Abstract
The ammonium permease Mep2 induces a switch from unicellular yeast to filamentous growth in response to nitrogen limitation in Saccharomyces cerevisiae and Candida albicans. In S. cerevisiae, the function of Mep2 and other ammonium permeases depends on the protein kinase Npr1. Mutants lacking NPR1 cannot grow on low concentrations of ammonium and do not filament under limiting nitrogen conditions. A G349C mutation in Mep2 renders the protein independent of Npr1 and results in increased ammonium transport and hyperfilamentous growth, suggesting that the signaling activity of Mep2 directly correlates with its ammonium transport activity. In this study, we investigated the role of Npr1 in ammonium transport and Mep2-mediated filamentation in C. albicans. We found that the two ammonium permeases Mep1 and Mep2 of C. albicans differ in their dependency on Npr1. While Mep1 could function well in the absence of the Npr1 kinase, ammonium transport by Mep2 was virtually abolished in npr1Δ mutants. However, the dependence of Mep2 activity on Npr1 was relieved at higher temperatures (37°C), and Mep2 could efficiently induce filamentous growth under limiting nitrogen conditions in npr1Δ mutants. Like in S. cerevisiae, mutation of the conserved glycine at position 343 in Mep2 of C. albicans to cysteine resulted in Npr1-independent ammonium uptake. In striking contrast, however, the mutation abolished the ability of Mep2 to induce filamentous growth both in the wild type and in npr1Δ mutants. Therefore, a mutation that improves ammonium transport by Mep2 under nonpermissible conditions eliminates its signaling activity in C. albicans.
INTRODUCTION
Microorganisms sense the availability of nutrients in their environment and express appropriate transporters and enzymes that are required for uptake and metabolization of these nutrients (11, 16). A preferred nitrogen source for many microorganisms is ammonium, which is transported into the cell by ammonium permeases of the Mep/Amt family (31). The yeast Saccharomyces cerevisiae possesses three ammonium permeases encoded by the MEP1 to MEP3 genes. Each of these transporters can support growth of S. cerevisiae on media containing low concentrations of ammonium as the only nitrogen source, but mutants lacking all three MEP genes are unable to grow on ammonium at concentrations below 5 mM (21). Expression of the MEP genes is induced under limiting nitrogen conditions and repressed at high ammonium concentrations (21). Under the latter conditions, sufficient ammonium may freely diffuse into the cell in the form of ammonia or be taken up by unspecific transporters to support growth.
In addition to its transport function, Mep2 is required for the transition from yeast to filamentous, pseudohyphal growth, which occurs under limiting nitrogen conditions on solid media (13, 19). It is believed that Mep2 is an ammonium sensor that induces pseudohyphal growth in response to the presence of extracellular ammonium. Current evidence suggests that the signaling activity of Mep2 is linked to ammonium transport, because amino acid substitutions that inhibit the transport activity of Mep2 also prevent pseudohyphal growth (20, 26, 30).
Ammonium transport by Mep1 to Mep3 requires the serine/threonine protein kinase Npr1 (nitrogen permease reactivator 1), which is important for the activity of several permeases mediating uptake of nitrogenous compounds (9, 14, 15, 29). S. cerevisiae npr1 mutants exhibit a growth defect on low-ammonium medium similar to that of mutants lacking the three ammonium permeases (10). Substitution of cysteine for the highly conserved glycine at position 349 in Mep2 results in a hyperactive transporter that is independent of Npr1 (4). The hyperactive Mep2 with the G349C mutation also induces filamentation more efficiently than does wild-type Mep2, further supporting a correlation between the ammonium transport and signaling activities of Mep2 (4).
The fungal pathogen Candida albicans also switches from budding yeast morphology to filamentous growth in response to different environmental signals, including nitrogen starvation (3). C. albicans has two ammonium permeases, Mep1 and Mep2, either of which is sufficient to enable growth in low ammonium concentrations (2). Similarly to the situation in S. cerevisiae, Mep2, but not Mep1, is required for filamentous growth of C. albicans in response to nitrogen limitation. The transport and signaling functions of Mep2 can be separated, as deletion of the C-terminal cytoplasmic tail of Mep2 abolishes filamentous growth without affecting ammonium uptake (2). Certain amino acid substitutions in Mep2 also disturb ammonium transport and/or filamentous growth (7). However, there is no direct correlation between the ammonium transport activity of mutated Mep2 proteins and their ability to stimulate filamentous growth. For example, mutation of the conserved residue W167 abolished filamentation without having a strong impact on ammonium transport. Vice versa, mutation of the conserved Y122, which is assumed to participate together with W167 in ammonium recruitment at the extracytosolic side of the cell membrane, reduced ammonium uptake more strongly than a W167A mutation but still allowed efficient filament formation (7). It is therefore possible that signaling by Mep2 is regulated in different ways in C. albicans and S. cerevisiae, and it has been proposed that a high transport activity of C. albicans Mep2 (CaMep2) in the presence of abundant extracellular ammonium may actually block its signaling activity and repress filamentous growth (2).
As the Npr1 kinase is essential for the function of Mep2 and the other ammonium permeases in S. cerevisiae, we investigated the role of Npr1 in ammonium uptake by Mep1 and Mep2 and in Mep2-mediated filamentous growth of C. albicans.
MATERIALS AND METHODS
Strains and growth conditions.
C. albicans strains used in this study are listed in Table 1. All strains were stored as frozen stocks with 15% glycerol at −80°C and subcultured on YPD agar plates (20 g peptone, 10 g yeast extract, 20 g glucose, 20 g agar per liter) at 30°C. Strains were routinely grown in YPD liquid medium at 30°C in a shaking incubator. For selection of nourseothricin-resistant transformants, 200 μg/ml nourseothricin (Werner Bioagents, Jena, Germany) was added to YPD agar plates. To obtain nourseothricin-sensitive derivatives in which the SAT1 flipper cassette was excised by FLP-mediated recombination, transformants were grown overnight in YPM medium (10 g yeast extract, 20 g peptone, 20 g maltose per liter) without selective pressure to induce the MAL2 promoter, which controls expression of the caFLP gene (“ca” indicates Candida-adapted gene) in the SAT1 flipper cassette. One hundred to two hundred cells were spread on YPD plates containing 10 μg/ml nourseothricin and grown for 2 days at 30°C. Nourseothricin-sensitive clones were identified by their small colony size and confirmed by restreaking on YPD plates containing 100 μg/ml nourseothricin as described previously (25). For growth assays, YPD overnight cultures of the strains were washed two times in water and the cell suspensions adjusted to an optical density of 2. Ten microliters of a 10-fold dilution series (100 to 10−5) was spotted on YPD and SD agar (1.7 g yeast nitrogen base without amino acids [YNB; BIO 101, Vista, CA], 20 g glucose, 15 g agar per liter) plates containing various concentrations of ammonium or other nitrogen sources (as indicated in the figures) and incubated at 30°C. Rapamycin sensitivity of the strains was tested on YPD agar plates containing 100 ng/ml rapamycin. Filamentation assays were performed by plating washed cells from a YPD overnight culture at a density of 30 to 50 cells per plate on SD 2% agar plates containing 100 μM ammonium or urea as described below. Colony phenotypes were recorded after 6 days of incubation at 37°C. Growth rates of the strains in SD medium containing 1 mM ammonium were determined at 30°C using a Bioscreen C analyzer (Growth Curves USA, Piscataway, NJ).
