Abstract
Eukaryotic phosphoinositide-specific phospholipases C (PI-PLC) specifically hydrolyze phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2], produce the Ca2+-mobilizing agent inositol 1,4,5-trisphosphate, and regulate signaling in multicellular organisms. Bacterial PtdIns-specific PLCs, also present in trypanosomes, hydrolyze PtdIns and glycosyl-PtdIns, and they are considered important virulence factors. All unicellular eukaryotes studied so far contain a single PI-PLC-like gene. In this report, we show that ciliates are an exception, since we provide evidence that Tetrahymena species contain two sets of functional genes coding for both bacterial and eukaryotic PLCs. Biochemical characterization revealed two PLC activities that differ in their phosphoinositide substrate utilization, subcellular localization, secretion to extracellular space, and sensitivity to Ca2+. One of these activities was identified as a typical membrane-associated PI-PLC activated by low-micromolar Ca2+, modestly activated by GTPγS in vitro, and inhibited by the compound U73122 [1-(6-{[17β-3-methoxyestra-1,3,5(10)-trien-17-yl]amino}hexyl)-1H-pyrrole-2,5-dione]. Importantly, inhibition of PI-PLC in vivo resulted in rapid upregulation of PtdIns(4,5)P2 levels, suggesting its functional importance in regulating phosphoinositide turnover in Tetrahymena. By in silico and molecular analysis, we identified two PLC genes that exhibit significant similarity to bacterial but not trypanosomal PLC genes and three eukaryotic PI-PLC genes, one of which is a novel inactive PLC similar to proteins identified only in metazoa. Comparative studies of expression patterns and PI-PLC activities in three T. thermophila strains showed a correlation between expression levels and activity, suggesting that the three eukaryotic PI-PLC genes are functionally nonredundant. Our findings imply the presence of a conserved and elaborate PI-PLC-Ins(1,4,5)P3-Ca2+ regulatory axis in ciliates.
INTRODUCTION
Phosphoinositides (PIs), i.e., phosphatidylinositol (PtdIns) and its phosphorylated derivatives, are ubiquitous constituents of eukaryotic membranes, with many and diverse functions in receptor signaling, membrane trafficking, and cytoskeleton organization. Among PI-metabolizing enzymes, phospholipases C (PLCs) that hydrolyze PtdIns and phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] are of particular importance because they generate second messengers such as inositol 1,4,5-trisphosphate [Ins(1,4,5)P3] and participate in the replenishment of the inositol pool available for phosphoinositide synthesis. These phospholipases fall into distinct families: (i) the well-established family of eukaryotic phosphoinositide-specific PLCs (PI-PLCs) (EC 3.1.4.11) (24, 39); (ii) the bacterial PtdIns-specific PLC (EC 4.6.1.13) family, members of which are secreted enzymes and are considered important virulence factors in pathogenic bacteria (15); and (iii) the trypanosomal glycosylPtdIns-PLCs (GPI-PLCs) (EC 4.6.1.14), members of which form a separate subfamily of bacterial PLCs (6). Eukaryotic and bacterial-trypanosomal PLCs share some homology in the N-terminal part of the catalytic PLC fold, i.e., the PLCXc domain, but they differ in regions that dictate Ca2+ sensitivity and substrate specificity. As such, PI-PLCs exhibit an exquisite dependence on μM Ca2+ levels for PtdIns(4,5)P2 hydrolysis and mM Ca2+ levels for PtdIns hydrolysis (39), while PtdIns-PLCs and GPI-PLCs are largely insensitive to mM Ca2+ levels and do not hydrolyze PtdIns(4,5)P2 (15, 17). PLC genes resembling those carried by bacteria have not been identified so far in eukaryotes, but several mammalian Ca2+-insensitive or Ca2+-inhibited PtdIns-PLC activities have long been known and have previously been rigorously characterized biochemically as cytosolic or membrane-lysosomal enzymes together with phospholipase A and lysophospholipase (PLA/lysoPLA) activities (22).
Eukaryotic PI-PLCs are considered ubiquitous enzymes, since most sequenced eukaryotic genomes contain PI-PLC-like gene(s) (34). Importantly, all unicellular organisms studied, including fungi (Saccharomyces cerevisiae), slime molds (Dictyostelium discoideum), kinetoplastids (Trypanosoma cruzi), and apicomplexa (Toxoplasma gondii), have a single PI-PLC gene (10, 12, 13, 34, 36), while Arabidopsis thaliana has at least nine PI-PLC genes (4, 43). In animals, six catalytically functional PI-PLC isoforms have been described, namely, δ, β, γ, ε, η, and ζ (24). PI-PLCs generate Ins(1,4,5)P3 and diacylglycerol (DAG) from PtdIns(4,5)P2 upon agonist-induced receptor activation. Ins(1,4,5)P3 and DAG serve as powerful second messengers by initiating Ca2+ mobilization and protein phosphorylation, respectively; the activation of this signaling pathway dictates cellular fate by regulating functions such as cellular activation, proliferation, differentiation, and apoptosis (39). Animal PI-PLC isoforms differ in their modes of activation; PI-PLCβ isoforms are coupled primarily to G-protein-coupled receptors (GPCRs) and PI-PLCγ isoforms are coupled to receptor tyrosine kinases (RTKs), whereas PI-PLCε isoforms are coupled to Ras GTPases. The acute signaling role, if any, of the rest of the isoforms is less well understood (24). Nonetheless, PI-PLCδ and -ζ exhibit a striking sensitivity to Ca2+, and they are widely considered Ca2+-activated enzymes, particularly PI-PLCζ, which is a sperm-specific isoform and participates in the Ca2+ waves during fertilization (35). Interestingly, members of a seventh group of PI-PLCs (the PLC-L [PLC-like] or PRIP [PLC-related inactive protein] group) share extended homology with PI-PLCδ but are inactive PLCs due to the lack of at least one catalytically active His residue (23, 24). PRIPs are conserved in Caenorhabditis elegans and Drosophila melanogaster, and extensive studies of mouse PRIP-1 have shown pleiotropic functions in Ins(1,4,5)P3-mediated Ca2+ signaling and neurotransmitter receptor functions (23). Importantly, all unicellular and plant PI-PLCs are closely related to PI-PLCδ and ζ isoforms, with no other isoform or PRIP-like PLC protein identified in nonanimal species so far (4, 24, 34).
In contrast to findings from studies of other unicellular organisms, specific evidence for PI-PLC-like genes and/or activity in ciliates is still missing. In fact, it has been argued that Paramecium and Tetrahymena species may not contain stringent homologs of eukaryotic PI-PLC genes (38). Nevertheless, a recent study of Paramecium PLC genes (25) suggested the presence of several PI-PLCs, but their relationship to PtdIns or PtdIns(4,5)P2-PLC activities and their involvement in signaling are unknown. In this report, we provide biochemical evidence for at least two distinct PLC activities that utilize PtdIns and PtdIns(4,5)P2 with different levels of Ca2+ sensitivity and localization that correspond to bacterial PtdIns-PLC and eukaryotic PI-PLC, respectively. Importantly, we have identified two bacterium-like PtdIns-PLC and three eukaryote-like PI-PLC genes in Tetrahymena, including a novel, Tetrahymena-specific, PRIP/PLC-L homolog. Furthermore, using pharmacological manipulation of Tetrahymena PI-PLC activity in vivo and comparative studies of expression patterns and catalytic activity of Tetrahymena PI-PLCs, we propose that Tetrahymena PI-PLCs have multiple nonredundant roles.
MATERIALS AND METHODS
Strains and culture conditions.
Tetrahymena pyriformis W and Tetrahymena thermophila CU438.1, obtained from P. Burns (Cornell University, Ithaca, NY), and MS-1 (lysosomal enzyme secretion mutant) (21) and SB281 (mucocyst secretion mutant) (5), both obtained from A. Tiedtke (University of Münster, Germany), were cultured at 25°C under conditions of constant shaking (75 to 100 rpm) in an enriched proteose peptone medium consisting of 2.0% (wt/vol) proteose peptone (Merck, Darmstadt, Germany), 0.2% (wt/vol) yeast extract (Serva, Heidelberg, Germany), and 0.5% (wt/vol) glucose (Panreac, Barcelona, Spain). Media were supplemented with 1% (vol/vol) ferrous sulfate-chelate solution (Sigma, Steinheim, Germany, or St. Louis, MO). Routinely, working cultures (10 to 25 ml) of late-logarithmic-phase cells (48 to 72 h) were used. Final cell densities ranged between 0.6 and 2 × 106 cells/ml for all strains, and cells were counted using a Neubauer hemocytometer.
Radioactive substrates.
