Abstract
CD82 is a member of the tetraspanin superfamily, whose physiological role is best described in the context of cancer metastasis. However, CD82 also associates with components of the class II major histocompatibility complex (MHC) antigen presentation pathway, including class II MHC molecules and the peptide-loading machinery, as well as CD63, another tetraspanin, suggesting a role for CD82 in antigen presentation. Here, we observe the dynamic rearrangement of CD82 after pathogen uptake by imaging CD82-mRFP1 expressed in primary living dendritic cells. CD82 showed rapid and specific recruitment to Cryptococcus neoformans-containing phagosomes compared to polystyrene-containing phagosomes, similar to CD63. CD82 was also actively recruited to phagosomes containing other pathogenic fungi, including Candida albicans and Aspergillus fumigatus. Recruitment of CD82 to fungal phagosomes occurred independently of Toll-like receptor (TLR) signaling. Recruitment was not limited to fungi, as bacterial organisms, including Escherichia coli and Staphylococcus aureus, also induced CD82 recruitment to the phagosome. CD82 intersected the endocytic pathway used by lipopolysaccharide (LPS), implicating CD82 in trafficking of small, pathogen-associated molecules. Despite its partial overlap with lysosomal compartments, CD82 recruitment to C. neoformans-containing phagosomes occurred independently of phagosome acidification. Kinetic analysis of fluorescence imaging revealed that CD82 and class II MHC simultaneously appear in the phagosome, indicating that the two proteins may be associated. Together, these data show that the CD82 tetraspanin is specifically recruited to pathogen-containing phagosomes prior to fusion with lysosomes.
Tetraspanins are a family of proteins that comprise 33 members in mammals and span cellular membranes four times (12, 15). These proteins are characterized by short, cytoplasmic amino and carboxyl termini and two extracellular loops of unequal size. Tetraspanins can be distinguished from other molecules with the same topology through conserved residues within both the transmembrane domains and the larger of the two extracellular loops. Their membrane topology and wide distribution throughout most tissues and cell types, as well as their ability to interact laterally with other proteins within membranes, have led to the hypothesis that tetraspanins play a structural role, organizing other proteins into complexes within membrane microdomains termed “tetraspanin webs” (16). Depending on the nature of the interacting proteins, tetraspanins have been implicated in the control of cellular migration, adhesion, and signaling, perhaps best described in cancer (22). A specific role has been difficult to attribute to individual tetraspanins, partly due to possible molecular redundancy. Since the same tetraspanin expressed in different tissues can associate with distinct partners, a given tetraspanin likely has unique functions that depend on its location. For example, CD81 associates with CD19 on the surfaces of B cells and contributes to signaling, whereas this same tetraspanin is found complexed with CD4 and CD8 on the surfaces of T cells (34).
In the context of cancer, CD82, also known as Kai1, associates with integrins on the surfaces of various tumor cells (3), and its expression is linked to metastasis suppression (26). CD82, along with several other tetraspanins, can also be found in cells of the immune system (23, 33). In dendritic cells (DCs), CD82 associates with class II major histocompatibility complex (MHC) and the tetraspanin CD63 (20), as well as with other components of the antigen-processing and presentation pathway, including the peptide-editing/loading mediators HLA-DM and HLA-DO (14, 29). CD82 and CD63 are enriched in class II MHC-positive compartments and exosomes (7, 11, 31), both compartments of central importance to antigen presentation and initiation of an effective immune response (46). The specific function of these tetraspanins in professional antigen-presenting cells (APCs) remains unclear. We have previously demonstrated that CD63 and class II MHC are specifically recruited to Cryptococcus neoformans phagosomes and that CD63's arrival at the phagosome is dependent on phagosomal acidification (2). C. neoformans is an opportunistic fungal pathogen that causes disease in immunocompromised patients, including those with AIDS and hematologic malignancies and transplant recipients (30, 32). Here, we demonstrate that CD82 was actively recruited to C. neoformans-containing phagosomes, but not to those containing size-matched polystyrene beads. In addition to C. neoformans, we observed CD82 recruitment to phagosomes with other fungal pathogens, namely, Candida albicans and Aspergillus fumigatus, but also to those containing either Gram-positive (Staphylococcus aureus) or Gram-negative (Escherichia coli) bacteria. Our data clearly imply a role for CD82 in the dynamic intracellular trafficking after pathogen uptake and support nonredundant functions for tetraspanins in these processes.
MATERIALS AND METHODS
Plasmids and antibodies.
CD82 cDNA was obtained by PCR amplification of a BALB/c mouse spleen cDNA library and is detailed elsewhere (37). After the cDNA encoding the monomeric red fluorescent protein (mRFP1) was appended to the 3′ end (a kind gift from Roger Tsien, University of California, San Diego, CA), the cDNA was inserted into the lentiviral vector pHAGE (27). We previously generated anti-mRFP1 rabbit polyclonal antiserum used for immunoprecipitation experiments. Immunofluorescence in transduced HeLa cells was done with mouse anti-human CD82 (Abcam, Cambridge, MA). The secondary antibody used was goat anti-mouse Alexa Fluor 488 (Molecular Probes, Eugene, OR).
Cell lines, viral transduction, and cell culture.
RAW 264.7 mouse macrophages, myd88−/− TRIF−/− immortalized macrophages (a gift from Douglas Golenbock, University of Massachusetts, Worcester, MA), HeLa cells, and HEK293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Grand Island, NY) supplemented with 10% fetal calf serum (FCS) (HyClone, Logan, UT) and penicillin/streptomycin (Gibco). Bone marrow-derived DCs from class II MHC-enhanced green fluorescent protein (eGFP) knock-in mice (5) and C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME) were prepared as previously described (2) and seeded into eight-well Labtek II chambered-coverglass wells (Nalge Nunc, Naperville, IL). Media were replaced every 48 h. HEK293T cells were used to produce CD82-mRFP1 lentivirus as described previously (2), and the virus was used to infect RAW 264.7 cells and DCs.
Fungal and bacterial growth.
