cells were grown for 16 h at 30° and left untreated or exposed to 0.125% propolis for 15 min, and cells were visualized by a fluorescence microscope. A positive control was made by incubating 16-h-old yeast cells with 0.4 mM H2O2 for 20 h at 30°C. Bar, 5 μm. DIC, differential interference contrast. Arrows indicate the nuclei.
High-throughput screen of the yeast deletion library for propolis sensitivity.
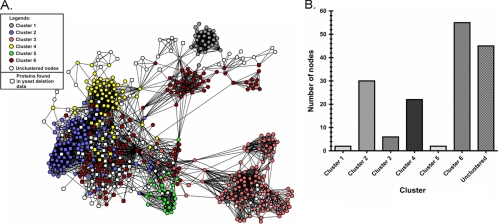
To elucidate the gene functions that may be required for propolis sensitivity in eukaryotes, the full collection of haploid S. cerevisiae deletion strains was screened for growth on YPD plates supplemented with 2.73% ethanol (as a control; 2.73% is the ethanol concentration used to dissolve 0.500% propolis) and 0.250 and 0.500% propolis for 5 days at 37°C. About approximately 4,800 different strains were involved in this screening, and after three confirming screenings, we were able to identify 138 deletion strains that have different degrees of propolis sensitivity compared to the corresponding wild-type strain (see Table S2 in the supplemental material). The survival data obtained from yeast deletion strains prompted us to ask how the absence of the determined proteins of propolis-sensitive yeast strains affects different biological processes that lead to cell death. In this sense, a search for potential proteins and/or mechanisms and their associated biological processes that are affected by propolis exposure was initiated. To achieve this goal, different PPPI networks using yeast deletion data were retrieved from the STRING database. Shared proteins and subnetworks present in the major PPPI network (Fig. 6) were identified and retrieved using the Cytoscape-associated plug-in MCODE and subjected to a Gene Ontology (GO) analysis in order to obtain information about the nature and number of subnetworks belonging to the network and their associated biological processes. Results obtained from MCODE and GO analysis showed that the final PPPI network (Fig. 6A) contains 553 nodes and 3,651 connectors and is composed of six heavily interconnected clusters, each comprising different biological processes. GO analyses revealed that these biological processes can be classified into the following categories: (i) translation/energy derivation by oxidation of organic compounds/mitochondrial genome maintenance, (ii) transcription/chromosome organization and biogenesis/G1 phase of mitotic cell cycle, (iii) carboxylic acid metabolic process/protein targeting to peroxisome/respiration metabolism, (iv) establishment and/or maintenance of chromatin architecture/histone deacetylation, (v) RNA catabolic process, and (vi) cell division mechanisms. As expected, proteins that could not be classified into any cluster were also identified in the network (unclustered protein subnetworks) (Fig. 6A and B).
Fig. 6.
System biology for the propolis sensitivity in S. cerevisiae. (A) A physical protein-protein interaction (PPPI) network obtained from yeast deletion data whose absence increases the sensitivity of yeast cells to propolis treatment. (B) Distribution of proteins (number of nodes) associated with propolis-induced sensitivity as observed from yeast deletion data in the different clusters and unclustered proteins of the major PPPI network. Cluster 1, translation/energy derivation by oxidation of organic compounds/mitochondrial genome maintenance; cluster 2, transcription/chromosome organization and biogenesis/G1 phase of mitotic cell cycle; cluster 3, carboxylic acid metabolic process/protein targeting to peroxisome/respiration metabolism; cluster 4, establishment and/or maintenance of chromatin architecture/histone deacetylation; cluster 5, RNA catabolic process; cluster 6, cell division mechanisms; and unclustered nodes.
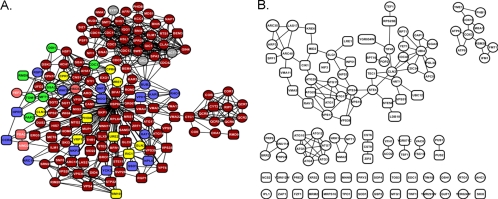
The distribution analysis of the proteins whose deletion increases the sensitivity of yeast strains to propolis indicated that both cluster 6 and unclustered protein subnetworks contain 61.73% of the total proteins observed in the deletion assay (Fig. 6B; see also Table S3 in the supplemental material). The other 38.27% proteins were found distributed among clusters 1 to 5 (Fig. 6B; see also Table S3), showing that cluster 6 and the unclustered protein subnetworks appear to be important for the pleiotropic responses observed when yeast cells are submitted to propolis treatment. When the proteins of the cluster 6 subnetwork (Fig. 7; see also Table S3) were subjected to GO analysis, the data indicated that the major biological processes found within this subnetwork were associated with cell division and reproduction, which includes the establishment of cell polarity, cell cycle, and budding (Table 1). Moreover, the proteins of the cluster 6 subnetwork appear to be important to the mitochondrial electron transport chain, vacuolar acidification, and negative regulation of transcription from RNA polymerase II promoter (Table 1). Interestingly, the GO analysis of the unclustered protein subnetworks (Fig. 7B; see also Table S3) showed that these proteins are important for the regulation of macroautophagy associated with protein targeting to vacuoles and the cellular response to starvation, among other processes (Table 2).
