Abstract
Streptococcus pneumoniae is a major cause of pneumonia and meningitis. Airway colonization is a necessary precursor to disease, but little is known about how the bacteria establish and maintain colonization. Carbohydrates are required as a carbon source for pneumococcal growth and, therefore, for colonization. Free carbohydrates are not readily available in the naso-oropharynx; however, N- and O-linked glycans are common in the airway. Sialic acid is the most common terminal modification on N- and O-linked glycans and is likely encountered frequently by S. pneumoniae in the airway. Here we demonstrate that sialic acid supports pneumococcal growth when provided as a sole carbon source. Growth on sialic acid requires import into the bacterium. Three genetic regions have been proposed to encode pneumococcal sialic acid transporters: one sodium solute symporter and two ATP binding cassette (ABC) transporters. Data demonstrate that one of these, satABC, is required for transport of sialic acid. A satABC mutant displayed significantly reduced growth on both sialic acid and the human glycoprotein alpha-1. The importance of satABC for growth on human glycoprotein suggests that sialic acid transport may be important in vivo. Indeed, the satABC mutant was significantly reduced in colonization of the murine upper respiratory tract. This work demonstrates that S. pneumoniae is able to use sialic acid as a sole carbon source and that utilization of sialic acid is likely important during pneumococcal colonization.
Streptococcus pneumoniae (pneumococcus) is responsible for over 1 million deaths per year worldwide (42). Although asymptomatic, colonization of the naso-oropharynx is required for the bacteria to cause disease (38). Relatively little is known about pneumococcal growth and colonization of the airway. Carbohydrates provide a carbon source to the bacteria and are therefore necessary for growth. Free carbohydrates are not readily available in the airway where initial colonization occurs (24). Instead, carbohydrates are found in the form of glycan modifications on human lipids and proteins.
S. pneumoniae strains express at least nine surface-associated glycosidases that modify host glycans (for a review, see reference 12). The ability to deglycosylate host glycans could contribute to bacterial survival by multiple mechanisms, including but not limited to providing a carbon source for growth, modifying function of host clearance molecules, competing with other bacteria for a niche, aiding in movement through the mucin layer, and promoting adherence to epithelial cells (3, 8, 14, 29, 33, 43). It was previously demonstrated that pneumococci sequentially deglycosylate both N- and O-linked glycan structures (14, 18). We have demonstrated that S. pneumoniae can grow on a human protein decorated with N-linked glycans and that this growth is dependent on the ability of the bacteria to sequentially deglycosylate this structure (3). The most common terminal carbohydrate present on N- and O-linked glycans is sialic acid, which is cleaved by pneumococcal neuraminidase NanA. As sialic acid is the initial sugar released during sequential deglycosylation, we hypothesize that it will contribute to pneumococcal growth in vivo.
Further supporting our hypothesis that sialic acid can be utilized for growth, S. pneumoniae is predicted to encode required proteins for the catabolism of sialic acid (2). While the catabolic enzymes appear to be conserved among many bacterial species, four distinct mechanisms of sialic acid transport have been identified: the major facilitator superfamily, sodium solute symporters, tripartite ATP-independent transporters, and ATP-binding cassette (ABC) transporters (1, 2, 25-27, 30, 36). Through a bioinformatics approach, a putative sodium solute symporter encoded by open reading frame SP1328 in strain TIGR4 was initially predicted to be the primary pneumococcal sialic acid transporter (32, 35). Based on their similarity to known sialic acid transporter components, it was later suggested that putative permease components of the ABC transporters SP1681-3 and SP1688-90 may also be involved in sialic acid uptake (2).
Here, we show that S. pneumoniae can utilize sialic acid as a carbon source for growth. Furthermore, using mutants in the three predicted transporters, we identify the ABC transporter encoded by SP1681-3 as the primary sialic acid transporter. This sialic acid transporter contributes to growth on a human glycoprotein and colonization in vivo. Together, these data suggest that sialic acid transport may be important for pneumococcal pathogenesis.
MATERIALS AND METHODS
Bacterial strains, culture media, and chemicals.
Parental and genetically modified strains of S. pneumoniae utilized in this study are described in Table 1. Broth cultures were routinely grown at 37°C in Todd-Hewitt broth (Becton Dickinson and Company) supplemented with 0.2% (wt/vol) yeast extract (Becton Dickinson and Company) (THY). C medium with 5% yeast extract (C+Y), pH 8.0, was used for transformations. Chemically defined medium (CDM) was used for growth analyses and was supplemented with no sugar, 12 mM glucose, 12 mM sialic acid, or 5 mg ml−1 alpha-1 glycoprotein (AGP), as indicated (15). S. pneumoniae was also grown at 37°C and 5% CO2 overnight on tryptic soy agar plates (Becton Dickinson and Company) spread with 5,000 U of catalase (Worthington Biochemical Corporation) prior to plating of bacteria, as well as on tryptic soy agar plates supplemented with 5% sheep blood (Becton Dickinson and Company). Bacteria were selected on tryptic soy agar plates that contained streptomycin (200 μg ml−1), kanamycin (500 μg ml−1), chloramphenicol (2.5 μg ml−1), or neomycin (20 μg ml−1), as appropriate.
TABLE 1.