C. neoformans H99 and C. neoformans cap 59Δ (acapsular strain) (13) (both serotype A) and C. albicans SC5314 and SC5314-GFP (41) (gifts from Gerald R. Fink, Whitehead Institute for Biomedical Research, Cambridge, MA) were grown in standard yeast extract-peptone-dextrose (YPD) medium. A. fumigatus (strain Af293) was a gift from Eleftherios Mylonakis (Massachusetts General Hospital, Boston, MA). A. fumigatus was plated on Sabouraud dextrose agar plates supplemented with 100 μg/ml ampicillin and grown at 30°C for 3 to 5 days. Conidia were dislodged from the plates by gentle scraping and resuspended in phosphate-buffered saline (PBS). A. fumigatus conidia were fluorescently labeled by selectively linking Alexa Fluor 647 succinimidyl ester reconstituted in dimethylformamide (100 mg/ml) to primary amines located on the conidial surface. This labeling procedure resulted in >95% viability of A. fumigatus conidia, as judged by growth on Sabouraud dextrose agar plates. After 3 PBS washes, the conidia were suspended in 500 μl of PBS, and 3 μg of dye was mixed with the pathogen at 37°C for 30 min with shaking for 10 s every 10 min. The dye-pathogen mixture was then washed again 3 times in PBS, kept on ice, and protected from light until imaging experiments were performed. S. aureus expressing cytosolic GFP was grown in Columbia medium (BD Biosciences, Franklin Lakes, NJ) supplemented with 100 μg/ml ampicillin. E. coli expressing cytosolic GFP was grown in LB medium supplemented with 100 μg/ml ampicillin. Both bacteria were gifts from Lynda Stuart (Massachusetts General Hospital, Boston, MA).
Phagocytosis and LPS-trafficking experiments.
Phagocytosis experiments using fungi were performed using live C. neoformans, C. albicans, or A. fumigatus. C. neoformans was introduced to APCs at a 100:1 ratio for 2 to 24 h as indicated, whereas C. albicans and A. fumigatus were used at a 20:1 ratio. Dragon green-labeled beads (5 μm; Bangs Laboratories, Fishers, IN) were washed three times in PBS and diluted to 0.01% (wt/vol). The beads were added in 40-μl aliquots to eight-well dishes containing 200 μl of medium per well for 2 to 24 h. Phagocytosis experiments with bacteria were performed using either live E. coli or S. aureus. The bacteria were washed three times in PBS and used at a ratio of 20:1. For lipopolysaccharide (LPS)-trafficking experiments, LPS-Alexa Fluor 488 (1 μg/ml) was diluted in prewarmed medium and added to the cells. After 8 min of incubation, the cells were washed twice, and images were acquired at the indicated times. Lysosomes were detected using Lysotracker Green (Molecular Probes) at a concentration of 50 nM. Phagosome acidification was detected with Lysosensor-Alexa 488 or Lysosensor-568 (Molecular Probes), which was added to cells at a concentration of 1 μM simultaneously with C. neoformans or beads for 10 min. Inhibition of acidification was done using the vacuolar ATPase inhibitor bafilomycin (AG Scientific, San Diego, CA) at a concentration of 1 μM, administered to the cells 2 h before imaging them.
Microscopy and image analysis.
Imaging was performed using a spinning-disk confocal microscope and Metamorph software as previously described (2). Image analysis was performed using Volocity 3.1 software (Improvision, Lexington, MA). The rate of acquisition of fluorescence intensity around individual phagosomes was determined by measuring the mean fluorescence of the phagosomes. Independent events were analyzed from time-lapse sequences. Each sequence consisted of 10 images measured every 30 s for 10 min; the image sequence started 5 min after C. neoformans was added. The change in the mean fluorescence of individual phagosomes was tracked manually in reverse chronological order. For a given phagosome present at the last time point of an image sequence, a two-dimensional ring was defined for measuring the mean intensity of that phagosome in its equatorial plane. Centered on a phagosome, an average ring was 5 μm in diameter and 1 μm thick. The mean intensity was measured at each preceding time point in the x-y plane where the equatorial position of the phagosome was apparent. The first time point in a sequence was defined as the point at which a dark mass was evident inside the cell but whose mean intensity was equal to that measured for the background of the images. Sequences from at least 10 separate events were collected. The intensities of fluorescence surrounding newly formed phagosomes were measured as a function of time for CD82-mRFP1. After background subtraction, the values were normalized to the maximum fluorescence intensity observed in each sequence. The average normalized intensity at each time point was plotted. Error bars indicate the standard deviations across the sequences.
Pulse-chase analysis.
Pulse-chase analysis was done as previously described (24). Briefly, RAW 264.7 cells expressing CD82-mRFP1 were metabolically labeled with [35S]cysteine/methionine for 15 min and chased for up to 3 h. The cells were lysed in NP-40 buffer, and CD82-mRFP1 was immunoprecipitated using anti-mRFP1 antiserum as described previously (2). Immunoprecipitated material was treated with endoglycosidase H (EndoH) or endoglycosidase F (EndoF) for 1 h prior to separation on SDS-PAGE. Polypeptides were visualized by autoradiography.
Immunofluorescence.
HeLa cells were seeded in eight-well dishes and infected with lentivirus to express CD82-mRFP1 as described above. Adherent cells were washed once in PBS and fixed with 4% paraformaldehyde for 10 min at room temperature. The cells were washed and submerged in blocking/permeabilization (BP) buffer (3% bovine serum albumin [BSA] and 0.5% Triton X-100 in PBS) for 10 min and exposed to antibody diluted 1:500 in BP buffer for 30 min at room temperature. The cells were washed three times in BP buffer and submerged in secondary antibody diluted 1:1,000 in the same buffer for 30 min at room temperature. The cells were washed three times in BP buffer, once in PBS, and once in distilled water and sealed with Fluoromount-G mounting medium (Southern Biotech, Birmingham, AL). The sealant was allowed to dry, and the cells were imaged in the 488- and 568-nm laser channels using a spinning-disk confocal microscope.
RESULTS
Expression of CD82 as a CD82-mRFP1 fusion protein did not affect its trafficking through the ER or its subcellular distribution.