Fig. 7.
(A) Subnetwork of proteins (cluster 6) associated with cell division, establishment of cell polarity, electron transport chain, and vacuolar acidification. (B) Unclustered protein subnetworks associated with macroautophagy, protein targeting to vacuoles, cellular response to starvation and stress, vacuolar transport, cell communication, and regulation of protein polymerization.
Table 2.
Specific GO classes derived from PPPI of unclustered protein subnetworks
| Biological process | GO identification no. | P valuea | Corrected P valueb | kc | fd |
|---|---|---|---|---|---|
| Macroautophagy | 34262 | 7.44 × 10−14 | 5.44 × 10−11 | 11 | 23 |
| CVT pathway | 32258 | 9.36 × 10−12 | 3.43 × 10−9 | 9 | 18 |
| Protein targeting to vacuole | 6623 | 7.37 × 10−11 | 1.80 × 10−8 | 13 | 62 |
| Cellular response to starvation | 9267 | 3.03 × 10−10 | 5.54 × 10−8 | 11 | 44 |
| Cellular response to stress | 33554 | 1.06 × 10−9 | 5.09 × 10−7 | 11 | 49 |
| Cellular response to nutrient levels | 31669 | 4.86 × 10−9 | 5.09 × 10−7 | 11 | 56 |
| Response to starvation | 42594 | 1.80 × 10−8 | 1.46 × 10−6 | 11 | 63 |
| Vacuolar transport | 7034 | 5.05 × 10−8 | 3.69 × 10−6 | 14 | 122 |
| Cell communication | 7154 | 1.44 × 10−6 | 6.21 × 10−5 | 20 | 317 |
| Regulation of protein polymerization | 32271 | 1.76 × 10−6 | 6.79 × 10−5 | 4 | 6 |
P values calculated by the hypergeometric distribution of one ontology class visualized in the network.
Calculated values based on P values obtained after FDR was applied.
Total number of proteins found in the network which belong to a gene ontology.
Total number of proteins that belong to a specific gene ontology.
In the next two sections, we show validation data about the role played by clusters related to the mitochondrial electron transport chain and vacuolar acidification.
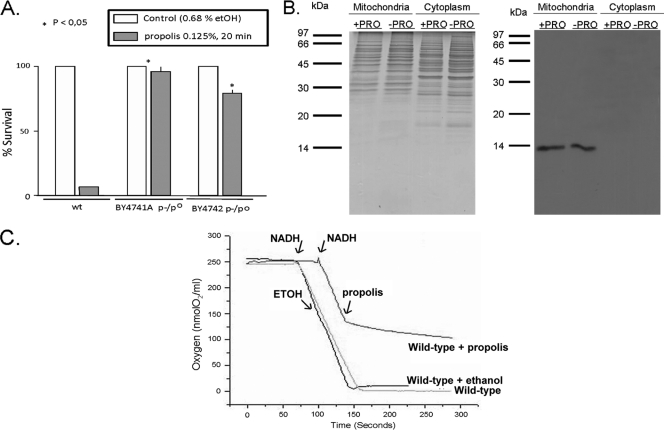
Petite strains are less sensitive to propolis.
It has recently been shown that during necrosis in S. cerevisiae, there is an inhibition of mitochondrial function with a corresponding reduction of ATP production (for a review, see reference 19). In this paper, there are several indications that propolis induces a secondary necrosis cell death mechanism in S. cerevisiae. In addition, as demonstrated in the previous section, we found that when several genes related to energy derivation by oxidation of organic compounds, mitochondrial genome maintenance, and the mitochondrial electron transport chain are deleted, there is an increase in S. cerevisiae propolis sensitivity. We decided to investigate a possible correlation between propolis sensitivity, mitochondria and S. cerevisiae necrotic cell death by isolating petite strains of S. cerevisiae and checking their sensitivity to propolis. In order to isolate BY4742 [rho0]/[rho−] cells, the respiratory-competent strain BY4742 (10) was mutagenized with ethidium bromide, and the resultant petite respiratory-deficient colonies were crossed to [rho0]/[rho−] tester strains (80) to confirm the nature of the isolated petite mutant respiratory incapacity. Two of these petite strains, BY4741A [rho0]/[rho−] and BY4742 [rho−]/[rho0], and the corresponding wild-type strain were exposed to 0.125% propolis for 20 min, and cell viability was determined (Fig. 8). The petite strains showed dramatic increased viability compared to the wild-type strain (compare 95 and 75% viability of BY4741A [rho−]/[rho0] and BY4742 [rho−]/[rho0] strains, respectively, against about 10% viability in the wild-type strain) (Fig. 8A). Accordingly, when cell protein extracts were fractionated, cytochrome c could not be observed in cytoplasmic protein extracts of petite cells exposed to propolis but only in mitochondrial protein extracts (Fig. 8B). We measured the effect of propolis on mitochondria isolated from yeast strain W303 by monitoring the oxygen consumption rate as a consequence of mitochondria NADH oxidase activity. In the presence of propolis the NADH oxidase activity is 1/10 of that observed before the addition of this compound, indicating its toxic effect on respiration (Fig. 8C). These results strongly suggest that S. cerevisiae propolis sensitivity is dependent on the mitochondrial function.