[3H]PtdIns(4,5)P2 (purchased from ARC, St. Louis, MO) (specific activity, 6.5 Ci/mmol) was used as a substrate in the phospholipase C assay after purification by thin-layer chromatography (TLC) using a chloroform-methanol-ammonium hydroxide-water (86:76:6:16 [by vol]) solvent system and oxalate-impregnated silica gel H plates (Merck) (30). Unlabeled PtdIns(4,5)P2 was isolated from a phosphoinositide mixture (Sigma) by the same procedure. [3H]PtdIns (specific activity, 3,200 to 4,500 cpm/nmol) was isolated from D-myo[2-3H]inositol-labeled Tetrahymena pyriformis cultures as previously described (30). For [3H]lysoPtdIns preparation, the isolated [3H]PtdIns was subjected to hydrolysis by the use of phospholipase A2 from Naja naja (Sigma). Briefly, dried [3H]PtdIns (80 nmol) was dispersed in 1 ml of CaCl2 (3.3 mM)–Tris (33 mM) (pH 6.8) in a bath sonicator for 15 min, and the reaction was started by addition of 1.8 ml of CaCl2 (3.3 mM)–Tris (33 mM) (pH 6.8)–0.2 ml of PLA2 solution (0.1 mg/ml in Tris [10 mM] [pH 7.4])–0.4 ml of diethyl ether). The mixture was incubated for 100 min at 37°C under conditions of vigorous shaking. The [3H]lyso-sn2-PtdIns produced was isolated by phase separation in a solvent system consisting of chloroform-methanol-water-isobutanol (2:1:4:1 [by vol]) and separated by TLC using silica gel G plates and an acidic solvent system, chloroform-methanol-acetic acid-water (75:45:12:6 [by vol]). Lipids were detected by exposure to iodine vapor, and the area corresponding to standard lysoPtdIns was scraped off and extracted with chloroform-methanol-water-isobutanol mixtures as described above. The specific activity of the isolated [3H]lysoPtdIns was 3,240 cpm/nmol. All radioactive lipid compounds were assayed for radioactivity by liquid scintillation counting using 5 to 10 ml of a toluene-based scintillation fluid after resuspension in 0.5 ml of methanol.
Subcellular fractionation.
Cells were harvested by centrifugation at 1,000 × g for 10 min, washed, resuspended in Tris (10 mM)–sucrose (0.2 M)–NaF (50 mM) (pH 7.4)–phenylmethylsulfonyl fluoride (PMSF) (0.5 mM)–leupeptin (5 μg/ml) and homogenized by probe sonication using a Vibra Cell Sonics and Materials device set at 40 Hz amplitude with 6 30-s bursts with 30-s intervals on ice. Homogenates were centrifuged at 1,000 × g for 10 min to remove unbroken cells and nuclei, and the supernatant was centrifuged at 25,000 × g for 20 min. The 25,000 × g pellet was resuspended in buffer and recentrifuged, and the final pellet was designated the heavy-membrane (HM) fraction. The 25,000 × g supernatant was further separated into a soluble fraction (cytosol) and a light-membrane (LM) fraction by ultracentrifugation at 100,000 × g for 1 h. The protein content of all fractions was measured according to the Lowry method, using bovine serum albumin as the standard.
Phospholipase assays.
The PLC activity of Tetrahymena cell supernatants, homogenates, and subcellular fractions was assayed at 37°C according to the method of Melin et al. (32). The standard reaction mixture consisted of Tris (50 mM) (pH 6.5), sodium deoxycholate (0.01%), 10 μl of radioactive substrate solution (final concentration of phosphoinositide, 30 to 60 μM), 8 to 16 μg of protein, and 5 μl of the appropriate EGTA, BAPTA [1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid], or Ca2+ concentrated solution in a total volume of 50 μl. Dried [3H]PtdIns(4,5)P2 (4,600 cpm/nmol), PtdIns (3,200 to 4,500 cpm/nmol), and lysoPtdIns (3,240 cpm/nmol) were solubilized by sonication in Tris 50 mM (pH 6.5) and sodium deoxycholate (0.05%) and used within 2 h. Ca2+/EGTA buffers covering a range of 0.1 to 100 μM free Ca2+ were prepared as described previously (20). Reactions were started by the addition of substrate solution and terminated after 15 min by the addition of 1 ml of ice-cold chloroform/methanol (2:1 [vol/vol]) and 0.25 ml of 1 N HCl. Linearity was observed for up to 20 μg of protein and up to 25 min of reaction time (data not shown). After phase separation by centrifugation at 3,000 × g for 3 min, aliquots of the aqueous phase were counted for radioactivity representing the total hydrolyzing activity. In all experiments, control reactions were performed in the absence of protein or with the addition of heat-inactivated protein preparation; the water-soluble radioactivity of these reaction mixtures (routinely, 1 to 2% of lipid substrate radioactivity) was used to correct all activity data. U73122 [1-(6-{[17β-3-methoxyestra-1,3,5(10)-trien-17-yl]amino}hexyl)-1H-pyrrole-2,5-dione] and GTPγS, where indicated, were added to the reaction mixtures from solutions concentrated 25-fold and were preincubated for 3 min at 37°C before the addition of radioactive substrate; equivalent amounts of vehicle (ethanol and water, respectively) were used for control reactions.
Analysis of inositol phosphates and lysoPtdIns.
Aqueous products of the PI-PLC assay were separated by anion-exchange chromatography using Dowex AG 1-x8 columns, formate form (Bio-Rad, Hercules, CA). The aqueous phases of the assays were diluted with water (1:8 [by vol]), neutralized with dilute NH4OH, and loaded onto the columns, and the products were eluted in batches with increasing concentrations of ammonium formate in formic acid. Glycerophosphoinositol was eluted with ammonium formate (60 mM)–sodium borate (5 mM), and inositol monophosphate, inositol bisphosphate, and inositol trisphosphate were eluted with 10 to 12 ml of ammonium formate (0.2 M)–formic acid (0.1 M), 12 to 14 ml of ammonium formate (0.4 M)–formic acid (0.1 M), and 10 to 12 ml of ammonium formate (1 M)–formic acid (0.1 M), respectively. For all samples subjected to chromatography, 2-ml fractions were collected and the radioactivity was assayed by liquid scintillation counting using a dioxane-naphthalene-water scintillation fluid. Standard [3H]Ins(1,4,5)P3 (Amersham, Buckinghamshire, England) and [3H]glycerophosphoinositol, derived from alkaline hydrolysis of [3H]PtdIns, were routinely used for the calibration of the columns. For quantification of [3H]lysoPtdIns, the chloroform phase was recovered after termination of the reactions and subjected to chromatography by TLC in a chloroform-methanol-acetic acid-water (75:45:12:6 [by vol]) solvent system. The areas corresponding to lysoPtdIns and PtdIns were scraped off the plate and counted to determine radioactivity levels.
U73122 treatment and analysis of phosphoinositides in vivo.
T. thermophila cells were labeled in vivo for 24 h under conditions of starvation in Tris (10 mM) (pH 7.4) with D-myo[2-3H]inositol (ICN MP Biomedicals, Irvine, CA) at a concentration of 1 μCi/ml. Aliquots of the cell suspension were treated with U73122 in the presence or absence of insulin (2.5 μM) for a total of 10 to 15 min. Incubations were terminated by the addition of ice-cold perchloric acid, and phosphoinositides were extracted and quantified by TLC or high-performance liquid chromatography (HPLC) as described previously (9, 29).
In silico analysis of Tetrahymena PLC genes.
The NCBI database and the Tetrahymena genome database (http://www.tigr.org/tdb/e2k1/ttg/) were queried in BLAST searches using the PLCXc domains of human PLCδ1 and Bacillus cereus PLC. Recovered Tetrahymena gene products were further investigated for identification of domains by analysis using the SMART (http://smart.embl-heidelberg.de/) and PFAM (http://pfam.sanger.ac.uk/) databases, and the domain structures were authenticated using the NCBI conserved domain search tool (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and sequence alignment with full-length human PLCδ1. Nuclear export and localization signals as well as signal peptide and putative transmembrane regions were analyzed by the use of Signal P (Signal Peptide cleavage sites) (http://www.cbs.dtu.dk/services/SignalP/), predictNLS (http://cubic.bioc.columbia.edu/predictNLS/), HMMTOP (http://www.enzim.hu/hmmtop/), and HMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/) software. The PLCXc domains of Tetrahymena PLC1 and BPLC1 (where “BPLC” represents “bacterium-like PLC”) were used in additional BLASTP searches of selected eukaryotic genomes available at NCBI to identify putative homologs. Accession numbers of all PLC genes analyzed are included in Table S1 and Table S2 in the supplemental material. All amino acid alignments were performed with ClustalW. For phylogenetic analyses, alignments of PLCXc domains were constructed and regions containing gaps were eliminated, resulting in a total of 135 positions in the final data set. In all cases, the bootstrap consensus tree inferred from 1,000 replicates was constructed using MEGA version 4.0 software (42).
Isolation of RNA and PCR.