Strains of Streptococcus pneumoniae used in this studya
| Strain name | Serotype | Characteristics/genotypeb | Site of isolationc | Source or reference |
|---|---|---|---|---|
| 1121 | 23F | Clinical isolate | McCool et al. (19a) | |
| 1121 Smr | 23F | Lys56→Thr in RpsL [rpsL(K56T)] conferring Smr | Marion et al. (18) | |
| 1121 ΔsatABC | 23F | ΔsatABC rpsL(K56T) (Smr) | This study | |
| 1121 ΔsatABC/satABC+ | 23F | ΔsatABC/satABC+ rpsL(K56T) (Smr) | This study | |
| 1121 Δ1688-90 | 23F | Δ1688-90 rpsL(K56T) (Smr) | This study | |
| 1121 Δ1688-90/1688-90+ | 23F | Δ1688-90/1688-90+rpsL(K56T) (Smr) | This study | |
| 1121 ΔsatABC Δ1688-90 | 23F | ΔsatABC Δ1688-90 rpsL(K56T) (Smr) | This study | |
| 1121 ΔsatABC Δ1688-90/satABC+ 1688-90+ | 23F | ΔsatABC Δ1688-90/satABC+ 1688-90+rpsL(K56T) (Smr) | This study | |
| 1121 ΔnanA | 23F | ΔnanA (Cmr) rpsL(K56T) (Smr) | This study | |
| TIGR4 | 4 | Clinical isolate | Tettelin et al. (32) | |
| TIGR4 Smr | 4 | Lys56→Thr in RpsL [rpsL(K56T)] conferring Smr | Bender and Weiser (2a) | |
| TIGR4 ΔsatABC | 4 | ΔsatABC rpsL(K56T) (Smr) | This study | |
| TIGR4 ΔsatABC/satABC+ | 4 | ΔsatABC/satABC+rpsL(K56T) (Smr) | This study | |
| TIGR4 Δ1328 | 4 | ΔSP1328 rpsL(K56T) (Smr) | This study | |
| TIGR4 ΔsatABC Δ1688-90 Δ1328 | 4 | ΔsatABC Δ1688-90 Δ1328 rpsL(K56T) (Smr) | This study | |
| C06_18 | 22F | Clinical isolate | Blood | Burnaugh et al. (3) |
| C06_29 | 15B/C | Clinical isolate | BAL | Burnaugh et al. (3) |
| C06_31 | 23F | Clinical isolate | BAL | Burnaugh et al. (3) |
| C06_57 | 6A/B | Clinical isolate | BAL | Burnaugh et al. (3) |
| C06_58 | 19A | Clinical isolate | Blood | Burnaugh et al. (3) |
+, strain with genetic mutation has been reconstituted to wild type.
Smr, resistance to streptomycin; Cmr, resistance to chloramphenicol.
BAL, bronchioalveolar lavage/aspirate.
Unless otherwise specified, all chemicals, substrates, and enzymes were purchased from Sigma Chemicals.
Construction of mutants.
Unmarked, in-frame deletions of the putative sialic acid transporters were generated using the Janus cassette selection system (31). This method requires two rounds of transformations. The first round introduces an engineered cassette conferring kanamycin resistance and streptomycin sensitivity (rpsL+) into a streptomycin-resistant (Smr) parental strain. Regions flanking the gene(s) of interest were amplified with primers 1 and 2 and primers 4 and 6 and then sequentially joined to the Janus cassette using modified splicing by overlap extension (SOE) (3, 10). A high-fidelity proofreading polymerase, Pfx-50 (Invitrogen), was used throughout to minimize PCR-generated errors, and all genomic DNA was prepared essentially as described previously (41). All Janus constructs were transformed into pneumococci, and transformants were selected on kanamycin and confirmed by PCR with primers 7 and 8, which flank the mutant construct. All primer sequences can be found in Table 2.
TABLE 2.
Primers used in this study
| Group | Number | Primer sequence (5′→3′) | Location (accession no.) |
|---|---|---|---|
| satABC | A.1 | CTATATGTTGTTCACGCATTC | 1505216-1505236 (AE007317) |
| A.2 | CATTATCAATTAAAAATCAAACGGTCATCCATCACTCTCCTCTGTa | 1504642-1504662 (AE007317) | |
| A.3 | GCATACCCTCCATTTTGTAGATCATCCATCACTCTCCTCTGTb | 1504642-1504662 (AE007317) | |
| A.4 | GGAAAGGGGCCCAGGTCTCTACAAAATGGAGGGTATGCc | 1501066-1501086 (AE007317) | |
| A.5 | TCTACAAAATGGAGGGTATGC | 1501066-1501086 (AE007317) | |
| A.6 | GAATGCTACTGCCTCGTCTTTGA | 1500702-1500724 (AE007317) | |
| A.7 | TTTGCAGGTCGTTTCTC | 1505403-1505419 (AE007317) | |
| A.8 | TATTCTCATTTCTCTACCTC | 1500451-1500470 (AE007317) | |
| A.9 | GCCTTTGAGGCGACAGC | 1652124-1652146 (AE007317) | |
| A.10 | ACACGATGCCCCACTTCTTTCTG | 1652408-1652433 (AE007317) | |
| SP1688-90 | B.1 | TGGTAATCGATTGTTTGGG | 1513786-1513804 (AE007317) |
| B.2 | CATTATCAATTAAAAATCAAACGGTTTTTCATCGTTCTTCTCTTTCa | 1513451-1513472 (AE007317) | |
| B.3 | CCTTCTTTCGTCTACTTCACAGTTTTTCATCGTTCTTCTCTTTCd | 1513451-1513472 (AE007317) | |
| B.4 | GGAAAGGGGCCCAGGTCCTGTGAAGTAGACGAAAGAAGGc | 1510314-1510335 (AE007317) | |
| B.5 | CTGTGAAGTAGACGAAAGAAGG | 1510314-1510335 (AE007317) | |
| B.6 | GAAGCTGGTCTAGAAAATAAATAA | 1509913-1509936 (AE007317) | |
| B.7 | GTTTAGGAACTTATGTGGGAGTA | 1513885-1513907 (AE007317) | |
| B.8 | GGATTTGATAAGGGAATAGTTGA | 1509625-1509647 (AE007317) | |
| B.9 | GTGTATTTTCAACTGCCTGTCC | 1509872-1509893 (AE007317) | |
| B.10 | ATGCGAATGAATTAAACTATGGTC | 1510202-1510225 (AE007317) | |
| SP1328 | C.1 | GCAACAATGCGTCAAAATCCTC | 1254113-1254134 (AE005672) |
| C.2 | CATTATCCATTAAAAATCAAACGGTAGATACCTGCAACCAACACa | 1253572-1253591 (AE005672) | |
| C.3 | AACTTGAATCCGCTTTAATTTCTAGATACCTGCAACCAACACe | 1253572-1253591 (AE005672) | |
| C.4 | AAGCATAAGGAAAGGGGCCCGAAATTAAAGCGGATTCAAGTTc | 1252127-1252148 (AE005672) | |
| C.5 | GAAATTAAAGCGGATTCAAGTT | 1252127-1252148 (AE005672) | |
| C.6 | TTCTTCAAAAGTCACCAACATA | 1251714-1251735 (AE005672) | |
| C.7 | CAGCTATCCCACCTATTTATTT | 1254259-1254280 (AE005672) | |
| C.8 | AGTTTTTTAACAGTTTCATCATT | 1251664-1251686 (AE005672) | |
| C.9 | GTGATTCTGATTAGTGGTGTC | 1253070-1253090 (AE005672) | |
| C.10 | ACAGCTGTTGGAGGAAGGA | 1252270-1252288 (AE005672) | |
| aroE | RT.F | TGCAGTTCARAAACATWTTCTAA | 1232187-1232203 (AE007317) |
| RT.R | TGCTAGCCCATCATATTCGTTTGTTG | 1231725-1231747 (AE007317) | |
| Janus | J.F | CCGTTTGATTTTTAATGGATAATG | 7-30 (AY334019) |
| J.R | GGGCCCCTTTCCTTATGCTT | 247511-247527 (AE005672) |
Underlining indicates reverse complement sequence of primer J.F.