Fig. 8.
Petite mutants are more tolerant to propolis. (A) Viability of 16-h yeast wild-type and petite mutant strains to 0.125% propolis (*, P < 0.05) was determined by plating appropriated cell concentrations and counting the number of colonies. The results are the means ± standard deviations of three independent experiments. (B) Exposure of yeast petite mutant cells to propolis does not induce cytochrome c release into the cytoplasm. Yeast cells were grown for 16 h at 30°C and left untreated or exposed to 0.125% propolis for 10 min at 30°C. The cells were harvested and fractionated (mitochondria and cytoplasm) as described in the Materials and Methods section, and proteins were run on a polyacrylamide gel. At left is a Coomassie-stained gel with the same amount of proteins that were transferred to a nitrocellulose filter (right panel). This membrane was probed with the antibody rabbit anti-cytochrome c. (C) NADH oxidase activity in wild-type yeast in the presence of propolis. Mitochondria (100 μg of protein) of the wild-type W303 strain were assayed for NADH-oxidase with a Clark electrode as described in the Materials and Methods section. Arrows indicate the addition of propolis (final concentration of 0.125%) or the identical volume of ethanol and KCN. These are representative traces of one experiment, which were reproduced with mitochondria isolated on three different days.
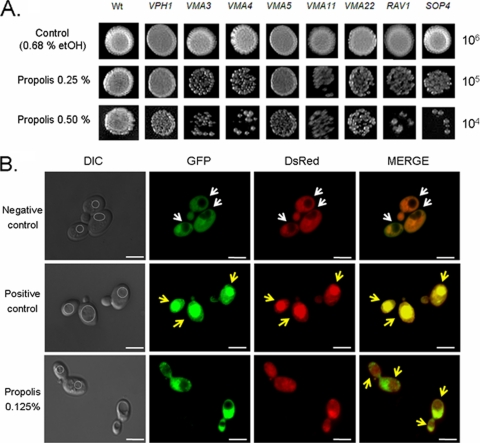
A role for the vacuole and the vacuolar H+-ATPase in propolis sensitivity.
Another set of genes important for propolis sensitivity observed in our screening were involved in vacuolar acidification and macroautophagy associated with protein targeting to vacuoles and cellular response to starvation (Table 2). Organelle acidification is implicated in protein sorting in the biosynthetic and endocytic pathways, proteolytic activation of zymogen precursors, and transmembrane transport of viral contents and toxins (for a review, see reference 32). In fungi, the lysosome-like vacuole also plays a role in storage of metabolic building blocks, calcium homeostasis, and osmotic control. Vacuolar acidification is essential for all these functions. The vacuolar proton-translocating ATPase (V-ATPase) is an essential enzyme to catalyze all these processes. Their primary role in eukaryotic cell is ATP-driven transport of protons from the cytosol into acidic organelles (32). When several genes (VPH1, VMA3, VMA4, VMA5, VMA11, VMA22, RAV1, and SOP4) involved in the assembly of the yeast V-ATPase were deleted, the corresponding yeast deletion strains became more sensitive to propolis (Fig. 9A). Recently, Rosado et al. (67) developed a method for monitoring autophagy using Rosella, a biosensor comprised of a fast-maturing pH-stable red fluorescent protein fused to a pH-sensitive green fluorescent protein variant. Its mode of action relies upon differences in pH between different cellular compartments and the vacuole. Rosella is a genetically encoded dual-color-emission biosensor, a product of a fusion of a relatively pH-insensitive fast-maturing variant of the red fluorescent protein, DsRed (6) with a pH-sensitive variant of GFP (56). The rationale of this assay relies on the pH difference between the vacuole (or lysosome; approximately pH 6.2 or 4.8, respectively) and other compartments of the cell (pH >7.0). The reporter is a dual-color emission biosensor comprising a relatively pH-stable DsRed and a pH-sensitive GFP variant (67). In yeast cells grown for 16 h, both red and green fluorescence were evenly distributed throughout the cytosol and absent from the vacuole (Fig. 9B, first row). Cells were then transferred to starvation medium (without nitrogen) to induce autophagy. After a 4-h incubation, red fluorescence, but not green, was observed to accumulate in the vacuole of approximately 40% of the cells (Fig. 9B, second row). Then, the cells were exposed to 0.125% propolis, and after a 90-min incubation, a subtle phenotype of red fluorescence, but not green, was observed to accumulate in the vacuole of approximately 60% of wild-type cells (Fig. 9B, second and third rows).