Table 1.
C. albicans strains used in this study
| Strain | Parent | Relevant characteristic or genotypea | Reference or source |
|---|---|---|---|
| SC5314 | Wild-type parental strain | 12 | |
| npr1Δ mutants and complemented strains | |||
| NPR1M1A | SC5314 | npr1-1::SAT1-FLIP/NPR1-2 | This study |
| NPR1M1B | SC5314 | NPR1-1/npr1-2::SAT1-FLIP | This study |
| NPR1M2A | NPR1M1A | npr1-1::FRT/NPR1-2 | This study |
| NPR1M2B | NPR1M1B | NPR1-1/npr1-2::FRT | This study |
| NPR1M3A | NPR1M2A | npr1-1::FRT/npr1-2::SAT1-FLIP | This study |
| NPR1M3B | NPR1M2B | npr1-1::SAT1-FLIP/npr1-2::FRT | This study |
| NPR1M4A | NPR1M3A | npr1-1::FRT/npr1-2::FRT | This study |
| NPR1M4B | NPR1M3B | npr1-1::FRT/npr1-2::FRT | This study |
| NPR1MK1A | NPR1M4A | NPR1-SAT1-FLIP/npr1-2::FRT | This study |
| NPR1MK1B | NPR1M4B | npr1-1::FRT/NPR1-SAT1-FLIP | This study |
| NPR1MK2A | NPR1MK1A | NPR1-FRT/npr1-2::FRT | This study |
| NPR1MK2B | NPR1MK1B | npr1-1::FRT/NPR1-FRT | This study |
| mep2Δ mutants | |||
| SCMEP2M4A | SC5314 | mep2-1::FRT/mep2-2::FRT | 6 |
| SCMEP2M4B | SC5314 | mep2-1::FRT/mep2-2::FRT | 6 |
| mep1Δ mep2Δ double mutants | |||
| SCMEP12M1A | SCMEP2M4A | MEP1-1/mep1-2::SAT1-FLIP | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| SCMEP12M1B | SCMEP2M4B | mep1-1::SAT1-FLIP/MEP1-2 | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| SCMEP12M2A | SCMEP12M1A | MEP1-1/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| SCMEP12M2B | SCMEP12M1B | mep1-1::FRT/MEP1-2 | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| SCMEP12M3A | SCMEP12M2A | mep1-1::SAT1-FLIP/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| SCMEP12M3B | SCMEP12M2B | mep1-1::FRT/mep1-2::SAT1-FLIP | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| SCMEP12M4A | SCMEP12M3A | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| SCMEP12M4B | SCMEP12M3B | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| mep1Δ mep2Δ mep3Δ triple mutants | |||
| SCMEP123M1A | SCMEP12M4A | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| mep3-1::SAT1-FLIP/MEP3-2 | |||
| SCMEP123M1B | SCMEP12M4B | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| MEP3-1/mep3-2::SAT1-FLIP | |||
| SCMEP123M2A | SCMEP123M1A | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| mep3-1::FRT/MEP3-2 | |||
| SCMEP123M2B | SCMEP123M1B | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| MEP3-1/mep3-2::FRT | |||
| SCMEP123M3A | SCMEP123M2A | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| mep3-1::FRT/mep3-2::SAT1-FLIP | |||
| SCMEP123M3B | SCMEP123M2B | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| mep3-1::SAT1-FLIP/mep3-2::FRT | |||
| SCMEP123M4A | SCMEP123M3A | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| mep3-1::FRT/mep3-2::FRT | |||
| SCMEP123M4B | SCMEP123M3B | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| mep3-1::FRT/mep3-2::FRT | |||
| mep1Δ mep2Δ npr1Δ triple mutants | |||
| Δmep12NPR1M1A | SCMEP12M4A | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| npr1-1::SAT1-FLIP/NPR1-2 | |||
| Δmep12NPR1M1B | SCMEP12M4B | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| NPR1-1/npr1-2::SAT1-FLIP | |||
| Δmep12NPR1M2A | Δmep12NPR1M1A | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| npr1-1::FRT/NPR1-2 | |||
| Δmep12NPR1M2B | Δmep12NPR1M1B | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| NPR1-1/npr1-2::FRT | |||
| Δmep12NPR1M3A | Δmep12NPR1M2A | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| npr1-1::FRT/npr1-2::SAT1-FLIP | |||
| Δmep12NPR1M3B | Δmep12NPR1M2B | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| npr1-1::SAT1-FLIP/npr1-2::FRT | |||
| Δmep12NPR1M4A | Δmep12NPR1M3A | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| npr1-1::FRT/npr1-2::FRT | |||
| Δmep12NPR1M4B | Δmep12NPR1M3B | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| npr1-1::FRT/npr1-2::FRT | |||
| Strains expressing MEP1, MEP2, or MEP2G343C in mep1Δ mep2Δ or mep1Δ mep2Δ npr1Δ backgrounds | |||
| SCΔmep12MEP1K1A | SCMEP12M4A | mep1-1::FRT/MEP1-caSAT1 | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| SCΔmep12MEP1K1B | SCMEP12M4A | MEP1-caSAT1/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| SCΔmep12MEP2K1A | SCMEP12M4A | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/MEP2-caSAT1 | |||
| SCΔmep12MEP2K1B | SCMEP12M4B | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/MEP2-caSAT1 | |||
| SCΔmep12MEP2K2A | SCMEP12M4A | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/MEP2G343C-caSAT1 | |||
| SCΔmep12MEP2K2B | SCMEP12M4B | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/MEP2G343C-caSAT1 | |||
| Δmep1Δmep2Δnpr1MEP1K1A | Δmep12NPR1M4A | mep1-1::FRT/MEP1-caSAT1 | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| npr1-1::FRT/npr1-2::FRT | |||
| Δmep1Δmep2Δnpr1MEP1K1B | Δmep12NPR1M4B | MEP1-caSAT1/mep1-2::FRT | This study |
| mep2-1::FRT/mep2-2::FRT | |||
| npr1-1::FRT/npr1-2::FRT | |||
| Δmep1Δmep2Δnpr1MEP2K1A | Δmep12NPR1M4A | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/MEP2-caSAT1 | |||
| npr1-1::FRT/npr1-2::FRT | |||
| Δmep1Δmep2Δnpr1MEP2K1B | Δmep12NPR1M4B | mep1-1::FRT/mep1-2::FRT | This study |
| MEP2-caSAT1/mep2-2::FRT | |||
| npr1-1::FRT/npr1-2::FRT | |||
| Δmep1Δmep2Δnpr1MEP2K2A | Δmep12NPR1M4A | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/MEP2G343C-caSAT1 | |||
| npr1-1::FRT/npr1-2::FRT | |||
| Δmep1Δmep2Δnpr1MEP2K2B | Δmep12NPR1M4B | mep1-1::FRT/mep1-2::FRT | This study |
| MEP2G343C-caSAT1/mep2-2::FRT | |||
| npr1-1::FRT/npr1-2::FRT | |||
| Strains expressing GFP-tagged MEP2 in mep1Δ mep2Δ or mep1Δ mep2Δ npr1Δ backgrounds | |||
| SCΔmep12MEP2G7A | SCMEP12M4A | mep1-1::FRT/mep1-2::FRT | This study |
| MEP2-GFP-caSAT1/mep2-2::FRT | |||
| SCΔmep12MEP2G7B | SCMEP12M4B | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/MEP2-GFP-caSAT1 | |||
| SCΔmep12Δnpr1MEP2G7A | Δmep12NPR1M4A | mep1-1::FRT/mep1-2::FRT | This study |
| MEP2-GFP-caSAT1/mep2-2::FRT | |||
| npr1-1::FRT/npr1-2::FRT | |||
| SCΔmep12Δnpr1MEP2G7B | Δmep12NPR1M4B | mep1-1::FRT/mep1-2::FRT | This study |
| mep2-1::FRT/MEP2-GFP-caSAT1 | |||
| npr1-1::FRT/npr1-2::FRT |
SAT1-FLIP denotes the SAT1 flipper cassette. The NPR1, MEP1, MEP2, and MEP3 alleles in strain SC5314 can be distinguished by HaeII, ClaI, EcoRI, and BamHI restriction site polymorphisms, respectively. The NPR1 allele containing the polymorphic HaeII site at position +1002 was arbitrarily designated NPR1-2, the MEP1 allele with the polymorphic upstream ClaI site MEP1-1, the MEP2 allele with the polymorphic upstream EcoRI site MEP2-1, and the MEP3 allele with the polymorphic downstream BamHI site MEP3-2.