Underlining indicates reverse complement sequence of primer A.5.
Underlining indicates reverse complement sequence of primer J.R.
Underlining indicates reverse complement sequence of primer B.5.
Underlining indicates reverse complement sequence of primer C.5.
The second round of transformation replaced the Janus cassette with a DNA construct consisting of the fragments flanking the region to be deleted; these fragments were generated with primers 1 and 3 and primers 5 and 6 and joined by SOE PCR. Introduction of this construct restores kanamycin sensitivity and streptomycin resistance. Transformants were confirmed by PCR with primers 7 and 8, and genetic sequencing of this same region confirmed that no spurious mutations had been introduced during generation of the mutants. As opacity can also affect pneumococcal colonization and glycosidase expression, all mutants were confirmed as being of the same opacity as their parental strains (13, 40).
All mutants have been genetically reconstituted by reintroducing the deleted region into the same genetic location in the same orientation through two additional rounds of genetic transformation: one to reintroduce the cassette and the second to restore the original parental region. Growth of all strains was tested in rich medium to ensure that the mutation did not introduce generalized growth defects.
The glycosidase NanA is predicted to be encoded in a single gene transcript (32). Therefore, an insertion-deletion mutation was constructed as previously described (13, 14).
RNA extraction and reverse transcriptase PCR.
The Janus system of mutant generation is designed to generate unmarked mutants; however, as the genes encoding the predicted ABC transporters are predicted to be in operons, we demonstrated the mutants had no effect on transcription of the distal genes by reverse transcriptase PCR. RNA extraction was conducted using a modified RNeasy extraction protocol (Qiagen). Bacteria were grown in THY to an optical density at 600 nm (OD600) of 0.3 ± 0.01. Each 5-ml culture was centrifuged and resuspended in 500 μl of 10 mM Tris-50 mM EDTA (pH 8), and cells were lysed by addition of 30 μl of sodium N-lauroyl sarcosinate (20%, wt/vol) with incubation for 10 min at 37°C. Concentrated samples were combined with 2 ml of Tri-Reagent (MRC Inc.) and allowed to stand at room temperature for 5 min. A 400-μl aliquot of chloroform was added to each sample, followed by gentle shaking for 15 s, and then the samples were allowed to stand at room temperature for an additional 3 min. After centrifugation at 4,000 × g at 4°C for 15 min, the aqueous layer was removed and 1 volume of 75% ethanol was added and mixed by pipetting. Samples were then processed on RNeasy Mini columns (Qiagen), eluted into 30 μl of RNase-free water, and quantified on a Nanodrop spectrophotometer.
After DNase I treatment (Invitrogen), 1 μg of RNA was used for cDNA synthesis with SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's specifications. Parallel samples lacking reverse transcriptase were processed and served as a negative control for each sample to ensure the purity of the RNA isolation. cDNA and reverse transcriptase-free negative-control samples were tested by PCR; primers 9 and 10 were designed in the gene immediately downstream to the relevant mutations, and primers RT.F and RT.R are within a control housekeeping gene, aroE (Table 2). Products were visualized by agarose gel electrophoresis and ethidium bromide staining.
Dialysis of human alpha-1 glycoprotein (AGP).
Dialysis was conducted in order to remove any free sugar from AGP as previously described (3). The Slide-A-Lyzer dialysis cassette (10,000-molecular-weight cutoff) was used according to the instructions of the manufacturer (Pierce). Human AGP was reconstituted at 10 mg ml−1 in distilled water (dH2O) and dialyzed against 2 liters of dH2O at 4°C for 2 h and then overnight in fresh, prechilled dH2O (2 liters). Samples were concentrated using Slide-A-Lyzer concentrating solution (Pierce) according to the instructions provided, and the protein was then extracted, diluted to the original volume, filter sterilized, and stored at 4°C.
Growth assays.
Growth assays were conducted essentially as described previously by our laboratory (3). S. pneumoniae strains were grown in THY to an OD600 of 0.6 ± 0.005, representative of exponential growth, and 1 ml was washed and resuspended in 130 μl of 1:1 phosphate-buffered saline (PBS)-catalase (3 × 104 U ml−1). A 20-μl portion of bacterial suspension or PBS-catalase (bacterium-free control) was added to 180 μl of CDM supplemented with the appropriate carbon source (12 mM glucose, 12 mM sialic acid, or 5 mg ml−1 AGP). Medium supplemented with glucose (12 mM) was used as a positive control in all experiments to confirm the viability of strains. Medium not supplemented with sugar served as a negative control in all experiments. Plates were incubated at 37°C in a BIO-TEK Synergy HT plate reader for at least 60 h, and the OD600 was measured every 20 min.
Data were corrected for path length, and the averages from triplicate sugar-free medium controls were subtracted from values for experimental wells. Results for triplicate wells were then averaged, and this value was considered one datum point for further analysis. Data from at least three independent experiments were averaged, and the 95% confidence interval was calculated for each time point.
To determine if the growth of bacterial inocula on rich medium contributed to the delay observed in growth on sialic acid compared to growth on glucose, we prepared inocula from bacteria grown on sialic acid. Bacteria were prepared and grown on sialic acid in a 96-well plate as described above. When strains reached a path length-corrected OD600 of 0.3, bacteria were harvested from the wells, pooled, washed in PBS, and resuspended in 1:1 PBS-catalase. To ensure a fair comparison of growth between conditioned and nonconditioned bacteria, it is important to use essentially identical bacterial inocula. To achieve this goal, a range of dilutions of the conditioned bacteria recovered from the plate was used to inoculate fresh sialic acid medium. Enumeration of inocula allowed the selection of wells containing inocula closest to but not exceeding the number of bacteria in the original inocula.
Neuraminidase activity assay.