Fig. 9.
Propolis induces vacuolar acidification in S. cerevisiae. (A) Spot dilution assays for YPD medium supplemented with different concentrations of propolis. Yeast cells were grown for 96 h at 30°C. (B) Rosella expressed in the yeast cytosol is delivered to the vacuole under cell exposure to propolis. DIC and fluorescence images are shown for wild-type cells under growing conditions (first row), after nitrogen starvation for 4 h (second row), and exposed to 0.125% propolis for 90 min at 30°C. The position of the vacuole in the DIC images is delineated by a dashed white line. Empty vacuoles are highlighted by white arrows. Filled vacuoles are highlighted by yellow arrows. Bar, 5 μm.
Autophagy can be involved in the turnover of long-lived proteins and whole organelles (for example, mitochondria in mitophagy and the endoplasmic reticulum in reticulophagy) (51). Autophagy is also an important process during starvation; i.e., by the catabolism of macromolecules autophagy generates metabolic substrates (51). Autophagy has been followed by various biochemical and morphological methods (for reviews, see references 35 and 36). Expression of GFP-Atg8p in yeast can be used to follow the localization or accumulation of the preautophagosomal structure, autophagosomes, and autophagic bodies. We have observed that ATG1 (encoding a protein serine/threonine kinase required for vesicle formation in autophagy and the cytoplasm-to-vacuole targeting [CVT] pathway), ATG10 (encoding a conserved E2-like conjugating enzyme that mediates formation of the Atg12p-Atg5p conjugate, which is a critical step in autophagy), and ATG20 (encoding a nexin family member required for the CVT pathway and for endosomal sorting) deletion strains are more sensitive to propolis (for reviews about yeast genes involved in autophagy, see references 33 and 39) (Fig. 10A). We have used mRNA accumulation of ATG14 and ATG8 and monitoring GFP-Atg8 as assays to verify if propolis induces an increase in autophagy. Propolis induced ATG8 and ATG14 mRNA accumulation about 4 times and 14 times, respectively (5- and 10-min exposure to 0.125% propolis) (Fig. 10B). The GFP-Atg8 translocated to vacuolar-like structures after exposure to 0.125% propolis for 20 min (Fig. 10C).
Fig. 10.
Propolis induces autophagy in S. cerevisiae. (A) Spot dilution assays for YPD medium supplemented with different concentrations of propolis. Yeast cells were grown for 96 h at 30°C. (B) Yeast wild-type cells were grown for 9 h in liquid YPD medium at 30°C and left untreated or exposed to 0.125% propolis for 5 or 10 min. Total RNA was extracted, and real-time reverse transcription-PCR (RT-PCR) was used to quantify mRNA accumulation. The measured quantity of the mRNA in each of the treated samples was normalized using the threshold cycle (CT) values obtained for the actin (ACT1) mRNA amplifications run in the same plate. The relative quantitation of ATG8, ATG14, and ACT1 gene expression was determined by a standard curve (i.e., CT values plotted against logarithm of the DNA copy number). The results are the means ± standard deviations of four sets of experiments. (C) Cells of yeast strains harboring GFP-Atg8 either untreated or exposed to 0.125% propolis for 20 min at 30°C. Bar, 5 μm.
Taken together, these data strongly indicate that vacuolar acidification and autophagy are important for yeast cell death caused by propolis.
DISCUSSION
The three main types of propolis around the world have different compositions: (i) caffeic acid phenethyl ester (CAPE)-based propolis in Europe, the Far East and New Zealand; (ii) artepillin C (ARC)-based Brazilian green propolis; and (iii) Brazilian red propolis. Recently, a screen of 1,266 compounds with known pharmaceutical activities was performed, and 15 compounds that prolonged survival of C. albicans-infected nematodes and inhibited in vivo filamentation of C. albicans were identified (11). One of these compounds, CAPE, exhibited antifungal activity in a murine model of candidiasis (11). CAPE is a poplar-type propolis while Brazilian green propolis is a Baccharis type. We along with other investigators have been unable to identify any traces of CAPE in the Brazilian green propolis (75, 79; also data not shown). Here, we have investigated molecular targets involved in cell lethality caused by the Brazilian green propolis. We were not able to see any cell death effect when S. cerevisiae was exposed to high concentrations of four of the major compounds that are present in the green propolis. Thus, we decided to verify the effects of propolis on S. cerevisiae cell lethality by using alcoholic propolis extracts. Cell death in S. cerevisiae is frequently followed by indicative apoptotic and/or necrotic markers such as externalization of phosphatidylserine to the outer leaflet of the plasma membrane, chromatin condensation, the generation of reactive oxygen species, propidium iodide accumulation, HMGB1/Nhp6p localization, detection of plasma membrane ruptures, and complete disintegration of subcellular structures (14, 19). In addition, ROS accumulation, mitochondrial fragmentation, cytochrome c release, the metacaspase Yca1p, the apoptosis-inducing factor Aif1p, the endonuclease G Nuc1p, the serine protease OMI, cytoskeleton perturbations, and chromatin epigenetic modification have also been observed (79). Necrosis has also been described during yeast chronological aging (20).