Plasmid constructions.
Oligonucleotide primers used in this study are listed in Table 2. An NPR1 deletion cassette was obtained by amplifying NPR1 upstream and downstream sequences from genomic DNA of strain SC5314 by PCR with the primer pairs NPR1-1/NPR1-2 and NPR1-3/NPR1-4, respectively, and substituting the ApaI/XhoI- and SacII/SacI-digested PCR products for the GAT1 flanking regions in the previously described plasmid pGAT1M2 (6) to generate pNPR1M2. For complementation of the npr1Δ mutants, a fragment containing the NPR1 coding region and upstream sequences was amplified with the primers NPR1-1 and NPR1-5, digested with ApaI/BglII, and ligated together with a BglII-SalI fragment containing the ACT1 transcription termination sequence (TACT1) from pMEP2K1 (2) into the ApaI/XhoI-digested pNPR1M2, yielding pNPR1K1. Plasmid pMEP1M5, which was used to delete the MEP1 gene in prototrophic C. albicans strains, was generated by substituting the SAT1 flipper cassette (25) for the URA3 flipper cassette in the previously described pMEP1M2 (2). A deletion cassette for orf19.4446, which has similarity to ammonium permease genes and was designated MEP3 for the purpose of the present study (although our results indicate that it does not encode a functional ammonium permease), was generated as follows. The orf19.4446 upstream and downstream sequences were amplified with the primer pairs MEP46/MEP47 and MEP48/MEP49, respectively, and the KpnI/XhoI- and BglII/SacI-digested PCR products substituted for the MEP1 flanking sequences in pMEP1M2 to obtain pMEP3M2. The URA3 flipper cassette was then replaced by the SAT1 flipper cassette to generate pMEP3M3. To introduce a wild-type MEP2 copy into mep1Δ mep2Δ double and mep1Δ mep2Δ npr1Δ triple mutants, a fragment containing the MEP2 coding region and upstream sequences was amplified with the primers MEP3 and MEP79, digested with KpnI/BglII, and cloned together with a BglII-PstI TACT1-caSAT1 fragment from pOPT1G22 (24) in the KpnI/PstI-digested pMEP2G6 (6), resulting in pMEP2K17. The MEP2G343C allele was obtained by an overlap PCR with the primer pairs MEP3/MEP108 and MEP107/ACT38 and substitution of the KpnI/BglII-digested PCR product for the corresponding fragment in pMEP2K17 to produce pMEP2K18. For reintroduction of MEP1 into the mep1Δ mep2Δ double and mep1Δ mep2Δ npr1Δ triple mutants, the BglII-PstI TACT1-caSAT1 fragment from pMEP2K17 was substituted for the corresponding fragment with the URA3 marker in pMEP1K1 (2), generating pMEP1K3.
Table 2.
Primers used in this study
| Primer name | Primer sequencea |
|---|---|
| ACT38 | 5′-ATATGGGCCCTGCAGACATTTTATGATGGAATGAATGGG-3′ |
| NPR1-1 | 5′-ATATGGGCCCTTCCAAGGCACGAGACC-3′ |
| NPR1-2 | 5′-ATATCTCGAGGCAGTTGACTTTGCTGCAGTG-3′ |
| NPR1-3 | 5′-ATATCCGCGGTGGGAATCATTACGAAGATC-3′ |
| NPR1-4 | 5′-ATATGAGCTCCCGTTGGTGAAGTGTCGTTG-3′ |
| NPR1-5 | 5′-ATATAGATCTATTTCTTCTTATTCTTTTCAAAAGC-3′ |
| MEP3 | 5′-TAAATACGGTACCCAAACGATTGGCTTGAATGTC-3′ |
| MEP46 | 5′-TACCACTGGTACCCGATTATCTAATTAC-3′ |
| MEP47 | 5′-TCAACATCTCGAGACATGGCTGACGG-3′ |
| MEP48 | 5′-ATAATACAGATCTTACATAGTACGATAC-3′ |
| MEP49 | 5′-CACTTATGAGCTCTTGATTGCCAGAG-3′ |
| MEP79 | 5′-AAAGAACAGATCTAATTTTTAGCTTCTCC-3′ |
| MEP107 | 5′-GTGTGGGCACTCCATTGTGTTGGTGG-3′ |
| MEP108 | 5′-CCACCAACACAATGGAGTGCCCACAC-3′ |
Restriction sites introduced into the primers are underlined; stop codons (reverse sequence) are highlighted in bold and mutated codons in italics.
Strain constructions.
C. albicans strains were transformed by electroporation (18) with the following gel-purified DNA fragments. The ApaI-SacI fragment from pNPR1M2 was used to delete NPR1 in the wild-type strain SC5314 and the mep1Δ mep2Δ double mutants. The ApaI-SacI fragment from pNPR1K1 was used to reintegrate an intact NPR1 copy into the npr1Δ mutants. The KpnI-SacI fragment from pMEP1M5 was used to delete MEP1 in the mep2Δ mutants, and the KpnI-SacI fragment from pMEP3M3 was used to delete MEP3 in the mep1Δ mep2Δ double mutants. The XhoI-SacI fragment from pMEP1K3 was used to reintegrate a wild-type MEP1 copy into the mep1Δ mep2Δ double mutants and the mep1Δ mep2Δ npr1Δ triple mutants. The KpnI-SacI fragments from pMEP2K17 and pMEP2K18 were used to reintegrate wild-type MEP2 and MEP2G343C, respectively, into the mep1Δ mep2Δ double mutants and the mep1Δ mep2Δ npr1Δ triple mutants. The KpnI-SacI fragment from pMEP2G7 (6) was used to integrate a GFP-tagged MEP2 copy into the mep1Δ mep2Δ double mutants and the mep1Δ mep2Δ npr1Δ triple mutants. The correct integration of all constructs was verified by Southern hybridization with gene-specific probes. During the course of this work, we noticed that one of the two independently constructed mep1Δ mep2Δ mutants and its derivatives (the B series) had become homozygous for chromosome R. However, in all assays described in this work, the two independently generated series of strains behaved identically, and the results obtained with these strains are therefore included.