Neuraminidase activity was measured using a fluorimetric assay (16). The fluorogenic substrate 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUAN) was resuspended in 0.25 M sodium acetate buffer (pH 7) at 0.35% (wt/vol) and stored in aliquots at −20°C. After cultures were grown to an OD600 of 0.2 ± 0.01 in CDM supplemented with 5 mg ml−1 AGP, bacteria were toluene lysed. A 10-μl sample of MUAN was mixed with 10 μl of bacterial lysate, and the mixture was incubated at 37°C for 5 min. The reaction was stopped by the addition of 1.5 ml of 50 mM sodium carbonate buffer, pH 9.6. The fluorescence was detected using a BIO-TEK Synergy HT plate reader at an excitation wavelength of 366 nm and an emission wavelength of 446 nm. The average fluorescence of controls with medium alone was subtracted from the sample readings. Experiments were conducted three times in triplicate; significance was determined by a two-tailed Student t test, and a P value below 0.05 was considered significant.
Southern blot.
Distribution of SP1328 was assessed via Southern blot analysis. Each 2-μg sample of genomic DNA was digested with the restriction enzyme EcoRV (Fisher Scientific) for 2 h at 37°C. Samples were resolved on a 1% agarose gel and visualized by ethidium bromide staining to ensure that DNA was digested and present in each lane. The Turboblotter rapid alkaline transfer system (Whatman) was used according to the manufacturer's instructions to transfer digested DNA to a polyvinylidine fluoride membrane. DNA was cross-linked to the membrane via UV radiation. The DNA probe was generated via PCR using primers C.9 and C.10 (Table 2) and labeled using the ECL direct nucleic acid labeling and detection system (GE Life Sciences). Hybridization was conducted per the manufacturer's instructions and followed by autoradiography to detect DNA binding.
Sialic acid transport assay.
Cultures of parental and mutant S. pneumoniae strains were grown in THY to an OD600 of 0.6 ± 0.01. Each sample was pelleted, washed in PBS, and concentrated in CDM without sugar to an OD600 of 2.0. CDM containing sialic acid was prepared such that [3H]sialic acid (American Radiolabeled Chemicals Inc.) accounted for 1% total sialic acid. Triplicate 90-μl aliquots were first incubated for 3 min at 37°C to acclimate. CDM containing sialic acid was then added to the culture to achieve a final concentration of 2.5 μM. After incubation at 37°C for the designated time, reactions were terminated by filtration through 0.22-μm-pore-size filters (Millipore). Filters were washed three times with 2 ml CDM without sugar. After the filters were air dried, 10 ml of 30% ScintiSafe LCS Cocktail (Fisher Scientific) was added to each sample. Incorporated sialic acid was measured in a Packard Tri-Carb liquid scintillation analyzer. Average measurements conducted with heat-killed bacteria were subtracted from each value. Data were normalized to the amount of label incorporated per 107 cells. Time course experiments were conducted twice in triplicate. All other transport experiments were conducted three times in triplicate. Where applicable, significance was determined by a two-tailed Student t test, and a P value below 0.05 was considered significant.
Murine colonization.
Nasopharyngeal colonization was performed as described previously (19). Groups of 10 6- to 8-week-old female C57BL/6 mice (Jackson Laboratory) were inoculated intranasally with 1 × 108 mid-log-phase organisms of 1121 Smr, 1121 ΔsatABC, 1121 ΔSP1688-90, 1121 ΔsatABC ΔSP1688-90, or 1121 ΔnanA. After 5 days, the density of colonization was assessed by upper respiratory tract lavage and quantitative culture of recovered organisms on tryptic soy agar supplemented with neomycin. The animal data are presented as the mean CFU ml−1 ± standard error of the mean (SEM). Significance was assessed by a two-tailed Student t test; a P value below 0.05 was considered significant.
RESULTS
S. pneumoniae can utilize sialic acid as a sole carbon source.
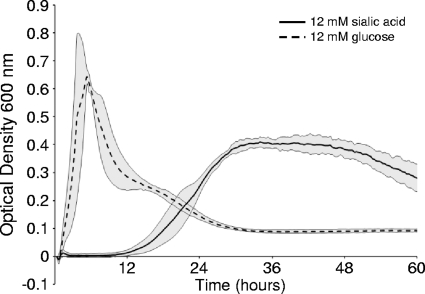
We have previously demonstrated the ability of S. pneumoniae to remove sialic acid from both N- and O-linked glycans via neuraminidase cleavage (3, 14, 18). As sialic acid is found terminally positioned on glycoconjugates, S. pneumoniae likely encounters sialic acid frequently in the airway (7). Release of sialic acid may thus provide S. pneumoniae with a source of carbohydrates necessary for growth. To first test whether sialic acid could be used as a sole carbon source, bacteria were grown in CDM supplemented with sialic acid. Sialic acid was able to support significant pneumococcal growth (OD600 = 0.4), although this growth was delayed and to a lower final optical density than growth in glucose at the same molar concentration (Fig. 1). Conditioning bacteria in sialic acid medium resulted in a reduction, but not elimination, of the time taken to achieve the maximal optical density (data not shown). These data suggest that the delay in growth on sialic acid is in part due to the prior growth of the bacteria in rich medium.
FIG. 1.
Sialic acid supports S. pneumoniae growth. Strain 1121 Smr was grown on chemically defined medium for 60 h supplemented with 12 mM sialic acid or 12 mM glucose as the sole carbon source. Growth was measured by the optical density at 600 nm. Data points are the means of three independent experiments performed in triplicate. Gray shading indicates a 95% confidence interval.
To determine whether sialic acid can support the growth of diverse pneumococcal strains, recent clinical isolates of different serotypes and genetic backgrounds (Table 1) were tested for their ability to utilize sialic acid as a sole carbon source (data not shown). While growth rate and maximal OD600 achieved varied, each strain tested grew on sialic acid, suggesting that the ability to utilize sialic acid is conserved. In order for S. pneumoniae to utilize sialic acid as a nutrient source, the bacteria must have a mechanism for import of the carbohydrate; however, no pneumococcal sialic acid transporter has yet been identified.
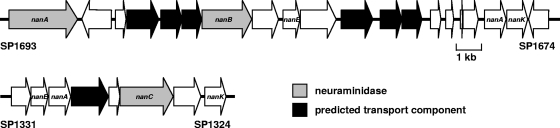
Proposed pneumococcal sialic acid transporters.