We have noticed that apoptosis and necrosis markers are induced by propolis, but increased time exposure to propolis intensifies the number of cells with necrotic markers, such as PI and nucleo-cytosolic translocation of the Nhp6Ap-GFP. Our data indicate a dual role for propolis treatment as an agent that induces apoptosis and secondary necrosis. These effects are partially mediated by YCA1 metacaspase since the YCA1Δ mutant is more resistant to propolis. In S. cerevisiae overexpression of YCA1 caused cell death while YCA1 deletion protected against cell death caused by reactive oxygen species or chronological aging (49). Additionally, we have also observed that Nuc1p does not translocate to the nucleus upon propolis cell death induction. Interestingly, Büttner et al. (13) observed that apoptotic death mediated by Nuc1p does not require YCA1. More interestingly, simultaneous markers for apoptosis and necrosis have also been noticed during propolis cell death induction in Trypanosoma cruzi (55).
There are several conditions where mitochondria-produced ROS have been associated with yeast apoptosis (for reviews, see references 19, 59, and 62). Propolis can induce ROS formation, and it is more lethal when S. cerevisiae grows in the presence of glycerol and ethanol as carbon sources (data not shown), suggesting that respiration increases propolis lethality. Actually, we were able to demonstrate that propolis can inhibit respiration in S. cerevisiae. We have observed that propolis cell death induction stimulates the cytochrome c release and that also [rho0] cells are more tolerant to propolis. Cytochrome c is a mitochondrial protein with a function in the respiratory chain and an additional function as an activator of caspase-9 in the intrinsic pathway of mammalian apoptosis (40). Treatment of yeast cells with acetic acid leads to mitochondrial cytochrome c release (45). Disruption of cytochrome c partially prevents acetic acid-induced cell death, and accordingly [rho0] cells show resistance against acetic acid-induced cell death (45). However, it is not very clear, as it is in mammals, if cytochrome c release can lead to the formation of an apoptosome-like structure and activation of caspases. In contrast, Sripriya et al. (77) have observed loss of mitochondrial membrane potential and absence of cytochrome c release during necrotic cell death of S. cerevisiae induced by expression of a proteinaceous elicitor hairpin (Pss) from Pseudomonas syringae. However, the deficiency of functional mitochondrial DNA, and consequent inability to respire from petite mutants, conferred resistance to death.
We have applied the power of genomics by screening the full collection of haploid S. cerevisiae deletion strains to comprehend how propolis, a complex phytotherapeutic compound, affects cell metabolism. This strategy has already been extensively used for understanding how other compounds affect S. cerevisiae cell metabolism (8, 15, 18, 27, 71, 86). By applying systems biology, we observed that most of the proteins whose deletion increases the sensitivity of yeast strains to propolis are involved in cell division mechanisms, the mitochondrial electron transport chain, vacuolar acidification, regulation of macroautophagy associated with protein targeting to vacuoles, cellular response to starvation, and negative regulation of transcription from the RNA polymerase II promoter. We have investigated and validated in more detail the mitochondrial electron transport chain and vacuolar acidification pathways. We have shown that propolis induces vacuolar acidification and translocation of Atg8p to the vacuoles, one of the hallmarks of autophagy. In S. cerevisiae, the vacuole is very important for maintaining cellular homeostasis comprising the regulation of intracellular pH and degradation mainly during nutrient limitation of proteins and organelles by autophagy (for reviews, see references 2, 29, and 46). Cell death induced by acetic acid is increased in S. cerevisiae VPS gene deletion mutants (VPS genes are involved in homotypic vacuole fusion and vacuolar protein sorting and are essential for normal vacuolar function) (70). It has been observed that the intracellular pH was acidified in VPS mutant cells upon treatment with acetic acid (70). It is possible that the disturbance of the homeostatic pH control may trigger necrosis by release of pronecrotic proteases, which would find an optimal pH for their enzymatic activity in the acidified cytosol (19).
Our work emphasizes the importance of S. cerevisiae as a model system to understand at a molecular level the mechanism of action of a phytotherapeutic compound. There is a multitude of studies describing the most diverse effects of propolis on different biological systems, ranging from cancer chemoprevention (78) to cariostatic activity on streptococci (42) to an influence on immunomodulatory action and lymphocytes and antibody production (73). Our study is the first one that investigates systematically by using functional genomics how propolis influences the metabolism of an organism. This opens new possibilities for understanding and validating propolis as an alternative therapeutic agent against fungal infections.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil.
We thank R. J. Devenish for providing us with the yeast vectors containing Rosella, the four anonymous reviewers for their suggestions, and Rogelio Lopes Brandão for helping us with the yeast deletion library. We also thank D. J. Klionsky for providing the ATG8::GFP construct.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 30 December 2010.
