Abstract
Uropathogenic Escherichia coli (UPEC), the predominant cause of uncomplicated urinary tract infection (UTI), utilizes an array of outer membrane iron receptors to facilitate siderophore and heme import from within the iron-limited urinary tract. While these systems are required for UPEC in vivo fitness and are assumed to be functionally redundant, the relative contributions of specific receptors to pathogenesis are unknown. To delineate the relative roles of distinct UPEC iron acquisition systems in UTI, isogenic mutants in UPEC strain CFT073 or 536 lacking individual receptors were competed against one another in vivo in a series of mixed infections. When combinations of up to four mutants were coinoculated using a CBA/J mouse model of ascending UTI, catecholate receptor mutants (ΔfepA, Δiha, and ΔiroN mutants) were equally fit, suggesting redundant function. However, noncatecholate siderophore receptor mutants, including the ΔiutA aerobactin receptor mutant and the ΔfyuA yersiniabactin receptor mutant, were frequently outcompeted by coinoculated mutants, indicating that these systems contribute more significantly to UPEC iron acquisition in vivo. A tissue-specific preference for heme acquisition was also observed, as a heme uptake-deficient Δhma ΔchuA double mutant was outcompeted by siderophore receptor mutants specifically during kidney colonization. The relative contribution of each receptor to UTI only partially correlated with in vivo levels of receptor gene expression, indicating that other factors likely contributed to the observed fitness differences. Overall, our results suggest that UPEC iron receptors provide both functional redundancy and niche specificity for this pathogen as it colonizes distinct sites within the urinary tract.
Both pathogenic and nonpathogenic bacteria encode complex systems for acquisition of environmental iron, an essential metal that is insoluble under neutral, aerobic conditions and protein bound within mammalian hosts. Uptake of iron in bacteria is achieved primarily through the synthesis, export, and uptake of small iron-chelating molecules termed siderophores. In Gram-negative bacteria, uptake of ferrisiderophores and heme iron is facilitated by specific outer membrane receptors. Ligand transport by these structurally conserved beta-barrel proteins is an energy-dependent process, requiring transduction of the proton motive force via the inner membrane complex TonB-ExbBD.
An essential role for iron acquisition by Escherichia coli, the causative agent of most community-acquired urinary tract infections (UTIs), has been well established. The genome of pyelonephritis E. coli strain CFT073 encodes 14 characterized outer membrane iron compound receptors, three siderophore biosynthesis systems, and a number of putative TonB-dependent receptors that may also be involved in iron acquisition (51). The genes encoding these iron acquisition systems were among the most highly upregulated genes in strain CFT073 during experimental infection of the murine urinary tract (46). Similarly, most of these systems, including the yersiniabactin and heme uptake loci, were highly expressed by clinical strains of E. coli in urine collected directly from women with cystitis (14), implying a role for iron uptake during human UTI. Indeed, obtaining iron is a critical process for uropathogenic E. coli (UPEC) survival in vivo, as a tonB mutant was severely attenuated in a mouse pyelonephritis model, indicating that TonB-dependent receptors are required for UTI (49). Interestingly, asymptomatic bacteriuria strains, while lacking or containing mutations in classical UPEC virulence factors such as fimbriae, express the full complement of iron acquisition systems, providing further evidence for the necessity of iron uptake during successful urinary tract colonization (40).
UPEC strains produce up to four different siderophores: catecholates enterobactin and salmochelin, the hydroxamate aerobactin, and yersiniabactin, a mixed-type siderophore (20). Salmochelin, a glucosylated form of enterobactin that is resistant to binding and inactivation by the mammalian protein lipocalin-2 (11), also represents an immune evasion strategy. While enterobactin is produced and utilized by nearly all E. coli strains, genes encoding aerobactin, salmochelin, and yersiniabactin receptors are found more frequently among pathogens (5, 23, 25). Furthermore, increased yersiniabactin and salmochelin synthesis was detected in urinary E. coli isolates, compared to fecal isolates, of patients with recurrent UTI (20).
Although iron acquisition in general is required for UTI, no single uptake system that is necessary for this function has been identified. Although able to colonize the urinary tract during independent challenge, mutants defective for ChuA- or Hma-mediated heme uptake (15, 49), IroN-mediated salmochelin uptake (43), IutA-mediated aerobactin uptake (49), or Iha-mediated enterobactin uptake (22) were outcompeted by a wild-type strain during competitive infection. Similarly, UPEC strain CFT073 maintained the ability to colonize the murine urinary tract when either enterobactin or aerobactin synthesis was disrupted, but an iucB entD double mutant, defective for synthesis of both siderophores, was attenuated (49). Thus, functional redundancy exists among these iron acquisition systems.
Although both heme and siderophore iron acquisition contribute to the ability of UPEC to infect the urinary tract, evidence for distinct roles of specific systems exists. Heme uptake via ChuA was shown to play a role in bladder and kidney colonization (49), as well as in the formation of intracellular bacterial communities within superficial bladder epithelial cells (39), contributing to virulence more significantly than the heme receptor Hma (15). Mutants lacking the salmochelin receptor IroN, which were defective for urothelial cell invasion (10), were outcompeted by the wild-type strain in the bladders of infected mice, but kidney colonization was unaltered (43). Conversely, a mutant deficient in both enterobactin and aerobactin synthesis was only modestly attenuated in the bladder but showed a 1-log reduction in kidney colonization (49). Consequently, different iron acquisition systems appear to have differing contributions to bladder and kidney infection. Moreover, there is evidence for the unequal contributions of distinct iron acquisition systems to the virulence of other bacterial pathogens (29, 47, 50).
While there is substantial evidence for functional redundancy among UPEC iron acquisition systems, we hypothesized that specific systems may have a greater impact on urinary tract colonization. In this study, we used a series of mixed infections, competing mutants defective for specific iron receptors against one another to directly assess the relative contributions of heme and siderophore receptors to UTI. The data presented here support a model of UPEC pathogenesis in which uptake of heme and uptake of noncatecholate siderophores play dominant roles at specific sites within the urinary tract.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli strains used in this study are listed in Table 1. Bacteria were routinely cultured in Luria broth (10 g/liter tryptone, 5 g/liter yeast extract, 0.5 g/liter NaCl) with appropriate antibiotics (100 μg/ml ampicillin, 25 μg/ml kanamycin, 20 μg/ml chloramphenicol) at 37°C with aeration.
TABLE 1.