Based on annotation of the TIGR4 genome, three distinct genetic regions have been suggested to encode sialic acid transporters: a predicted sodium solute symporter, SP1328, and two predicted ABC transporters, SP1681-3 and SP1688-90 (Fig. 2) (2, 27, 32, 35). Of these, the predicted sodium solute symporter (SP1328) was previously proposed to be the primary transporter (35). However, open reading frame SP1328 is absent in at least half of pneumococcal strains (23, 35). By Southern blot analysis, strain 1121 and four of five recent clinical isolates tested were shown to lack homologs to SP1328, and yet all were able to maintain growth upon sialic acid (Fig. 1 and data not shown). These data suggest that SP1328 is not the primary sialic acid transporter; therefore, initial efforts focused on determining the role of the two putative ABC transporters.
FIG. 2.
Schematic of regions encoding known and putative neuraminidases and transporters as found in S. pneumoniae strain TIGR4. Arrows indicate open reading frames. Neuraminidases are depicted in gray, and predicted transporter components are depicted in black. Open reading frames predicted to be involved in sialic acid utilization are labeled as follows: nanK, sugar kinase; nanA (note that this is distinct from neuraminidase nanA), N-acetylneuraminate lyase; and nanE, N-acetylmannosamine-6-phosphate 2-epimerase.
The open reading frames SP1681-3 and SP1688-90 are predicted to encode components of ABC transporters (Fig. 2) (32). Unlike the predicted symporter, both loci are considered to be part of the pneumococcal core genome (20). The presence of open reading frames SP1681-3 and SP1688-90 was confirmed in strain 1121 and five recent clinical isolates by PCR amplification (Table 1 and data not shown). These genes exist in proximity to the genes encoding the two characterized neuraminidases, NanA and NanB, as well as a predicted sugar kinase (SP1675), a predicted N-acetylneuraminate lyase (SP1676), and a predicted N-acetylmannosamine-6-phosphate 2-epimerase (SP1685) (32). Together, this evidence suggests that one or both of these predicted ABC transporters are involved in sialic acid utilization.
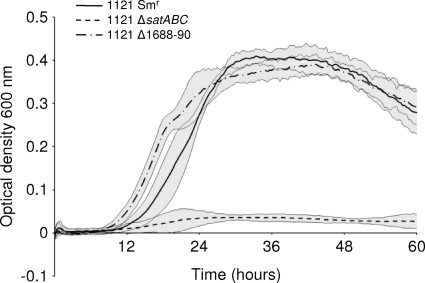
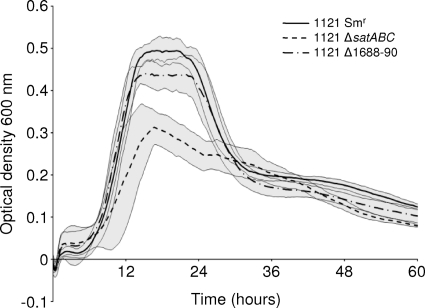
SP1681-3 (satABC) is required for growth on sialic acid.
In order to test whether SP1681-3 or SP1688-90 contributes to growth on sialic acid, unmarked deletions were constructed, resulting in strains 1121 Δ1681-3, 1121 Δ1688-90, and 1121 Δ1681-3 Δ1688-90. Mutations were confirmed by both PCR and sequencing and determined to lack polar effects by reverse transcriptase PCR (data not shown). Data suggest that only one of these regions, SP1681-3, contributed to growth on sialic acid (Fig. 3). As such, 1121 Δ1681-3, but not 1121 ΔSP1688-90, showed a significant reduction in growth (maximum OD600, <0.1) compared to that of the parental strain 1121 Smr. The double mutant showed no additional reduction in growth, suggesting that no additive effect of the mutation of the two transporters existed (data not shown). All mutant strains were genetically reconstituted, and each reconstituted strain grew as efficiently as the parent on sialic acid (data not shown). These data suggest that SP1681-3 encodes the primary sialic acid transporter in S. pneumoniae, and henceforth it is referred to as the sialic acid transporter satABC. In this locus, satA (SP1683) is predicted to encode a substrate binding protein, while satB (SP1682) and satC (SP1681) are predicted to encode permease components.
FIG. 3.
satABC (SP1681-3) is required for efficient growth on sialic acid. Strain 1121 Smr and mutants were grown for 60 h on chemically defined medium supplemented with 12 mM sialic acid as the sole carbon source. Growth was measured by the optical density at 600 nm. Data points are the means of three independent experiments performed in triplicate. Gray shading indicates a 95% confidence interval.
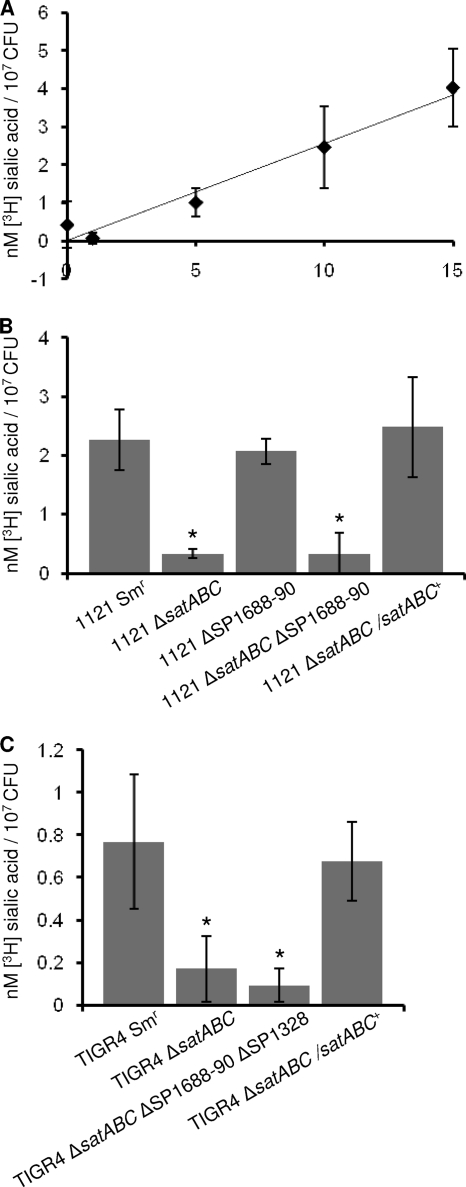
satABC is required for transport of sialic acid.