Isolation of genomic DNA and Southern hybridization.
Genomic DNA from C. albicans strains was isolated as described previously (25). DNA was digested with appropriate restriction enzymes, separated on a 1% agarose gel, and, after ethidium bromide staining, transferred by vacuum blotting onto a nylon membrane and fixed by UV cross-linking. Southern hybridization with enhanced-chemiluminescence-labeled probes was performed with an Amersham ECL direct nucleic acid labeling and detection system (GE Healthcare, Braunschweig, Germany) according to the instructions of the manufacturer.
Fluorescence microscopy.
C. albicans strains expressing green fluorescent protein (GFP)-tagged Mep2 were grown overnight in SD medium containing 100 μM proline. Two milliliters of the overnight cultures was washed three times in water, resuspended in 10 ml SD medium containing 100 μM ammonium chloride, and incubated for 6 h at 30°C or 37°C. Fluorescence of the cells was detected using a Zeiss Observer Z1 microscope with a Zeiss HXP120C illuminator. Images were taken successively with filter settings for GFP and transmission images. Cells were observed with a 100× immersion oil objective.
Ammonium uptake assay.
To determine ammonium uptake rates of the strains, an ammonium removal assay was used. Cells were grown overnight in 50 ml SD medium with 0.1% proline at 30°C, washed two times in water, and resuspended in SD medium with 1 mM ammonium chloride at an optical density of 2. The cultures were incubated with shaking (200 rpm) at 30°C in 30-ml volumes, and 1-ml samples were taken at 10 min, 30 min, 60 min, and then every 60 min until 6 h. The cells were pelleted, and 40 μl of the supernatant was added to 760 μl OPA solution (540 mg o-phthaldialdehyde, 10 ml ethanol, 50 μl β-mercaptoethanol, 0.2 M phosphate buffer, pH 7.3, at 100 ml) to quantify the remaining ammonium (1). After 20 min of incubation in the dark, the extinction at 420 nm was measured. As a reference, 760 μl OPA plus 40 μl water was used. The system was calibrated with ammonium chloride concentrations from 0 to 2 mM.
RESULTS
Requirement of the Npr1 kinase for growth of C. albicans on ammonium.
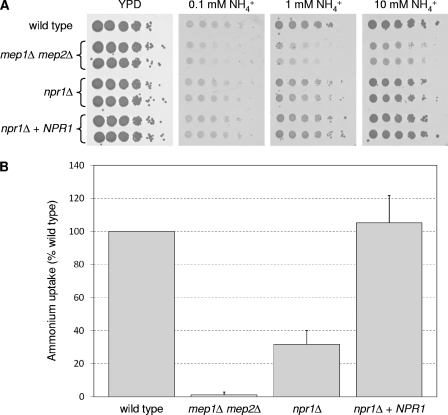
To investigate whether the Npr1 kinase is required for ammonium uptake in C. albicans, we deleted the NPR1 gene (orf19.6232) from the genome of the wild-type strain SC5314 by using the SAT1-flipping strategy (25). Two independent series of homozygous npr1Δ mutants and complemented strains (A and B series) were generated and tested for their ability to grow on agar plates containing different concentrations of ammonium as the only nitrogen source. As can be seen in Fig. 1A, the npr1Δ mutants had a slight growth defect on these media. However, the growth defect was not as severe as that of mutants lacking the ammonium permeases Mep1 and Mep2, indicating that ammonium can still be taken up into the cells in the absence of Npr1, albeit not as efficiently as in the wild-type strain. A reduced growth rate of the npr1Δ mutants was also observed in liquid medium containing 1 mM ammonium (Table 3). Ammonium uptake assays demonstrated that ammonium uptake by the npr1Δ mutants was reduced to ca. 30% of wild-type uptake rates (Fig. 1B).
Fig. 1.
(A) Growth of the wild-type strain SC5314, mep1Δ mep2Δ double mutants, and npr1Δ mutants and complemented strains on different concentrations of ammonium as the sole nitrogen source. Tenfold dilution series of the strains were spotted on a YPD control plate or on SD plates containing the indicated ammonium concentrations and incubated for 1 day (YPD) or 3 days (SD) at 30°C. The following strains were used: SC5314 (wild type), SCMEP12M4A and -B (mep1Δ mep2Δ), NPR1M4A and -B (npr1Δ), and NPR1MK2A and -B (npr1Δ + NPR1). (B) Ammonium uptake by the same strains. Uptake rates were determined in the presence of 1 mM ammonium as described in Materials and Methods. Ammonium uptake by the wild-type strain SC5314 was set to 100%, and uptake rates by the mutants are given as percentages of the wild-type uptake rate. Shown are the means and standard deviations from three independent experiments (one with the A series and two with the B series of mutants).
Table 3.
Doubling times of C. albicans strains in liquid medium with 1 mM NH4+
| Strain | Description or genotype | Doubling time (min)a |
|---|---|---|
| SC5314 | Wild type | 111 |
| NPR1M4 | npr1Δ | 229 ± 14 |
| NPR1MK2 | npr1Δ + NPR1 | 125 ± 12 |
| SCMEP12M4 | mep1Δ mep2Δ | 1,215 ± 70 |
| SCΔmep12MEP1K1 | mep1Δ mep2Δ + MEP1 | 113 ± 5 |
| SCΔmep12MEP2K1 | mep1Δ mep2Δ + MEP2 | 145 ± 27 |
| SCΔmep12MEP2K2 | mep1Δ mep2Δ + MEP2G343C | 170 ± 17 |
| Δmep12NPR1M4 | mep1Δ mep2Δ npr1Δ | 1,305 ± 154 |
| Δmep1Δmep2Δnpr1MEP1K1 | mep1Δ mep2Δ npr1Δ + MEP1 | 266 ± 21 |
| Δmep1Δmep2Δnpr1MEP2K1 | mep1Δ mep2Δ npr1Δ + MEP2 | 1,056 ± 64 |
| Δmep1Δmep2Δnpr1MEP2K2 | mep1Δ mep2Δ npr1Δ + MEP2G343C | 193 ± 4 |
Means ± standard deviations (SD) from four biological replicates (two each for the A and B series). The doubling time of strain SC5314 is the means from two biological replicates (106 min and 116 min).
Differential requirement of Npr1 for ammonium transport by Mep1 and Mep2.