E. coli strains and plasmids
| Strain or plasmid | Descriptiona | Reference(s) or source |
|---|---|---|
| Strains | ||
| CFT073 | E. coli pyelonephritis isolate (Ent+ Sal+ Aer+) | 31 |
| 536 | E. coli UTI isolate (Ent+ Sal+ Ybt+) | 2 |
| ΔfepA mutant | CFT073 ΔfepA::cat Camr | This study |
| Δiha mutant | CFT073 Δiha::scaR Kans; kan cassette removed | This study |
| Δiha ΔfepA mutant | CFT073 Δiha::scaR ΔfepA::kan Kanr; kan cassette removed | This study |
| ΔiroN mutant | CFT073 ΔiroN::kan Kanr or ΔiroN::scaR Kans; kan cassette removed | This study |
| ΔcirA mutant | CFT073 ΔcirA::kan Kanr | This study |
| Δfiu mutant | CFT073 Δfiu::cat Camr | This study |
| ΔiutA mutant | CFT073 ΔiutA::cat Camr or ΔiutA::scaR Kans; kan cassette removed | This study |
| ΔfhuA mutant | CFT073 ΔfhuA::kan Kanr | This study |
| ΔfhuE mutant | CFT073 ΔfhuE::cam Camr | This study |
| chuA mutant | CFT073 chuA::cat Camr | 49 |
| Δhma chuA mutant | CFT073 Δhma::kan chuA::cat Kanr Camr | 15 |
| ΔfepA536 mutant | 536 ΔfepA::cat Camr | This study |
| ΔiroN536 mutant | 536 ΔiroN::scaR Cams; cat cassette removed | This study |
| ΔfhuA536 mutant | 536 ΔfhuA::scaR Cams; cat cassette removed | This study |
| ΔfhuE536 mutant | 536 ΔfhuE::cat Camr | This study |
| ΔfyuA536 mutant | 536 ΔfyuA::kan Kanr | This study |
| Δhma ΔchuA536 mutant | 536 Δhma::kan ΔchuA::cat Kanr Camr | This study |
| ΔentF536 mutant | 536 ΔentF::kan Kanr | This study |
| ΔybtS536 mutant | 536 ΔybtS::kan Kanr | This study |
| Plasmids | ||
| pKD4 | λ Red template vector; Kanr Ampr | 7 |
| pKD3 | λ Red template vector; Camr Ampr | 7 |
| pKD46 | Red recombinase helper plasmid, temp sensitive; Ampr | 7 |
| pCP20 | Flp recombinase helper plasmid, temp sensitive; Ampr | 7 |
| pGEN | pGEN-MCS, promoterless expression vector, p15A ori (copy no. ∼15); par hok sok mok parM parR Ampr | 13, 27 |
| pnativefyuA | fyuA with native promoter (290 bp upstream) in pGEN; Ampr | This study |
| pnativechuA | chuA with native promoter (360 bp upstream) in pGEN; Ampr | This study |
| pnativehma | hma with native promoter (900 bp upstream) in pGEN; Ampr | 15 |
Ent, enterobactin; Sal, salmochelin; Aer, aerobactin; Ybt, yersiniabactin; Kan, kanamycin; Cam, chloramphenicol; Amp, ampicillin.
Mutant construction and cloning.
Isogenic mutants were generated in E. coli CFT073 and 536 (Table 1) using the λ Red recombinase system (7). Briefly, kanamycin or chloramphenicol resistance cassettes were PCR amplified from the template plasmid pKD4 or pKD3, respectively, using primers containing regions identical to the 5′ and 3′ ends of the gene to be deleted (Table 2). The resulting product was used to replace >80% of the target gene by homologous recombination in bacteria expressing Red recombinase from pKD46. Flp recombinase expression from pCP20 was used to remove antibiotic resistance markers from the strains. All mutants were confirmed by PCR.
TABLE 2.
Primers for mutant construction and complementationa
| Gene or plasmid | Forward primer | Reverse primer |
|---|---|---|
| fepA | GCCCGCGATGTATCGGAGATCATTCGTACCATGCCTGGCGGTGTAGGCTGGAGCTGCTTC | ACGTCCCGGCTCGTTATAGGTATACGCACCGGCACCGGCGATGGGAATTAGCCATGGTCC |
| iha | CCACTCTGGCTTCCGTAGTCATTCCCTGTCTCGGATTTTCGTGTAGGCTGGAGCTGCTTC | GTTGATGATCCCGTCTGGAAGTAATCACCGGCATACAGCGATGGGAATTAGCCATGGTCC |
| iroN | CTCCGACGATGATAATGACGAGACTCTGGTGGTGGAAGCCGTGTAGGCTGGAGCTGCTTC | CGGCCTGGCTCGTTATAGGTATTCGCCCCTTCAGAAGATCATGGGAATTAGCCATGGTCC |
| cirA | CCGTACGCAAGGGGACGTAAAGAAGATGTGAGCGATACCCGTGTAGGCTGGAGCTGCTTC | CGTTATAGCTGTAGTCGTCACGACTGAGATCCTTGTCGCCATGGGAATTAGCCATGGTCC |
| fiu | GCCGGTCTTTGTATTGGTATTACGCCTGTGGCACAGGCACGTGTAGGCTGGAGCTGCTTC | CTGTGAGCAAGAAGGTTCTTGGCTCGCCCGGGTGATAACGATGGGAATTAGCCATGGTCC |
| iutA | CCCGGGCTCTTGGTCCGCTGCTTCTTGTCGTGCTGTCACCGTGTAGGCTGGAGCTGCTTC | CCGGCCTTTGTAGTCGTACAGTGAAGCAGGGCCGTAACCCGATGGGAATTAGCCATGGTCC |
| fhuA | GTTGTAGTAGCCACAGCGGTTAGCGGCATGTCTGTTTATGGTGTAGGCTGGAGCTGCTTC | CTGACGTTCTGCGCCCCAGAAGCAACCATAAGTGTTAAAGATGGGAATTAGCCATGGTCC |
| fhuE | ACATCAAGCTATCATCAAACCGTCACTCCTTGCCGGTTGCGTGTAGGCTGGAGCTGCTTC | CGGTAATGCTGAAATTACGCGGTGCGCCGTAGACGATAGATGGGAATTAGCCATGGTCC |
| chuA | CCGCAATTTACCTCGTTGCGTTTGAGTTTGTTGGCTTTGGGTGTAGGCTGGAGCTGCTTC | CGTTACGACCATCCTGTGGGATGCCTTGCGGCGACCAGTAATGGGAATTAGCCATGGTCC |
| hma | GTCGAATTTATATCCTTTCCACTCTGTCATTATGCATTTCGTGTAGGCTGGAGCTGCTTC | CAGAGATTATGTTGGCCTGACCCATCACAAGAGTATGTTTATGGGAATTAGCCATGGTCC |
| fyuA | GGCTTTATCCTCTGGCCTTGGGGGGATTATTGCTCCCCGCCGTGTAGGCTGGAGCTGCTTC | CGTATTGATACCGACGGTGCGACCCATATTGACCTGCGCGATGGGAATTAGCCATGGTCC |
| entF | GCCAGCATTTACCTTTGGTCGCCGCACAGCCCGGCATCTGGTGTAGGCTGGAGCTGCTTC | CGCCACCGGAGAGATAATATCCACATGCGCACAATCCTGACATGGGAATTAGCCATGGTCC |
| ybtS | GAATTTCTACATCTGGCGTTACCAGAGGAACAATGGCTACGTGTAGGCTGGAGCTGCTTC | ACCATTAAATAGGGCGCAATGCTCGCTAATTTCTCCCGGGATGGGAATTAGCCATGGTCC |
| pnativefyuA | CCTGCAGGTGCCAACTGACTATC | CCATGGTGTAAAAGGGATACCTTTTCGGT |
| pnativechuA | CCGTACGAATTCTTATTGTGTTAATGGCGG | GCTTTCCGGCCGTTACCATTGATAACTCACG |
Primer sequences are listed 5′ to 3′. Underlined sequences in mutant construction primers are identical to regions at the 5′ and 3′ ends of the gene to be disrupted.
For complementation, the fyuA and chuA open reading frames (ORFs) plus ∼300 bp upstream sequence were PCR amplified from E. coli 536 genomic DNA using EasyA polymerase (Agilent). Fragments containing fyuA and chuA were cloned into the SbfI-NcoI and EcoRI-EagI sites of pGEN (Table 1), respectively. The resulting constructs were verified by DNA sequencing.
In vitro growth.
CFT073 and 536 wild-type strains and isogenic mutants were cultured overnight in LB in the absence of antibiotics. Cultures were washed once in phosphate-buffered saline (PBS) and standardized to an optical density at 600 nm (OD600) of ∼1.0, and approximately 105 CFU was inoculated into LB or LB containing 200 μM 2,2′-dipyridyl (DPD). Growth curves were determined using a Bioscreen C growth curve analyzer (Growth Curves) at 37°C with aeration.
Murine cochallenge model of ascending UTI.