To directly test whether satABC encodes a sialic acid transporter, uptake assays utilizing radiolabeled sialic acid were conducted. A time course experiment with the parental strain was conducted first to demonstrate a temporal increase in the uptake of [3H]sialic acid (Fig. 4 A). From this experiment, a 10-minute incubation time was selected for subsequent experiments. Compared to the parent strain, 1121 ΔsatABC was significantly reduced in uptake of [3H]sialic acid (Fig. 4B). While SP1688-90 showed no growth defect on sialic acid, it remained possible that this predicted transporter was still responsible for some uptake of sialic acid. However, the SP1688-90 mutant strain showed no statistically significant reduction in transport of sialic acid under these conditions, and the double mutant showed no significant reduction in transport compared to that of the satABC single mutant (Fig. 4B). These data demonstrate that satABC encodes the primary transporter for sialic acid under these conditions.
FIG. 4.
satABC is required for transport of sialic acid. Strain 1121 Smr and mutants were incubated for the designated amount of time in chemically defined medium supplemented with 2.5 μM sialic acid containing 1% [3H]sialic acid. Following incubation, bacteria were filtered and washed, and radioactivity associated with cells was measured using a scintillation counter. (A) Data are representative of two time course experiments with strain 1121 Smr conducted in triplicate. Values on the x axis show time (in minutes). (B and C) Data are averages from parental and mutant strains from three independent experiments performed in triplicate. Standard deviation is depicted. Statistical significance was tested by a two-tailed Student t test; *, P ≤ 0.05.
The SP1328 predicted symporter does not contribute to sialic acid transport.
Although 1121 lacks the predicted sodium solute symporter, it remained possible that SP1328 contributes to transport of sialic acid when present. To test this, we measured the uptake of [3H]sialic acid in the TIGR4 strain, which contains all three candidate transporters. TIGR4 ΔsatABC showed a significant reduction in sialic acid transport which could be restored in the genetically reconstituted strain, TIGR4 ΔsatABC/satABC+ (Fig. 4C). A TIGR4 ΔsatABC ΔSP1688-90 ΔSP1328 triple mutant had no greater reduction compared to the satABC mutant, which suggests that neither SP1688-90 nor SP1328 contributes to sialic acid import under these assay conditions. Overall transport in the TIGR4 background was lower than for 1121; this is consistent with the delayed growth and lower maximal OD achieved when TIGR4 is grown on sialic acid as a sole carbon source (data not shown). Together, these data suggest that satABC encodes the primary sialic acid transporter in multiple strain backgrounds.
satABC contributes to growth on human glycoprotein.
While the data presented demonstrate that SatABC transports sialic acid, we wanted to test whether this transport contributed to growth on human glycoprotein. We have previously shown that S. pneumoniae can grow on a model human glycoprotein, AGP (3). AGP is decorated with complex N-linked glycans containing terminal sialic acid, galactose, and N-acetylglucosamine. These three carbohydrates can be sequentially cleaved by pneumococcal exoglycosidases (neuraminidase NanA, beta-galactosidase BgaA, and N-acetylglucosaminidase StrH) and used for growth. Mutation of any of these exoglycosidases results in a reduction in growth (3). Thus, sialic acid released by neuraminidase and transported by SatABC likely contributes to growth of S. pneumoniae. We therefore hypothesized that the inability to transport sialic acid would result in a reduction in, but not elimination of, growth on AGP.
When 1121 Smr and the satABC mutant were grown on AGP, a significant reduction in growth on AGP was seen for 1121 ΔsatABC (Fig. 5); growth was restored in the 1121 ΔsatABC/satABC+ reconstituted strain (data not shown). As was the case with growth on free sialic acid, 1121 Δ1688-90 was not significantly reduced in growth on AGP and the double mutant showed no further reduction compared to 1121 ΔsatABC (Fig. 5 and data not shown), suggesting that SP1688-90 does not contribute to growth on AGP under these experimental conditions.
FIG. 5.
satABC contributes to growth on a human glycoprotein. Strain 1121 Smr and mutants were grown for 60 h on chemically defined medium supplemented with 5 mg ml−1 alpha-1 glycoprotein as the sole carbon source. Growth was measured by the optical density at 600 nm. Data points are the means from three independent experiments performed in triplicate. Gray shading indicates a 95% confidence interval.
As the transport of a substrate into the cell often upregulates utilization pathways, it was possible that 1121 ΔsatABC may express lower neuraminidase levels than 1121 Smr (6, 11). A reduction in neuraminidase expression would reduce cleavage of sialic acid, which would restrict other pneumococcal glycosidases from releasing underlying carbohydrates for growth. To determine if less efficient cleavage of carbohydrates from AGP significantly contributed to the reduced growth of 1121 ΔsatABC, the neuraminidase activity of bacteria growing on AGP (OD600 = 0.3) was determined. No significant difference in neuraminidase activity between the mutant and parental strain was observed (data not shown). This supports the original hypothesis that the inability to utilize sialic acid results in a reduction in growth on AGP. As glycoconjugates are likely used as a carbon source during colonization, these data suggest that sialic acid utilization contributes to growth in the airway.
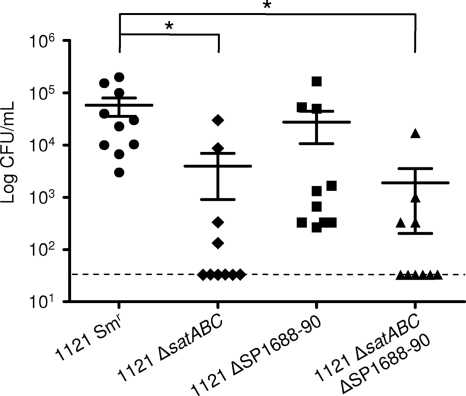
satABC contributes to airway colonization.
Although no reduction in colonization was observed in a nanA mutant compared to the parental strain, it remained possible that transport of sialic acid may still contribute to colonization (data not shown). Indeed, significantly fewer bacteria were recovered from the upper respiratory tract of mice inoculated with 1121 ΔsatABC than from those inoculated with 1121 Smr, suggesting that sialic acid transport contributes to colonization (Fig. 6). In contrast, 1121 Δ1688-90 showed no significant attenuation in vivo and there was no statistically significant difference in colonization between 1121 ΔsatABC and 1121 ΔsatABC Δ1688-90. Although we cannot rule out that SP1688-90 is important at other times during infection or that its role is human specific, these data suggest that SP1688-90 does not contribute to airway colonization. Collectively, these data demonstrate that satABC, and presumably transport of sialic acid, is important for colonization of the naso-oropharynx.
FIG. 6.
satABC contributes to pneumococcal colonization. Groups of 10 mice received intranasal inoculation with 1 × 108 CFU of mutant or parental strain (1121 Smr). Five days after inoculation, mice were sacrificed by carbon dioxide asphyxiation. Nasal lavage fluid was collected, and bacterial titers were assessed. The gray dashed line indicates the limit of detection. The mean and standard error of the mean are depicted. Statistical significance was tested by a two-tailed Student t test; *, P ≤ 0.05.