Total RNA from Tetrahymena thermophila or Tetrahymena pyriformis cells was isolated from 8 × 106 cells from each of the strains grown at equivalent time points of their growth curves by the use of RNAwiz reagent (Ambion, Austin, TX), resuspended in RNase-free water, and treated with DNase I at a ratio of 1 U per 50 μl of isolated RNA. For cDNA, 0.5 μg of total RNA was reverse transcribed using 5 U of AMV reverse transcriptase (Finzyme, Espoo, Finland) (RNase-free; 20 U/μl), deoxynucleoside triphosphates (dNTPs), and 2.5 μM random hexameric primers (Promega, Madison, WI). For amplification of PLCs, cDNA was amplified using GoTaq DNA polymerase (Promega) (5 U/μl), in the presence of the following gene-specific primers: for PLC1 forward, 5′-AAAAGTTGTCCGCATCCAAA-3′; for PLC1 reverse, 5′-TCATTGCAAGCTCAGGACAT-3′; for PLC2 forward, 5′-GACGACGAGACTGACAGCAA-3′; for PLC2 reverse, 5′-TCTACTCTCGAACCCTTTGGA-3′; for PLC3a forward, 5′-GCAGCCAGGAAATAGCAAAG-3′; for PLC3a reverse, 5′-TTTGTGAGATAGCCATT-3′; PLC4a forward, 5′-TACCTGCAAGCCCTCAGATT-3′; for PLC4a reverse, 5′-AACTCTCTTCGCTGCCGTTA-3′; for ACT1 (actin-1) forward, 5′-TCGACTCTGGTGATGGTGTT-3′; for ACT1 reverse, 5′-CGACGTAGCAGAGCTTTTCC-3′; for ATU1 (α-tubulin-1) forward, 5′-CTGATGGTCAAATGCCCTCT-3′; and for ATU1 reverse, 5′-CCTCTGGCGAAGTTGTTAGC-3′ (additional primers for PLC3 and PLC4 are listed in Fig. S3C in the supplemental material). All primers were designed using primer 3 software (http://frodo.wi.mit.edu/primer3/). PCRs consisted of the following steps: 95°C for 105 s, 35 cycles of 95°C for 15 s, 56°C for 30 s, 72°C for 30 s, and 72°C for 7 min. Samples were analyzed by 1.5% agarose gel electrophoresis, and PCR products were visualized using a UV transilluminator following staining with ethidium bromide and compared to molecular size markers (100 to 3,000 bp; Fermentas, Vilnius, Lithuania). Band intensities were quantified by the use of ImageJ software (http://rsbweb.nih.gov/ij/) and were normalized with respect to ATU1 levels.
Nucleotide sequence accession numbers.
Sequences were deposited in GenBank under accession numbers HQ317216 and HQ317217.
RESULTS
Multiple Tetrahymena PLC activities can be distinguished by their phosphoinositide substrate utilization, localization, Ca2+ sensitivity, and secretion to extracellular space.
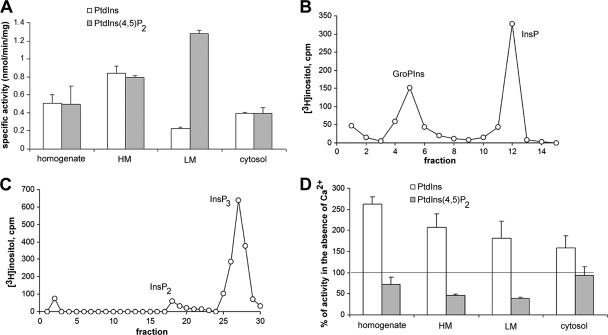
In previous experiments, we utilized myristate-rich, [3H]inositol-labeled PtdIns isolated from Tetrahymena pyriformis cultures as a suitable exogenous substrate for phospholipase assays and established that it is hydrolyzed into three products: lysoPtdIns, inositol monophosphate (InsP), and glycerophosphoinositol (GroPIns) (31). To study the potential relationship of these phospholipase activities to PI-PLC, T. pyriformis cells were homogenized by sonication and homogenates were fractionated by differential centrifugation into heavy-membrane (HM), light-membrane (LM), and cytosolic fractions. Phospholipase activity was assayed using [3H]PtdIns or [3H]PtdIns(4,5)P2 as a substrate in mixed deoxycholate micelles as described in Materials and Methods. As shown in Fig. 1A, hydrolysis of PtdIns and PtdIns(4,5)P2 was readily detected in all fractions. PtdIns-hydrolyzing activity at 1 mM Ca2+ was higher in the HM fraction and crude homogenate, while PtdIns(4,5)P2-hydrolyzing activity at 10 μM Ca2+ was higher in the LM and HM fractions. Analysis of water-soluble products showed the presence of various amounts of InsP and GroPIns when PtdIns was used as a substrate (Fig. 1B), whereas InsP3 was the major product formed by hydrolysis of PtdIns(4,5)P2 (Fig. 1C). These results suggested that PtdIns is utilized by both PLC and PLA/lysoPLA activities and that PtdIns(4,5)P2 is utilized by PLC activities. We next tested the Ca2+ sensitivities of both activities in all fractions. We found that PtdIns(4,5)P2-PLC activity was decreased in the absence of Ca2+ in the homogenate and HM and LM but not the cytosol fractions and that PtdIns-PLC plus PLA/lysoPLA activity was invariably increased in all fractions (Fig. 1D; note that in Fig. 1D, activity in the presence of EGTA is plotted as a percentage of the activity in the presence of Ca2+). Similar results were obtained with a second Ca2+ chelator, BAPTA (data not shown). These results suggested that, under these conditions, PtdIns and PtdIns(4,5)P2 are hydrolyzed by apparently different PLC activities with distinct localization and Ca2+ specificity characteristics.
Fig. 1.
Phosphoinositide-hydrolyzing activities of Tetrahymena. (A) [3H]PtdIns and [3H]PtdIns(4,5)P2-hydrolyzing activities were determined in homogenate and subcellular fractions (HM, heavy membrane; LM, light membrane) as described in Materials and Methods. For PtdIns, Ca2+ was adjusted to 1 mM, whereas for PtdIns(4,5)P2, free Ca2+ was adjusted to 10 μM. Data represent means ± standard deviations (SD) of the results from three independent experiments, each assayed in duplicate. (B and C) [3H]inositol-labeled water-soluble products of PtdIns- and PtdIns(4,5)P2-hydrolyzing activities of HM and LM fractions, respectively, were subjected to chromatography on Dowex AG-1x8 columns, and inositol phosphates were eluted with ammonium formate-formic acid solutions as described in Materials and Methods. InsP (fractions 3 to 8) and GroPIns (fractions 10 to 14) are the products of PtdIns hydrolysis, indicating PLC and PLA/lysoPLA activities, respectively, while InsP3 (fractions 25 to 30) is the major product of PtdIns(4,5)P2 hydrolysis, indicating a PLC activity. (D) PtdIns- and PtdIns(4,5)P2-hydrolyzing activities exhibit different sensitivities to Ca2+. [3H]PtdIns- and [3H]PtdIns(4,5)P2-hydrolyzing activities were determined as described for panel A but in the presence of EGTA (3 mM). Data represent means ± SD of the results from three independent experiments and are expressed as percentages of the specific activity determined in the presence of Ca2+ [1 mM for PtdIns and 10 μM for PtdIns(4,5)P2] represented by the horizontal line.
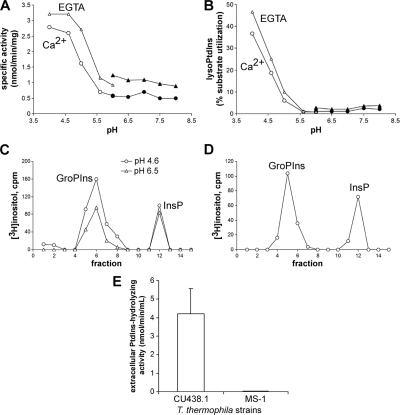
We further investigated the PtdIns-PLC activity by analyzing the pH dependency of homogenate and HM activities. As shown in Fig. 2A, for the homogenate fractions, PtdIns-hydrolyzing activity was maximal at pH 4 to 5 and was slightly increased by Ca2+ depletion at all pH values. Interestingly, parallel analysis of lysoPtdIns production indicated a robust PtdIns-PLA activity at acidic but not neutral pH values (Fig. 2B) that was accompanied by increased GroPIns production, presumably via a lysoPLA activity (data not shown). Since PtdIns-PLC may also have PLC activity against lysoPtdIns, we directly assessed this possibility by utilizing [3H]lyso-sn2-PtdIns produced by Naja naja PLA2 as a substrate (see Materials and Methods). As shown in Fig. 2C, lyso-sn2-PtdIns was hydrolyzed at both acidic and neutral pH to GroPIns and InsP in homogenates (in a Ca2+-insensitive manner; data not shown), indicating the presence of both lysoPLA1 and lysoPLC activities. Overall, we could not identify a Ca2+-activated PtdIns-PLC activity due to the predomination of Ca2+-insensitive or -inhibited PtdIns-PLC (and PLA/lysoPLA activities). This was not for technical reasons, since the same methodology (including radioactive substrates) was previously used in the characterization of Ca2+-sensitive human PLC activities (41). We reasoned that this Ca2+-insensitive or -inhibited PtdIns-PLC activity could represent a bacterium-like PLC activity in T. pyriformis. Since Tetrahymena cells constitutively secrete a variety of phospholipases (14), we tested whether such an activity can be detected in the extracellular medium. Indeed, PtdIns-hydrolyzing activity was detected in the medium of logarithmic-phase T. pyriformis cultures (12.7 ± 1.2 nmol/min/ml; n = 3) and both PtdIns-PLC and PLA/lysoPLA were present, as indicated by analysis of products (Fig. 2D). Similar results were also obtained for extracellular PtdIns-hydrolyzing activities of the T. thermophila CU438.1 strain (Fig. 2E; total activity, 4.2 ± 1.2 nmol/min/ml; n = 3). Importantly, all extracellular PtdIns-hydrolyzing activities were abolished in the T. thermophila MS-1 strain (21), which does not secrete lysosomal enzymes (Fig. 2E).