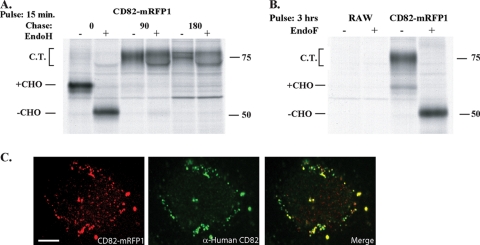
CD82 was fluorescently tagged with a C-terminal monomeric red fluorescent protein (mRFP1) to enable its visualization in real time. To ensure that this modification did not interfere with normal folding and trafficking out of the endoplasmic reticulum (ER), the fusion protein was subjected to pulse-chase analysis. Mouse RAW 264.7 macrophages were lentivirally transduced to stably express CD82-mRFP1. The cells were metabolically labeled with [35S]cysteine/methionine for 15 min and chased in medium for up to 3 h. Immunoprecipitation with anti-mRFP1 antibody and digestion with EndoH allowed the visualization of the fusion protein's maturation by autoradiography (Fig. 1 A). CD82 is predicted to contain three glycans, which explains the gradual appearance of a smear characteristic of terminally glycosylated proteins. The addition of these glycans illustrates its proper translation, folding, and traversal of the ER and Golgi apparatus. In the trans-Golgi, glycans are further modified, rendering them resistant to cleavage by EndoH, a point demonstrated by the inability of the enzyme to affect the smear observed for the later chase points. To show that proper folding and trafficking pertains to all translated CD82-mRFP1, cells were labeled to steady state and glycans were removed by digestion with EndoF, an enzyme capable of removing these modifications irrespective of a protein's maturation stage. The complete collapse of the glycosylated fusion to a discrete band demonstrates this point (Fig. 1B).
FIG. 1.
Mouse CD82-mRFP1 matures normally and demonstrates intracellular distribution similar to that of human CD82. RAW 264.7 cells were transduced to stably express CD82-mRFP1. The cells were pulse-labeled for 15 min with [35S]cysteine/methionine and chased for 0, 90, or 180 min (A) or labeled for 3 h (B). The cells were lysed in 1% NP-40, and proteins were immunoprecipitated with anti-mRFP1 antisera. At each time point, half of the sample was treated with endoglycosidase H (A) or F (B) for 1 h at 37°C. Samples were run on a 10% SDS-polyacrylamide gel, and polypeptides were visualized by autoradiography. The “CHO” labels indicate the presence of N-linked glycans, and complex-type sugars (C.T.) were present. Molecular sizes (in kilodaltons) are given at right. (C) HeLa cells expressing mouse CD82-mRFP1 were fixed, permeabilized, and stained with an anti-human CD82 antibody. Shown is an image of a single cell illustrating mouse CD82-mRFP1 (left, red), human CD82 (center, green), and a merged image (right). Bar, 5 μm.
As an added control, we sought to establish the proper subcellular localization of CD82-mRFP1. In the absence of anti-mouse CD82 antibodies, we took advantage of the high degree of sequence identity between human and mouse tetraspanins by testing for colocalization of endogenous human CD82—for which antibodies exist—with the mouse CD82 fusion protein. Lentiviral transduction of HeLa cells with CD82-mRFP1 allowed detection by immunofluorescence of the endogenous CD82 within the same compartments. The overlay of the human and the mouse forms shows colocalization of these tetraspanin orthologues in small vesicles, indicating the presence of the proteins in endosomes (Fig. 1C). Therefore, we conclude that CD82-mRFP1 can reliably be used as a proxy to study the behavior of this tetraspanin.
CD82 is selectively recruited to C. neoformans-containing phagosomes.
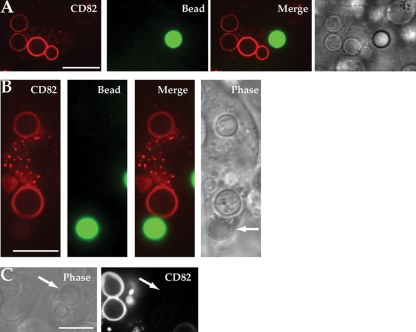
As we have previously demonstrated recruitment of CD63 to C. neoformans-containing phagosomes (2), we now wished to investigate if the CD63-associated tetraspanin CD82 (20, 44) was also specifically recruited to phagosomes containing the yeast. Primary DCs were lentivirally transduced to express CD82-mRFP1 and exposed to a combination of C. neoformans H99 and size-matched green fluorescently labeled polystyrene beads. The cells were allowed to ingest these particles for 30 min and subsequently imaged using spinning-disk confocal microscopy. As with CD63, we observed selective recruitment of CD82 within the same cell to phagosomes containing yeast, but not to phagosomes containing polystyrene beads (Fig. 2 A and B). Bright-field images clearly differentiated intracellular yeast from ingested polystyrene beads. To ensure that CD82 recruitment was present throughout the phagosome, serial images in the z dimension demonstrated consistent CD82-mRFP1 recruitment (see movies S1 and S2 in the supplemental material). CD82 could be seen on the phagosomal membrane within 30 min following C. neoformans H99 ingestion, and its presence persisted for several hours (data not shown). Conversely, polystyrene beads consistently failed to induce recruitment despite prolonged incubation (24 h) (data not shown). To ensure that the fluorescent ring visualized was not a result of autofluorescence by C. neoformans, cells devoid of CD82-mRFP1 expression were imaged adjacent to DCs expressing CD82-mRFP1. Phase images confirmed phagocytosis of the yeast. Despite its intracellular location, no fluorescence could be seen in cells without CD82-mRFP1, whereas robust recruitment of CD82 to the fungus-containing phagosomes in cells expressing CD82-mRFP1 was seen (Fig. 2C). These data show that CD82 is selectively recruited to the C. neoformans-containing phagosome, similar to CD63.
FIG. 2.
CD82 is recruited selectively to C. neoformans H99 phagosomes. (A) DCs expressing CD82-mRFP1 were exposed to fluorescent polystyrene beads (green) and C. neoformans (unlabeled). Confocal images were taken 30 min after exposure of particles to DCs. (B) Selective recruitment of CD82 to fungal phagosomes in DCs that had ingested both bead and yeast. The arrow indicates the location of the polystyrene bead. (C) DCs not expressing CD82-mRFP1 that had taken up C. neoformans do not show any signal in the red channel, indicating that autofluorescence from the yeast does not contribute to this signal. The arrows point to intracellular yeast in cells. Scale bars, 10 μm.