DISCUSSION
The data here demonstrate that sialic acid can be used as a sole carbon source for growth. Several bacteria that are able to colonize the human airway or gut mucosa have been shown to utilize sialic acid as a carbon source, including Haemophilus influenzae and multiple streptococcal and bifidobacterial strains (4, 5, 28, 34, 37). Growth of S. pneumoniae on free sialic acid appeared less efficient than growth on the same molar concentration of glucose under our experimental conditions. While the reason for this remains unknown, it may relate to the differences in the carbohydrate structures or mechanisms of transport. Even though growth on sialic acid may be less efficient than that on other carbohydrates, because it is often the first carbohydrate encountered on glycoconjugates it is likely used as a carbon source in the airway.
Growth on sialic acid is dependent upon satABC, which encodes an ABC transporter. The absence of satABC resulted in reduced bacterial growth on a human glycoprotein and reduced colonization in a mouse model, suggesting that this genetic locus and therefore transport of sialic acid are important for human airway colonization. Previous studies have suggested coregulation of the operon containing satABC and nanA; as nanA encodes the neuraminidase that cleaves terminal sialic acid from host glycoconjugates, these data suggest that the cleavage and utilization of sialic acid is a coordinated process (13).
Despite this, a nanA mutant showed no reduction in colonization in the same model, although a role for NanA in murine colonization has previously been demonstrated (17, 21). The reason for this discrepancy is unclear, but data here are consistent with our previous colonization modeling with nanA mutants in other strain backgrounds (13, 14). These results may indicate that SatABC has additional unidentified substrates or that sialic acid is being made available to pneumococcus by some other mechanism. It is possible that host protein turnover or neuraminidases from other sources, including the host, other bacterial cohabitants of the niche, or pneumococcus itself (NanB), may each contribute to sialic acid availability that is independent of NanA.
The SatABC transporter is a member of the ABC transport superfamily. Although other ABC transporters are predicted to import sialic acid, the only other characterized ABC transporter responsible for sialic acid import is SatABCD in Haemophilus ducreyi (25). ABC importers require two nuclear binding domains (ATPases), two transmembrane domains (permeases), and a substrate-binding domain that confers specificity (9, 22). The genetic organization of satABC in pneumococcus notably lacks predicted ATPases. While transport components are typically organized within a single operon, there are other examples of streptococcal species lacking ATPases in operon organization (39). Our data show that the SatABC transporter is functional for the uptake and utilization of sialic acid and thus require that the ATPases be encoded elsewhere.
The functions of the other two predicted sialic acid transporters remain unknown. Despite SP1688-90 and SP1328 being located within the same operons as nanB and nanC, respectively, we could demonstrate no role for either in sialic acid transport under our assay conditions. These data suggest that the additional predicted transporters either are specific for other substrates or contribute to sialic acid transport under other conditions. SP1688-90 is considered part of the core pneumococcal genome and as such is likely essential for some aspect of pathogenesis. In contrast, the gene encoding the predicted symporter (SP1328) is not part of the core genome (20). SP1328 is encoded within a 20-kb region that correlates with invasive disease; however, how each gene product within this region contributes to this is unknown (20). The absence of this region in some strains demonstrates that SP1328 cannot be essential for human colonization (23). Furthermore, our data demonstrate that this locus is not essential for invasive disease, as neither of our clinical isolates recovered from blood possess this genomic region (Table 1).
The human glycoprotein AGP contains N-linked glycans with sialic acid, galactose, and N-acetylglucosamine that can be sequentially cleaved by pneumococcal glycosidases (14). Here we show that sialic acid released from AGP is imported via SatABC and utilized as a carbon source for growth. For these experiments, a limiting concentration of AGP was used; given the solubility of AGP, higher concentrations of AGP could not be tested. This leaves open the possibility that sialic acid is not utilized for growth in vivo, where pneumococcus presumably encounters an excess of glycoconjugates. However, the significant reduction of the satABC mutant in murine colonization supports the hypothesis that sialic acid is used as a carbon source in vivo.
In summary, we have demonstrated that S. pneumoniae is able to utilize sialic acid as a sole carbon source and have identified satABC as encoding an ABC transporter specific for sialic acid. Furthermore, we have shown that import of sialic acid by SatABC contributes to colonization in the airway.
Acknowledgments
This work was supported by American Heart Association Predoctoral Fellowship 10PRE3490014 (C.M.) and National Institute of Allergy and Infectious Diseases grant 1R01AI076341 (S.J.K.).
We thank Robert Munson, Jr., for his assistance with design of the transport assays.
Editor: A. Camilli
Footnotes
Published ahead of print on 28 December 2010.