Six- to 8-week-old female CBA/J mice were inoculated transurethrally as previously described (21), with modification. Bacteria were cultured overnight in LB without antibiotics prior to inoculation. Each strain was harvested by centrifugation (3,500 × g, 30 min, 4°C) and resuspended in PBS to an OD600 of about 4.0 (∼109 CFU/ml). Three to four strains were mixed in equal ratios, and 50 μl of this suspension (108 CFU) was delivered to each mouse via a sterile 0.28-mm polyethylene catheter attached to an infusion pump (Harvard Apparatus). To quantify each strain in the inoculum, dilutions were plated onto LB agar with and without appropriate antibiotics to differentiate antibiotic-sensitive and -resistant mutants. Seventy-two hours postinoculation (hpi), organs were removed from euthanized mice and homogenized in 3 ml PBS using a GLH homogenizer (Omni International). Using an Autoplate 4000 spiral plater (Spiral Biotech), tissue homogenate was plated onto LB agar containing appropriate antibiotics. Colonies were enumerated with a QCount automated plate counter (Spiral Biotech) and CFU/g tissue determined (output). For cochallenge experiment (two inoculated strains), competitive indices were calculated by dividing the ratio of ΔfepA to Δiha mutant in the output by the ratio of the two mutants in the inoculum [(CFUfepA/CFUiha)output/(CFUfepA/CFUiha)input]. For multistrain coinfections, fitness indices (FI) for each organ sample were calculated by dividing the fraction of each mutant in the output by the fraction of each mutant in the inoculum [(CFUmutant/CFUtotal)output/(CFUmutant/CFUtotal)input].
RNA isolation and qPCR.
For in vitro measurements, strains were cultured with aeration to late exponential phase (OD600 of 0.5 to 0.6) in 100 ml LB with or without 200 μM DPD and 10 μM hemin. Culture samples (200 μl) were mixed with 25 μl cold 5% phenol-ethanol stop solution, pelleted (1 min, 10,000 × g, 4°C), and stored at −80°C for RNA isolation. For in vivo samples, CBA/J mice (n = 15) were infected with E. coli strain CFT073 or 536 as described above. From 6 to 72 hpi, urine was collected periodically and pooled from each cage of mice (5 animals). Immediately after collection, cold 5% phenol-ethanol stop solution was added to each pool (0.125 μl solution per μl urine), and samples were pelleted (1 min, 10,000 × g, 4°C) and stored at −80°C. For RNA isolation, bacterial pellets (collected from five mice) were pooled from all time points. Thawed pellets were resuspended in 100 μl RNase-free Tris-EDTA (TE) containing 1 mg/ml lysozyme and RNA isolated using the RNeasy protocol (Qiagen). Samples were DNase treated according to the Turbo DNA-Free procedure (Ambion) and cDNA synthesized using SuperScript II first-strand synthesis reagents (Invitrogen) according to the manufacturer's instructions. Real-time quantitative PCR (qPCR) was performed using 30 ng cDNA template and Brilliant SYBR green reagents (Agilent). Data were normalized to the gapA transcript and analyzed using MxPro 4.0 software (Agilent).
In vitro competition assay.
Cocultures were performed as previously described (28). Overnight cultures of CFT073 ΔfhuA, ΔfhuE, and ΔiutA mutants were diluted 1:100 in LB and cultured to late exponential phase. Culture samples (1 ml) were pelleted (1 min, 10,000 × g, room temperature), resuspended in 1 ml PBS, and standardized to an OD600 of 1.0. Cell suspensions were mixed 1:1:1, diluted 1:100 into 5 ml LB with or without 200 μM DPD, and incubated at 37°C. Every 8 or 16 h, cultures were passaged 1:100 or 1:500 into 5 ml fresh medium, respectively. At 24, 48, and 72 h, cultures were plated on LB agar and LB containing kanamycin or chloramphenicol to enumerate each mutant.
Iron source utilization assays.
The ability of complemented mutants to utilize various iron sources was determined as previously described (48), with modifications. Supernatants from the ΔentF536 or ΔybtS536 mutant, cultured overnight with 100 μM DPD, were filter sterilized and concentrated 10-fold using a vacuum centrifuge. Overnight cultures of wild-type 536 and ΔfyuA536 and Δhma ΔchuA536 mutants carrying pGEN constructs were diluted 1:100 into LB containing 200 μM DPD and cultured for ∼6 h at 37°C. One-milliliter samples of iron-limited cultures were pelleted (1 min, 10,000 × g, room temperature), resuspended in 1 ml PBS, and adjusted to an OD600 of 1.0. Bacterial suspension was added to molten sorbitol-MacConkey agar containing 350 or 375 μM DPD to ∼5 × 104 CFU/ml. Using the ends of pipette tips, ∼8-mm diameter wells were punched into solidified plates, and 80 μl of 100 μM FeCl2, 100 μM hemin, or concentrated supernatant was added to each well (6). Plates were imaged, and diameter of growth surrounding each well was measured after 72 h at 37°C.
Statistical analyses.
Statistical analyses were performed using GraphPad Prism 5. For competitive infections, significance was determined using the Wilcoxon rank sum test on log10-transformed fitness indices or competitive indices with a hypothetical median of 0 (log101 = 0). The Mann-Whitney test was used to compare fitness indices for the same mutant between different organs. Correlations were determined using the Spearman rank test.
RESULTS
Heterologous iron receptor gene expression is unaltered in specific receptor mutant strains.
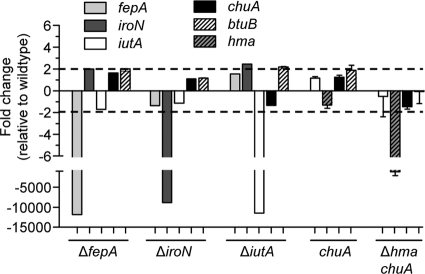
To examine the contributions of heme and siderophore uptake to UTI, deletion mutations in individual iron receptor genes were made in UPEC strain CFT073. To simplify analysis, only receptors having defined substrates were included (Table 3). Growth of all mutants was examined during culture in LB and in the presence of 200 μM DPD. All mutants had growth rates similar to that of the isogenic parent strain both in rich medium and under iron limitation (data not shown). Previous work from our laboratory has shown that expression of iron receptor genes in CFT073 is upregulated under iron-limiting conditions, both in vitro (1) and in vivo (46). As expression of these genes is likely Fur regulated, they are predicted to be expressed independently of one another. Nonetheless, to determine whether receptor gene expression was dysregulated in the receptor mutants, transcripts isolated from selected mutants, cultured under iron limitation, were measured by qPCR. With the exception of the deleted genes themselves, none of the receptor genes examined differed by more than 2-fold from wild-type expression levels (Fig. 1). Lack of chuA “downregulation” by the chuA and Δhma chuA mutants is due to the fact that, rather than gene deletion, these constructs contain an insertion mutation in chuA (Table 1) and retain the entire chuA coding sequence. Although expression of iroN appeared slightly elevated in the ΔiutA mutant, expression of btuB, the vitamin B12 receptor included as a control, was also slightly elevated, suggesting that this difference is not biologically significant. For heme receptor Δhma (data not shown), chuA, and Δhma chuA mutants, heterologous receptor expression was unaltered in both the absence (Fig. 1) and the presence (data not shown) of excess hemin. Thus, deletion of one or more outer membrane iron receptor genes does not affect transcript abundance of the remaining receptor genes, indicating that any defects in mutant fitness are not compensated for by upregulation of heterologous receptor genes.
TABLE 3.