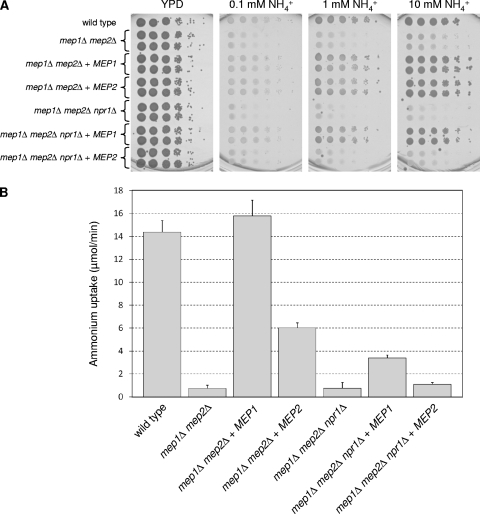
The phenotype of the npr1Δ mutants suggested that, in contrast to the situation in S. cerevisiae, one or both of the C. albicans ammonium permeases Mep1 and Mep2 can function in a partially Npr1-independent fashion. To address the specific requirement of Npr1 for Mep1 and/or Mep2 activity, we deleted NPR1 in a mep1Δ mep2Δ double mutant background and then reintroduced a functional copy of MEP1 or MEP2 into the double and triple mutants. Reinsertion of MEP1 into the mep1Δ mep2Δ double mutants restored growth and ammonium uptake to wild-type levels (Fig. 2 and Table 3). MEP2 also complemented the growth defect of the mep1Δ mep2Δ double mutants on agar plates containing limiting ammonium concentrations (Fig. 2A), but ammonium uptake was still reduced (Fig. 2B), in agreement with a slightly slower growth of the strains in liquid medium (Table 3).
Fig. 2.
(A) Growth of the wild-type strain SC5314, mep1Δ mep2Δ double mutants, mep1Δ mep2Δ npr1Δ triple mutants, and strains in which a functional copy of MEP1 or MEP2 was reinserted on different concentrations of ammonium as the sole nitrogen source. Tenfold dilution series of the strains were spotted on a YPD control plate or on SD plates containing the indicated ammonium concentrations and incubated for 1 day (YPD) or 3 days (SD) at 30°C. The following strains were used: SC5314 (wild type), SCMEP12M4A and -B (mep1Δ mep2Δ), SCΔmep12MEP1K1A and -B (mep1Δ mep2Δ + MEP1), SCΔmep12MEP2K1A and -B (mep1Δ mep2Δ + MEP2), Δmep12NPR1M4A and -B (mep1Δ mep2Δ npr1Δ), Δmep1Δmep2Δnpr1MEP1K1A and -B (mep1Δ mep2Δ npr1Δ + MEP1), and Δmep1Δmep2Δnpr1MEP2K1A and -B (mep1Δ mep2Δ npr1Δ + MEP2). (B) Ammonium uptake by the same strains. Uptake rates were determined in the presence of 1 mM ammonium as described in Materials and Methods. Shown are the means and standard deviations from three independent experiments (two with the A series and one with the B series of mutants).
A more striking difference between MEP1 and MEP2 was seen in strains lacking the Npr1 kinase. Reintegration of MEP1 into the mep1Δ mep2Δ npr1Δ triple mutants strongly improved growth both on solid and in liquid medium, although ammonium uptake was only partially restored. In contrast, expression of MEP2 in the same strains only slightly ameliorated ammonium uptake and growth (Fig. 2 and Table 3), indicating that Mep2 does not function efficiently in the absence of Npr1.
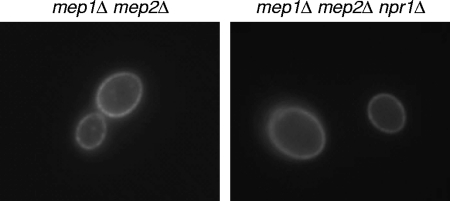
To evaluate whether expression or localization of Mep2 was impaired in the absence of Npr1, we expressed a GFP-tagged Mep2 in the same strains. As can be seen in Fig. 3, Mep2 was localized at the cell periphery and expressed at similar levels in the presence or absence of Npr1, demonstrating that Npr1 is not required for expression and correct localization of Mep2. Altogether, these results argue that ammonium transport by Mep2 strongly depends on Npr1, while Mep1 can efficiently support growth in the absence of the kinase.
Fig. 3.
Expression of GFP-tagged Mep2 in mep1Δ mep2Δ double mutants (strains SCΔmep12MEP2G7A and -B) and mep1Δ mep2Δ npr1Δ triple mutants (strains SCΔmep12Δnpr1MEP2G7A and -B). Cells were grown in SD medium containing 100 μM ammonium at 30°C and observed by fluorescence microscopy. The two independently generated series of mutants behaved identically, and only one of them is shown in each case.
Npr1 is required for ammonium uptake by unspecific transporters.
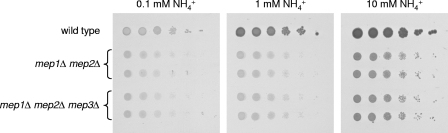
Of note, deletion of NPR1 in a mep1Δ mep2Δ double mutant background resulted in a further growth impairment (Fig. 2A and Table 3), although no difference between mep1Δ mep2Δ double mutants and mep1Δ mep2Δ npr1Δ triple mutants could be detected in ammonium uptake assays (Fig. 2B). These results indicated that Npr1 is required for the activity of other proteins that contribute to ammonium uptake/utilization. The most obvious candidate for an Npr1-dependent ammonium transporter was orf19.4446, which encodes a putative protein with 34% amino acid identity to Mep1 and Mep2. To test whether this protein, which we designated Mep3 for the purpose of the present study, is responsible for the residual ammonium uptake in the absence of Mep1 and Mep2, we deleted the corresponding gene in a mep1Δ mep2Δ double mutant background. However, no growth differences between mep1Δ mep2Δ double mutants and mep1Δ mep2Δ mep3Δ triple mutants were observed at a range of ammonium concentrations (Fig. 4), indicating that orf19.4446 does not encode a functional ammonium permease. It is likely that Npr1 is required for the activity of unspecific cation transporters that allow some ammonium uptake into the cells in the absence of the specific ammonium transporters Mep1 and Mep2, especially at increased ammonium concentrations.
Fig. 4.
Growth of the wild-type strain SC5314, mep1Δ mep2Δ double mutants, and mep1Δ mep2Δ mep3Δ triple mutants on different concentrations of ammonium as the sole nitrogen source. Tenfold dilution series of the strains were spotted on SD plates containing the indicated ammonium concentrations and incubated for 3 days at 30°C. The following strains were used: SC5314 (wild type), SCMEP12M4A and -B (mep1Δ mep2Δ), and SCMEP123M4A and -B (mep1Δ mep2Δ mep3Δ).
Npr1 is required for growth of C. albicans on other nitrogen sources.