REFERENCES
- 1.Allen, S., A. Zaleski, J. W. Johnston, B. W. Gibson, and M. A. Apicella. 2005. Novel sialic acid transporter of Haemophilus influenzae. Infect. Immun. 73:5291-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almagro-Moreno, S., and E. F. Boyd. 2009. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol. Biol. 9:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Bender, M. H., and J. N. Weiser. 2006. The atypical amino-terminal LPNTG-containing domain of the pneumococcal human IgA1-specific protease is required for proper enzyme localization and function. Mol. Microbiol. 61:526-543. [DOI] [PubMed] [Google Scholar]
- 3.Burnaugh, A. M., L. J. Frantz, and S. J. King. 2008. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 190:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byers, H. L., K. A. Homer, and D. Beighton. 1996. Utilization of sialic acid by viridans streptococci. J. Dent. Res. 75:1564-1571. [DOI] [PubMed] [Google Scholar]
- 5.Byers, H. L., K. A. Homer, E. Tarelli, and D. Beighton. 1999. N-acetylneuraminic acid transport by Streptococcus oralis strain AR3. J. Med. Microbiol. 48:375-381. [DOI] [PubMed] [Google Scholar]
- 6.Chan, P. F., et al. 2003. Characterization of a novel fucose-regulated promoter (PfcsK) suitable for gene essentiality and antibacterial mode-of-action studies in Streptococcus pneumoniae. J. Bacteriol. 185:2051-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, X., and A. Varki. 2010. Advances in the biology and chemistry of sialic acids. ACS Chem. Biol. 5:163-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalia, A. B., A. J. Standish, and J. N. Weiser. 2010. Three surface exoglycosidases from Streptococcus pneumoniae, NanA, BgaA, and StrH, promote resistance to opsonophagocytic killing by human neutrophils. Infect. Immun. 78:2108-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holland, I. B., and M. A. Blight. 1999. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 293:381-399. [DOI] [PubMed] [Google Scholar]
- 10.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 11.Iyer, R., N. S. Baliga, and A. Camilli. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187:8340-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King, S. J. 2010. Pneumococcal modification of host sugars: a major contributor to colonization of the human airway? Mol. Oral Microbiol. 25:15-24. [DOI] [PubMed] [Google Scholar]
- 13.King, S. J., et al. 2004. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol. Microbiol. 54:159-171. [DOI] [PubMed] [Google Scholar]
- 14.King, S. J., K. R. Hippe, and J. N. Weiser. 2006. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol. Microbiol. 59:961-974. [DOI] [PubMed] [Google Scholar]
- 15.Kloosterman, T. G., J. J. Bijlsma, J. Kok, and O. P. Kuipers. 2006. To have neighbour's fare: extending the molecular toolbox for Streptococcus pneumoniae. Microbiology 152:351-359. [DOI] [PubMed] [Google Scholar]
- 16.Lock, R. A., J. C. Paton, and D. Hansman. 1988. Purification and immunological characterization of neuraminidase produced by Streptococcus pneumoniae. Microb. Pathog. 4:33-43. [DOI] [PubMed] [Google Scholar]
- 17.Manco, S., et al. 2006. Pneumococcal neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect. Immun. 74:4014-4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marion, C., et al. 2009. Identification of a pneumococcal glycosidase that modifies O-linked glycans. Infect. Immun. 77:1389-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCool, T. L., and J. N. Weiser. 2004. Limited role of antibody in clearance of Streptococcus pneumoniae in a murine model of colonization. Infect. Immun. 72:5807-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.McCool, T. L., T. R. Cate, G. Moy, and J. N. Weiser. 2002. The immune response to pneumococcal proteins during experimental human carriage. J. Exp. Med. 195:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obert, C., et al. 2006. Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect. Immun. 74:4766-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orihuela, C. J., G. Gao, K. P. Francis, J. Yu, and E. I. Tuomanen. 2004. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J. Infect. Dis. 190:1661-1669. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen, P. L. 2005. Transport ATPases: structure, motors, mechanism and medicine: a brief overview. J. Bioenerg. Biomembr. 37:349-357. [DOI] [PubMed] [Google Scholar]
- 23.Pettigrew, M. M., K. P. Fennie, M. P. York, J. Daniels, and F. Ghaffar. 2006. Variation in the presence of neuraminidase genes among Streptococcus pneumoniae isolates with identical sequence types. Infect. Immun. 74:3360-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips, N. J., et al. 1990. Structural models for the cell surface lipooligosaccharides of Neisseria gonorrhoeae and Haemophilus influenzae. Biomed. Environ. Mass Spectrom. 19:731-745. [DOI] [PubMed] [Google Scholar]
- 25.Post, D. M., R. Mungur, B. W. Gibson, and R. S. Munson, Jr. 2005. Identification of a novel sialic acid transporter in Haemophilus ducreyi. Infect. Immun. 73:6727-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy, S., C. W. Douglas, and G. P. Stafford. 2010. A novel sialic acid utilization and uptake system in the periodontal pathogen Tannerella forsythia. J. Bacteriol. 192:2285-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Severi, E., A. H. Hosie, J. A. Hawkhead, and G. H. Thomas. 2010. Characterization of a novel sialic acid transporter of the sodium solute symporter (SSS) family and in vivo comparison with known bacterial sialic acid transporters. FEMS Microbiol. Lett. 304:47-54. [DOI] [PubMed] [Google Scholar]
- 28.Severi, E., et al. 2005. Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol. Microbiol. 58:1173-1185. [DOI] [PubMed] [Google Scholar]
- 29.Shakhnovich, E. A., S. J. King, and J. N. Weiser. 2002. Neuraminidase expressed by Streptococcus pneumoniae desialylates the lipopolysaccharide of Neisseria meningitidis and Haemophilus influenzae: a paradigm for interbacterial competition among pathogens of the human respiratory tract. Infect. Immun. 70:7161-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steenbergen, S. M., et al. 2005. Sialic acid metabolism and systemic pasteurellosis. Infect. Immun. 73:1284-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sung, C. K., H. Li, J. P. Claverys, and D. A. Morrison. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tettelin, H., et al. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 33.Uchiyama, S., et al. 2009. The surface-anchored NanA protein promotes pneumococcal brain endothelial cell invasion. J. Exp. Med. 206:1845-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vimr, E., C. Lichtensteiger, and S. Steenbergen. 2000. Sialic acid metabolism's dual function in Haemophilus influenzae. Mol. Microbiol. 36:1113-1123. [DOI] [PubMed] [Google Scholar]
- 35.Vimr, E. R., K. A. Kalivoda, E. L. Deszo, and S. M. Steenbergen. 2004. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68:132-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vimr, E. R., and F. A. Troy. 1985. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J. Bacteriol. 164:845-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward, R. E., M. Ninonuevo, D. A. Mills, C. B. Lebrilla, and J. B. German. 2007. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol. Nutr. Food Res. 51:1398-1405. [DOI] [PubMed] [Google Scholar]
- 38.Watt, J. P., et al. 2004. Nasopharyngeal versus oropharyngeal sampling for detection of pneumococcal carriage in adults. J. Clin. Microbiol. 42:4974-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webb, A. J., K. A. Homer, and A. H. Hosie. 2008. Two closely related ABC transporters in Streptococcus mutans are involved in disaccharide and/or oligosaccharide uptake. J. Bacteriol. 190:168-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiser, J. N., R. Austrian, P. K. Sreenivasan, and H. R. Masure. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62:2582-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whatmore, A. M., V. A. Barcus, and C. G. Dowson. 1999. Genetic diversity of the streptococcal competence (com) gene locus. J. Bacteriol. 181:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.World Health Organization. 2008. The global burden of disease: 2004 update. http://www.who.int/healthinfo/global_burden_disease/2004_report_update/en/index.html.
- 43.Yesilkaya, H., S. Manco, A. Kadioglu, V. S. Terra, and P. W. Andrew. 2008. The ability to utilize mucin affects the regulation of virulence gene expression in Streptococcus pneumoniae. FEMS Microbiol. Lett. 278:231-235. [DOI] [PubMed] [Google Scholar]