REFERENCES
- 1. Aguero M. B., et al. 2010. Argentinean propolis from Zuccagnia punctata Cav. (Caesalpinieae) exudates: phytochemical characterization and antifungal activity. J. Agric. Food Chem. 58:194–201 [DOI] [PubMed] [Google Scholar]
- 2. Ariño J., Ramos J., Sychrová H. 2010. Alkali metal cation transport and homeostasis in yeasts. Microbiol. Mol. Biol. Rev. 74:95–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bader G. D., Hogue C. W. 2003. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Basnet P., Matsuno T., Neidlein R. Z. 1997. Potent free radical scavenging activity of propol isolated from Brazilian propolis. Z. Naturforsch. C 52:828–833 [DOI] [PubMed] [Google Scholar]
- 5. Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289–300 [Google Scholar]
- 6. Bevis B. J., Glick B. S. 2002. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 20:83–87 [DOI] [PubMed] [Google Scholar]
- 7. Bharucha N., Kumar A. 2007. Yeast genomics and drug target identification. Comb. Chem. High Throughput Screen. 10:618–634 [DOI] [PubMed] [Google Scholar]
- 8. Blackburn A. S., Avery S. V. 2003. Genome-wide screening of Saccharomyces cerevisiae to identify genes required for antibiotic insusceptibility of eukaryotes. Antimicrob. Agents Chemother. 47:676–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boone C., Bussey H., Andrews B. J. 2007. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 8:437–449 [DOI] [PubMed] [Google Scholar]
- 10. Brachmann C. B., et al. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132 [DOI] [PubMed] [Google Scholar]
- 11. Breger J., et al. 2007. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 3:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burhans W., Weinberger M. 2007. Yeast endonuclease G: complex matters of death, and of life. Mol. Cell 25:323–325 [DOI] [PubMed] [Google Scholar]
- 13. Büttner S., et al. 2007. Endonuclease G regulates budding yeast life and death. Mol. Cell 25:233–246 [DOI] [PubMed] [Google Scholar]
- 14. Carmona-Gutierrez D., et al. 2010. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 17:763–773 [DOI] [PubMed] [Google Scholar]
- 15. Chan T-F., Carvalho J., Riles L., Zheng X. F. S. 2000. A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR). Proc. Natl. Acad. Sci. U. S. A. 97:13227–13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Costanzo M., et al. 2010. The genetic landscape of a cell. Science 327:425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dalben-Dota K. F., et al. 2010. Antifungal activity of propolis extract against yeasts isolated from vaginal exudates. J. Altern. Complement. Med. 16:285–290 [DOI] [PubMed] [Google Scholar]
- 18. Desmoucelles C., Pinson B., Saint-Marc C., Daignan-Fornier B. 2002. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J. Biol. Chem. 277:27036–27044 [DOI] [PubMed] [Google Scholar]
- 19. Eisenberg T., Carmona-Gutierrez D., Büttner S., Tavernarakis N., Madeo F. 2010. Necrosis in yeast. Apoptosis 15:257–268 [DOI] [PubMed] [Google Scholar]
- 20. Eisenberg T., et al. 2009. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 11:1305–1314 [DOI] [PubMed] [Google Scholar]
- 21. Elmore S. 2007. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35:45–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ghisalberti E. L. 1979. Propolis: a review. Bee World 60:59–84 [Google Scholar]
- 23. Glick B., Schatz G. 1991. Import of proteins into mitochondria. Annu. Rev. Genet. 25:21–44 [DOI] [PubMed] [Google Scholar]
- 24. Greenaway W., Scaysbrook T., Whatley F. R. 1990. The composition and plant origins of propolis. Bee World 71:107–118 [Google Scholar]
- 25. Gullo A. 2009. Invasive fungal infections: the challenge continues. Drugs 69(Suppl. 1):65–73 [DOI] [PubMed] [Google Scholar]
- 26. Gulshan K., Moye-Rowley W. C. 2007. Multidrug resistance in fungi. Eukaryot. Cell 6:1933–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hillenmeyer M. E., et al. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320:362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hoon S., St. Onge R. P., Giaever G., Nislow C. 2008. Yeast chemical genomics and drug discovery: an update. Trends Pharmacol. Sci. 29:499–504 [DOI] [PubMed] [Google Scholar]
- 29. Inoue Y., Klionsky D. J. 2010. Regulation of macroautophagy in Saccharomyces cerevisiae. Semin. Cell Dev. Biol. 21:664–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ito J., et al. 2001. Anti-AIDS agents. 48.(1) Anti-HIV activity of moronic acid derivatives and the new melliforone-related triterpenoid isolated from Brazilian propolis. J. Nat. Prod. 64:1278–1281 [DOI] [PubMed] [Google Scholar]
- 31. Kanafani Z., Perfect J. 2008. Antimicrobial resistance: resistance to antifungal agents: mechanisms and clinical impact. Clin. Infect. Dis. 46:120–128 [DOI] [PubMed] [Google Scholar]