CD82 redistributes to phagosomes containing other pathogenic yeasts, including C. albicans and A. fumigatus.
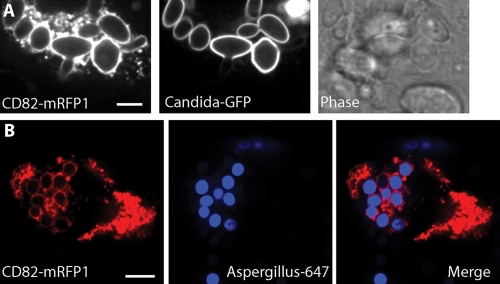
While CD82 exhibited exquisite specificity when DC phagosomes contained either C. neoformans or polystyrene beads, we wanted to see if recruitment was due to the specific surface chemistry of the fungal pathogen. C. albicans can cause invasive fungal infections in patients whose immune systems are compromised or in immunocompetent patients possessing indwelling vascular catheters (28). The external polysaccharide coats of C. neoformans H99 and C. albicans differ significantly. The surface of C. neoformans is composed primarily of glucuronoxylomannan and, to a lesser extent, galactoxylomannan (9, 45). In contrast, the surface of C. albicans is composed of mannoproteins that serve in part to prevent exposure of β-glucans to cells of the immune system (40). Likewise, A. fumigatus is another opportunistic pathogenic fungus that infects primarily immunocompromised patients. The A. fumigatus cell wall is comprised of galactomannan and β-1,3-1,4-glucan, which are absent on C. albicans (21). To determine if CD82 recruitment required specific carbohydrate moieties found on C. neoformans H99, we exposed CD82-mRFP1-expressing DCs to fluorescent C. albicans or A. fumigatus labeled with Alexa 647. CD82 recruitment was robust on phagosomes containing either C. albicans (Fig. 3 A) or A. fumigatus (Fig. 3B). The kinetics of CD82 delivery to these phagosomes was similar to what we observed with C. neoformans (data not shown). These data exclude a dependence on specific surface chemistry of C. neoformans for CD82 recruitment and extend our observations to include an assortment of polysaccharides and proteins on the pathogen surface.
FIG. 3.
CD82 is recruited to phagosomes containing either C. albicans or A. fumigatus. DCs were transduced with mouse CD82-mRFP1 and allowed to phagocytose either C. albicans (A) or A. fumigatus (B). To facilitate the identification of organisms, we used pathogens expressing surface GFP (C. albicans) or organisms labeled with Alexa 647 (A. fumigatus). The left image in each panel represents CD82, and the middle image demonstrates fluorescence from the fungus. A bright-field image of C. albicans taken up by DC expressing CD82-mRFP1 is shown. The merged image shows an overlay of CD82-mRFP1 signal and the organism. Scale bar, 5 μm.
CD82 recruitment to fungal phagosomes is independent of TLR signaling.
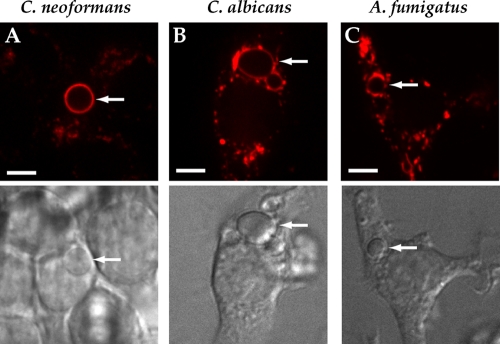
Toll-like receptor (TLR) signaling is critical in the host defense against fungal organisms. TLR2 and TLR4 are readily recruited from the plasma membrane to microbial phagosomes (36), and we have shown that TLR9 is recruited to phagosomes containing A. fumigatus (18). We wished to determine if CD82 recruitment to fungal phagosomes required signaling from any TLR family members. To evaluate this possibility, we expressed CD82-mRFP1 in immortalized bone marrow macrophage cell lines deficient in both MyD88 and TRIF. These cells are completely incapable of all TLR signaling. Despite this, we still observed robust recruitment of CD82 to phagosomes containing either C. neoformans (Fig. 4 A), C. albicans (Fig. 4B), or A. fumigatus (Fig. 4C). Thus, our results indicate that CD82 redistribution to the fungal phagosomes is independent of TLR signaling.
FIG. 4.
CD82 recruitment to fungal phagosomes is independent of TLR signaling. Immortalized macrophages from myd88−/− TRIF−/− mice were lentivirally transduced to express CD82-mRFP1. These APCs were exposed to C. neoformans H99 (A), C. albicans (B), or A. fumigatus (C), and cells that had taken up fungal organisms were imaged. Differential interference contrast (DIC) images of cells are shown to facilitate identification of fungal organisms. The arrows indicate intracellular locations of fungi. Scale bar, 5 μm.
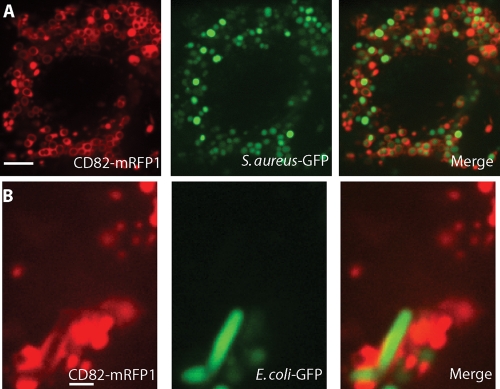
CD82 is recruited to phagosomes containing either Gram-positive or Gram-negative bacteria.
To determine if prokaryotic pathogens also possess the capacity to recruit CD82 to the phagosome, we examined DCs expressing CD82-mRFP1 after they had taken up either S. aureus, a Gram-positive bacterium of clinical importance, or E. coli, a Gram-negative bacterium. Again, we utilized microorganisms that expressed GFP to confirm that these organisms had been ingested by the professional APCs. Recruitment of CD82 to phagosomes containing S. aureus was observed (Fig. 5 A). A minority of S. aureus cells failed to express GFP in sufficient quantities to be visualized by fluorescence microscopy, but these microorganisms still recruited CD82 to their phagosomes. Since CD82 recruitment is not immediate upon phagosome formation, a small percentage of intracellular bacteria can be seen without saturating amounts of CD82 on the phagosomal membrane. Likewise, CD82-mRFP1-expressing DCs exposed to E. coli also underwent phagosomal recruitment of CD82, displaying the characteristic rod-shaped form of the bacteria (Fig. 5B). Taken together, these data indicate that CD82 recruitment is not specific to fungal pathogens and that CD82 localizes to phagosomal compartments containing a variety of organisms, but notably, not to polystyrene beads.
