Abstract
Like all ciliates, Paramecium tetraurelia is a unicellular eukaryote that harbors two kinds of nuclei within its cytoplasm. At each sexual cycle, a new somatic macronucleus (MAC) develops from the germ line micronucleus (MIC) through a sequence of complex events, which includes meiosis, karyogamy, and assembly of the MAC genome from MIC sequences. The latter process involves developmentally programmed genome rearrangements controlled by noncoding RNAs and a specialized RNA interference machinery. We describe our first attempts to identify genes and biological processes that contribute to the progression of the sexual cycle. Given the high percentage of unknown genes annotated in the P. tetraurelia genome, we applied a global strategy to monitor gene expression profiles during autogamy, a self-fertilization process. We focused this pilot study on the genes carried by the largest somatic chromosome and designed dedicated DNA arrays covering 484 genes from this chromosome (1.2% of all genes annotated in the genome). Transcriptome analysis revealed four major patterns of gene expression, including two successive waves of gene induction. Functional analysis of 15 upregulated genes revealed four that are essential for vegetative growth, one of which is involved in the maintenance of MAC integrity and another in cell division or membrane trafficking. Two additional genes, encoding a MIC-specific protein and a putative RNA helicase localizing to the old and then to the new MAC, are specifically required during sexual processes. Our work provides a proof of principle that genes essential for meiosis and nuclear reorganization can be uncovered following genome-wide transcriptome analysis.
INTRODUCTION
Among unicellular eukaryotes, ciliates exhibit a characteristic nuclear dimorphism. Their germ line nucleus, the micronucleus (MIC), is transcriptionally silent during vegetative growth but undergoes meiosis during sexual processes and transmits the germ line genome to the following generation. In contrast, the somatic macronucleus (MAC) is responsible for gene expression but is destroyed at each sexual cycle (29). Two modes of sexual reproduction have been described and are induced mostly in response to starvation. Conjugation involves reciprocal exchange of haploid gametic nuclei between two mating partners. In contrast, autogamy is a self-fertilization process described in some ciliate species, during which the zygotic nucleus is formed by the fusion of two identical gametic nuclei from a single cell. Conjugation and autogamy involve complex biological processes, including MIC meiosis, mitotic division of one haploid meiotic product to yield the gametic nuclei, fertilization and karyogamy, two successive rounds of mitosis of the zygotic nucleus, and the differentiation of new MICs and MACs. Thus, the formation of a new MAC is part of a highly regulated succession of nuclear events that extend over two noncanonical cell cycles and involve a particular type of cell division, called the karyonidal division.
Within the developing new MAC, also called anlagen, a complex program of genome amplification takes place, concurrently with the programmed elimination of germ line sequences (17, 39). In Paramecium tetraurelia, an estimated 60,000 single-copy, short, and noncoding internal eliminated sequences (IESs) are precisely excised from the genome during the assembly of functional genes (reviewed in reference 9). Moreover, imprecise loss of generally repetitive germ line DNA is associated with chromosome fragmentation (21). Maternal inheritance of rearrangement patterns from the old to the new MAC involves a trans-nuclear genome comparison mediated by different populations of noncoding RNAs (reviewed in reference 13), including constitutive maternal transcripts originating from the old MAC (22) and 25-nucleotide (nt) scan RNAs (scnRNAs) produced during MIC meiosis by a specialized RNA interference (RNAi) machinery (23).
Only a few proteins involved in developmentally programmed nuclear reorganization have been identified in Paramecium. Spo11, a protein conserved in all eukaryotes, was also shown to be essential for meiosis in P. tetraurelia (6). The PiggyMac domesticated transposase, produced specifically during MAC development, is a likely candidate for the introduction of the double-strand breaks that initiate all DNA elimination events (6). Previous screens in P. tetraurelia uncovered the essential role of a developmentally regulated SUMO pathway likely to operate in the developing new MAC (24) and of Die5p, a nuclear protein of unknown function acting at a late step during DNA rearrangements (25). Essential genes encoding proteins involved in scnRNA biogenesis or transport were shown to be expressed specifically during sexual processes (10, 23, 26).
To gain broader insight into which biological functions participate in the correct progression of sexual processes and in the formation of a functional new MAC, we started a large-scale analysis of the transcriptome based on the available annotation of the P. tetraurelia MAC genome sequence (5). In this study, we focused on the genes carried by the Megabase chromosome, which was the first macronuclear chromosome to be annotated manually (40). We used dedicated DNA microarrays to monitor transcription profiles during vegetative growth and throughout autogamy, and we identified four major differential expression patterns during sexual processes: (i) an early induction peak, (ii) a late induction peak, (iii) gradual induction, and (iv) strong repression. We picked 15 confirmed induced genes for further in vivo functional studies and knocked down their expression by RNA interference (14). Four genes were found to be essential during vegetative growth and were not tested further during autogamy. Two other genes were shown to encode differentially localized nuclear proteins, and their expression was required for the recovery of viable sexual progeny.
MATERIALS AND METHODS
Autogamy time course and RNA extraction.
Experiments were carried out with the entirely homozygous P. tetraurelia strains 51new, here also designated strain 51 (15), and d4-2, which is essentially genetically identical to 51 but carries the A29 allele of the surface antigen A gene (34). Cells were grown in a wheatgrass powder (WGP) infusion medium inoculated with Klebsiella pneumoniae the day before use. Cultivation and autogamy were carried out at 27°C, and total RNA was extracted using TRIzol as described in reference 5. For Northern blots, 20 μg of total RNA were denatured and loaded on 1% agarose gels. Electrophoresis in 1× Tris-borate-EDTA (31), transfer to Hybond N+ membranes (GE Healthcare), and hybridization with 32P-labeled DNA probes were performed as described in reference 22.
Design of dedicated Megabase DNA microarrays.
A total of 439 PCR fragments from the Megabase chromosome were synthesized using plasmid templates from the library constructed for the Megabase sequencing project (40) and purified on customized G50 plates (Sigma). Among these probes, 218 were internal to single genes while 221 encompassed more that one gene. In total, 484 out of the 558 predicted Megabase genes (Community Annotation Project, version 1.47 [http://paramecium.cgm.cnrs-gif.fr/parawiki/Community_Annotation_Project]) were covered by at least one probe. A total of 42 PCR fragments external to the Megabase chromosome were also spotted. In particular, we used control probes from known autogamy-induced genes DNAPKcs (S. Malinsky, personal communication), PtSPO11 (6), NOWA2 (26), and NOWA2bis and D7 (E. Meyer, personal communication). A detailed description of the probes can be found in the GEO database (number GPL7296). All probes can also be visualized on the Megabase sequence using the GBrowse interface from ParameciumDB (1, 3). Each array was designed to contain three (sometimes six) replicates of each probe. The procedures used for spotting, hybridization, and data analysis are described in the supplemental material.