- 32. Kane P. 2006. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 70:177–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kanki T., Klionsky D. J. 2010. The molecular mechanism of mitochondria autophagy in yeast. Mol. Microbiol. 75:795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kimoto T., et al. 1998. Apoptosis and suppression of tumor growth by artepillin C extracted from Brazilian propolis. Cancer Detect. Prev. 22:506–515 [DOI] [PubMed] [Google Scholar]
- 35. Klionsky D. J., Cuervo A. M., Seglen P. O. 2007. Methods for monitoring autophagy from yeast to human. Autophagy 3:181–206 [DOI] [PubMed] [Google Scholar]
- 36. Klionsky D. J., et al. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4:151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kontoyiannis D. P., Lewis R. E. 2002. Antifungal drug resistance of pathogenic fungi. Lancet 359:1135–1144 [DOI] [PubMed] [Google Scholar]
- 38. Korua O., et al. 2007. Oral and dental bacteriology and infection. In vitro antimicrobial activity of propolis samples from different geographical origins against certain oral pathogens. Anaerobe 13:140–145 [DOI] [PubMed] [Google Scholar]
- 39. Kourtis N., Tavernarakis N. 2009. Autophagy and cell death in model organisms. Cell Death Differ. 16:21–30 [DOI] [PubMed] [Google Scholar]
- 40. Kushnareva Y., Newmeyer D. D. 2010. Bioenergetics and cell death. Ann. N. Y. Acad. Sci. 1201:50–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li J., Yuan J. 2008. Caspases in apoptosis and beyond. Oncogene 27:6194–6206 [DOI] [PubMed] [Google Scholar]
- 42. Libério S. A., et al. 2009. The potential use of propolis as a cariostatic agent and its actions on mutans group streptococci. J. Ethnopharmacol. 125:1–9 [DOI] [PubMed] [Google Scholar]
- 43. Lowry O. H., Rosebrough N. J., Lewis Farr A., Randall R. J. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 44. Ludovico P., Sousa M. J., Silva M. T., Leão C., Corte-Real M. 2001. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology 147:2409–2415 [DOI] [PubMed] [Google Scholar]
- 45. Ludovico P., et al. 2002. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2598–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lynch-Day M. A., Klionsky D. J. 2010. The Cvt pathway as a model for selective autophagy. FEBS Lett. 584:1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Madeo F., Fröhlich E., Fröhlich K.-U. 1997. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 139:729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Madeo F., et al. 1999. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 145:757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Madeo F., et al. 2002. A caspase-related protease regulates apoptosis in yeast. Mol. Cell 9:911–917 [DOI] [PubMed] [Google Scholar]
- 50. Maere S., Heymans K., Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21:3448–3449 [DOI] [PubMed] [Google Scholar]
- 51. Maiuri M. C., Zalckvar E., Kimchi A., Kroemer G. 2007. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 8:741–752 [DOI] [PubMed] [Google Scholar]
- 52. Manavathu E. K., Culright J. L., Chandrasekar P. H. 1998. Organism-dependent fungicidal activities of propolis. Antimicrob. Agents Chemother. 42:3018–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Matsuno T., Matsumoto Y., Saito N., Morikawa J. 1997. Isolation and characterization of cytotoxic diterpenoid isomers from propolis. Z. Naturforsch. C 52:702–704 [DOI] [PubMed] [Google Scholar]
- 54. Mazzoni C., Falcone C. 2008. Caspase-dependent apoptosis in yeast. Biochim. Biophys. Acta 1783:1320–1327 [DOI] [PubMed] [Google Scholar]
- 55. Menna-Barreto R. F. S., et al. 2009. Different cell death pathways induced by drugs in Trypanosoma cruzi: an ultrastructural study. Micron 40:157–168 [DOI] [PubMed] [Google Scholar]
- 56. Miesenböck G., De Angelis D. A., Rothman J. E. 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394:192–195 [DOI] [PubMed] [Google Scholar]
- 57. Morschhäuser J. 2010. Regulation of multidrug resistance in pathogenic fungi. Fungal Genet. Biol. 47:94–106 [DOI] [PubMed] [Google Scholar]
- 58. Osiewacz H. D., Scheckhuber C. Q. 2006. Impact of ROS on ageing of two fungal model systems: Saccharomyces cerevisiae and Podospora anserine. Free Radic. Res. 40:1350–1358 [DOI] [PubMed] [Google Scholar]
- 59. Ota C., Unterkircher C., Fantinato V., Shimizu M. T. 2001. Antifungal activity of propolis on different species of Candida. Mycoses 44:375–378 [DOI] [PubMed] [Google Scholar]
- 60. Park Y. K., et al. 2005. Suppressive effects of ethanolic extracts from propolis and its main botanical origin on dioxin toxicity. J. Agric. Food Chem. 53:10306–10309 [DOI] [PubMed] [Google Scholar]