FIG. 5.
CD82 is recruited to phagosomes containing either Gram-positive or Gram-negative bacteria. DCs were transduced with mouse CD82-mRFP1 and exposed to S. aureus (A) or E. coli (B). The left image in each panel represents CD82, and the middle image demonstrates GFP fluorescence in order to facilitate the identification of organisms. The merged images show overlays of CD82-mRFP1 signals and bacteria. Scale bar, 5 μm.
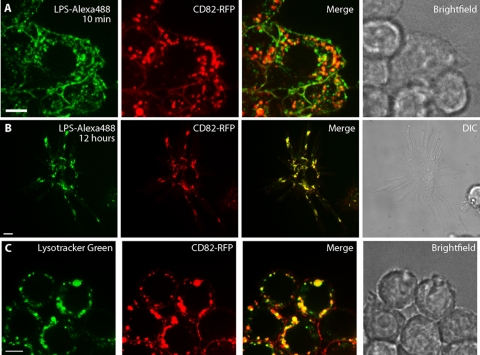
LPS intersects CD82 upon endocytosis.
Since CD82 was recruited to phagosomes containing Gram-negative organisms, we wished to determine if CD82 was also involved in trafficking of small pathogen-associated molecules or limited to recruitment to phagosomes. LPS is a component of the Gram-negative cell wall and a potent TLR4 agonist; its fate within the endocytic compartment of professional APCs has been well studied (17, 19). In hematopoietic cells, CD82 can be found on the plasma membrane, as well as in intracellular vesicles (14, 44). CD82 internalization from the cell surface requires both dynamin and clathrin (44). To determine if CD82 participated in the endocytosis of LPS in macrophages, we incubated RAW cells expressing CD82-mRFP1 with fluorescent LPS. Ten minutes after the addition of the fluorescent ligand, the distributions of LPS and CD82-mRFP1 showed partial overlap both on the cell surface and within small intracellular vesicles (Fig. 6 A), indicating that LPS intersects the CD82-positive compartment at the time of cell entry. Strikingly, while the overlap between LPS and CD82 was partial at early time points, after prolonged incubation we observed nearly complete colocalization (Fig. 6B). Since LPS traffics into acidic compartments, we wished to determine if CD82 was also found in lysosomes of APCs. To evaluate this possibility, CD82-mRFP1-expressing RAW cells were incubated with Lysotracker, an acidophilic dye that accumulates in the lysosomes. CD82-mRFP1 and Lysotracker appeared to overlap in most, but not all, compartments (Fig. 6C). These data indicate that CD82 intersects the endocytic pathway used by LPS and is also found in lysosomes of professional APCs.
FIG. 6.
LPS intersects CD82 in the endocytic pathway, and CD82 is found in acidic compartments. RAW cells expressing CD82-mRFP1 were incubated with 1 mg/ml LPS conjugated to Alexa 488, and serial images were taken. (A) After 10 min, LPS had been internalized and showed partial overlap with CD82. (B) After 12 h, there was nearly complete colocalization between CD82 and LPS, as shown in the activated macrophage. (C) To determine if CD82 resides in acidic compartments, 50 nM Lysotracker was added to RAW cells expressing CD82-mRFP1 and imaged 30 min later. Scale bar, 5 μm.
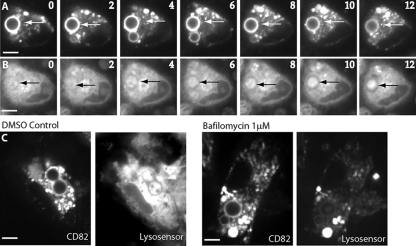
CD82 recruitment is independent of phagosome acidification.
When probing the conditions under which CD63 is recruited, we demonstrated that its appearance on the phagosomal membrane is dependent upon, and shortly follows, phagosomal acidification. To determine whether this sequence of events is also the case for CD82, we exposed CD82-mRFP1-expressing DCs to Lysosensor, a cell-permeable dye that fluoresces in acidic environments, such as the maturing phagosome (2). Treated DCs were allowed to ingest C. neoformans H99, and maturing phagosomes were imaged in real time. Unlike the recruitment kinetics seen for CD63, phagosomal acquisition of CD82 preceded Lysosensor fluorescence (Fig. 7 A and B). To further distinguish CD82 recruitment from phagosome acidification, we blocked acidification and tracked the behavior of CD82. Intracellular acidification was impeded by treating cells with bafilomycin, a potent inhibitor of the H+-pumping vacuolar ATPase, which mediates transport of protons across vacuolar membranes. Cells expressing CD82-mRFP1 treated with bafilomycin were exposed to yeast, and acidification of subcellular compartments was monitored using Lysosensor. Again, unlike what we observed for CD63, blocking acidification had no effect on the recruitment of CD82, which readily occurred despite acidification of the phagosome (Fig. 7C). These data indicate that CD82 recruitment occurs prior to acidification of the phagosomal compartment and is not affected by inhibition of the vacuolar H+ ATPase.
FIG. 7.
Phagosomal recruitment of CD82 is independent of phagosomal acidification. DCs were transduced to express CD82-mRFP1 and incubated with C. neoformans and the pH indicator Lysosensor. Time-lapse imaging of CD82-mRFP1 recruitment (A) and phagosome acidification as determined by Lysosensor fluorescence (B) are shown. The images were taken at 2-min intervals. The arrows indicate C. neoformans-containing phagosomes. (C) DCs were lentivirally transduced with CD82-mRFP1 and incubated with C. neoformans in the absence or presence of 1 μM bafilomycin. The DCs were serially imaged, and CD82-mRFP1 recruitment and acidification as determined by Lysosensor were followed. The images shown were taken after 30 min of incubation with C. neoformans. Scale bars, 5 μm.