Gene inactivation by RNAi.
Gene inactivation was performed in P. tetraurelia strain 51 following the previously published “feeding” procedure (14, 26). Silencing media were prepared by inoculating a single colony of Escherichia coli HT115 (37) harboring the appropriate silencing plasmid into 1× WGP containing 0.1 mg/ml ampicillin and 0.01 mg/ml tetracycline. After overnight shaking at 37°C, the culture was diluted 10-fold into WGP with ampicillin. Following 6 to 8 h of shaking at 37°C, the culture was diluted 6- to 10-fold into the same medium and IPTG was added to a final concentration of 0.5 mM for overnight induction of double-stranded RNA (dsRNA) synthesis at 37°C. Silencing medium was supplemented with 0.8 mg/liter β-sitosterol before use.
Screening procedure.
All genes were silenced in at least three independent experiments. For those genes that exhibited RNAi phenotypes in this first screen, experiments were repeated 10 times on the average, either by transferring single cells to each silencing medium or by starting from small populations of cells.
To monitor RNAi phenotypes during vegetative growth, 6 to 24 cells were placed individually into ∼200 μl of freshly induced silencing medium. As a control, the same number of cells were transferred to standard Klebsiella medium or, alternatively, to control silencing medium containing induced E. coli harboring either the empty L4440 vector or ND7- or ICL7-silencing plasmids, which target nonessential genes (see supplemental material). The next day, each clone was replicated by transferring a single cell to 200 μl of fresh medium. The cells were counted in each microculture to evaluate their growth rate. For all replicate experiments, the global percentage of cell lines for which we observed phenotypic defects after a 48-hour incubation period in each silencing medium (shown in Table 1) was calculated, using the data obtained for 50 cell lines on average.
Table 1.
Screening for RNAi phenotypes for selected genes from clusters I and IIa
| Megabase gene | Human curation | Induction on Northern blot (fold induction) | Induction on microarrays (fold induction) | Predicted gene product | RNAi vegetative phenotype | Progeny of RNAi-treated cells |
|---|---|---|---|---|---|---|
| PTMB.08c | PTETG100018001 | Early (5.0) | Early (2.8) | Hypothetical protein | 100% lethality | NT |
| PTMB.96 | PTETG100036001 | NT | Early (1.8) | Microtubule-binding protein, putative | Normal growth | Normal |
| PTMB.104 | PTETG100063001 | NT | Late (20.5) | Hypothetical protein | Normal growthb | Normalb |
| PTMB.157c | PTETG100110001 | NT | Late (2.1) | Hypothetical protein | Normal growth | Normal |
| PTMB.170c | PTETG100119001 | Early (>8.7)c | Early (3.1) | Hypothetical protein | Normal growth | 19% survivors |
| PTMB.182 | PTETG100055001 | NT | Early (1.8) | Hypothetical protein with coiled-coil domains | Partial phenotyped (10-15%): small cells (do not feed),e slower growth, lethality | NT |
| PTMB.186c | PTETG100031001 | Early (5.7) | Early (1.9) | Cyclin-dependent kinase regulatory subunit, putative | 1 division less per day | 63% survivors |
| PTMB.220 | PTETG100042001 | Early (93.0) | Early (5.7) | Nucleic acid helicase, putative | Normal growth | 1% survivors |
| PTMB.236c | PTETG100023001 | NT | Early (3.2) | Microtubule-associated protein, putative | Normal growth | Normal |
| PTMB.238c | PTETG100024001 | NT | Early (5.1) | Hypothetical protein | Normal growth | Normal |
| PTMB.239c | PTETG100035001 | NT | Early (1.8) | Hypothetical protein with coiled-coil domains | Partial phenotyped (30-50%): arrested growth, lethality | NT |
| PTMB.285c | PTETG100068001 | Early (3.3) | Not significant (1.4) | Hypothetical protein | Normal growth | Normal |
| PTMB.422c | PTETG100014001 | Early (9.0) | Early (1.5) | Guanylate nucleotide binding protein, putative | 100% lethality | NT |
| PTMB.344c | PTETG100152001 | Early (3.1) | Early (2.5) | DNA mismatch repair protein Msh2, putative | Normal growth | Normal |
| PTMB.443c | PTETG100004001 | Late (>19.0)c | Late (27.1) | DNA ligase, putative | Normal growth | Normal |
NT, not tested.
Ohnolog may not be silenced.
Vegetative band was not visible on Northern blot; fold induction was calculated relative to the weakest visible band.
Only the indicated fraction of RNAi-treated cell lines showed the phenotype.
Cells were incubated in India ink-containing medium to visualize food vacuoles.
To observe survival of sexual progeny, cells grown for 12 to 15 vegetative divisions in silencing medium were starved for 3 days to induce autogamy. Then, ∼30 postautogamous cells were transferred individually to Klebsiella medium and each microculture was monitored for a few days to check cell survival and growth. Only cell lines exhibiting a normal fission rate were considered postautogamous survivors, while those in which cell lethality or slow growth was observed were counted as the progeny of RNAi-affected cells. For each of the three genes exhibiting phenotypes in sexual progeny, we present the data calculated for more than 350 postautogamous cells in all experiments taken together (Table 1).
Immunolabeling of P. tetraurelia cells.
Immunolabeling of cytoskeletal structures was performed as described in reference 19. Basically, permeabilized and fixed cells were incubated for 1 h in a 1:10 to 1:400 dilution of monoclonal antibody (MAb) 12G9, which was raised against a cytoskeleton preparation from Tetrahymena thermophila but labels the basal bodies and the fission line specifically during morphogenesis in dividing Paramecium cells (36). Cells were then incubated for 1 h in a 1:250 to 1:500 dilution of tetramethyl rhodamine isocyanate (TRITC)-conjugated goat anti-mouse secondary antibody (Jackson Immunoresearch Laboratories Inc., West Grove, PA) and washed twice in Tris-buffered saline with Tween–bovine serum albumin (TBST-BSA). For the staining of nuclei, DAPI (4′,6-diamidino-2-phenylindole; Sigma) was added to the last wash to a final concentration of 1 μg/ml. Cells were mounted in Citifluor (Citifluor Ltd., London) before observation with a Nikon or Zeiss epifluorescence microscope, using filters to distinguish TRITC and DAPI staining.
Complementation of a lethal CKS1 mutation in Saccharomyces cerevisiae.