Fig. 2.
Characterization of intracellular and extracellular PtdIns-PLC and PLA activities in Tetrahymena. (A) pH dependency of PtdIns-hydrolyzing activities. Homogenate activity was assayed at the indicated pH values with acetate-based buffers (open symbols) and Tris-based buffers (closed symbols) in the presence of 3 mM EGTA (triangles) or 1 mM Ca2+ (circles). (B) pH dependency of PtdIns-PLA activity. [3H]LysoPtdIns was quantified after separation of lipids by TLC and scintillation counting as described in Materials and Methods. (C) LysoPLA and lysoPLC activities against lysoPtdIns. Homogenate activities against [3H]lysoPtdIns were assayed as described in Materials and Methods at the indicated pH values, and water-soluble products were separated as described for Fig. 1B. LysoPLA activity was substantially increased at acidic pH. (D) Extracellular PtdIns-hydrolyzing activities. Aliquots of cell supernatant after 48 h of growth were incubated with [3H]PtdIns as described for Fig. 1A, and water-soluble products were separated as described for Fig. 1B. Addition of 1 mM Ca2+ instead of EGTA did not significantly affect InsP or GroPIns production (not shown in the panel). (E) Absence of extracellular PtdIns-hydrolyzing activities in the T. thermophila MS-1 strain. Equal amounts from T. thermophila CU438.1 and MS-1 cell supernatants were assayed for PtdIns-hydrolyzing activities. Data represent the means ± SD of the results from three independent experiments, each assayed in duplicate.
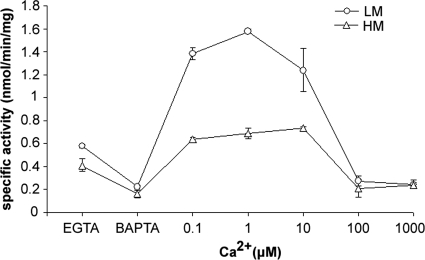
Despite the apparent lack of a readily detectable PI-PLC-like activity against PtdIns, characterization of the T. pyriformis PtdIns(4,5)P2-PLC activity in LM and HM fractions revealed an exquisite dependence on low-micromolar Ca2+. As shown in Fig. 3, depletion of endogenous Ca2+ by addition of either BAPTA or EGTA resulted in reduced PtdIns(4,5)P2-PLC activity in both fractions and addition of 0.1 to 10 μM free Ca2+ resulted in increased activity against PtdIns(4,5)P2. Notably, further increasing the Ca2+ concentration above 100 μM caused an abrupt decrease of activity. This type of Ca2+ sensitivity is typical of all of the mammalian PI-PLC isoforms that have been studied (39) and provided definite evidence for the presence of a eukaryote-like PI-PLC in T. pyriformis that utilizes PtdIns(4,5)P2. In contrast to the results seen with the bacterium-like secreted PtdIns-PLC, PtdIns(4,5)P2-PLC activity was not detected in the growth medium of T. pyriformis cells, indicating that it is strictly an intracellular activity (data not shown). Importantly, biochemical characterization of PLC activities of T. thermophila strain CU438.1 again suggested the presence of two PLC activities with different localizations, one of which is a membrane-associated, micromolar Ca2+-sensitive, eukaryote-like, PI-PLC activity hydrolyzing PtdIns(4,5)P2 (see Fig. S1 in the supplemental material).
Fig. 3.
PtdIns(4,5)P2-PLC is activated by low-micromolar Ca2+ concentrations. PtdIns(4,5)P2-PLC activity was assayed in HM and LM fractions in the presence of EGTA and BAPTA or in the presence of 0.1 to 1,000 μM free Ca2+ concentrations as indicated. Data represent the means ± SD of the results from three independent experiments, each assayed in duplicate.
Pharmacological manipulation of PI-PLC activity by U73122 and GTPγS suggests high turnover of PtdIns(4,5)P2 in vivo and regulation by GTPases.
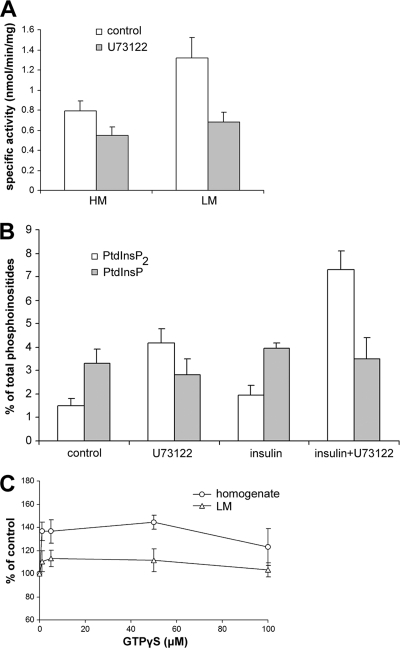
The compound U73122 [1-(6-{[17β-3-methoxyestra-1,3,5(10)-trien-17-yl]amino}hexyl)-1H-pyrrole-2,5-dione] is a general inhibitor of animal, plant, and protist PI-PLCs, and it has proven valuable in dissection of PI-PLC functions (11, 12, 18). Having established the basic biochemical properties of Tetrahymena PI-PLC, we used U73122 in in vitro inhibition experiments. Preincubation of a T. pyriformis or T. thermophila HM or LM fraction with U73122 at 10 μM resulted in 30 to 50% inhibition of PtdIns(4,5)P2-PLC activity (Fig. 4A; results shown for T. pyriformis), suggesting a common basis for catalysis between mammalian, plant, and Tetrahymena PI-PLC activity. In order to gain insight into the functional role of PI-PLC in regulating Tetrahymena PtdIns(4,5)P2 turnover in vivo, [3H]inositol-labeled starved T. thermophila cells were incubated with U73122 and phosphoinositides were extracted and analyzed by TLC (see Materials and Methods). Interestingly, within 10 min of incubation, we observed a significant increase of PtdInsP2 levels (approximately 3-fold compared to control samples) without any changes in PtdInsP levels (Fig. 4B). HPLC analysis of deacylated phosphoinositides according to the method of Leondaritis et al. (29) verified that this increase was primarily due to the presence of PtdIns(4,5)P2 and not to that of other PtdInsP2 isomers (data not shown). Importantly, under conditions of accelerated PtdIns(4,5)P2 synthesis, i.e., treatment of starved T. thermophila cells with insulin (T. Prevedoros and D. Galanopoulou, unpublished data), pretreatment with U73122 resulted in even more pronounced increases in PtdIns(4,5)P2 levels (approximately 5-fold compared to nontreated control samples; Fig. 4B). These results firmly establish that PI-PLC is a direct regulator of PtdIns(4,5)P2 levels in Tetrahymena.
Fig. 4.
Pharmacological manipulation of PI-PLC activity and PtdIns(4,5)P2 levels in Tetrahymena. (A) U73122 inhibits PI-PLC in vitro. HM and LM fraction PI-PLC activity against 60 μM PtdIns(4,5)P2 at 1 μM Ca2+ was assayed in the absence or presence of U73122 (10 μM). Data represent the means ± SD of the results from two independent experiments, each assayed in duplicate. (B) Effect of U73122 on T. thermophila phosphoinositide levels in vivo. [3H]inositol-labeled cells were preincubated with U73122 (10 μM) or vehicle (dimethylsulfoxide [DMSO]) for 10 min in the absence or presence of insulin (2.5 μM) for an additional 5 min, and [3H]phosphoinositides were extracted and subjected to chromatography using TLC as described in Materials and Methods. Data are expressed as percentages of total [3H]inositol-labeled phosphoinositides and represent the means ± standard errors of the means (SEM) of the results from two independent experiments. [3H]PtdInsP2 levels were increased 3- to 5-fold by U73122 pretreatment, suggesting effective PI-PLC inhibition in vivo. (C) Effect of GTPγS on PI-PLC activity in vitro. Homogenate and LM fraction PI-PLC activity against 60 μM PtdIns(4,5)P2 in the presence of 10 μM Ca2+ and 250 μM MgCl2 was assayed in the absence or presence of the indicated concentrations of GTPγS. Data represent the means ± SEM of the results from two independent experiments, each assayed in duplicate, and are expressed as percentages of the specific activity in the absence of GTPγS.
Several previous studies have suggested that a purported PI-PLC-like activity may be coupled to receptor activation via G proteins in Tetrahymena (40); however, in vitro data are missing and most experiments have mainly relied on pharmacological agents of uncertain or untested ciliate specificity (38). Interestingly, while there are several GPCR-like genes in the T. thermophila genome, there are no recognizable trimeric G proteins (1), and this further challenges the results obtained by analysis using classical mammalian G-protein pharmacological agents. To provide a firm link between PI-PLC and G proteins or small GTPases in Tetrahymena, we used a nonhydrolyzable GTP analogue, GTPγS, in in vitro assays (45). Preincubation of T. thermophila homogenate fractions with 1 to 100 μM GTPγS in the presence of 250 μM Mg2+ and 10 μM Ca2+ resulted in activation of PI-PLC activity at all GTPγS concentrations, with a maximum of a 45% increase at 50 μM (Fig. 4C). A similar trend was observed with LM fractions but with marginal activation of PI-PLC, suggesting substantial loss of factors necessary for GTPγS-induced activation (Fig. 4C). Although the extent of T. thermophila PI-PLC activation is smaller than that found in previous studies examining mammalian and D. discoideum PI-PLCs (3, 44), these results provide the first evidence that GTPase activity, either direct or indirect, may regulate PI-PLC in Tetrahymena.