UPEC iron receptors and summary of receptor mutant fitness during competitive mixed infection
| Gene | Locus tag | Receptor substrate | Substrate type | Reference | Gene presence |
No. of FI <1a |
||
|---|---|---|---|---|---|---|---|---|
| CFT073 | 536 | Bladder | Kidney | |||||
| fepA | c0669 | Enterobactin | Catecholate | 54 | + | + | 0/5 | 1/5 |
| iha | c3610 | Enterobactin | Catecholate | 30 | + | − | 0/5 | 0/5 |
| iroN | c1250 | Salmochelin | Catecholate | 19 | + | + | 1/4 | 0/4 |
| fiu | c1374 | DBS, DHBb | Catecholate | 17 | + | + | 0/1 | 0/1 |
| cirA | c2690 | DBS, DHB | Catecholate | 17 | + | + | 0/1 | 0/1 |
| iutA | c3623 | Aerobactin | Hydroxamate | 8 | + | − | 2/3 | 3/3 |
| fhuA | c0185 | Ferrichrome | Hydroxamate | 24 | + | + | 0/2 | 0/2 |
| fhuE | c1374 | Coprogen, rhodotorulic acid | Hydroxamate | 18 | + | + | 0/2 | 0/2 |
| fyuA | c2436 | Yersiniabactin | Mixed | 38 | +c | + | 2/2 | 1/2 |
| chuA | c4308 | Heme | NAd | 48 | + | + | 1/3 | 3/3 |
| hma | c2482 | Heme | NA | 15 | + | + | 1/3 | 3/3 |
Number of times a mutant defective for the indicated gene was outcompeted (fitness index significantly less than 1.0) per number of mixed infections in which it was included (both CFT073 and 536 backgrounds). Values for fepA and iha both include results for the Δiha ΔfepA double mutant, and values for chuA and hma reflect data from the Δhma ΔchuA double mutant.
DBS, dihydroxybenzolyserine; DHB, dihyroxybenzoate.
CFT073 carries an intact fyuA gene, but nonsense mutations and a 711-bp insertion element in genes encoding yersiniabactin biosynthesis machinery (irp1 and irp2) prevent siderophore production.
NA, not applicable.
FIG. 1.
Heme and siderophore receptor expression levels by qPCR in receptor mutants. RNA was isolated from wild-type CFT073 and receptor mutants (indicated on x axis) cultured in LB containing 200 μM DPD. Transcript levels were measured in cDNA preparations from each strain and normalized to the gapA level, and results are shown as fold change relative to wild-type levels. Dashed lines represent 2-fold change and bars show the means.
Catecholate siderophore receptor mutants are equally fit in the murine urinary tract.
Previous work from our laboratory demonstrated that a chuA heme receptor mutant was outcompeted by an hma heme receptor mutant in a murine model of UTI (15), indicating that receptors with similar substrate specificities can have differing contributions to UPEC colonization. Because E. coli CFT073 encodes multiple receptors that import the catecholate siderophore enterobactin, we hypothesized that they may contribute unequally to UTI. To test this, mice were challenged transurethrally with a 1:1 mixture of a ΔfepA mutant and a mutant lacking the IrgA homolog adhesin gene iha, which is carried outside the ent fep locus and transports both enterobactin and its iron-chelating linear degradation product dihydroxybenzoylserine (DBS) (30). Competitive indices, calculated at 72 hpi, indicated that both mutants were equally fit in the murine urinary tract (Fig. 2 A), although the ΔfepA mutant tended to be outcompeted by the Δiha mutant in the kidney (P = 0.0502).
FIG. 2.
Fitness of E. coli CFT073 catecholate siderophore receptor mutants during mixed infections. (A) Competitive indices from cochallenge in CBA/J mice transurethrally challenged with a mixture of equal amounts of ΔfepA and Δiha mutants (n = 7). The total inoculum equaled 1 × 108 CFU/mouse. (B to D) Fitness indices calculated from the cochallenge results shown in panel A (B) or from mixed infections in mice inoculated with a mixture of ΔfepA, Δiha, and ΔiroN mutants (n = 10) (C) or wild-type CFT073 (WT) and ΔcirA and Δfiu mutants (n = 9) (D). Fitness indices at 72 hpi were calculated by dividing the fraction of each mutant in the output (CFU/g tissue) by the fraction of each mutant in the input (CFU/ml inoculum). Bars represent the medians, and the dashed line indicates the theoretical fitness index of 1.0 (log101 = 0). *, P = 0.0333.
Salmochelin, imported by the receptor IroN, is a glucosylated form of enterobactin shown to resist sequestration by the host protein lipocalin-2 (11) and contribute to UPEC pathogenesis (43). In addition to salmochelin, IroN was demonstrated to transport enterobactin and DBS (37). Therefore, we speculated that salmochelin uptake via the receptor IroN may contribute more significantly to UPEC pathogenesis than FepA or Iha, the substrates of which are susceptible to lipocalin-2 inactivation. Overall, we reasoned that, while mutants defective for each of these receptors displayed fitness defects when competed against a wild-type parent strain (22, 43), the receptors may have unequal contributions to in vivo fitness that would not be apparent during traditional cochallenge experiments. To examine this possibility directly, a three-way in vivo competition assay was performed with CFT073 ΔfepA, Δiha, and ΔiroN mutants. Mutants were mixed at a 1:1:1 ratio and coinoculated transurethrally into CBA/J mice. However, because more than two strains were inoculated, we reasoned that calculation of traditional competitive indices would not take into account the competition occurring among all three mutant strains. Therefore, we developed a modified competitive index, which we termed the fitness index (FI), that compared the fraction of each mutant recovered from each organ to the fraction of the same mutant in the inoculum [(CFUmutant/CFUtotal)output/(CFUmutant/CFUtotal)input]. When the cochallenge data presented in Fig. 2A were reanalyzed by this new measure, similar results were obtained (Fig. 2B). However, FI values for the ΔfepA, Δiha, and ΔiroN mutants were not significantly different from 1.0 (log101.0 = 0) (Fig. 2C), indicating that all three mutants were equally fit in the murine urinary tract when placed in direct competition.
Two additional outer membrane receptors, CirA and Fiu, have been shown to transport the enterobactin breakdown products DBS and dihydroxybenzoate (DHB) (17). While roles for iha, iroN, and fepA (ent) in UPEC (22, 43, 49) have been demonstrated, the potential contributions of cirA and fiu are unknown. Before their relative contributions to UPEC colonization could be assessed, we first sought to test whether they played any role in UTI. Thus, isogenic ΔcirA and Δfiu mutants were coinoculated with wild-type CFT073. Surprisingly, neither mutant displayed a significant decrease in fitness when competed against the wild type (Fig. 2D), indicating that neither cirA nor fiu contributes significantly to UTI in this model. Consequently, they were not tested for their ability to compete against the other catecholate receptors. Together, these data demonstrate that the catecholate siderophore receptors FepA, Iha, and IroN contribute equally to UPEC colonization of the murine urinary tract.
Aerobactin receptor mutant is outcompeted in vivo but not in vitro.
While E. coli encodes several outer membrane receptors for the import of catecholate siderophores, a single receptor, IutA, is responsible for uptake of the hydroxamate siderophore aerobactin. Most E. coli strains, including CFT073, can also utilize the fungal hydroxamate siderophores ferrichrome, coprogen, and rhodotorulic acid, via the receptors FhuA and FhuE (18). Although we predicted that these xenosiderophore receptors would not be functional in the urinary tract in the absence of fungal species, we used the in vivo competition assay to measure the relative contributions of hydroxamate siderophore receptors to infection. As expected, the ΔiutA mutant was significantly outcompeted in both the bladder (P = 0.0020) and the kidneys (P = 0.0371) (Fig. 3 A), indicating that IutA is the most significant hydroxamate receptor during UTI. Interestingly, although the ΔiutA mutant was outcompeted in both organs, its bladder fitness indices were significantly lower than those in the kidney (P = 0.0232), suggesting that this receptor contributes most during bladder colonization and that nonaerobactin iron sources compensate for the loss of iutA during kidney infection. In contrast to in vivo competition, all hydroxamate receptor mutants were maintained at nearly equivalent ratios when cocultured in an in vitro competition assay (28), both during growth under iron-limited conditions (Fig. 3B) and in rich medium (data not shown). This suggests that the multimutant competition infection approach is capable of specifically identifying mutants with in vivo fitness defects.