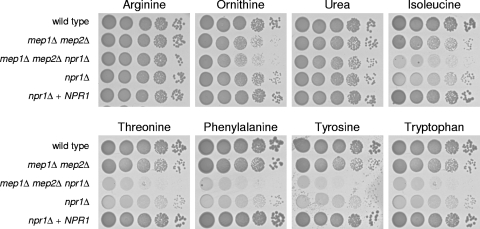
In S. cerevisiae, Npr1 is required for growth on other nitrogen sources in addition to ammonium, in part because the ammonium permeases contribute to growth under these conditions by retrieval of excreted ammonium (4). We therefore tested growth of the C. albicans npr1Δ mutants on a panel of these alternative nitrogen sources (Fig. 5). Similarly to S. cerevisiae npr1Δ mutants, C. albicans mutants lacking NPR1 exhibited a growth defect on isoleucine, tyrosine, and tryptophan. However, there were also notable differences in the phenotypes of npr1Δ mutants of the two species. While Npr1 has been shown to be required for growth of S. cerevisiae on arginine, ornithine, and urea, little or no growth defect of the C. albicans npr1Δ mutants was observed on these nitrogen sources. Vice versa, inactivation of NPR1 in C. albicans severely affected the ability of the mutants to utilize threonine and phenylalanine, whereas Npr1 is not required for growth of S. cerevisiae on these amino acids. In C. albicans, the ammonium permeases Mep1 and Mep2 contributed only marginally to growth on most of the tested alternative nitrogen sources, except for ornithine and isoleucine, where a relatively strong growth defect of the mep1Δ mep2Δ mutants was observed. These results suggest that the activity of certain amino acid transporters or enzymes involved in the metabolization of the corresponding amino acids depends on Npr1. However, the exacerbated growth defect of the mep1Δ mep2Δ npr1Δ triple mutants compared to that of the npr1Δ single mutants demonstrates that ammonium retrieval by Mep1 is important for the residual growth of npr1Δ mutants on various amino acids.
Fig. 5.
Growth of the wild-type strain SC5314 and different mutants on SD agar plates containing the indicated amino acids (10 mM) as the sole nitrogen source. Tenfold dilution series of the strains were spotted on the plates and incubated for 3 days at 30°C. The following strains were used: SC5314 (wild type), SCMEP12M4A and -B (mep1Δ mep2Δ), Δmep12NPR1M4A and -B (mep1Δ mep2Δ npr1Δ), NPR1M4A and -B (npr1Δ), and NPR1MK2A and -B (npr1Δ + NPR1). The two independently generated series of mutants behaved identically, and only one of them is shown in each case.
Mep2-mediated ammonium transport and filamentous growth become independent of Npr1 at elevated temperatures.
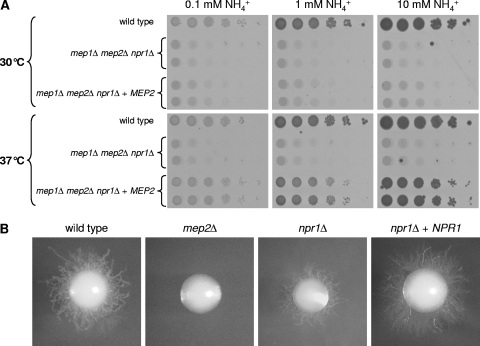
In both S. cerevisiae and C. albicans, Mep2 also acts as a signaling protein and stimulates filamentous growth in response to nitrogen limitation. As explained in the introduction, the ability of S. cerevisiae Mep2 (ScMep2) to induce filamentation correlates with its ammonium transport activity. Consequently, S. cerevisiae npr1Δ mutants have a filamentation defect, because Npr1 is required for Mep2 activity (4, 19). As our previous experiments demonstrated that Npr1 is important for ammonium transport by Mep2 also in C. albicans, we investigated whether the npr1Δ mutants had a filamentation defect under limiting nitrogen conditions. Filamentous growth is usually stimulated at elevated temperatures in C. albicans. Surprisingly, we observed that Npr1 is much less important for Mep2-dependent growth at 37°C than at 30°C, as growth of mep1Δ mep2Δ npr1Δ triple mutants expressing a single copy of MEP2 was largely restored at the elevated temperature (Fig. 6A). Analysis of strains expressing a GFP-tagged Mep2 showed that Mep2 was expressed at comparable levels at 30°C and 37°C, both in the absence and in the presence of Npr1 (data not shown). In line with the Npr1-independent activity of Mep2 at 37°C, the C. albicans npr1Δ mutants exhibited filamentous growth on media containing limiting ammonium concentrations, although filamentation was slightly reduced in the absence of Npr1 (Fig. 6B).
Fig. 6.
(A) Mep2-dependent growth becomes independent of the Npr1 kinase at elevated temperatures. Tenfold dilution series of the strains were spotted on SD plates containing the indicated ammonium concentrations and incubated for 3 days at 30°C or 37°C. The following strains were used: SC5314 (wild type), Δmep12NPR1M4A and -B (mep1Δ mep2Δ npr1Δ), and Δmep1Δmep2Δnpr1MEP2K1A and -B (mep1Δ mep2Δ npr1Δ + MEP2). (B) Filamentous growth of the wild-type strain SC5314, mep2Δ mutants, and npr1Δ mutants and complemented strains on SD agar plates containing 100 μM ammonium as the sole nitrogen source. The plates were incubated for 6 days at 37°C. The following strains were used: SC5314 (wild type), SCMEP2M4A and -B (mep2Δ), NPR1M4A and -B (npr1Δ), and NPR1MK2A and -B (npr1Δ + NPR1). The two independently generated series of mutants behaved identically, and only one of them is shown in each case.
A mutation that restores Mep2-mediated ammonium transport in the absence of Npr1 abolishes the signaling activity of Mep2.
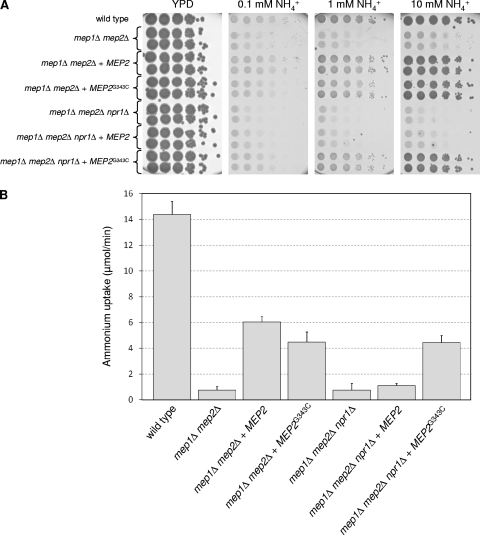
As a G349C mutation in ScMep2 results in a hyperactive, Npr1-independent ammonium transporter, we investigated the effect of a corresponding G343C mutation in CaMep2. Similarly to the situation in S. cerevisiae, the MEP2G343C allele restored growth of C. albicans mep1Δ mep2Δ npr1Δ triple mutants on media containing ammonium as the sole nitrogen source (Fig. 7A and Table 3), and ammonium uptake rates by the mutated Mep2 were similar in the presence and absence of Npr1 (Fig. 7B). Therefore, the G343C mutation resulted in Npr1-independent ammonium transport activity of Mep2 also in C. albicans. When expressed in a mep1Δ mep2Δ mutant, MEP2G343C restored ammonium uptake and growth slightly less efficiently than did wild-type MEP2.
Fig. 7.