- 61. Parrish J., et al. 2001. Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature 412:90–94 [DOI] [PubMed] [Google Scholar]
- 62. Perrone G. G., Tan S. X., Dawes I. W. 2008. Reactive oxygen species and yeast apoptosis. Biochim. Biophys. Acta 1783:1354–1368 [DOI] [PubMed] [Google Scholar]
- 63. Ramsdale M. 2008. Programmed cell death in pathogenic fungi. Biochim. Biophys. Acta 1783:1369–1380 [DOI] [PubMed] [Google Scholar]
- 64. Reers M., et al. 1995. Mitochondrial membrane potential monitored by JC-1 dye. Methods Enzymol. 260:406–417 [DOI] [PubMed] [Google Scholar]
- 65. Ribeiro G. F., Côrte-Real M., Johansson B. 2006. Characterization of DNA damage in yeast apoptosis induced by hydrogen peroxide, acetic acid, and hyperosmotic shock. Mol. Biol. Cell 17:4584–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rivals I., Personnaz L., Taing L., Potier M. C. 2007. Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23:401–407 [DOI] [PubMed] [Google Scholar]
- 67. Rosado C. J., Mijaljica D., Hatzinisiriou I., Prescott M., Devenish R. J. 2008. Rosella: a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy 4:205–213 [DOI] [PubMed] [Google Scholar]
- 68. Salomão K., et al. 2008. Brazilian propolis: correlation between chemical composition and antimicrobial activity. Evid. Based Complement. Alternat. Med. 5:317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Scaffidi P., Misteli T., Bianchi M. E. 2002. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418:191–195 [DOI] [PubMed] [Google Scholar]
- 70. Schauer A., et al. 2009. Vacuolar functions determine the mode of cell death. Biochim. Biophys. Acta 1793:540–545 [DOI] [PubMed] [Google Scholar]
- 71. Scherens B., Goffeau A. 2004. The uses of genome-wide yeast mutant collections. Genome Biol. 5:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Semighini C. P., Marins M., Goldman M. H. S., Goldman G. H. 2002. Quantitative analysis of the relative transcript levels of ABC transporter Atr genes in Aspergillus nidulans by real-time reverse transcription-PCR assay. Appl. Environ. Microbiol. 68:1351–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sforcin J. M. 2007. Propolis and the immune system: a review. J. Ethnopharmacol. 113:1–14 [DOI] [PubMed] [Google Scholar]
- 74. Shannon P., et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shimazawa M., et al. 2005. Neuroprotection by Brazilian green propolis against in vitro and in vivo ischemic neuronal damage. Evid. Based Complement. Alternat. Med. 2:201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Siqueira A. B. S., et al. 2009. Trichophyton species susceptibility to green and red propolis from Brazil. Lett. Appl. Microbiol. 48:90–96 [DOI] [PubMed] [Google Scholar]
- 77. Sripriya P., Vedantam L. V., Podile A. R. 2009. Involvement of mitochondria and metacaspase elevation in hairpin Pss-induced cell death of Saccharomyces cerevisiae. J. Cell Biochem. 107:1150–1159 [DOI] [PubMed] [Google Scholar]
- 78. Szliszka E., Krol W. 2011. The role of dietary polyphenols in tumor necrosis factor-related apoptosis inducing ligand (TRAIL)-induced apoptosis for cancer chemoprevention. Eur. J. Cancer Prev. 20:63–69 [DOI] [PubMed] [Google Scholar]
- 79. Teixeira E. W., Negri G., Meira R. M. S. A., Message D., Salatino A. 2005. Plant origin of green propolis: bee behavior, plant anatomy and chemistry. Evid. Based Complement. Alternat. Med. 2:85–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tzagoloff A., Akai A., Needleman R. B., Zulch G. 1975. Assembly of the mitochondrial membrane system. Cytoplasmic mutants of Saccharomyces cerevisiae with lesions in enzymes of the respiratory chain and in the mitochondrial ATPase. J. Biol. Chem. 250:8236–8242 [PubMed] [Google Scholar]
- 81. Uren A. G., et al. 2000. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 6:961–967 [DOI] [PubMed] [Google Scholar]
- 82. Viuda-Martos M., Ruiz-Navajas Y., Fernandez-López J., Pérez-Álvarez J. A. 2008. Functional properties of honey, propolis, and royal jelly. J. Food Sci. 73:R117–R124 [DOI] [PubMed] [Google Scholar]
- 83. Vynograd N., Vynograd I., Sosnowski Z. 2000. A comparative multicenter study of the efficacy of propolis, acyclovir and placebo in the treatment of genital herpes (HSV). Phytomedicine 7:1–6 [DOI] [PubMed] [Google Scholar]
- 84. Winzeler E. A., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906 [DOI] [PubMed] [Google Scholar]
- 85. Wuster A., Madan Babu M. 2008. Chemogenomics and biotechnology. Trends Biotechnol. 26:252–258 [DOI] [PubMed] [Google Scholar]
- 86. Zewail A., et al. 2003. Novel functions of the phosphatidylinositol metabolic pathway discovered by a chemical genomics screen with wortmannin. Proc. Natl. Acad. Sci. U. S. A. 100:3345–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.