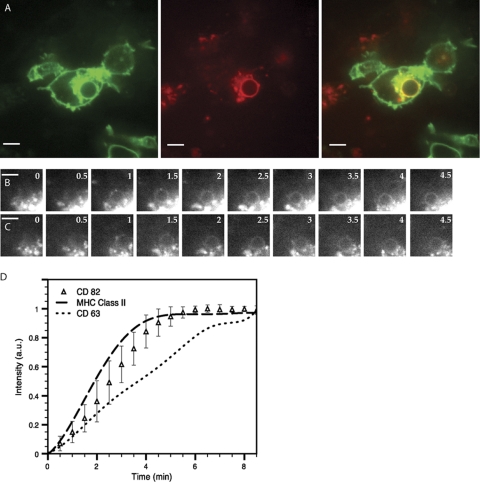
CD82 and class II MHC recruitment to the phagosome occur with similar kinetics.
Both class II MHC and CD82 recruitment to C. neoformans-containing phagosomes do not require acidification. To determine if class II MHC and CD82 have similar kinetics of appearance on C. neoformans phagosomes, DCs generated from the class II MHC-eGFP mice (5) were transduced with CD82-mRFP1-encoding lentivirus in order to track the dynamic movements of both fluorescent proteins. C. neoformans was added, and the DCs were imaged. Phagosomes formed after introduction of the organism demonstrated strong recruitment of both class II MHC-eGFP and CD82-mRFP1 (Fig. 8 A). To determine if the timing of class II MHC and CD82 appearances on the phagosome coincided, we obtained serial images of a nascently formed phagosome in a DC expressing both class II-eGFP (Fig. 8B) and CD82-mRFP1 (Fig. 8C). These images demonstrate that the appearances of the two proteins on the phagosome coincided temporally.
FIG. 8.
The kinetics of CD82 recruitment are similar to those of class II MHC recruitment to phagosomes containing C. neoformans. (A) Within 10 min after the addition of C. neoformans to DCs grown from class II MHC-eGFP mice and transduced to express CD82-mRFP1, phagosomes acquire both CD82-mRFP1 and class II MHC (left, class II MHC; center, CD82-mRFP1; right, merged image). Scale bar, 5 μm. (B and C) Time-lapse images in both red and green channels show that class II MHC-eGFP (B) appears around the phagosomes with kinetics similar to those of CD82-mRFP1 (C). Scale bar, 5 μm. (D) Kinetic analysis of fluorescence acquisition around phagosomes. Each data point (▵) represents the average intensity measured at each time point for at least 10 independent C. neoformans phagocytic events for CD82-mRFP1 over time (in minutes). The curves for class II MHC and CD63 are derived from identical experiments as described previously (2). a.u., arbitrary units. The error bars represent standard deviations.
To further probe the timing of arrival on the phagosomal membrane of CD82 in relation to class II MHC and CD63, DCs from C57BL/6 mice transduced to express CD82-mRFP1 were exposed to Cap59Δ C. neoformans and imaged over 30 min. The acapsular form of C. neoformans was chosen to facilitate the collection of a large number of phagocytic events. Cap59Δ C. neoformans induced the same endosomal rearrangements as wild-type C. neoformans H99, the only difference being that terminal levels of CD82 were slightly lower, a point that did not interfere with resolution of the recruitment kinetics (data not shown). Additionally, we chose to do this analysis in multiple DCs expressing only one type of fluorescent protein to eliminate any contribution of pixel intensity from other fluorophores. Phagosomes that had recruited maximal amounts of CD82 (as judged by pixel density) were followed back in time to the initial point of yeast entry. The pixel density was then plotted going forward to generate a curve representing the kinetics of recruitment. This curve was overlaid with curves generated previously representing the kinetics of recruitment for CD63 and class II MHC performed under identical conditions (2). The resulting graph demonstrates that CD82 has recruitment kinetics similar to those of class II MHC, arriving slightly before CD63 on the phagosomal membrane (Fig. 8D). These data show that CD63 and CD82 traffic independently to phagosomes and further support the notion that CD82 and CD63 serve nonredundant functions and that CD82 may indeed serve to escort class II MHC to the site of the antigen.
DISCUSSION
Antigen processing and presentation are multistep processes whose many components are progressively being characterized and defined (38). Phagocytosed microbes enter phagosomal compartments that subsequently undergo fusion and fission events that modify their immediate environment. This dynamic delivery and removal of material is necessary to ensure proper peptide processing, loading of class II MHC molecules, and transport to the cell surface, where these antigens are used to activate T cells. Although the roles of many components involved in this process have been defined, such as HLA-DO, DM, and CLIP in peptide loading, the activation of proteases at low pH for processing, and the tubulation of endocytic compartments for delivery of loaded class II MHC to the cell surface, there are many other components present at various stages of this pathway whose roles have yet to be described. Tetraspanins fall into the latter category. In this study, we presented data showing that CD82 was involved in the endolysosomal pathway and that the content of the phagosome specifically triggered CD82 rearrangement to the phagosome. Finally, we showed that CD82 recruitment was independent of phagosomal acidification and appeared on the phagosome with kinetics similar to those of class II MHC. To our knowledge, this report is the first to implicate CD82 in intracellular trafficking after pathogen uptake, and it supports nonredundant roles for the two tetraspanins CD82 and CD63.
We have previously described the selective recruitment of CD63 to yeast-containing phagosomes (2), and we now extend this observation to include CD82. Not only is CD82 rapidly recruited to the membrane of nascent C. neoformans-containing phagosomes, but its recruitment is selective to the microorganism, with polystyrene bead-containing phagosomes within the same cell effectively devoid of the tetraspanin. This observation mirrors that seen with CD63, indicating that the cargo determines the composition of proteins found on the phagosomal membrane, a point supported by other studies (4, 35, 39). The ability of other microorganisms to recruit CD82 suggests that the component required to trigger tetraspanin redistribution to the phagosome is not linked to a specific structure found exclusively on yeast. Although the selective and rapid recruitment of CD82 is reminiscent of that seen for CD63, we did observe two important differences that serve to separate these two tetraspanins and pinpoint the timing of their involvement in the class II MHC pathway. Unlike CD63, whose appearance on the phagosome is strictly dependent on prior acidification, the addition of bafilomycin to DCs had no bearing on CD82 recruitment. Furthermore, careful examination of recruitment kinetics revealed that CD82 arrives at the phagosome before CD63, indicating that if CD82 and CD63 assemble into tetraspanin microdomains to associate with class II MHC molecules, as suggested by other studies (10, 20, 29), this association must come about following arrival at the phagosome. Once there, these tetraspanins may serve to escort peptide-loaded class II MHC to the cell surface, a hypothesis supported by the presence of both CD82 and CD63 in endocytic tubules of LPS-stimulated primary dendritic cells (37).