PCR-amplified PTMB.186c cDNA was introduced downstream of the GAL1 promoter into the pYES2 vector carrying the URA3 gene (Invitrogen). Two consecutive internal stop codons, TAG-TAA, present in the Paramecium sequence (bp 151 to 156 starting from the ATG) were replaced by CAG-CAA codons encoding Gln in S. cerevisiae to give plasmid pRG186. The diploid strain Y23274 (BY4743 MAT a/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/lys2Δ0 MET15/met15Δ0 ura3Δ0/ura3Δ0 YBR135w::kanMX4/YBR135w) from EUROSCARF was transformed with pRG186, and transformants were selected on W0-Ura plates. Sporulation was induced at 28°C as described previously (33), and tetrads were dissected using a Singer MSM System micromanipulator. Individual spores were grown on YP Gal medium prior to spotting onto YP plates supplemented with 2% glucose (Glu) or 2% galactose (Gal), whenever indicated. G418 was used at a final concentration of 200 μg/ml.
Injection of GFP fusion transgenes.
Linearized plasmids carrying green fluorescent protein (GFP) fusion transgenes (see the supplemental material) were microinjected into the MAC of vegetative 51 cells, as described previously (26). Briefly, Paramecium cells were microinjected in mineral water (Volvic, France) or Dryl solution containing 0.2% bovine serum albumin, under an oil film (Nujol), while they were visualized with a phase-contrast inverted microscope. GFP fluorescence was observed using a Zeiss or Leica microscope.
RESULTS AND DISCUSSION
Identification of a set of 15 Megabase genes specifically induced during autogamy.
For the functional analysis of Paramecium genes that exhibit variable expression levels throughout the sexual cycle, we focused our pilot study on the Megabase chromosome, the largest P. tetraurelia somatic chromosome (40), designated scaffold_1 in the assembly of the whole-genome sequence (5). Automatic annotation of the genome revealed a uniform gene distribution along MAC chromosomes (5), and the currently annotated Megabase genes represent around 1% of the total number of genes in the MAC genome (Community Annotation Project, version 1.47). To follow gene expression during sexual events, we focused on autogamy because it allows the preparation of large amounts of total RNA from large-scale cultures, even though the synchrony of cells is not perfect (see Fig. S1A in the supplemental material).
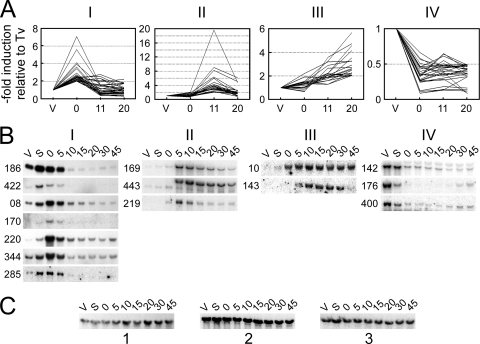
In a first approach, total RNA was extracted from vegetative d4-2 cells and during autogamy, at three characteristic time points with respect to the timing of IES excision (see Fig. S1 in the supplemental material). Fluorescently labeled cDNAs from each time point were hybridized to dedicated arrays carrying PCR fragments covering most Megabase genes (see Materials and Methods). Statistical analysis of microarrays (see the supplemental material) revealed four major transcription profiles that showed significant variations (at least 2-fold) throughout autogamy (Fig. 1A; see Tables S2 to S5 in the supplemental material): an early induction peak (cluster I), a late induction peak (cluster II), gradual induction (cluster III), and repression (cluster IV). This represented a total of 104 probes out of 439, the remaining ones showing only small variations in their transcription profiles or no specific expression pattern.
Fig. 1.
Four characteristic gene expression patterns during autogamy in P. tetraurelia. (A) Graphic representation of the four major expression profiles identified during the analysis of Megabase microarrays. For each probe, the graph shows the variations of the average normalized microarray hybridization signal obtained throughout autogamy for stock d4-2 (0, 11, and 20, time in hours), relative to the value calculated for the same probe for vegetative cells (V or Tv). At time zero (T0), 50% of cells had fragmented old MACs and amplification of germ line DNA had started in the developing new MACs but no products of IES excision were detected (see Fig. S1 in the supplemental material). At T11, double-strand breaks were readily detected at IES ends but only a few excised molecules were observed. At T20, IES elimination from chromosomes and accumulation of excised circles had become conspicuous. Probes with the largest induction or repression factors were selected within each cluster by applying the following filters: T0/TV > 2 for cluster I (30 probes), T11/TV > 2 for cluster II (23 probes), T20/TV > 2 for cluster III (20 probes), and T0/TV < 0.5 for cluster IV (31 probes). The full data set can be found in Tables S2 (cluster I), S3 (cluster II), S4 (cluster III), and S5 (cluster IV) in the supplemental material. (B) Validation of clusters by Northern blot hybridization of total RNA from stock 51 during autogamy (V, vegetative cells; S, starved cells; 0 to 45, autogamy time points in hours; see Fig. S2A in the supplemental material). Three identical blots were used in parallel for the successive hybridization of individual 32P-labeled gene probes. (PTMB gene numbers are shown on the left of each panel. For clarity, gene orientation relative to Megabase sequence is not indicated.) Details for all hybridization probes are listed in Table S6 in the supplemental material. (C) Loading control for the Northern blots shown in panel B using a 32P-labeled 17S ribosomal DNA probe. Blot 1 was used for PTMB.186c, PTMB.422c, PTMB.08c, PTMB.170c, PTMB.220, and PTMB.344c. Blot 2 was used for PTMB.285c, PTMB.169c, PTMB.443c, PTMB.143c, PTMB.142c, PTMB.176c, and PTMB.400c. Blot 3 was used for PTMB.219 and PTMB.10c.
Almost 50% of the probes spotted on our custom arrays encompassed more than one gene, making it difficult to provide directly an exhaustive list of individual genes that followed the profiles shown in Fig. 1A. To identify differentially expressed genes, we therefore used two complementary strategies. We first hybridized Northern blots of total RNAs extracted during an autogamy time course of strain 51, with 32P-labeled probes from genes picked from each expression cluster (Fig. 1B). In parallel, we used whole-genome oligonucleotide microarrays (2) hybridized with cDNAs from two other independent autogamy time course experiments performed with strain 51 (see Fig. S2B and C in the supplemental material). We extracted the data obtained for all genes covered by the probes listed in clusters I to IV and selected those that showed significant variations in their expression levels and followed the transcription profile of their respective expression cluster (see Tables S7 to S10 in the supplemental material).