(A) Growth of the wild-type strain SC5314, mep1Δ mep2Δ double mutants, mep1Δ mep2Δ npr1Δ triple mutants, and strains in which a wild-type copy of MEP2 or the MEP2G343C allele was reinserted on different concentrations of ammonium as the sole nitrogen source. Tenfold dilution series of the strains were spotted on a YPD control plate or on SD plates containing the indicated ammonium concentrations and incubated for 1 day (YPD) or 3 days (SD) at 30°C. The following strains were used: SC5314 (wild type), SCMEP12M4A and -B (mep1Δ mep2Δ), SCΔmep12MEP2K1A and -B (mep1Δ mep2Δ + MEP2), SCΔmep12MEP2K2A and -B (mep1Δ mep2Δ + MEP2G343C), Δmep12NPR1M4A and -B (mep1Δ mep2Δ npr1Δ), Δmep1Δmep2Δnpr1MEP2K1A and -B (mep1Δ mep2Δ npr1Δ + MEP2), and Δmep1Δmep2Δnpr1MEP2K2A and -B (mep1Δ mep2Δ npr1Δ + MEP2G343C). (B) Ammonium uptake by the same strains. Uptake rates were determined in the presence of 1 mM ammonium as described in Materials and Methods. Shown are the means and standard deviations from three independent experiments (two with the A series and one with the B series of mutants). Data are from the same experiments with results shown in Fig. 2, and the values of the control strains are included for comparison.
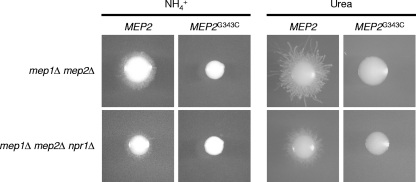
We then tested the ability of the wild-type MEP2 and MEP2G343C alleles to complement the filamentation defect of mep1Δ mep2Δ mutants in the presence and absence of Npr1. Unexpectedly, the MEP2G343C allele was unable to induce filamentous growth under limiting nitrogen conditions, in contrast to wild-type MEP2, which induced morphogenesis regardless of the presence of Npr1 (Fig. 8). Therefore, a mutation in Mep2 that restores the ammonium transport capacity in the absence of the Npr1 kinase abolishes the ability to induce filamentous growth.
Fig. 8.
Filamentous growth of mep1Δ mep2Δ double mutants and mep1Δ mep2Δ npr1Δ triple mutants expressing wild-type MEP2 or the MEP2G343C allele on SD agar plates containing 100 μM ammonium or urea as the sole nitrogen source. The plates were incubated for 6 days at 37°C. The following strains were used: SCΔmep12MEP2K1A and -B (mep1Δ mep2Δ + MEP2), SCΔmep12MEP2K2A and -B (mep1Δ mep2Δ + MEP2G343C), Δmep1Δmep2Δnpr1MEP2K1A and -B (mep1Δ mep2Δ npr1Δ + MEP2), and Δmep1Δmep2Δnpr1MEP2K2A and -B (mep1Δ mep2Δ npr1Δ + MEP2G343C). The two independently generated series of mutants behaved identically, and only one of them is shown in each case.
DISCUSSION
The results presented in this work demonstrate that the importance of the Npr1 kinase for ammonium permease function in S. cerevisiae is different from that in C. albicans. The transport activity of all three ammonium permeases of S. cerevisiae strongly depends on Npr1, and none of the Mep proteins can support growth at low ammonium concentrations in the absence of this kinase (4, 10). In contrast, the two ammonium permeases of C. albicans differ in their dependency on Npr1. While Mep1 was sufficiently active in cells lacking Npr1 to support growth under low-ammonium conditions, ammonium transport by Mep2 was largely abolished in npr1Δ mutants at the standard growth temperature of 30°C. Interestingly, however, at 37°C Mep2 functioned well in the absence of Npr1 and enabled growth on ammonium as the sole nitrogen source.
In S. cerevisiae, Npr1 is required to maintain the general amino acid permease Gap1, which is expressed when the cells are grown in a nitrogen-poor medium, at the cytoplasmic membrane by preventing ubiquitination-dependent endocytosis. In the absence of Npr1, Gap1 is endocytosed and targeted to the vacuole for degradation, and newly synthesized Gap1 is directly sorted to the vacuole and never reaches the plasma membrane (8, 28). There is evidence that Npr1 does not phosphorylate Gap1 directly but acts in an indirect fashion by phosphorylating the α-arrestin Aly2, thereby interfering with Gap1 sorting to the vacuole (23). On the other hand, the nitrate transporter Ynt1 of Hansenula polymorpha is protected from ubiquitinylation-mediated sorting to the vacuole by direct, Npr1-dependent phosphorylation in response to nitrogen deprivation, allowing its delivery to the plasma membrane (22). Npr1 itself is inhibited under nutrient-rich conditions by the TOR (target of rapamycin) kinase signaling pathway and activated upon nitrogen limitation by Sit4-dependent dephosphorylation (17, 27). Inactivation of NPR1 confers increased resistance to rapamycin in S. cerevisiae (27), and we observed the same phenotype for the C. albicans npr1Δ mutants (data not shown), indicating that Npr1 activity is controlled by TOR also in C. albicans.
We found that the role of Npr1 in maintaining the transport activity of Mep2 in C. albicans is different from its role in stabilizing Gap1 in S. cerevisiae, as Mep2 was properly expressed and localized in C. albicans npr1Δ mutants at the restrictive temperature of 30°C. Similar results have been reported for S. cerevisiae, where Mep2 was also correctly targeted to the plasma membrane in the absence of Npr1 (26). Therefore, instead of preventing endocytosis and degradation of Mep2, Npr1 seems to enable Mep2 to adopt a transport-competent conformation. Apparently, CaMep2 can attain its active conformation at higher temperatures also in an Npr1-independent fashion. Additional observations suggest that the role of Npr1 in maintaining ammonium permease function in S. cerevisiae is not protection from ubiquitination-dependent degradation. Gap1 degradation in the absence of Npr1 is prevented when ubiquitination is inhibited by an rsp5 mutation (8). In contrast, nitrogen catabolite repression (NCR)-sensitive genes were still expressed at normally repressing ammonium concentrations in an npr1 mutant even when RSP5 was also inactivated, indicating that ammonium permease activity in the npr1 mutant was not restored by the rsp5 mutation (10). These findings support the idea that the role of Npr1 in maintaining Mep function is different from its role in stabilizing Gap1 at the plasma membrane.
Our results indicate that ammonium permeases can attain a transport-competent state by different mechanisms in C. albicans. Mep2 becomes transport proficient in the presence of a functional Npr1 kinase, at elevated temperatures, or by a G343C mutation. Interestingly, Mep1 also contains the conserved glycine, mutation of which to cysteine in Mep2 of S. cerevisiae and C. albicans renders these transporters Npr1 independent. Therefore, the ability of CaMep1 to efficiently transport ammonium in the absence of Npr1 must be caused by some other feature of this permease in which it differs from Mep2.