DCs capture C. neoformans delivered to a mouse intranasally and present peptides to antigen-specific T cells using an in vivo model of cryptococcosis (43). Using zipper phagocytosis, DCs place C. neoformans in phagosomes that acquire markers of early endocytic compartments and eventually fuse with the lysosome (42). Our data support the following model of C. neoformans phagosomal maturation. Upon phagocytosis of C. neoformans by DCs, the organism is placed into a phagosome, which then initially recruits CD82 and class II MHC with nearly identical kinetics. Shortly after this event, the compartment becomes acidified. This event then permits the subsequent recruitment of CD63 to the phagosome. Interestingly, C. neoformans can escape from phagosomes within macrophages without killing the host cell by means of an unusual expulsive mechanism that may represent a mode of dissemination (1, 25). Detailed molecular analysis of the fate of C. neoformans phagosomes may assist in elucidating this pathway.
We provide the first evidence that CD82 is actively recruited to the phagosome after uptake of a pathogen. In addition to delivery of specialized proteins in the antigen-processing and presentation machinery, CD82 may also be linked with proteins involved in membrane fusion required for phagosomal maturation. TI-VAMP/VAMP7 (tetanus neurotoxin-insensitive vesicle-associated membrane protein) is involved in heterotypic fusion between endosomes and lysosomes and directly transports CD82 in HeLa cells (8). Similar to our observations for CD82, TI-VAMP7 is found in endosomal and lysosomal compartments and is recruited to phagosomes (6). It remains to be determined, though, whether TI-VAMP7 will show similar specificities for phagosomes containing microorganisms and those with polystyrene beads. Moreover, TI-VAMP7 is required for optimal phagocytosis in macrophages, as depletion of TI-VAMP7 by small interfering RNA (siRNA) led to a defect in FcR-mediated phagocytosis in RAW cells. It is possible that CD82 and TI-VAMP7 both escort phagosomes to lysosomes and promote phagosomal acidification. It is noteworthy that TI-VAMP7 is also involved in exocytosis and appears to be associated with vesicles released by cells during phagosome formation (6). It is tempting to speculate that TI-VAMP7 and CD82 may be involved in the release of C. neoformans phagosomes from living professional APCs (1, 25).
Supplementary Material
Acknowledgments
K.A.-T. was supported by a National Research Service Award 1F32CA105862-01 from the NCI. P.V.K., M.L.C., and N.S.K. were supported by DOM start up funds from the MGH Department of Medicine. E.P. was an MIT Poitras Fellow. This work was supported by grant 5R01AI034893-17 from the NIAID awarded to H.L.P. J.M.V. was supported by grant 5K08AI57999 from the NIAID and startup funds from the MGH Department of Medicine.
We thank Roger Tsien, Douglas Golenbock, Gerald Fink, Eleftherios Mylonakis, and Lynda Stuart for sharing their reagents.
We declare no conflict of interest.
Editor: G. S. Deepe, Jr.
Footnotes
Published ahead of print on 13 December 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Alvarez, M., and A. Casadevall. 2006. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr. Biol. 16:2161-2165. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas, K., J. C. Love, H. L. Ploegh, and J. M. Vyas. 2006. Recruitment of CD63 to Cryptococcus neoformans phagosomes requires acidification. Proc. Natl. Acad. Sci. U. S. A. 103:15945-15950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandyopadhyay, S., et al. 2006. Interaction of KAI1 on tumor cells with DARC on vascular endothelium leads to metastasis suppression. Nat. Med. 12:933-938. [DOI] [PubMed] [Google Scholar]
- 4.Blander, J. M., and R. Medzhitov. 2006. Toll-dependent selection of microbial antigens for presentation by dendritic cells. Nature 440:808-812. [DOI] [PubMed] [Google Scholar]
- 5.Boes, M., et al. 2002. T-cell engagement of dendritic cells rapidly rearranges MHC class II transport. Nature 418:983-988. [DOI] [PubMed] [Google Scholar]
- 6.Braun, V., et al. 2004. TI-VAMP/VAMP7 is required for optimal phagocytosis of opsonised particles in macrophages. EMBO J. 23:4166-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chairoungdua, A., D. L. Smith, P. Pochard, M. Hull, and M. J. Caplan. 2010. Exosome release of {beta}-catenin: a novel mechanism that antagonizes Wnt signaling. J. Cell Biol. 190:1079-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danglot, L., et al. 2010. Role of TI-VAMP and CD82 in EGFR cell-surface dynamics and signaling. J. Cell Sci. 123:723-735. [DOI] [PubMed] [Google Scholar]
- 9.De Jesus, M., et al. 2009. Galactoxylomannan-mediated immunological paralysis results from specific B cell depletion in the context of widespread immune system damage. J. Immunol. 183:3885-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engering, A., and J. Pieters. 2001. Association of distinct tetraspanins with MHC class II molecules at different subcellular locations in human immature dendritic cells. Int. Immunol. 13:127-134. [DOI] [PubMed] [Google Scholar]
- 11.Escola, J. M., et al. 1998. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273:20121-20127. [DOI] [PubMed] [Google Scholar]
- 12.Fedor Berditchevski, E. O. 2007. Tetraspanins as regulators of protein trafficking. Traffic 8:89-96. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Rivera, J., Y. C. Chang, K. J. Kwon-Chung, and A. Casadevall. 2004. Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot. Cell 3:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond, C., et al. 1998. The tetraspan protein CD82 is a resident of MHC class II compartments where it associates with HLA-DR, -DM, and -DO molecules. J. Immunol. 161:3282-3291. [PubMed] [Google Scholar]
- 15.Hemler, M. E. 2008. Targeting of tetraspanin proteins—potential benefits and strategies. Nat. Rev. Drug Discov. 7:747-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hemler, M. E. 2003. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19:397-422. [DOI] [PubMed] [Google Scholar]