For cluster I, whole-genome microarrays identified 14 genes exhibiting an early peak of activation (see Tables S2 and S7 in the supplemental material), 6 of which were also confirmed by Northern blot hybridization. One additional gene (PTMB.285c), with an early activation peak detected on Northern blots, was not recovered from microarray analysis because its expression level varied only slightly. It was nevertheless added to our set of validated genes: taken together, 15 Megabase genes were confirmed to follow the cluster I expression profile. For cluster II, 10 genes were found to be activated as a late peak according to whole-genome microarrays (see Tables S3 and S8 in the supplemental material), and this expression profile was confirmed on Northern blots for 3 of them. Cluster III includes both early- and late-induced genes, whose mRNA levels remain high at later time points following induction. Whole-genome microarrays revealed a significant increase in mRNA levels throughout autogamy for 18 genes from this cluster (Tables S4 and S9), among which 2 were confirmed by Northern blot hybridization (Fig. 1B). Finally, the expression profile of cluster IV was confirmed for 32 genes, which are repressed early according to whole-genome microarrays (Tables S5 and S10). Interestingly, some genes within this subset participate in the metabolism of stable RNAs or in mRNA translation, both of which are expected to be turned down in starved cells.
Induction of a gene during autogamy may reflect the participation of its encoded protein in the progression of sexual processes. For further functional analysis, we picked 12 early-induced genes from our confirmed cluster I set. We added gene PTMB.443c from later-induced cluster II, because it was annotated as a DNA ligase gene, and two genes covered by cluster I probes (PTMB.104 and PTMB.157c), which were activated at later time points according to whole-genome microarrays. This set of 15 specifically induced genes was used to systematically screen for essential functions during vegetative growth and during autogamy.
Screening for essential induced genes.
Expression of each gene was silenced individually by feeding Paramecium cultures dsRNA-overproducing bacteria to trigger RNA interference (14). The effect of each RNAi was first examined during vegetative growth (see Materials and Methods). Paramecium cells were grown for 12 to 15 vegetative divisions in the presence of bacteria overproducing dsRNA homologous to each target gene. The phenotypes examined were cell shape, growth rate, and survival (Table 1). Out of the 15 genes tested, 2 were found to be essential for vegetative growth (PTMB.08c and PTMB.422c) and 2 gave a partial phenotype, leading to growth arrest and eventually to cell death in a fraction of the population (PTMB.182 and PTMB.239c). Our experimental procedure did not allow us to study the effect of silencing these four genes on the outcome of sexual processes, and future work should address this question. The remaining 11 genes were silenced during autogamy, and the survival of sexual progeny was monitored following transfer of autogamous cells to standard rich medium (Table 1). RNAi against PTMB.170c and PTMB.220 gave rise to only 19% and 1% surviving cells with a normal fission rate in the progeny, respectively, indicative of an essential function of these two genes during autogamy. Although PTMB.170c does not have an ohnolog from the last whole genome duplication (WGD), we found three paralogs originating from older WGDs, which probably were not silenced by our RNAi constructs (Table S11). This might explain the residual 19% survival observed in the postautogamous progeny of silenced cells (Table 1). A partial and variable phenotype was observed for PTMB.186c, the silencing of which led to 63% wild-type postautogamous progeny.
All 15 genes analyzed here had been selected for functional analysis because their expression levels were found to increase specifically during autogamy. In our screen, however, the silencing of only 7 resulted in significant cell death, either during normal vegetative growth or following autogamy. It is, of course, possible that a few other genes were not completely silenced by the RNAi method used in our study and, therefore, that no phenotype was revealed in our screen. However, this relatively small number of established essential genes might also be explained in part by the three rounds of WGD that shaped the P. tetraurelia genome during evolution (5). For the genes examined in this study (except for PTMB.104), the two paralogs from the last WGD (also called ohnologs), whenever still present in the genome, generally exhibit a high level of sequence identity and were probably cosilenced with the same silencing construct (see Table S11 in the supplemental material). However, paralogs from previous WGDs or from other duplication events are often more divergent: they may have escaped RNAi and still produce proteins of redundant function. This might be the case for genes PTMB.96, PTMB.157, PTMB.236c, PTMB.285c, and PTMB.443c.
Six genes (PTMB.08c, PTMB.422c, PTMB.186c, PTMB.344c, PTMB.170c, and PTMB.220) were chosen for further analyses and are described in more detail below.
The PTMB.08c and PTMB.422c gene products are required for vegetative growth.
The expression of PTMB.08c and PTMB.422c is not restricted to autogamy, since their respective mRNAs can be detected in vegetative cells (Fig. 1B, panel I). In our initial screen, 100% lethality was observed in vegetative populations less than 48 h after transfer to PTMB.08c- or PTMB.422c-silencing medium (Table 1). Northern blot hybridization confirmed that after ∼24 h of incubation, the amount of each mRNA was ∼70% lower in silenced cells than in control cells (see Fig. S3A in the supplemental material). To further characterize their phenotype, cells submitted to each RNAi were monitored during the first 10 h of incubation in silencing medium.
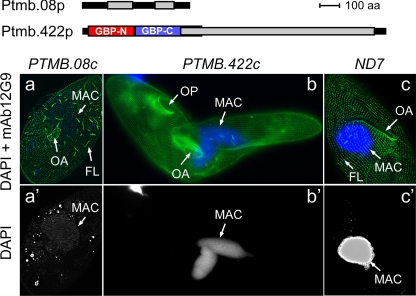
(i) PTMB.08c is essential for the maintenance of macronuclear integrity.
PTMB.08c encodes a hypothetical protein of 566 amino acids (aa) with no known homolog in other organisms and harbors two predicted coiled-coil domains (Fig. 2). After 4 h in PTMB.08c-silencing medium, DNA started to disappear from the MAC even before cells divided, as judged by a very faint DAPI staining of residual MAC structures (Fig. 2a and a′) relative to that of control cells (Fig. 2c and c′). Following a 10-hour incubation, around 40 cells were fixed and stained with Protargol silver protein staining reagent to document both the loss of MAC DNA and the absence of a nuclear protein scaffold (18). We observed that the whole MAC structure had disappeared from 46% of silenced cells while 54% still contained apparently normal MACs (see Fig. S4 in the supplemental material). The MICs were not examined in this experiment.
Fig. 2.
Vegetative RNAi phenotypes of PTMB.08c- and PTMB.422c-silenced cells. In panels a to c, fixed cells were labeled by MAb 12G9-TRITC (green) and DAPI (blue) to reveal basal bodies and nuclei, respectively. Panels a′ to c′ show DAPI staining of the same cells. The structure of Ptmb.08p and Ptmb.422p putative proteins is displayed on top. Predicted coiled coils are represented by gray boxes, and the GBP-like N- and C-terminal domains within Ptmb.422p are drawn as red and blue boxes, respectively. (a and a′) Silencing of PTMB.08c interferes with MAC integrity. The cell shown was fixed during the first division following transfer to PTMB.08c silencing medium. It has a normal newly formed oral apparatus (OA) and the beginning of the fission line (FL). The residual macronucleus (MAC) is visible as a faint round shape. The exact nature of the surrounding small dots detected in the cytoplasm using the DAPI filter (a′) has not been clearly established. These could either be dispersed DNA-containing structures or artifactual fluorescence signals due to overexposure. (b and b′) Silencing of PTMB.422c impairs cytokinesis. In the cell shown here, the two divided MACs are located in the anterior component of the boomerang structure. (c and c′) Control ND7-silenced cell at a division stage comparable to the one shown in panels a and a′. Note the bright blue labeling of the MAC. ND7 is a nonessential gene involved in trichocyst discharge, and its silencing has no effect on cell division (14).