The Tetrahymena thermophila genome contains two sets of genes encoding bacterial and eukaryotic PLCs, including a novel PRIP homolog.
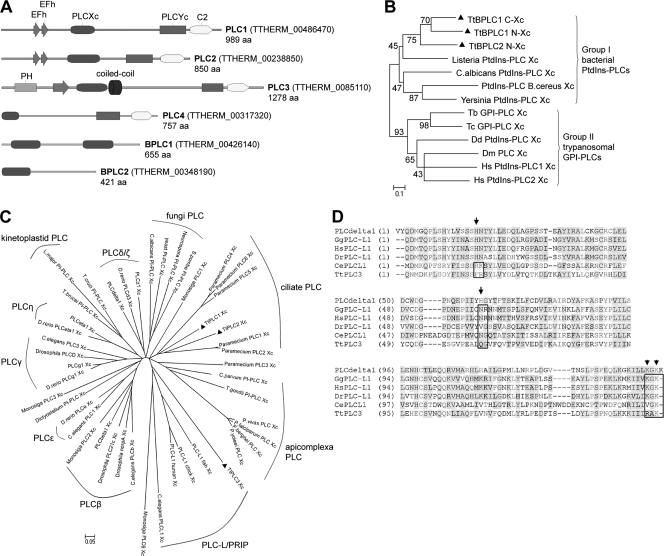
Our biochemical characterization indicated the presence of both bacterium-like and eukaryote-like PLC enzymes in Tetrahymena species. We sought to identify putative genes for these enzymes in order to study their expression patterns and thus gain further insight into their possible roles in Tetrahymena functions. Interrogation of the Tetrahymena thermophila genome and NCBI database with the human PI-PLCδ1 sequence revealed the presence of four putative T. thermophila PI-PLC genes, and similar searches performed with the B. cereus PtdIns-PLC sequence resulted in identification of two putative T. thermophila PtdIns-PLC genes (Fig. 5A; see also Table S1 in the supplemental material). All gene products had a PLCXc-like domain that forms the N-terminal part of the catalytic PLC fold and is common in both eukaryotic and bacterial PLCs (17). Analysis of the domain structure of T. thermophila PI-PLCs (designated TtPLC1-4) revealed a typical eukaryotic PLC domain structure consisting of EF-hand motifs, the PLC catalytic Xc-Yc module separated by a variable X/Y linker, and a C-terminal C2 domain (Fig. 5A; see also Table S1 in the supplemental material). Multiple alignment of the C2 domain indicated conservation of several Ca2+-interacting residues that are responsible for Ca2+-induced association of PI-PLCs with membrane phospholipids (see Fig. S2A in the supplemental material). An N-terminal PH domain was detected in PLC3 but was absent from the rest of the TtPLCs (Fig. 5A; see also Table S1 in the supplemental material). PLC4 had N-terminally truncated PLCXc and Yc domains with low e-value scores in BLAST searches and no recognizable EF-hand motifs (see Table S1 in the supplemental material) and was excluded from subsequent multiple sequence alignments. A consensus nuclear export sequence (NES) was identified in PLC1-3 that corresponded to the NES of PLCδ1 (amino acids 164 to 177) (45), while two putative nuclear localization signals were identified in PLC3 (amino acids 193 to 228 and 550 to 557), suggesting functional nucleocytoplasmic shuttling. Interestingly, PLC2, in contrast to PLC1 and despite their common domain organization and gene structures (see Table S1 in the supplemental material), contained a much shorter X/Y linker resembling that of most mammalian PLC isoforms. Analysis of the domain structure of the PtdIns-PLC TtBPLC1 and TtBPLC2 revealed the presence of two tandem PLCXc domains separated by a 200-amino-acid linker in the former, whereas the latter had an N-terminal PLCXc domain (Fig. 5A; see also Table S1 in the supplemental material). In order to functionally categorize these proteins, we performed multiple alignments of the PLCXc domains of T. thermophila PLCs, eukaryotic PI-PLCs, and bacterial and trypanosomal PLCs, including their putative homologs from unicellular, invertebrate, and mammalian species (Fig. 5B and C; the accession numbers of the sequences used for analysis are listed in Table S2 in the supplemental material, while the alignments for bacterial and eukaryotic PLCs are shown in Fig. S2B and S2C in the supplemental material).
Fig. 5.
Domain architecture and homology of T. thermophila bacterial and eukaryotic PLCs. (A) Domain architecture of PLC1-4 and BPLC1-BPLC2. EFh, EF-hands; PLCXc and Yc, PLC catalytic domains; C2, PKC C2-homology domain; PH, pleckstrin-homology domain; aa, amino acid. (B) Unrooted neighbor-joining tree for bacterium-like PtdIns-PLC PLCXc domains. T. thermophila BPLC1-BPLC2 PLCXc domains (TtBPLC1 C and TtBPLC1 N correspond to the C- and N-terminal PLCXc domains, respectively) are indicated by black triangles. Bootstrap values from 100 replicates are indicated near the corresponding branches. Group I and group II PtdIns-PLCs corresponding to bacterial and trypanosome-like enzymes, respectively, are indicated by brackets. Tb, T. brucei; Tc, T. cruzi; Dd, D. discoideum; Dm, D. melanogaster; Hs, H. sapiens. (C) Unrooted neighbor-joining tree for eukaryotic PI-PLC PLCXc domains from mammals, invertebrates, and unicellular eukaryotes. Tetrahymena PLC1-3 PLCXc domains are indicated by black triangles. Groups corresponding to specific mammalian PI-PLC isoforms or kinetoplastid, fungal, ciliate, and apicomplexa PI-PLCs are indicated. Trees constructed using parsimony and maximum-likelihood methods largely corroborated the overall topology shown, with the exception of those representing Paramecium PLC3, Dictyostelium PI-PLC, Monosiga PLC1, -3, and -6 and C. elegans PLC-L; notably, TtPLC3 was situated in the ciliate PLC group (see text). (D) Sequence alignment of PLCXc domains from PLCδ1, animal and invertebrate PLC-L/PRIP (Hs, H. sapiens; Gg, G. gallus; Dr, D. rerio; Ce, C. elegans), and TtPLC3/PRIP. Arrows indicate the positions of catalytic His residues (H311/356 in PLCδ1) and arrowheads the position of Lys residues that interact with the D-4/5 phosphates of the polar head of PtdIns(4,5)P2 (K438/440 in PLCδ1). Boxed regions highlight the lack of one (HsPLC-L, GgPLC-L, and DrPLC-L) or both (TtPLC3/PRIP and CePLC-L) catalytic His residues and preservation of one or both substrate-interacting Lys residues in PLC-L/PRIP proteins. Similar sequence alignments of the PLCYc domains also indicated preservation of the PLCδ1 Arg549 residue that dictates a preference for PtdIns(4,5)P2 (not shown). Accession numbers for all PLCs in panels B and C are given in Table S2 in the supplemental material.
As shown in Fig. 5B, PLCXc domains of TtBPLC1 and TtBPLC2 are closely related to PtdIns-PLCXc domains from B. cereus, Listeria monocytogenes, Yersinia mollaretii, and a Candida albicans PtdIns-PLC. In contrast, PtdIns-PLCXc domains from other eukaryotic species, including Homo sapiens, D. melanogaster, D. discoideum, and Entamoeba dispar, seem to be more closely related to the trypanosomal GPI-PLCXc domain. This degree of diversification is apparent in the sequence alignment shown in Fig. S2B in the supplemental material, where all group II PtdIns-PLCs have trypanosome-like insertions between the β-strand I and α-helix 1′ of the PLCXc structure (17). Our analysis substantiates the idea that T. thermophila possesses bacterium-like PtdIns-PLCs, which were characterized biochemically in the previous section, and, furthermore, provides the first report of bacterium-like PtdIns-PLC genes, distinct from trypanosomal GPI-PLCs, in lower eukaryotes. Since Tetrahymena PtdIns-PLC is apparently a secreted activity (Fig. 2D) and since TtBPLC1 (but not BPLC2) codes for an active PLC (see legend in Fig. S2B in the supplemental material), we searched for putative signal peptide sequences and/or transmembrane regions in the TtBPLC1 coding sequence but with negative results. However, this may have been due to incorrect annotation of the BPLC1 gene; we found an alternative methionine start codon 228 bp upstream of the annotated start codon of BPLC1, and interrogation of the translated amino acid sequence with the SignalP program resulted in identification of a putative signal peptide sequence and a cleavage site between new positions 24 and 25. Thus, whether TtBPLC1 is indeed a classical Tetrahymena-secreted phospholipase responsible for extracellular digestion of PtdIns remains to be determined.