FIG. 3.

In vivo and in vitro competitive fitness of E. coli CFT073 hydroxamate receptor mutants. (A) In vivo competition in CBA/J mice transurethrally challenged with a mixture of equal amounts of ΔfhuA, ΔfhuE, and ΔiutA mutants (n = 10). Fitness indices at 72 hpi were calculated by dividing the fraction of each mutant in the output (CFU/g tissue) by the fraction of each mutant in the input (CFU/ml inoculum). Bars represent the medians, and the dashed line indicates the theoretical fitness index of 1.0. The P value displayed indicates significance by the Mann-Whitney test. *, P = 0.0371, and **, P = 0.0020, by the Wilcoxon signed-rank test. (B) In vitro culture competition of ΔfhuA, ΔfhuE, and ΔiutA mutants cultured in LB containing 200 μM DPD. After inoculation, cultures were passaged into fresh medium every 8 (1:100 dilution) or 16 (1:500 dilution) h, indicated by arrows. Means of triplicate cultures are plotted and error bars show standard errors of the means. Fitness indices, calculated as described for panel A, are shown.
Aerobactin and heme acquisition contributes most significantly to UTI.
To directly assess the relative contributions of aerobactin, salmochelin, and enterobactin siderophore uptake, ΔiroN, ΔfepA, and ΔiutA mutants were competed in a mixed infection of the murine UTI model. As observed in Fig. 2C, neither the ΔiroN mutant nor the ΔfepA mutant was significantly underrepresented in the total bacterial output of the bladder or kidneys, suggesting that they contribute relatively equally to virulence (Fig. 4 A). In contrast, in both the bladder and the kidneys, the ΔiutA mutant was significantly outcompeted (P = 0.0059 and P = 0.0098, respectively), indicating that iutA contributes more to UPEC iron uptake during urinary tract colonization than either iroN or fepA.
FIG. 4.

Fitness of E. coli CFT073 siderophore and heme receptor mutants during mixed infection. In vivo competition in CBA/J mice transurethrally challenged with a mixture of ΔiutA, ΔiroN, and ΔfepA mutants (n = 10) (A) or ΔiroN, Δiha ΔfepA, ΔiutA, and Δhma chuA mutants (n = 17) (B). Fitness indices at 72 hpi were calculated by dividing the fraction of each mutant in the output (CFU/g tissue) by the fraction of each mutant in the input (CFU/ml inoculum). Bars represent the medians, and the dashed line indicates the theoretical fitness index of 1.0. *, P < 0.02; **, P < 0.01.
In addition to receptors for ferrisiderophore uptake, E. coli CFT073 encodes two TonB-dependent heme receptors, ChuA and Hma, and requires at least one for full virulence (15). To establish the role for heme acquisition in the context of siderophore utilization, a heme uptake-deficient Δhma chuA double mutant was included in a four-way competition infection with the aerobactin and catecholate receptor mutants. To control for potential redundancy effects that may decrease the fitness of a mutant lacking two outer membrane iron receptors, a Δiha ΔfepA double mutant lacking the two known enterobactin receptors was constructed and included in the mixed infection. In both the bladders and the kidneys of infected mice, the Δhma chuA mutant was outcompeted by the coinoculated mutants (P = 0.0121 and P = 0.0052, respectively), while the Δiha ΔfepA mutant was not (Fig. 4B), demonstrating that Hma and ChuA contribute more to UPEC colonization than Iha and FepA. Similarly to our previous observations, the ΔiutA mutant was significantly underrepresented in the kidney after a 72-h infection (P = 0.0060), again suggesting the relative importance of aerobactin utilization. However, in contrast to the fitness of the ΔiroN mutant during cochallenge with the ΔiutA and ΔfepA mutants (Fig. 4A), here the ΔiroN mutant was outcompeted in the bladder (P = 0.0140), indicating that the magnitude of the competitive advantage provided by iroN depends on the relative fitness of the coinoculated mutants. These data taken together suggest that heme utilization via Hma and ChuA and aerobactin uptake via IutA contribute most significantly to UPEC iron acquisition during UTI.
Yersiniabactin and heme uptake is most critical for colonization by E. coli 536.
While E. coli CFT073 produces three siderophores, enterobactin, salmochelin, and aerobactin, the majority of UPEC strains produce a fourth siderophore, yersiniabactin, often instead of aerobactin (20). Because CFT073 does not produce yersiniabactin, UPEC strain 536 was used to assess the relative contribution of this mixed-type siderophore to UTI. Isogenic receptor mutants were constructed in strain 536 and iron source utilization assays performed to confirm their ability to mediate heme- and siderophore-dependent growth. As expected, while wild-type 536 was capable of using FeCl2, hemin, enterobactin/salmochelin (supernatants from a yersiniabactin-deficient ΔybtS536 mutant), or yersiniabactin (supernatants from an enterobactin- and salmochelin-deficient ΔentF536 mutant) as a sole iron source (Fig. 5 A), a ΔfyuA536 mutant could not grow utilizing supernatants from a ΔentF536 mutant (Fig. 5B). This defect in yersiniabactin utilization could be complemented by expression of fyuA in trans. Similarly to our findings with CFT073 (15), a Δhma ΔchuA536 double mutant was defective for hemin utilization and could be complemented with plasmids carrying either hma or chuA (Fig. 5A).
FIG. 5.
Relative contribution of yersiniabactin uptake to E. coli UTI. (A) Growth of the E. coli 536 wild type carrying pGEN vector (top left) or the Δhma ΔchuA536 mutant carrying pGEN vector (top right), pnativechuA (bottom left), or pnativehma (bottom right) around wells containing (clockwise from top) 100 μM FeCl2, concentrated supernatant from the ΔentF536 mutant, concentrated supernatant from the ΔybtS536 mutant, or 100 μM hemin. Bars indicate zones of growth. (B) Growth of the E. coli ΔfyuA536 mutant carrying pGEN vector (left) or pnativefyuA (right) around wells containing 100 μM FeCl2 or concentrated supernatants from the ΔybtS536 or the ΔentF536 mutant. Bars indicate zones of growth. (C and D) In vivo competition in CBA/J mice transurethrally challenged with a mixture of E. coli 536 ΔfhuA536, ΔfhuE536, ΔfyuA536, and Δhma ΔchuA536 mutants (n = 16) (C) or ΔiroN536, ΔfepA536, ΔfyuA536, and Δhma ΔchuA536 mutants (n = 13) (D). Fitness indices at 72 hpi were calculated by dividing the fraction of each mutant in the output (CFU/g tissue) by the fraction of each mutant in the input (CFU/ml inoculum). Bars represent the medians, and the dashed line indicates the theoretical fitness index of 1.0. *, P < 0.05; **, P < 0.01.
Results obtained from strain CFT073 suggest that aerobactin and heme utilization contributes most significantly to UTI (Fig. 4), and so the relative role of the yersiniabactin receptor FyuA was first examined in the context of heme and hydroxamate receptor function. Mice were challenged transurethrally with a mixture of equal amounts of ΔfhuA536, ΔfhuE536, ΔfyuA536, and Δhma ΔchuA536 mutants, and the fitness of each mutant was examined at 72 hpi. As expected, the ΔfhuA536 and ΔfhuE536 fungal hydroxamate receptor mutants had fitness indices that did not significantly differ from 1.0 (Fig. 5C), confirming the observations made with CFT073, suggesting that these receptors are relatively dispensable for UTI (Fig. 3A). In the bladder, only the ΔfyuA536 mutant was outcompeted (P = 0.0020), indicating that, of the four mutants tested, the yersiniabactin receptor represented the most contributive iron receptor to UPEC 536 colonization. However, both the ΔfyuA536 and Δhma ΔchuA536 mutants were outcompeted in the kidney (P = 0.0098 and P = 0.0273, respectively), suggesting that yersiniabactin and heme receptors contributed relatively equally to infection of this organ.