Our data suggest that the putative Ptmb.08p protein is involved in the control of DNA content and integrity of the vegetative MAC. Future studies, including the determination of Ptmb.08p intracellular localization and a close inspection of the fate of all types of nuclei in RNAi-treated cells, will allow a better understanding of the biological significance of PTMB.08c overexpression during autogamy. One simple hypothesis would be that early de novo synthesis of Ptmb.08p during autogamy is required to maintain selectively the integrity of new MICs or MACs, while other nuclei get degraded. Indeed, several new types of nuclei are formed during sexual processes (haploid products of MIC meiosis, gametic nuclei, zygotic nucleus, and new MICs and MACs). However, while new MICs and MACs differentiate, selective degradation of seven out of eight meiotic products is observed at early stages and, under prolonged starvation, old MAC fragments undergo autolysis at later stages (8).
(ii) Ptmb.422p: a putative GTP-binding protein involved in cell division.
The N terminus of the 1,602-aa Ptmb.422p protein has a high degree of similarity to human guanylate-binding proteins GBP1 to GBP5 (see Fig. S5 in the supplemental material). Its long C-terminal extension is predicted to adopt a coiled-coil structure (Fig. 2). The function of GBP-related proteins, which are not conserved in yeast, has not been established clearly, but they are related to dynamins, a superfamily of proteins shown to participate in various cellular processes, such as the budding of transport vesicles, the division of organelles, cytokinesis, or pathogen resistance (16, 28).
PTMB.422c has a close ohnolog from the latest WGD, with 92% identity at the nucleic acid level (96% identity at the amino acid level), which should be silenced efficiently in our experiments (see Table S11 in the supplemental material). A strong vegetative phenotype was readily observed at the first cell division following cell transfer to PTMB.422c silencing medium. Morphogenesis started normally, but daughter cells failed to separate and exhibited a characteristic “boomerang” phenotype (Fig. 2b). After 8 h of RNAi, Protargol silver staining revealed that up to 46% of the PTMB.422c-silenced population exhibited the “boomerang” phenotype (data not shown), and a strong defect in MAC segregation was observed in 51% of cells blocked in cytokinesis: 27% had both MACs in the anterior prospective daughter cell, 2% had both MACs in the posterior prospective daughter cell, and 22% presented two divided MACs blocked in the middle of the boomerang structure. Both daughter cells were sometimes able to enter the next cell cycle and divide again without separation, resulting in the formation of “monsters” (not shown) and eventually in cell death.
Expression of PTMB.422c is essential for cell morphogenesis and/or cytokinesis during vegetative growth. As proposed for GTPases from the dynamin superfamily, it could be involved in membrane tubulation or fission (28). Because the silencing of this gene could not be performed during autogamy, we can only speculate on the significance of the increase in PTMB.422c mRNA levels during sexual processes. It could for instance be required for karyonidal division, during which the two developing MACs originating from the zygotic nucleus are segregated to the two daughter cells. A study of the localization of Ptmb.422p in Paramecium should provide more insight into the function of this protein during autogamy.
The product of gene PTMB.186c complements a null cks1 mutation in S. cerevisiae.
Sequence analysis reveals that PTMB.186c encodes a protein homologous to yeast cyclin-dependent kinase regulatory subunit Cks1p (see Fig. S6 in the supplemental material), which is involved in the control of cell cycle progression through its physical association with cyclin-dependent kinases (CDKs) and the stabilization of CDK-cyclin complexes (27).
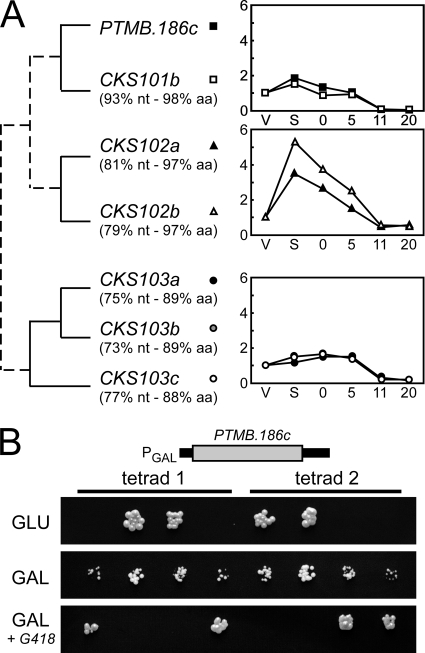
PTMB.186c has 1 ohnolog from the latest WGD. In addition, five other paralogs were found at other genomic loci not related by any obvious WGD relationship (Fig. 3A). The proteins encoded by all seven CKS1-like genes are highly conserved at the amino acid level, including the residues involved in CDK binding in other organisms (see Fig. S6 in the supplemental material). Reciprocally, 13 putative CDK proteins are encoded by the Paramecium genome (not shown), some of which may interact with Cks (11), and at least one Paramecium CDK was shown by others to bind in vitro to the Schizosaccharomyces pombe Cks homolog (41). Thus, the function of Paramecium Cks proteins is likely to be conserved. To confirm this hypothesis, we expressed P. tetraurelia Ptmb.186p under the control of the yeast GAL1 promoter in a diploid S. cerevisiae strain heterozygous for a null cks1 mutation. Following sporulation, we observed that Ptmb.186p, like its human homologs (30), could complement a lethal cks1 deletion in yeast (Fig. 3B).
Fig. 3.