The analysis of eukaryote-like TtPLC1-3 PLCXc domains, together with analysis of PLCXc domains of selected isoforms from H. sapiens, Danio rerio, D. melanogaster, C. elegans, the choanoflagellate Monosiga brevicollis, and unicellular PI-PLCs (Fig. 5C), revealed two interesting aspects. First, PI-PLC isoforms are clustered together; for example, our analysis identified Monosiga PLC2 as a β-isoform and all known or suspected PLCγ-, ε-, and β-isoforms from the invertebrates included in the analysis. In contrast, PI-PLCXc domains from fungi, apicomplexa, ciliates, and kinetoplastids occupied distinct groups that were largely unrelated to each other or to a specific PI-PLC isoform (Fig. 5C). Our analysis placed TtPLC1/ TtPLC2 together with Paramecium PLCs (25) and grouped all apicomplexa PI-PLCs from T. gondii, Cryptosporidium parvum, and Plasmodia species. Second, the PLCXc domains of PRIP proteins from H. sapiens, Gallus gallus, and D. rerio, which overall display a PLCδ-like structure (23), formed a distinct group together with a putative PRIP homolog from C. elegans, an M. brevicollis PLC, and, surprisingly, TtPLC3 (Fig. 5C). It should be noted, however, that TtPLC3 is a ciliate-specific gene and is not evolutionarily related to vertebrate PRIP genes, since phylogenetic trees based on parsimony and maximum-likelihood methods placed TtPLC3 within the ciliate group (see Fig. S2D in the supplemental material). Similar discrepancies were also observed for the C. elegans PLC-L, Paramecium PLC3, and some M. brevicollis PLCs (discussed in the Fig. S2D legend in the supplemental material). These discrepancies probably reflect the complex phylogenetic relationships of the eukaryotic PI-PLC family, as has been previously suggested by several researchers in the field (4, 24, 43). Nevertheless, in all aspects studied, TtPLC3 is most likely an inactive PI-PLC and a true PRIP homolog, since it lacks the two critical His residues necessary for catalysis (Fig. 5D, arrows) (17), it retains crucial amino acids for efficient recognition of PtdIns(4,5)P2 (Fig. 5D, arrowheads), and it has an N-terminal PH domain (Fig. 5A); therefore, we have renamed TtPLC3 as TtPRIP. To our knowledge, this is the first description of a putative PRIP homolog in nonanimal species.
Comparative analyses of PI-PLC activity and expression patterns of PLC1, PLC2, and PRIP in T. thermophila strains indicate that they are functional genes.
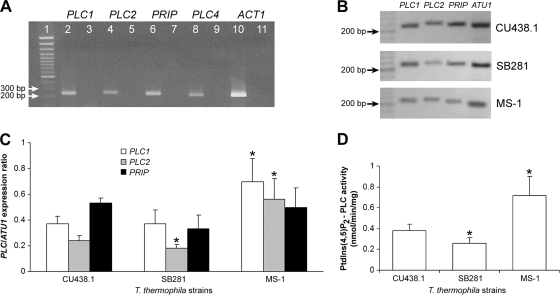
Recent data from a study of Tetrahymena expressed-sequence-tag (EST) clones (8) suggested that PLC1, PLC2, PRIP, and BPLC1 are likely to be functional, constitutively expressed genes. We thus focused our study on PLC1-PLC2 and PRIP, for the additional reason that they are likely to represent important signaling enzymes, and generated primers that were used to amplify the respective transcripts from total RNA isolated from vegetative T. thermophila CU438.1 cells (see Materials and Methods). In semiquantitative reverse transcription-PCRs (RT-PCRs) optimized in the logarithmic phase of amplification and utilizing ACT1 (actin-1) or ATU1 (α-tubulin-1) expression levels for normalization, we found that all genes were indeed expressed; in fact, expression of PRIP was highest, followed by that of PLC1 and PLC2 (Fig. 6A). In these experiments, we also amplified PLC4 transcripts and found that it is also expressed (Fig. 6A). Importantly, the same set of primers was used to amplify the respective transcripts from a cDNA preparation obtained from T. pyriformis mRNAs with positive results, suggesting the presence, conservation, and expression of all PLC genes in T. pyriformis as well (see Fig. S3A to C in the supplemental material). Indeed, direct sequencing of the T. pyriformis PLC1 and PRIP PCR products (GenBank accession numbers HQ317216 and HQ317217) revealed a perfect match at the nucleotide level, suggesting strong conservation in Tetrahymena species (see Fig. S3 in the supplemental material).
Fig. 6.
Expression and comparative analysis of Tetrahymena PLC1, PLC2, PRIP, and PI-PLC activity. (A) Expression of Tetrahymena PLCs and ACT1. Total RNA was isolated from T. thermophila CU438.1 cells and subjected to reverse transcription, and cDNA was amplified with gene-specific primers as described in Materials and Methods. Lane 1 shows the results of electrophoresis of DNA size markers, and lanes 3, 5, 7, 9, and 11 show the results of reactions in the absence of cDNA. All PCR products were of the expected size (233, 239, 228, 208, and 201 bp for PLC1, PLC2, PRIP, PLC4, and ACT1, respectively). (B and C) Expression of PLC1, PLC2, PRIP, and ATU1 genes in the T. thermophila strains SB281 and MS-1. RNA was isolated from CU438.1, SB281, and MS-1 cells in the same exponential phase of growth, and cDNA was amplified as described above. Reactions were optimized for linear amplification of PRIP and ATU1 genes (not shown). Expression of each PLC gene was normalized for ATU1 expression, and data representing the results from three different cultures were plotted as shown in panel C. Asterisks denote statistical significance (P < 0.05 [Student's t test]). (D) PI-PLC activity in T. thermophila strains. Activity was determined as described for Fig. 3 in the presence of 60 μM PtdIns(4,5)P2 and 10 μM Ca2+; data represent means ± SD of the results from three experiments. Asterisks denote statistical significance (P < 0.05 [Student's t test]).
Since PRIP is likely to code for an inactive PLC (Fig. 5D), the bulk of PI-PLC activity characterized in the previous sections was primarily due to PLC1 and PLC2. Of note, PLC1 and PLC2 may contribute differentially to total PLC activity, at least under basal conditions, due to their differences in X/Y linker regions (Fig. 5A). Indeed, a recent study proposed that the structure of the X/Y linker is very important for constitutive activity of most mammalian PLC isoforms (19). As a first step toward understanding the contribution of these proteins to PLC activity, we proceeded to perform a comparative study of their expression patterns and PI-PLC activities by the use of a collection of T. thermophila strains. We therefore assessed the expression levels of PLC1-PLC2 and PRIP in CU438.1 (wild type) and two well-characterized and -studied mutant strains, SB281 (5) and MS-1 (21). In parallel, we assayed PI-PLC activities in the same strains as described in previous sections. A typical profile of PLC1-PLC2, PRIP, and ATU1 expression in all strains is shown in Fig. 6B and the quantification of results obtained from three different cultures of each strain in Fig. 6C. Interestingly, expression levels of PLC1 and/or PLC2 correlated perfectly with PI-PLC activity in mutant strains compared to the wild-type CU438.1 strain. As such, reduced PLC activity in the SB281 strain (30% reduction compared to CU438.1) correlated with reduced expression of PLC2 (25% decrease compared to CU438.1), while increased PLC activity in the MS-1 strain (2-fold compared to CU438.1) correlated with similar increases in expression of both PLC1 and PLC2 (2-fold compared to CU438.1) (Fig. 6C and D). Considering the differences between the CU438.1 and SB281 strains, we observed that, during starvation, SB281 cells exhibited approximately half of the activity of CU438.1 cells (0.13 ± 0.04 and 0.25 ± 0.02 nmol/min/mg, respectively; n = 3). Interestingly, expression of PLC2 was also differentially regulated during starvation, since it was reduced by 40% in starved CU438.1 and SB281 cells compared to cells in growth medium (not shown). These results suggest a high level of coordination between the expression levels of PLC2 and (probably) PLC1 and total PI-PLC activity.
DISCUSSION
Phospholipases C that specifically hydrolyze PtdIns and/or PtdIns(4,5)P2 are important regulatory enzymes in both prokaryotic and eukaryotic functions. In this report, comparative biochemical characterization of PLC activities as well as in silico and molecular analysis of PLC-like genes in Tetrahymena provide compelling evidence for the presence of distinct PtdIns-PLC and PtdIns(4,5)P2-PLC activities that correspond to two sets of genes coding for bona fide bacterial and eukaryotic PLCs, respectively.
Earlier attempts to identify a PI-PLC activity in Tetrahymena were hindered by significant degradation of substrates [i.e., PtdIns or PtdIns(4,5)P2] by PLA/lysoPLA activities. In fact, with a different fractionation protocol (31) or by the use of nonpurified [3H]PtdIns(4,5)P2 as the substrate, robust PtdIns(4,5)P2-hydrolyzing activity that is insensitive to Ca2+ and results in production of glycerol derivatives of phosphoinositides is detected primarily in the cytosol (our unpublished data). Accordingly, efforts to purify PtdIns and PtdIns(4,5)P2-PLC activities from cytosolic fractions were unsuccessful, since the purified activity displayed insensitivity to micromolar Ca2+ and U73122 and corresponded to a PLA/lysoPLA activity (data not shown). Interestingly, a similar situation was described earlier in a report from a study performed with the slime mold D. discoideum, in which exogenous PtdIns(4,5)P2 is hydrolyzed preferentially to glyceroderivatives (3), and also in a report from a study using S. cerevisiae, in which robust production of phosphoinositide glyceroderivatives was detected in vivo (16). In the latter two cases, detection of a PI-PLC activity was achieved only by monitoring hydrolysis of endogenous substrates and overexpression of the PI-PLC gene, respectively (3, 16). These data collectively suggest that, in these eukaryotes, the bulk of phosphoinositide hydrolysis, at least under certain conditions, occurs primarily via deacylating and not via PLC activities. Although the biological meaning of this observation is unclear, it should be noted that the glyceroderivatives of phosphoinositides have been described as putative signaling molecules, at least in studies of mammalian cells (7).