- 17.Husebye, H., et al. 2006. Endocytic pathways regulate Toll-like receptor 4 signaling and link innate and adaptive immunity. EMBO J. 25:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasperkovitz, P. V., M. L. Cardenas, and J. M. Vyas. 2010. TLR9 is actively recruited to Aspergillus fumigatus phagosomes and requires the N-terminal proteolytic cleavage domain for proper intracellular trafficking. J. Immunol. 185:7614-7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitchens, R. L., P. Wang, and R. S. Munford. 1998. Bacterial lipopolysaccharide can enter monocytes via two CD14-dependent pathways. J. Immunol. 161:5534-5545. [PubMed] [Google Scholar]
- 20.Kropshofer, H., et al. 2002. Tetraspan microdomains distinct from lipid rafts enrich select peptide-MHC class II complexes. Nat. Immunol. 3:61-68. [DOI] [PubMed] [Google Scholar]
- 21.Latge, J. P. 2010. Tasting the fungal cell wall. Cell Microbiol. 12:863-872. [DOI] [PubMed] [Google Scholar]
- 22.Lazo, P. A. 2007. Functional implications of tetraspanin proteins in cancer biology. Cancer Sci. 98:1666-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy, S., and T. Shoham. 2005. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 5:136-148. [DOI] [PubMed] [Google Scholar]
- 24.Lorenzo, M. E., J. U. Jung, and H. L. Ploegh. 2002. Kaposi's sarcoma-associated herpesvirus K3 utilizes the ubiquitin-proteasome system in routing class major histocompatibility complexes to late endocytic compartments. J. Virol. 76:5522-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma, H., J. E. Croudace, D. A. Lammas, and R. C. May. 2006. Expulsion of live pathogenic yeast by macrophages. Curr. Biol. 16:2156-2160. [DOI] [PubMed] [Google Scholar]
- 26.Miranti, C. K. 2009. Controlling cell surface dynamics and signaling: how CD82/KAI1 suppresses metastasis. Cell Signal. 21:196-211. [DOI] [PubMed] [Google Scholar]
- 27.Mostoslavsky, G., et al. 2005. Efficiency of transduction of highly purified murine hematopoietic stem cells by lentiviral and oncoretroviral vectors under conditions of minimal in vitro manipulation. Mol. Ther. 11:932-940. [DOI] [PubMed] [Google Scholar]
- 28.Peter, G. P. 2006. Invasive candidiasis. Infect. Dis. Clin. N. Am. 20:485-506. [DOI] [PubMed] [Google Scholar]
- 29.Poloso, N. J., L. K. Denzin, and P. A. Roche. 2006. CDw78 defines MHC class II-peptide complexes that require II chain-dependent lysosomal trafficking, not localization to a specific tetraspanin membrane microdomain. J. Immunol. 177:5451-5458. [DOI] [PubMed] [Google Scholar]
- 30.Pukkila-Worley, R., and E. Mylonakis. 2008. Epidemiology and management of cryptococcal meningitis: developments and challenges. Exp. Opin. Pharmacother. 9:551-560. [DOI] [PubMed] [Google Scholar]
- 31.Schorey, J. S., and S. Bhatnagar. 2008. Exosome function: from tumor immunology to pathogen biology. Traffic 9:871-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh, N., F. Dromer, J. Perfect, and O. Lortholary. 2008. Immunocompromised hosts: cryptococcosis in solid organ transplant recipients: current state of the science. Clin. Infect. Dis. 47:1321-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tarrant, J. M., L. Robb, A. B. van Spriel, and M. D. Wright. 2003. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol. 24:610-617. [DOI] [PubMed] [Google Scholar]
- 34.Todd, S. C., S. G. Lipps, L. Crisa, D. R. Salomon, and C. D. Tsoukas. 1996. CD81 expressed on human thymocytes mediates integrin activation and interleukin 2-dependent proliferation. J. Exp. Med. 184:2055-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torchinsky, M. B., J. Garaude, A. P. Martin, and J. M. Blander. 2009. Innate immune recognition of infected apoptotic cells directs TH17 cell differentiation. Nature 458:78-82. [DOI] [PubMed] [Google Scholar]
- 36.Underhill, D. M., et al. 1999. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature 401:811-815. [DOI] [PubMed] [Google Scholar]
- 37.Vyas, J. M., et al. 2007. Tubulation of class II MHC compartments is microtubule dependent and involves multiple endolysosomal membrane proteins in primary dendritic cells. J. Immunol. 178:7199-7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vyas, J. M., A. G. Van der Veen, and H. L. Ploegh. 2008. The known unknowns of antigen processing and presentation. Nat. Rev. Immunol. 8:607-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West, M. A., et al. 2008. TLR ligand-induced podosome disassembly in dendritic cells is ADAM17 dependent. J. Cell Biol. 182:993-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wheeler, R. T., and G. R. Fink. 2006. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheeler, R. T., D. Kombe, S. D. Agarwala, and G. R. Fink. 2008. Dynamic, morphotype-specific Candida albicans beta-glucan exposure during infection and drug treatment. PLoS Pathog. 4:e1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wozniak, K. L., and S. M. Levitz. 2008. Cryptococcus neoformans enters the endolysosomal pathway of dendritic cells and is killed by lysosomal components. Infect. Immun. 76:4764-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wozniak, K. L., J. M. Vyas, and S. M. Levitz. 2006. In vivo role of dendritic cells in a murine model of pulmonary cryptococcosis. Infect. Immun. 74:3817-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu, C., et al. 2009. CD82 endocytosis and cholesterol-dependent reorganization of tetraspanin webs and lipid rafts. FASEB J. 23:3273-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yauch, L. E., J. S. Lam, and S. M. Levitz. 2006. Direct inhibition of T-cell responses by the Cryptococcus capsular polysaccharide glucuronoxylomannan. PLoS Pathog. 2:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zitvogel, L., et al. 1998. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat. Med. 4:594-600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.