PTMB.186c encodes a homolog of cyclin-dependent kinase regulatory subunit Cks1p. (A) CKS-like genes in P. tetraurelia and their transcription profiles during autogamy. Paralogs of gene PTMB.186c (CKS101a) are CKS101b (PTETG800015001), CKS102a (GSPATG00005766001), CKS102b (GSPATG00007882001), CKS103a (GSPATG00004184001), CKS103b (GSPATG00010146001), and CKS103c (GSPATG00024666001). The percentages of nucleotide sequence identity (nt) and amino acid identity (aa) of the predicted gene product relative to PTMB.186c are indicated below each paralog. WGD paralogs are linked by solid lines; other evolutionary relationships are indicated by dotted lines. Transcription profiles were determined following hybridization of whole genome microarrays with cDNAs from two independent autogamy time course experiments performed with strain 51 (see the supplemental material). For each gene, mean hybridization signals obtained at each time point throughout autogamy were normalized relative to the value calculated for vegetative cells. (B) Complementation of a null cks1 mutation by the product of PTMB.186c in S. cerevisiae. A diploid S. cerevisiae strain carrying both a wild-type CKS1 and a null cks1::kanMX4 allele was made by inserting a G418 resistance cassette in the CKS1 coding sequence. It was transformed with a plasmid carrying the Paramecium PTMB.186c gene under the control of the S. cerevisiae GAL1 promoter, which is strongly induced by galactose (GAL) and repressed by glucose (GLU). Therefore, growing cells on galactose or glucose controls the expression of the P. tetraurelia CKS1 homolog. The transformed diploid strain was sporulated in order to segregate each allele into haploid spores. From each tetrad, only spores carrying a wild-type CKS1 gene were viable under repressive conditions (GLU). Under inducing conditions (GAL), normal-sized colonies were obtained from spores harboring a wild-type endogenous CKS1 allele, and smaller-sized colonies were obtained if a null background was complemented by exogenous PTMB.186c. In the latter case, the presence of the cks1::kanMX4 allele was confirmed by spotting on YP GAL + G418 medium, selective for the antibiotic resistance phenotype.
Although not distinguishable by Northern blot hybridization, three different gene expression profiles are revealed within the Paramecium CKS multigene family by the analysis of whole-genome microarrays (Fig. 3A). PTMB.186c and its close ohnolog CKS101b are only modestly induced in starved cells, while CKS102a and CKS102b exhibit a larger induction peak. Based on nucleotide sequence comparisons, CKS102a and CKS102b would be only partially knocked down by our PTMB.186c RNAi construct (see Table S11 in the supplemental material) and residual levels of Cks proteins may account for the 63% wild-type postautogamous survivors obtained in the progeny of PTMB.186c-silenced cells (Table 1).
The accumulation of some Cks-encoding mRNAs during autogamy, and the prominent decrease in all CKS transcript levels observed under prolonged starvation might reflect the involvement of Cks homologs in controlling the progression through the sexual cycle. To investigate this further, it would be informative to silence, independently or together, the three families of Paramecium CKS genes shown in Fig. 3A and follow the successive stages of autogamy in the different knockdowns.
The products of PTMB.344c, PTMB.170c, and PTMB.220 localize in different nuclei in vegetative cells and during sexual processes.
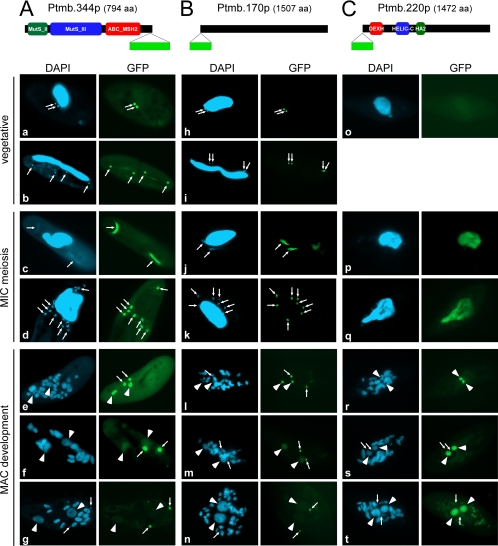
The products of PTMB.170c, PTMB.220, and PTMB.344c were predicted to have a nuclear localization using the Nucpred package (12). GFP fusions were constructed for each protein, and their localization was monitored in vegetative cells and during autogamy (Fig. 4). All three proteins were confirmed to be nuclear but did not localize to the same nuclei.
Fig. 4.
Localization of GFP fusion proteins for Ptmb.344p, Ptmb.170p, and Ptmb.220p. Predicted protein domains within Ptmb.344p and Ptmb.220p are indicated as colored boxes. The position of the GFP insertion is drawn as a light green box. (a, h, and o) Vegetative cells. (b and i) Dividing vegetative cells. All other panels show successive stages of autogamy. (c and j) First meiotic division. (d and k) Cells with eight haploid nuclei resulting from meiosis II. (p and q) Meiotic cells. (e, l, and r) Early MAC development. (f, m, and s) Intermediate stage of MAC development. (g, n, and t) Late MAC development. In all panels, white arrows point at MICs (some were omitted when MICs were not clearly distinguishable by DAPI staining), and arrowheads indicate new MACs. (A) Ptmb.344p-GFP fusion localizes to MICs during vegetative growth and autogamy. (B) GFP-Ptmb.170p fusion localizes to MICs during vegetative growth and autogamy. (C) GFP-Ptmb.220p fusion localizes to old and then new MACs specifically during autogamy.
(i) PTMB.170c and PTMB.344c encode micronuclear proteins.
PTMB.170c encodes a hypothetical protein of 1,507 aa with no significant homology to other known proteins except to one putative ortholog in another ciliate, Tetrahymena thermophila (BLASTP E value = 1e−7). In contrast, sequence analysis indicates that the PTMB.344c product is the sole putative ortholog of the DNA mismatch repair protein Msh2 encoded in the P. tetraurelia genome (see Fig. S7 in the supplemental material): it shares 32% identity with S. cerevisiae Msh2p (BLASTP E value = 1e−112) and 34% with its human ortholog (BLASTP E value = 1e−138). PTMB.344c is transcribed in vegetative cells, whereas low transcription levels do not allow the detection of a vegetative transcript of PTMB.170c by Northern blot hybridization. During autogamy, both genes clearly show the same early induction pattern (Fig. 1B). We studied the localization of their corresponding protein products by injecting GFP fusion transgenes into the MAC of vegetative cells and following the GFP fluorescence during vegetative growth and throughout autogamy (Fig. 4). The Ptmb.344p-GFP C-terminal fusion was expressed under the control of the endogenous PTMB.344c promoter. To increase the fluorescence yield, the GFP-Ptmb.170p N-terminal fusion was expressed from a constitutive Paramecium promoter: injected cells exhibited a normal vegetative growth phenotype and no dramatic effect was observed on the survival of postautogamous progeny (not shown). Both GFP fusion proteins were found in MICs during vegetative growth and meiosis. During MAC development, fluorescence was also found in early anlagen (Fig. 4, panels e and l), but it disappeared from new MACs at later stages (panels f and g and m and n). GFP fluorescence merged with DAPI-stained DNA for Ptmb.170p, except at meiosis I (panel j), while it consistently occupied a larger space for Ptmb.344p (compare panels a to g with h to n). In addition, GFP-Ptmb344p also appeared to stain the mitotic fission furrow during MIC division (panel b).