In general, the signaling activity of mutated Mep2 proteins in S. cerevisiae correlates with their transport activity, supporting the model that ammonium transport is required for signaling by Mep2 (4, 20, 26, 30). On the other hand, an H194E mutation abolished pseudohyphal growth despite the fact that it increased ammonium transport by Mep2 (5). Therefore, the possibility that the signaling activity of Mep2 is in fact repressed when it is engaged in ammonium transport, especially in the presence of relatively high ammonium concentrations, cannot be ruled out. So far, no mutations in Mep2 that prevent ammonium transport and result in constitutive signaling, which would be in favor of such an alternative model for the regulation of Mep2 signaling activity, have been described. Nevertheless, our finding that one mechanism by which Mep2 achieves a transport-competent state, namely, the G343C mutation, abolishes its signaling activity is compatible with this hypothesis. However, the ammonium uptake rate of Mep2 was slightly reduced by the G343C mutation in a wild-type background, which may be the reason for the filamentation defect. In S. cerevisiae, the analogous G349S mutation has a different effect in that it increases both the transport and signaling activities of Mep2 above those of the wild-type protein. It is therefore possible that the signaling activity of Mep2 is controlled in different ways by ammonium availability in the two yeast species.
ACKNOWLEDGMENTS
This study was supported by the Deutsche Forschungsgemeinschaft (DFG grants MO 846/4 and SFB 630).
Footnotes
Published ahead of print on 28 January 2011.
REFERENCES
- 1. Benson J. R., Hare P. E. 1975. Ortho-phthalaldehyde–fluorogenic detection of primary amines in picomole range—comparison with fluorescamine and ninhydrin. Proc. Natl. Acad. Sci. U. S. A. 72:619–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Biswas K., Morschhäuser J. 2005. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 56:649–669 [DOI] [PubMed] [Google Scholar]
- 3. Biswas S., Van Dijck P., Datta A. 2007. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71:348–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boeckstaens M., Andre B., Marini A. M. 2007. The yeast ammonium transport protein Mep2 and its positive regulator, the Npr1 kinase, play an important role in normal and pseudohyphal growth on various nitrogen media through retrieval of excreted ammonium. Mol. Microbiol. 64:534–546 [DOI] [PubMed] [Google Scholar]
- 5. Boeckstaens M., Andre B., Marini A. M. 2008. Distinct transport mechanisms in yeast ammonium transport/sensor proteins of the Mep/Amt/Rh family and impact on filamentation. J. Biol. Chem. 283:21362–21370 [DOI] [PubMed] [Google Scholar]
- 6. Dabas N., Morschhäuser J. 2007. Control of ammonium permease expression and filamentous growth by the GATA transcription factors GLN3 and GAT1 in Candida albicans. Eukaryot. Cell 6:875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dabas N., Schneider S., Morschhäuser J. 2009. Mutational analysis of the Candida albicans ammonium permease Mep2p reveals residues required for ammonium transport and signaling. Eukaryot. Cell 8:147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Craene J. O., Soetens O., Andre B. 2001. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 276:43939–43948 [DOI] [PubMed] [Google Scholar]
- 9. Dubois E., Grenson M. 1979. Methylamine/ammonia uptake systems in Saccharomyces cerevisiae: multiplicity and regulation. Mol. Gen. Genet. 175:67–76 [DOI] [PubMed] [Google Scholar]
- 10. Feller A., Boeckstaens M., Marini A. M., Dubois E. 2006. Transduction of the nitrogen signal activating Gln3-mediated transcription is independent of Npr1 kinase and Rsp5-Bul1/2 ubiquitin ligase in Saccharomyces cerevisiae. J. Biol. Chem. 281:28546–28554 [DOI] [PubMed] [Google Scholar]
- 11. Forsberg H., Ljungdahl P. O. 2001. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40:91–109 [DOI] [PubMed] [Google Scholar]
- 12. Gillum A. M., Tsay E. Y., Kirsch D. R. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 13. Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077–1090 [DOI] [PubMed] [Google Scholar]
- 14. Grenson M. 1983. Study of the positive control of the general amino-acid permease and other ammonia-sensitive uptake systems by the product of the NPR1 gene in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 133:141–144 [DOI] [PubMed] [Google Scholar]
- 15. Grenson M., Dubois E. 1982. Pleiotropic deficiency in nitrogen-uptake systems and derepression of nitrogen-catabolic enzymes in npr-1 mutants of Saccharomyces cerevisiae. Eur. J. Biochem. 121:643–647 [DOI] [PubMed] [Google Scholar]
- 16. Holsbeeks I., Lagatie O., Van Nuland A., Van de Velde S., Thevelein J. M. 2004. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 29:556–564 [DOI] [PubMed] [Google Scholar]
- 17. Jacinto E., Guo B., Arndt K. T., Schmelzle T., Hall M. N. 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 8:1017–1026 [DOI] [PubMed] [Google Scholar]
- 18. Köhler G. A., White T. C., Agabian N. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179:2331–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lorenz M. C., Heitman J. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17:1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marini A. M., Boeckstaens M., Benjelloun F., Cherif-Zahar B., Andre B. 2006. Structural involvement in substrate recognition of an essential aspartate residue conserved in Mep/Amt and Rh-type ammonium transporters. Curr. Genet. 49:364–374 [DOI] [PubMed] [Google Scholar]
- 21. Marini A. M., Soussi-Boudekou S., Vissers S., Andre B. 1997. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4282–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Navarro F. J., Martin Y., Siverio J. M. 2008. Phosphorylation of the yeast nitrate transporter Ynt1 is essential for delivery to the plasma membrane during nitrogen limitation. J. Biol. Chem. 283:31208–31217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. O'Donnell A. F., Apffel A., Gardner R. G., Cyert M. S. 2010. Alpha-arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol. Biol. Cell 21:3552–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reuß O., Morschhäuser J. 2006. A family of oligopeptide transporters is required for growth of Candida albicans on proteins. Mol. Microbiol. 60:795–812 [DOI] [PubMed] [Google Scholar]
- 25. Reuß O., Vik Å., Kolter R., Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127 [DOI] [PubMed] [Google Scholar]
- 26. Rutherford J. C., Chua G., Hughes T., Cardenas M. E., Heitman J. 2008. A Mep2-dependent transcriptional profile links permease function to gene expression during pseudohyphal growth in Saccharomyces cerevisiae. Mol. Biol. Cell 19:3028–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schmidt A., Beck T., Koller A., Kunz J., Hall M. N. 1998. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17:6924–6931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soetens O., De Craene J. O., Andre B. 2001. Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J. Biol. Chem. 276:43949–43957 [DOI] [PubMed] [Google Scholar]
- 29. Vandenbol M., Jauniaux J. C., Grenson M. 1990. The Saccharomyces cerevisiae NPR1 gene required for the activity of ammonia-sensitive amino acid permeases encodes a protein kinase homologue. Mol. Gen. Genet. 222:393–399 [DOI] [PubMed] [Google Scholar]
- 30. Van Nuland A., et al. 2006. Ammonium permease-based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol. Microbiol. 59:1485–1505 [DOI] [PubMed] [Google Scholar]
- 31. von Wirén N., Merrick M. 2004. Regulation and function of ammonium carriers in bacteria, fungi, and plants. Top. Curr. Genet. 9:95–120 [Google Scholar]