Importantly, by using a simple fractionation scheme we were able to distinguish biochemically between two distinct Tetrahymena phosphoinositide-specific PLC activities: Ca2+-insensitive or -inhibited PtdIns-PLC and a membrane-associated Ca2+-sensitive PtdIns(4,5)P2-PLC (Fig. 1 and 2). Further characterization of the PtdIns(4,5)P2-PLC activity revealed sensitivity to low-micromolar Ca2+, inhibition by U73122, and modest activation by GTPγS, the nonhydrolyzable analogue of GTP (Fig. 3 and 4). These characteristics, and in particular the typical low-micromolar Ca2+sensitivity shown in Fig. 3, are common to all unicellular, plant, and mammalian PI-PLC enzymes studied so far (4, 10, 12, 13, 36, 39). U73122 has been used in previous studies reporting the involvement of a purported PLC activity in several Tetrahymena functions (2, 40), and our in vitro data justify the rationales of these and other reports. Most importantly, in vivo treatment of cells with U73122 revealed that PI-PLC is constitutively active in Tetrahymena and that it is involved in the regulation of phosphoinositide flux through PtdIns(4,5)P2 toward Ins(1,4,5)P3 and other inositol phosphates. In principle, PtdIns(4,5)P2 is subjected to continuous cycling by PtdInsP kinases (PIPKs) and 5-phosphatases and degradation by PI-PLC. Of note, the T. thermophila genome contains at least nine distinct PIPK genes that code for low-abundance proteins; comparison of their catalytic domains to those of mammalian PIPKs suggests that they may display reduced catalytic efficiency (Leondaritis and Galanopoulou, unpublished data). Collectively, these data point to the interesting possibility that PI-PLC, in addition to its apparent role in production of Ins(1,4,5)P3, a known Ca2+-mobilizing agent, is concurrently the principal regulator of steady-state PtdIns(4,5)P2 levels in Tetrahymena.
Analysis of the genes coding for bacterium-like PtdIns-PLC and eukaryote-like PI-PLCs in Tetrahymena revealed that both are present and likely functional, as suggested also by biochemical characterization. Overall, it appears that bacterial and eukaryotic PLCs are restricted to bacteria and eukaryotes, respectively, with a few known exceptions: trypanosomes, which have a distant relative of bacterial PtdIns-PLCs named GPI-PLC (6); the dimorphic yeast C. albicans, which has two bacterial PtdIns-PLC genes (26), and Tetrahymena (this study). It is likely that many eukaryotic unicellular and multicellular organisms also have trypanosome-like PLC genes, as suggested by our nonexhaustive search (Fig. 5B), but we were unable to detect bacterium-like PLCs in other eukaryotes; this implies that the acquisition of a bacterium-like PLC in eukaryotes may be due to lateral gene transfer events in a species-specific manner and may not be a generalized trait. Apparently, TtBPLC1-TtBPLC2 may have a lysosomal origin, since the extracellular PtdIns-PLC activity was found to be abolished in the well-characterized secretion mutant strain T. thermophila MS-1 (Fig. 2D and E). The biological function of TtBPLC1-TtBPLC2 is at present unclear, although it might be involved in extracellular digestion of phospholipids together with many other phospholipases that have long been known to be actively secreted by Tetrahymena cells (14).
Among the most important findings of this study were the identification and characterization of an expanded set of PI-PLC genes in Tetrahymena (Fig. 5 and 6). Our experiments suggested that PLC1, PLC2, and PRIP are functional expressed genes and identified a conspicuous relationship of PLC2 (and PLC1) expression to total PI-PLC activity in T. thermophila strains (Fig. 6). The changes in expression and activity observed in SB281 and MS-1 strains should not be taken as directly correlating to the specific defects in those mutant strains, although this would be an interesting aspect to study in the future. Nevertheless, the results of these experiments complement a recent Tetrahymena genome-wide expression study by Miao et al. (33). Interestingly, those researchers provided evidence that expression of PLC1 is unrelated to that of PLC2 and PRIP throughout vegetative growth, starvation, and conjugation (see Fig. S4 in the supplemental material; see also the TGED database [http://tged.ihb.ac.cn/]). Taken together, these data indicate that catalytically active PLC1 and PLC2 serve probably nonredundant functions. The putative functions of the Tetrahymena PRIP homolog are currently obscure. It seems to be a Tetrahymena-specific innovation, since we have been unable so far to identify any other PRIP homolog in the close relative Paramecium or in other alveolates. In animal cells, a major role of PRIP1 appears to be the control of the extent of Ins(1,4,5)P3-induced Ca2+ mobilization by active PLCs (23). Interestingly, Miao et al. identified TtPRIP as a gene coexpressed with a putative transposase at late stages of conjugation (33), and TtPRIP and PLC2 expression patterns are closely related, at least during conjugation (see Fig. S4 in the supplemental material and the TGED database). Obviously, a major goal of future studies would be to dissect genetically the roles of PLC1, PLC2, and PRIP. Our ongoing studies have shown specific changes and fluctuations of PI-PLC activity during chemotaxis, phagocytosis, and cilium regeneration, which are accompanied by timely changes in PtdIns(4,5)P2 levels (our unpublished data). These studies may point to a specific role of PLC1-PLC2 in these cellular processes, some of which are also sensitive to U73122 in vivo.
From an evolutionary aspect, the results of this study and a recent study on Paramecium PLCs (25) suggest a ciliate-specific expansion of PI-PLC genes among all other unicellular organisms studied so far, which invariably have a single PI-PLC gene (34). Eukaryotic PI-PLCs have been extensively studied in animals, where they have been shown to regulate acute receptor signaling in an isoform-specific manner (24, 34). In plants, nine PI-PLC genes have been similarly identified, and they are involved in gravistimulation and growth (4, 18). Furthermore, S. cerevisiae genetic studies have shown a pleiotropic function of its single PI-PLC gene (13, 37). Since the most prominent function of animal PI-PLC is mobilization of Ca2+ via Ins(1,4,5)P3-sensitive Ca2+ channels (IP3R), it is surprising that this canonical Ins(1,4,5)P3/Ca2+/IP3R system is not conserved in plants or yeasts, as indicated by the apparent absence of IP3R-related homologs in their genomes (4, 34). In this evolutionary context, ciliates like Tetrahymena and Paramecium represent unique cases. Besides the expansion of PI-PLC genes, which are likely to have nonredundant roles, at least in Tetrahymena (this study), they apparently express members of an expanded family of IP3R-related Ca2+ channels, the functionality of which has been previously demonstrated, at least in Paramecium studies (27, 28). Collectively, these findings imply that the regulation of Ca2+ homeostasis via a PI-PLC-Ins(1,4,5)P3-IP3R axis is likely to be evolutionarily conserved and elaborated in ciliates; accordingly, future studies aimed at unraveling the role of Tetrahymena PLC1-PLC2 (and PRIP) may focus primarily on well-established Ca2+-regulated processes.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Tiedtke (University of Münster, Germany) for the T. thermophila strains, T. Prevedoros for his help with in vivo phosphoinositide analysis, I. Siokos for his help with RNA isolation, and A. Lykidis (U.S. Department of Energy-Joint Genome Institute) for critically reading the manuscript.
This work was partially supported by a University of Athens grant (KA 70/4/8779).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 17 December 2010.