The localization of Ptmb.344p and its homology to Msh2 proteins indicate that it might play a MIC-specific role, and its early upregulation during autogamy suggests that it is involved in meiosis. A participation of the mismatch repair system during meiotic recombination has been documented in other organisms (reviewed in references 32 and 38). Msh2 proteins participate in the repair of isolated mismatched base pairs and favor gene conversion (35). They also prevent homologous recombination between heavily mismatched DNA duplexes and therefore contribute to maintaining the integrity of repeated DNA sequences (e.g., 20). In our experiments, the silencing of PTMB.344c gave no phenotype during vegetative growth and no phenotype following autogamy (Table 1). We cannot exclude that the residual amounts of full-length PTMB.344c mRNA detected in silenced cells (see Fig. S3B in the supplemental material) account for this lack of effect. Furthermore, we found nine more distant MutS-like genes in the P. tetraurelia genome, some of which are upregulated during autogamy (not shown) and might complement a PTMB.344c knockdown. Additional experiments are clearly required to investigate the function of Msh2-related proteins during meiosis in Paramecium.
Nevertheless, based on the MIC-restricted localization of the GFP fusion protein, we can propose that Ptmb.170p is involved in some essential micronuclear function that would be revealed following autogamy. DAPI staining or immunolabeling of cells with anti-γ-tubulin antibodies failed to detect any significant cytological defect in the development of new MICs and MACs in autogamous cells (see Fig. S8 in the supplemental material). We cannot rule out, however, that a depletion in Ptmb.170p induces MIC genomic or chromosomal defects, during either vegetative growth or meiosis, that would result in the formation of a nonfunctional new MAC and cause cell death in sexual progeny.
(ii) Ptmb.220p, a putative DEAH-box helicase, localizes to old and then to new MAC.
PTMB.220 has one ohnolog from the last WGD (90% nucleotide [nt] identity, 93% aa identity), which should be silenced efficiently in our RNAi experiments (see Table S11 and Fig. S3C in the supplemental material). The corresponding 1,472-aa protein, Ptmb.220p, harbors conserved domains that are characteristic of DEAH-box helicases in its N-terminal region (Fig. 4; see Fig. S9 in the supplemental material). In contrast to the findings for PTMB.170c and PTMB.344c, no fluorescence was observed in vegetative cells injected with a PTMB.220-GFP fusion transgene transcribed under the control of the endogenous PTMB.220 promoter (Fig. 4C, panel o). During autogamy, the fusion protein was first detected in the parental MAC at the onset of meiosis (panel p) and during MAC fragmentation (panels p and q). At later stages, bright staining of the new MACs became conspicuous, whereas fluorescence disappeared from the old MAC (panels r to t). No staining of the MICs was detected at any stage.
The closest Ptmb.220p homolog is the Ema1p RNA helicase recently shown to be involved in programmed DNA elimination during MAC development in Tetrahymena (4). Ptmb.220p and Ema1p follow the same localization pattern during sexual processes. In addition, Ema1p interacts with the Tetrahymena Piwi protein Twi1p, together with two GW-repeat proteins that were also found to promote genome rearrangements (7). In P. tetraurelia, the GW-rich protein Nowa1p exhibits the same localization as Ptmb.220p: it accumulates in the old MAC during meiosis and is transferred to the developing new MACs at the time DNA rearrangements take place (26). As for PTMB.220, the silencing of NOWA1 resulted in 100% lethality in postautogamous progeny and Nowa1p was shown to be essential for the noncoding RNA-mediated control of DNA elimination. Taken together, these observations suggest that Ptmb.220p is also involved in the trans-nuclear cross talk that regulates genome rearrangements in Paramecium, possibly through an interaction with Nowa1p and/or noncoding RNAs.
Conclusion.
The 484 genes (out of 558) covered by the Megabase microarray probes used in this study represent 1.2% of the total number of genes that are currently annotated in the P. tetraurelia genome. Our search for specifically expressed genes during autogamy revealed the existence of two successive peaks of transcription, which were confirmed at a genome-wide scale by an independent microarray analysis (2). The earliest peak (expression cluster I) coincides with meiosis and the beginning of old MAC fragmentation and includes at least 15 Megabase genes. A second peak, including at least 10 Megabase genes, was detected 11 h later, after IES excision had started in the developing new MACs (expression cluster II). Two different explanations may be proposed for this second peak of transcription and need to be investigated further: either the switch-on of late genes within the old MAC or the start of transcription of newly rearranged genes from the anlagen.
Specific induction of a gene during autogamy in Paramecium is indicative of a putative role during this process. Indeed, previously described genes involved in the control or the catalysis of programmed DNA rearrangements in the new MAC were shown to be induced during sexual reproduction (6, 10, 23–26). Our functional analysis, performed on a subset of 15 genes from expression clusters I and II, revealed that one gene (PTMB.186c), likely to be involved in the control of the cell cycle, is required for a normal outcome of autogamy. Furthermore, we uncovered an essential function for six other genes (PTMB.08c, PTMB.170c, PTMB.182, PTMB.220, PTMB.239c, and PTMB.422c). Closer observation of the phenotypes of cells that have been inactivated for expression of each gene may lead to the identification of new essential functions during autogamy, such as membrane trafficking or nuclear functions (either in the MIC or in the MAC). In conclusion, our work indicates that the study of the transcriptome of P. tetraurelia during autogamy can pave the way for the identification of essential genes during meiosis and nuclear reorganization.
Supplementary Material
ACKNOWLEDGMENTS
We thank Laurence Amar, Delphine Gogendeau, France Koll, Anne Le Mouël, Sophie Malinsky, and Mariusz Nowacki for the gift of PCR fragments spotted as controls on our dedicated microarrays. We are also grateful to Sophie Malinsky and Eric Meyer for the kind gift of GFP plasmids and to Catherine Klotz for antibodies. Thanks to all members of the Bétermier and Cohen labs for very helpful discussions and to the staff of the GODMAP facility for their technical support.
This work was supported by the Polish Ministry of Sciences and Higher Education (grants 3 PO4A 006 25 and N303 075 32/2520), the CNRS (ATIP-Plus grant) and the Agence Nationale pour la Recherche (grant BLAN08-3_310945). J.K.N., R.G., M.J., K.M., C.J.H., M.Z., and M.B. received exchange grants from the CNRS and the Polish Academy of Sciences in the framework of the CNRS GDRE Paramecium Genomics.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 21 January 2011.