In addition to yersiniabactin, pyelonephritis strain 536 produces enterobactin and salmochelin. To examine the role of FyuA in the context of these siderophores, ΔfepA536, ΔiroN536, ΔfyuA536, and Δhma ΔchuA536 mutants were coinoculated, and their relative fitness levels were measured in a mixed-infection competition. Because E. coli 536 does not carry iha (4), the ΔfepA536 single mutant is equivalent to the CFT073 Δiha ΔfepA double mutant, at least with respect to known enterobactin receptors. As observed for CFT073, the FepA receptor appeared to play a lesser role in E. coli 536 colonization; the fitness index of the ΔfepA536 mutant was not different from 1.0 (Fig. 5D). The ΔiroN536 salmochelin receptor mutant was also able to compete well in both the bladder and the kidney. While the ΔfyuA536 mutant was outcompeted in the bladder (P = 0.0057) but not the kidney, the opposite pattern was observed for the Δhma ΔchuA536 mutant, as it was outcompeted only in the kidney (P = 0.0046). Together, these data demonstrate that FyuA-mediated yersiniabactin uptake and heme acquisition via Hma and ChuA are most critical for murine urinary tract colonization by UPEC strain 536.
Heme and noncatecholate siderophore receptors contribute most significantly to UTI.
As expected, the relative competitive fitness of each iron receptor mutant depends, to some extent, on the other mutants included in the mixed infection. However, comparison of all fitness indices of both CFT073 and 536 mutants from the array of mixed-infection competitions conducted revealed several patterns. First, the salmochelin and enterobactin receptors appeared individually to contribute the least to urinary tract colonization, as mutants lacking these genes were rarely outcompeted (Table 3). In contrast, the ΔiutA, ΔfyuA, and Δhma ΔchuA mutants were the most frequently outcompeted. All had fitness indices less than 1.0 at least half of the times they were tested, regardless of coinoculated mutants, suggesting that these receptors contribute most significantly to UTI by either E. coli strain CFT073 or 536. Finally, tissue specificity was also observed, as a Δhma ΔchuA mutant displayed defective bladder fitness in only 1 of 3 inoculum mixtures, while it was always outcompeted during kidney infection (Table 3). Thus, it appears that heme and noncatecholate receptors contribute most significantly to UPEC iron acquisition during colonization of the murine urinary tract.
Relative receptor contribution to virulence partially correlates with in vivo expression.
Previous work from our laboratory indicated that, for hma and chuA, receptor expression correlated with relative contribution to heme uptake and virulence (15). To determine whether differences in siderophore receptor gene expression may account for the observed differences in competitive fitness, receptor transcript levels were measured by qPCR from bacteria isolated from the urine of infected mice. All iron receptor genes examined were upregulated by CFT073 or 536 during murine UTI or culture in iron-limited medium, compared to culture in rich medium (data not shown). The relative transcript levels of the receptors varied, though. In CFT073, transcripts for chuA and iroN were the most abundant, followed by that for iutA, while iroN, fyuA, and chuA transcripts had the highest levels in E. coli 536 (Fig. 6 A). While expression of chuA and fyuA appears to correlate with the relatively high contribution of these genes to UTI, expression of iroN does not, as the iroN mutant was outcompeted in only 1 of 4 mixed-infection competitions (Table 1). When the median fitness index of each mutant was compared to the relative transcript abundance of the corresponding iron receptor gene, a significant correlation (P = 0.0307) was found in the kidneys but not the bladder (Fig. 6B). Iron receptor genes with higher expression levels tended to contribute more significantly to kidney infection than those with lower expression levels (Fig. 4, 5, and 6B). However, there were notable exceptions to this tendency, including iroN, suggesting that additional factors also contribute to the relative importance of each iron receptor to UPEC pathogenesis.
FIG. 6.
Expression of iron receptor genes by UPEC during murine experimental UTI. (A) Relative quantities of UPEC iron receptor transcripts, normalized to that of the gapA transcript, present in the urine of mice inoculated transurethrally with 1 × 108 CFU of wild-type CFT073 or 536. Bars represent the means of duplicate or triplicate samples, with each sample pooled from five mice over 6 to 72 hpi, and error bars show the standard errors of the means. (B) Mean relative transcript quantity shown in panel A plotted against the median fitness index of the corresponding mutant during bladder or kidney infection by CFT073 or 536. The FI value for each gene/mutant is the median of all mixed infections shown. For the CFT073 ΔfepA mutant, the FI value is the median of the ΔfepA and Δiha ΔfepA mutant FIs combined. Best-fit lines, Spearman correlation coefficients (r), and corresponding P values are shown.
DISCUSSION
Here we demonstrate nonequivalent roles for distinct iron acquisition systems during urinary tract colonization by UPEC and provide evidence for a functional hierarchy of these systems. Enterobactin and salmochelin receptors FepA, Iha, and IroN generally supplied redundant functions and contributed less to virulence and, presumably, iron uptake in vivo than noncatecholate siderophore and heme receptors. This hierarchy correlated with receptor gene expression for kidney, but not bladder, colonization. Overall, our data suggest a model for UPEC pathogenesis in which heme receptors Hma and ChuA and the noncatecholate siderophore receptor IutA or FyuA provide the most critical iron acquisition functions in the urinary tract.
In contrast to traditional coinfection studies, which compete a mutant against its wild-type parent strain, this study utilized a series of mixed infections that competed groups of mutants against one another, directly evaluating their relative roles in infection. We reasoned that mutants lacking iron receptors that contributed more significantly to uropathogenesis would be outcompeted by mutants lacking receptors whose functions were less important. To our knowledge, this is the first report to utilize direct competition between groups of mutants to examine the utility or redundancy of distinct iron acquisition systems in E. coli or other bacterial species; however, in vivo competition between two mutants has been used by our and other laboratories to examine the contribution of heme receptors (15, 49) and TonB systems (45) to UPEC and Vibrio cholerae pathogenesis, respectively. Related approaches using up to three coinoculated strains, often including double or triple mutants, have also been used in other infection models to assess the roles of Proteus mirabilis fimbriae (55) and Streptococcus pneumoniae oligopeptide transporters (26). We propose that this mixed mutant coinfection approach may be useful to dissect other redundant or partially redundant systems in UPEC or other bacterial pathogens.
While the approach used here is novel, several groups have investigated the relative importance of different iron acquisition systems to bacterial virulence. Ornibactin was concluded to be the most important siderophore for Burkholderia cenocepacia, as an ornibactin uptake mutant was cleared from a rat lung model, while a mutant lacking pyochelin uptake was maintained at wild-type levels (50). Similarly to our results, yersiniabactin was found to contribute more substantially than enterobactin to murine intranasal infection with Klebsiella pneumoniae (29). Miller and colleagues speculated that acquisition of nonenterobactin siderophore systems may be a critical component of enterobacterial pathogenesis, a hypothesis supported by our findings with UPEC.