REFERENCES
- 1. Anantharaman V., Iyer L. M., Aravind L. 2007. Comparative genomics of protists: new insights into the evolution of eukaryotic signal transduction and gene regulation. Annu. Rev. Microbiol. 61:453–475 [DOI] [PubMed] [Google Scholar]
- 2. Bartholomew J., et al. 2008. GTP avoidance in Tetrahymena thermophila requires tyrosine kinase activity, intracellular calcium, NOS, and guanylyl cyclase. Purinergic Signal. 4:171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bominaar A. A., Kesbeke F., Van Haastert P. J. 1994. Phospholipase C in Dictyostelium discoideum. Cyclic AMP surface receptor and G-protein-regulated activity in vitro. Biochem. J. 297:181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boss W. F., Davis A. J., Im Y. J., Galvão R. M., Perera I. Y. 2006. Phosphoinositide metabolism: towards an understanding of subcellular signaling. Subcell. Biochem. 39:181–205 [DOI] [PubMed] [Google Scholar]
- 5. Bowman G. R., Turkewitz A. P. 2001. Analysis of a mutant exhibiting conditional sorting to dense core secretory granules in Tetrahymena thermophila. Genetics 159:1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carrington M., et al. 1998. The properties and function of the glycosylphosphatidylinositol-phospholipase C in Trypanosoma brucei. Mol. Biochem. Parasitol. 91:153–164 [DOI] [PubMed] [Google Scholar]
- 7. Corda D., Zizza P., Varone A., Filippi B. M., Mariggio S. 2009. The glycerophosphoinositols: cellular metabolism and biological functions. Cell. Mol. Life Sci. 66:3449–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coyne R. S., et al. 2008. Refined annotation and assembly of the Tetrahymena thermophila genome sequence through EST analysis, comparative genomic hybridization, and targeted gap closure. BMC Genomics 9:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deli D., Leondaritis G., Tiedtke A., Galanopoulou D. 2008. Deficiency in lysosomal enzyme secretion is associated with upregulation of phosphatidylinositol 4-phosphate in Tetrahymena. J. Eukaryot. Microbiol. 55:343–350 [DOI] [PubMed] [Google Scholar]
- 10. Drayer A. L., van Haastert P. J. 1992. Molecular cloning and expression of a phosphoinositide-specific phospholipase C of Dictyostelium discoideum. J. Biol. Chem. 267:18387–18392 [PubMed] [Google Scholar]
- 11. Dupont G., McGuinness O. M., Johnson M. H., Berridge M. J., Borgese F. 1996. Phospholipase C in mouse oocytes: characterization of beta and gamma isoforms and their possible involvement in sperm-induced Ca2+ spiking. Biochem. J. 316:583–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fang J., Marchesini N., Moreno S. N. J. 2006. A Toxoplasma gondii phosphoinositide phospholipase C (TgPI-PLC) with high affinity for phosphatidylinositol. Biochem. J. 394:417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flick J. S., Thorner J. 1993. Genetic and biochemical characterization of a phosphatidylinositol-specific phospholipase C in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:5861–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Florin-Christensen J., Florin-Christensen M., Knudsen J., Rasmussen L. 1986. Phospholipases and phosphonolipids in a ciliate: an attack and defence system? Trends Biochem. Sci. 11:354–355 [Google Scholar]
- 15. Griffith O. H., Ryan M. 1999. Bacterial phosphatidylinositol-specific phospholipase C: structure, function, and interaction with lipids. Biochim. Biophys. Acta 1441:237–254 [DOI] [PubMed] [Google Scholar]
- 16. Hawkins P. T., Stephens L. R., Piggott J. R. 1993. Analysis of inositol metabolites produced by Saccharomyces cerevisiae in response to glucose stimulation. J. Biol. Chem. 268:3374–3383 [PubMed] [Google Scholar]
- 17. Heinz D. W., Essen L. O., Williams R. L. 1998. Structural and mechanistic comparison of prokaryotic and eukaryotic phosphoinositide-specific phospholipases C. J. Mol. Biol. 275:635–650 [DOI] [PubMed] [Google Scholar]
- 18. Helling D., Possart A., Cottier S., Klahre U., Kost B. 2006. Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. Plant Cell 18:3519–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hicks S. N., et al. 2008. General and versatile autoinhibition of PLC isozymes. Moll. Cell 31:383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Homma Y., Emori Y. 1997. Purification and assay of PLCδ, p. 99–116 In Shears S. B. (ed.), Signaling by inositides. IRL Press, Oxford, United Kingdom [Google Scholar]
- 21. Hünseler P., Scheidgen-Kleyboldt G., Tiedtke A. 1987. Isolation and characterization of a mutant of Tetrahymena thermophila blocked in secretion of lysosomal enzymes. J. Cell Sci. 88:47–55 [DOI] [PubMed] [Google Scholar]
- 22. Irvine R. F., Hemington N., Dawson R. M. 1978. The hydrolysis of phosphatidylinositol by lysosomal enzymes of rat liver and brain. Biochem. J. 176:475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanematsu T., Takeuchi H., Terunuma M., Hirata M. 2005. PRIP, a novel Ins(1,4,5)P3 binding protein: functional significance in Ca2+ signaling and extension to neuroscience and beyond. Mol. Cells 20:305–314 [PubMed] [Google Scholar]
- 24. Katan M. 2005. New insights into the families of PLC enzymes: looking back and going forward. Biochem. J. 391:e7–e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klöppel C., Muller A., Marker S., Simon M. 2009. Two isoforms of eukaryotic phospholipase C in Paramecium affecting transport and release of GPI-anchored proteins in vivo. Eur. J. Cell Biol. 88:577–592 [DOI] [PubMed] [Google Scholar]
- 26. Kunze D., et al. 2005. Functional analysis of the phospholipase C gene CaPLC1 and two unusual phospholipase C genes, CaPLC2 and CaPLC3, of Candida albicans. Microbiology 151:3381–3394 [DOI] [PubMed] [Google Scholar]
- 27. Ladenburger E. M., Sehring I. M., Korn I., Plattner H. 2009. Novel types of Ca2+-release channels participate in the secretory cycle of Paramecium cells. Mol. Cell. Biol. 29:3605–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ladenburger E. M., Korn I., Kasielke N., Wassmer T., Plattner H. 2006. An Ins(1,4,5)P3 receptor in Paramecium is associated with the osmoregulatory system. J. Cell Sci. 119:3705–3717 [DOI] [PubMed] [Google Scholar]
- 29. Leondaritis G., Tiedtke A., Galanopoulou D. 2005. D-3 phosphoinositides of the ciliate Tetrahymena: characterization and study of their regulatory role in lysosomal enzyme secretion. Biochim. Biophys. Acta 1745:330–341 [DOI] [PubMed] [Google Scholar]
- 30. Leondaritis G., Galanopoulou D. 2000. Characterization of inositol phospholipids and identification of a mastoparan-induced polyphosphoinositide response in Tetrahymena pyriformis. Lipids 35:525–532 [DOI] [PubMed] [Google Scholar]
- 31. Leondaritis G., Kapetaniou E., Galanopoulou D. 2000. Study of phosphatidylinositol-hydrolyzing activities in a unicellular eukaryote, p. 265–276 In Kokotos G., Constantinou-Kokotou V. (ed.), Lipases and lipids: structure, function and biotechnological applications. Crete University Press, Crete, Greece [Google Scholar]
- 32. Melin P. M., Sundler R., Jergil B. 1986. Phospholipase C in rat liver plasma membranes. Phosphoinositide specificity and regulation by guanine nucleotides and calcium. FEBS Lett. 198:85–88 [DOI] [PubMed] [Google Scholar]
- 33. Miao W., et al. 2009. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One 4:e4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Michell R. H. 2008. Inositol derivatives: evolution and functions. Nat. Rev. Mol. Cell Biol. 9:151–161 [DOI] [PubMed] [Google Scholar]
- 35. Nomikos M., et al. 2005. Role of phospholipase C-zeta domains in Ca2+-dependent phosphatidylinositol 4,5-bisphosphate hydrolysis and cytoplasmic Ca2+ oscillations. J. Biol. Chem. 280:31011–31018 [DOI] [PubMed] [Google Scholar]
- 36. Nozaki T., Toh-e A., Fujii M., Yagisawa H., Nakazawa M., Takeuchi T. 1999. Cloning and characterization of a gene encoding phosphatidylinositol-specific phospholipase C from Trypanosoma cruzi. Mol. Biochem. Parasitol. 102:283–295 [DOI] [PubMed] [Google Scholar]
- 37. Odom A. R., Stahlberg A., Wente S. R., York J. D. 2000. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287:2026–2029 [DOI] [PubMed] [Google Scholar]
- 38. Plattner H., Sehring I. M., Schilde C., Ladenburger E. M. 2009. Pharmacology of ciliated protozoa-drug (in)sensitivity and experimental drug (ab)use. Int. Rev. Cell Mol. Biol. 273:163–218 [DOI] [PubMed] [Google Scholar]
- 39. Rhee S. G. 2001. Regulation of phosphoinositide-specific phospholipase C. Annu. Rev. Biochem. 70:281–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ryals P. E. 2009. Inositols and phosphoinositides in Tetrahymena. Acta Protozool. 48:191–202 [Google Scholar]
- 41. Spyridakis S., Leondaritis G., Nakos G., Lekka M. E., Galanopoulou D. 2010. A specific phospholipase C activity regulates phosphatidylinositol levels in lung surfactant of patients with acute respiratory distress syndrome. Am. J. Respir. Cell Mol. Biol. 42:357–362 [DOI] [PubMed] [Google Scholar]
- 42. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 43. Tasma I. M., Brendel V., Whitham S. A., Bhattacharyya M. K. 2008. Expression and evolution of the phosphoinositide-specific phospholipase C gene family in Arabidopsis thaliana. Plant Physiol. Biochem. 46:627–637 [DOI] [PubMed] [Google Scholar]
- 44. Wu D. Q., Lee C. H., Rhee S. G., Simon M. I. 1992. Activation of phospholipase C by the alpha subunits of the Gq and G11 proteins in transfected Cos-7 cells. J. Biol. Chem. 267:1811–1817 [PubMed] [Google Scholar]
- 45. Yamaga M. M., Fujii, Kamata H., Hirata H., Yagisawa H. 1999. Phospholipase C-delta1 contains a functional nuclear export signal sequence. J. Biol. Chem. 274:28537–28541 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.