REFERENCES
- 1. Arnaiz O., Cain S., Cohen J., Sperling L. 2007. ParameciumDB: a community resource that integrates the Paramecium tetraurelia genome sequence with genetic data. Nucleic Acids Res. 35:D439–D444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arnaiz O., et al. 2010. Gene expression in a paleopolyploid: a transcriptome resource for the ciliate Paramecium tetraurelia. BMC Genomics 11:547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arnaiz O., Sperling L. 2011. ParameciumDB in 2011: new tools and new data for functional and comparative genomics of the model ciliate Paramecium tetraurelia. Nucleic Acids Res. 39:D632–D636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aronica L., et al. 2008. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 22:2228–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aury J. M., et al. 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444:171–178 [DOI] [PubMed] [Google Scholar]
- 6. Baudry C., et al. 2009. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev. 23:2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bednenko J., et al. 2009. Two GW repeat proteins interact with Tetrahymena thermophila argonaute and promote genome rearrangement. Mol. Cell. Biol. 29:5020–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berger J. D. 1974. Selective autolysis of nuclei as a source of DNA precursors in Paramecium aurelia exconjugants. J. Protozool. 21:145–152 [DOI] [PubMed] [Google Scholar]
- 9. Bétermier M. 2004. Large-scale genome remodelling by the developmentally programmed elimination of germ line sequences in the ciliate Paramecium. Res. Microbiol. 155:399–408 [DOI] [PubMed] [Google Scholar]
- 10. Bouhouche K., Gout J. F., Kapusta A., Bétermier M., Meyer E. Functional specialization of Piwi proteins in Paramecium tetraurelia from post-transcriptional gene silencing to genome remodelling. Nucleic Acids Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bourne Y., et al. 1996. Crystal structure and mutational analysis of the human CDK2 kinase complex with cell cycle-regulatory protein CksHs1. Cell 84:863–874 [DOI] [PubMed] [Google Scholar]
- 12. Brameier M., Krings A., MacCallum R. M. 2007. NucPred—predicting nuclear localization of proteins. Bioinformatics 23:1159–1160 [DOI] [PubMed] [Google Scholar]
- 13. Duharcourt S., Lepere G., Meyer E. 2009. Developmental genome rearrangements in ciliates: a natural genomic subtraction mediated by non-coding transcripts. Trends Genet. 25:344–350 [DOI] [PubMed] [Google Scholar]
- 14. Galvani A., Sperling L. 2002. RNA interference by feeding in Paramecium. Trends Genet. 18:11–12 [DOI] [PubMed] [Google Scholar]
- 15. Gratias A., Bétermier M. 2003. Processing of double-strand breaks is involved in the precise excision of Paramecium IESs. Mol. Cell. Biol. 23:7152–7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heymann J. A., Hinshaw J. E. 2009. Dynamins at a glance. J. Cell Sci. 122:3427–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jahn C. L., Klobutcher L. A. 2002. Genome remodeling in ciliated protozoa. Annu. Rev. Microbiol. 56:489–520 [DOI] [PubMed] [Google Scholar]
- 18. Jerka-Dziadosz M. 1985. Mirror-image configuration of the cortical pattern causes modifications in propagation of microtubular structures in the hypotrich ciliate Paraurostyla weissei. Rouxs Arch. Dev. Biol. 194:311–324 [Google Scholar]
- 19. Jerka-Dziadosz M., et al. 1995. Cellular polarity in ciliates: persistence of global polarity in a disorganized mutant of Tetrahymena thermophila that disrupts cytoskeletal organization. Dev. Biol. 169:644–661 [DOI] [PubMed] [Google Scholar]
- 20. Kolas N. K., et al. 2005. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J. Cell Biol. 171:447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le Mouël A., Butler A., Caron F., Meyer E. 2003. Developmentally regulated chromosome fragmentation linked to imprecise elimination of repeated sequences in Paramecium. Eukaryot. Cell 2:1076–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lepère G., Bétermier M., Meyer E., Duharcourt S. 2008. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 22:1501–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lepère G., et al. 2009. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic Acids Res. 37:903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matsuda A., Forney J. D. 2006. The SUMO pathway is developmentally regulated and required for programmed DNA elimination in Paramecium tetraurelia. Eukaryot. Cell 5:806–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsuda A., Shieh A. W., Chalker D. L., Forney J. D. 2010. The conjugation-specific Die5 protein is required for development of the somatic nucleus in both Paramecium and Tetrahymena. Eukaryot. Cell 9:1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nowacki M., Zagorski-Ostoja W., Meyer E. 2005. Nowa1p and Nowa2p: novel putative RNA binding proteins involved in trans-nuclear crosstalk in Paramecium tetraurelia. Curr. Biol. 15:1616–1628 [DOI] [PubMed] [Google Scholar]
- 27. Pines J. 1996. Cell cycle: reaching for a role for the Cks proteins. Curr. Biol. 6:1399–1402 [DOI] [PubMed] [Google Scholar]
- 28. Praefcke G. J., McMahon H. T. 2004. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5:133–147 [DOI] [PubMed] [Google Scholar]
- 29. Prescott D. M. 1994. The DNA of ciliated protozoa. Microbiol. Rev. 58:233–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Richardson H. E., Stueland C. S., Thomas J., Russell P., Reed S. I. 1990. Human cDNAs encoding homologs of the small p34Cdc28/Cdc2-associated protein of Saccharomyces cerevisiae and Schizosaccharomyces pombe. Genes Dev. 4:1332–1344 [DOI] [PubMed] [Google Scholar]
- 31. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32. Schofield M. J., Hsieh P. 2003. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 57:579–608 [DOI] [PubMed] [Google Scholar]
- 33. Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21 [DOI] [PubMed] [Google Scholar]
- 34. Sonneborn T. M. 1974. Paramecium aurelia, p. 469–594 In King R. C. (ed.), Handbook of genetics: plants, plant viruses and protists, vol. 2 Plenum Press, New York, NY [Google Scholar]
- 35. Stone J. E., Petes T. D. 2006. Analysis of the proteins involved in the in vivo repair of base-base mismatches and four-base loops formed during meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 173:1223–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Strzyzewska-Jowko I., Jerka-Dziadosz M., Frankel J. 2003. Effect of alteration in the global body plan on the deployment of morphogenesis-related protein epitopes labeled by the monoclonal antibody 12G9 in Tetrahymena thermophila. Protist 154:71–90 [DOI] [PubMed] [Google Scholar]
- 37. Timmons L., Court D. L., Fire A. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263:103–112 [DOI] [PubMed] [Google Scholar]
- 38. Waldman A. S. 2008. Ensuring the fidelity of recombination in mammalian chromosomes. Bioessays 30:1163–1171 [DOI] [PubMed] [Google Scholar]
- 39. Yao M. C., Duharcourt S., Chalker D. L. 2002. Genome-wide rearrangements of DNA in ciliates, p. 730–758 In Craig N. L., Craigie R., Gellert M., Lambowitz A. M. (ed.), Mobile DNA II. ASM Press, Washington, DC [Google Scholar]
- 40. Zagulski M., et al. 2004. High coding density on the largest Paramecium tetraurelia somatic chromosome. Curr. Biol. 14:1397–1404 [DOI] [PubMed] [Google Scholar]
- 41. Zhang H., Berger J. D. 1999. A novel member of the cyclin-dependent kinase family in Paramecium tetraurelia. J. Eukaryot. Microbiol. 46:482–491 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.