The results presented here indicate that uptake of either aerobactin or yersiniabactin is more important to UPEC colonization of the urinary tract than enterobactin or salmochelin utilization. In strain CFT073, this disparity was so pronounced that the loss of the IutA aerobactin receptor was more detrimental to competitive fitness than the loss of two enterobactin receptors (FepA and Iha). While differences in receptor transcript abundance correlated with this hierarchy during kidney infection, this was not the case during bladder colonization, indicating that additional factors likely contribute to these observations. The difference cannot be explained simply by siderophore affinity, as the affinity of enterobactin for ferric iron (Kd [dissociation constant] = 10−52 M) is substantially higher than that of aerobactin (Kd = 10−23 M) or yersiniabactin (Kd = 10−36 M) (33, 35). Furthermore, while the low contribution of enterobactin to UPEC virulence may be explained by its susceptibility to sequestration by lipocalin-2 (12), salmochelin is resistant to this inactivation (11) and would therefore be expected to play a greater role in urinary tract colonization.
Previous biochemical studies examining the properties of aerobactin and enterobactin may provide insight into these results. Aerobactin was shown to be more effective than enterobactin at delivering iron to E. coli at low siderophore concentrations (53), as would be expected in a dynamic urinary tract environment. Aerobactin was also found to be excreted from E. coli more rapidly than enterobactin, and each siderophore's efficiency varied with the type and amount of iron chelator used in the culture medium (9). Moreover, aerobactin, which has long been associated with invasive E. coli (52), demonstrated a preference for chelating and delivering host cell-associated, rather than transferrin-bound, iron (3). Thus, it appears that the urinary tract environment and the intimate association of UPEC with the urothelium, perhaps during invasion and formation of intracellular bacterial communities (32), may favor aerobactin (and perhaps yersiniabactin) function over enterobactin activity. It remains to be tested whether these properties of enterobactin are also characteristic of salmochelin, which is structurally identical to enterobactin at the chelation site. Although not examined in this study, the ferrous iron transporter encoded by sitABCD, which is induced by Shigella flexneri during intracellular growth (41), may also contribute to UPEC iron acquisition during invasive UTI.
Our findings are generally consistent with previous studies examining the role of individual UPEC iron receptors in UTI. Our results indicated that in strain CFT073 heme receptors ChuA and Hma and aerobactin receptor IutA contributed most significantly to UTI. Similarly, chuA, hma, and iutA single mutants were previously shown to be outcompeted 15-, 38-, and 125-fold, respectively, by wild-type CFT073 (15, 49). In contrast, mutants lacking the enterobactin or salmochelin receptor iha or iroN, which contributed only slightly to UPEC colonization in our studies, were outcompeted only ∼3-fold during cochallenge with the wild type (22, 43). This is the first report, however, to establish a role for yersiniabactin during UTI, as it was previously shown to be necessary for extraintestinal pathogenic E. coli virulence in a murine lethality model (44) and UPEC biofilm formation (16).
Although aerobactin, yersiniabactin, and heme receptor mutants were most often outcompeted by coinoculated mutants, the relative competitive fitness of each iron receptor mutant depended, to some extent, on the other mutants included in the mixed infection. We speculate that this may be due to the relative fitness of the coinoculated mutants, as well as inherent variability in the experimental model. For example, the ΔiutA mutant had reduced fitness in the bladder when competed against the relatively fit ΔfhuE and ΔfhuA fungal siderophore receptor mutants (Fig. 3A) but not when competed against the likely less fit ΔiroN, Δiha ΔfepA, and Δhma chuA mutants (Fig. 4B). While these differences indeed complicate our results, the use of independent replicates, as well as multiple combinations of mutants, with generally consistent results yields confidence in our findings.
Receptor cross-specificity, especially among the catecholate receptors, and the presence of undefined receptors in UPEC strains CFT073 and 536 complicate our findings. For example, while IroN is the predominant receptor for salmochelin, low uptake of this siderophore was also observed for Cir and FepA of Salmonella (19). Conversely, both FepA and IroN were demonstrated to transport enterobactin in Salmonella (37), and FepA, IroN, Cir, and Fiu all transport the enterobactin breakdown product DBS (17, 36). This overlapping substrate specificity likely contributed to the redundancy we observed among the catecholate receptors and may have diminished the relative importance of the individual receptors tested here. Consequently, while the relative role of salmochelin, for which a single receptor (IroN) exists, can be assessed by our studies, the relative role of enterobactin requires additional investigation. Future studies directly examining the relative role of catecholate siderophores in general will allow dissection of these redundancies as they relate to UPEC pathogenesis.
Additionally, several iron-responsive putative TonB-dependent receptors for which substrates have yet to be identified, including IreA and FitA, have been described in UPEC strains (34, 42). It is possible, and perhaps likely, that these receptors transport one of the siderophores examined here, thus adding complexity to an already complex system. These undefined receptors are expected to contribute to iron acquisition by UPEC during UTI, and future studies examining them within the context of siderophore and heme uptake will allow construction of a more complete hierarchy of UPEC iron uptake in vivo.
The entire length of the urinary tract is iron limiting, but the dramatic anatomical differences from urethra to kidneys likely offer substantial differences in iron source availability. Our results suggest that not only do certain iron acquisition systems appear more important than others for UPEC virulence but differences in the importance of these systems in specific urinary tract organs exist. Heme uptake, for example, was important consistently for kidney colonization and only rarely for bladder infection. In contrast, IutA-mediated aerobactin utilization was occasionally, but not always, found to contribute more significantly to bladder colonization than kidney infection. Variations in substrate availability or siderophore activity at disparate locations in the urinary tract may have contributed to these observed differences.
While the diverse iron acquisition systems of UPEC indeed provide redundancy for an essential biological function, our data establish that specific loci are more critical than others, often at distinct anatomical sites, indicating that a degree of specificity exists. Further delineation of these interactions will have implications for our overall understanding of UPEC virulence and pathogen evolution, as well as for the development of vaccines and therapeutics that target iron acquisition systems.
Acknowledgments
We thank Sara N. Smith for assistance with animal studies.
This work was supported in part by Public Health Service grant AI43363 from the National Institutes of Health.
Editor: S. M. Payne
Footnotes
Published ahead of print on 10 January 2011.
REFERENCES
- 1.Alteri, C. J., and H. L. Mobley. 2007. Quantitative profile of the uropathogenic Escherichia coli outer membrane proteome during growth in human urine. Infect. Immun. 75:2679-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, H., J. Hacker, A. Juarez, C. Hughes, and W. Goebel. 1982. Cloning of the chromosomal determinants encoding hemolysin production and mannose-resistant hemagglutination in Escherichia coli. J. Bacteriol. 152:1241-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brock, J. H., P. H. Williams, J. Liceaga, and K. G. Wooldridge. 1991. Relative availability of transferrin-bound iron and cell-derived iron to aerobactin-producing and enterochelin-producing strains of Escherichia coli and to other microorganisms. Infect. Immun. 59:3185-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brzuszkiewicz, E., et al. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. U. S. A. 103:12879-12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carbonetti, N. H., et al. 1986. Aerobactin-mediated iron uptake by Escherichia coli isolates from human extraintestinal infections. Infect. Immun. 51:966-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuiv, P. O., P. Clarke, D. Lynch, and M. O'Connell. 2004. Identification of rhtX and fptX, novel genes encoding proteins that show homology and function in the utilization of the siderophores rhizobactin 1021 by Sinorhizobium meliloti and pyochelin by Pseudomonas aeruginosa, respectively. J. Bacteriol. 186:2996-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lorenzo, V., A. Bindereif, B. H. Paw, and J. B. Neilands. 1986. Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J. Bacteriol. 165:570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Der Vartanian, M. 1988. Differences in excretion and efficiency of the aerobactin and enterochelin siderophores in a bovine pathogenic strain of Escherichia coli. Infect. Immun. 56:413-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldmann, F., L. J. Sorsa, K. Hildinger, and S. Schubert. 2007. The salmochelin siderophore receptor IroN contributes to invasion of urothelial cells by extraintestinal pathogenic Escherichia coli in vitro. Infect. Immun. 75:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischbach, M. A., et al. 2006. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc. Natl. Acad. Sci. U. S. A. 103:16502-16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flo, T. H., et al. 2004. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432:917-921. [DOI] [PubMed] [Google Scholar]
- 13.Galen, J. E., et al. 1999. Optimization of plasmid maintenance in the attenuated live vector vaccine strain Salmonella typhi CVD 908-htrA. Infect. Immun. 67:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagan, E. C., A. L. Lloyd, D. A. Rasko, G. J. Faerber, and H. L. T. Mobley. 2010. Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog. 6:e1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagan, E. C., and H. L. Mobley. 2009. Haem acquisition is facilitated by a novel receptor Hma and required by uropathogenic Escherichia coli for kidney infection. Mol. Microbiol. 7:79-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock, V., L. Ferrieres, and P. Klemm. 2008. The ferric yersiniabactin uptake receptor FyuA is required for efficient biofilm formation by urinary tract infectious Escherichia coli in human urine. Microbiology 154:167-175. [DOI] [PubMed] [Google Scholar]
- 17.Hantke, K. 1990. Dihydroxybenzoylserine—a siderophore for E. coli. FEMS Microbiol. Lett. 55:5-8. [DOI] [PubMed] [Google Scholar]
- 18.Hantke, K. 1983. Identification of an iron uptake system specific for coprogen and rhodotorulic acid in Escherichia coli K12. Mol. Gen. Genet. 191:301-306. [DOI] [PubMed] [Google Scholar]
- 19.Hantke, K., G. Nicholson, W. Rabsch, and G. Winkelmann. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. U. S. A. 100:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson, J. P., et al. 2009. Quantitative metabolomics reveals an epigenetic blueprint for iron acquisition in uropathogenic Escherichia coli. PLoS Pathog. 5:e1000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, D. E., C. V. Lockatell, M. Hall-Craigs, H. L. Mobley, and J. W. Warren. 1987. Uropathogenicity in rats and mice of Providencia stuartii from long-term catheterized patients. J. Urol. 138:632-635. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, J. R., et al. 2005. The IrgA homologue adhesin Iha is an Escherichia coli virulence factor in murine urinary tract infection. Infect. Immun. 73:965-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 24.Kadner, R. J., K. Heller, J. W. Coulton, and V. Braun. 1980. Genetic control of hydroxamate-mediated iron uptake in Escherichia coli. J. Bacteriol. 143:256-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanamaru, S., et al. 2003. Distribution and genetic association of putative uropathogenic virulence factors iroN, iha, kpsMT, ompT and usp in Escherichia coli isolated from urinary tract infections in Japan. J. Urol. 170:2490-2493. [DOI] [PubMed] [Google Scholar]
- 26.Kerr, A. R., et al. 2004. The Ami-AliA/AliB permease of Streptococcus pneumoniae is involved in nasopharyngeal colonization but not in invasive disease. Infect. Immun. 72:3902-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lane, M. C., C. J. Alteri, S. N. Smith, and H. L. Mobley. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc. Natl. Acad. Sci. U. S. A. 104:16669-16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lane, M. C., et al. 2005. Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect. Immun. 73:7644-7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawlor, M. S., C. O'Connor, and V. L. Miller. 2007. Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun. 75:1463-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leveille, S., et al. 2006. Iha from an Escherichia coli urinary tract infection outbreak clonal group A strain is expressed in vivo in the mouse urinary tract and functions as a catecholate siderophore receptor. Infect. Immun. 74:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mobley, H. L., et al. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulvey, M. A., et al. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494-1497. [DOI] [PubMed] [Google Scholar]
- 33.Neilands, J. B. 1981. Microbial iron compounds. Annu. Rev. Biochem. 50:715-731. [DOI] [PubMed] [Google Scholar]
- 34.Ouyang, Z., and R. Isaacson. 2006. Identification and characterization of a novel ABC iron transport system, fit, in Escherichia coli. Infect. Immun. 74:6949-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perry, R. D., P. B. Balbo, H. A. Jones, J. D. Fetherston, and E. DeMoll. 1999. Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology 145:1181-1190. [DOI] [PubMed] [Google Scholar]
- 36.Rabsch, W., et al. 2003. Role of receptor proteins for enterobactin and 2,3-dihydroxybenzoylserine in virulence of Salmonella enterica. Infect. Immun. 71:6953-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabsch, W., W. Voigt, R. Reissbrodt, R. M. Tsolis, and A. J. Bäumler. 1999. Salmonella typhimurium IroN and FepA proteins mediate uptake of enterobactin but differ in their specificity for other siderophores. J. Bacteriol. 181:3610-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rakin, A., E. Saken, D. Harmsen, and J. Heesemann. 1994. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol. Microbiol. 13:253-263. [DOI] [PubMed] [Google Scholar]
- 39.Reigstad, C. S., S. J. Hultgren, and J. I. Gordon. 2007. Functional genomic studies of uropathogenic Escherichia coli and host urothelial cells when intracellular bacterial communities are assembled. J. Biol. Chem. 282:21259-21267. [DOI] [PubMed] [Google Scholar]
- 40.Roos, V., G. C. Ulett, M. A. Schembri, and P. Klemm. 2006. The asymptomatic bacteriuria Escherichia coli strain 83972 outcompetes uropathogenic E. coli strains in human urine. Infect. Immun. 74:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Runyen-Janecky, L. J., and S. M. Payne. 2002. Identification of chromosomal Shigella flexneri genes induced by the eukaryotic intracellular environment. Infect. Immun. 70:4379-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russo, T. A., U. B. Carlino, and J. R. Johnson. 2001. Identification of a new iron-regulated virulence gene, ireA, in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 69:6209-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russo, T. A., et al. 2002. IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 70:7156-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schubert, S., B. Picard, S. Gouriou, J. Heesemann, and E. Denamur. 2002. Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infect. Immun. 70:5335-5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seliger, S. S., A. R. Mey, A. M. Valle, and S. M. Payne. 2001. The two TonB systems of Vibrio cholerae: redundant and specific functions. Mol. Microbiol. 39:801-812. [DOI] [PubMed] [Google Scholar]
- 46.Snyder, J. A., et al. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72:6373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tashima, K. T., P. A. Carroll, M. B. Rogers, and S. B. Calderwood. 1996. Relative importance of three iron-regulated outer membrane proteins for in vivo growth of Vibrio cholerae. Infect. Immun. 64:1756-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torres, A. G., and S. M. Payne. 1997. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23:825-833. [DOI] [PubMed] [Google Scholar]
- 49.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Visser, M. B., S. Majumdar, E. Hani, and P. A. Sokol. 2004. Importance of the ornibactin and pyochelin siderophore transport systems in Burkholderia cenocepacia lung infections. Infect. Immun. 72:2850-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welch, R. A., et al. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams, P. H. 1979. Novel iron uptake system specified by ColV plasmids: an important component in the virulence of invasive strains of Escherichia coli. Infect. Immun. 26:925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams, P. H., and N. H. Carbonetti. 1986. Iron, siderophores, and the pursuit of virulence: independence of the aerobactin and enterochelin iron uptake systems in Escherichia coli. Infect. Immun. 51:942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wookey, P., and H. Rosenberg. 1978. Involvement of inner and outer membrane components in the transport of iron and in colicin B action in Escherichia coli. J. Bacteriol. 133:661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zunino, P., et al. 2007. Mannose-resistant Proteus-like and P. mirabilis fimbriae have specific and additive roles in P. mirabilis urinary tract infections. FEMS Immunol. Med. Microbiol. 51:125-133. [DOI] [PubMed] [Google Scholar]