Abstract
Vibrio vulnificus is the leading cause of reported deaths from infections related to consumption of seafood in the United States. Affected predisposed individuals frequently die rapidly from sepsis. Otherwise healthy people can experience severe wound infection, which can lead to sepsis and death. A question is why, with so many people consuming contaminated raw oysters, the incidence of severe V. vulnificus disease is low. Molecular typing systems have shown associations of V. vulnificus genotypes and the environmental or clinical source of the strains, suggesting that different genotypes possess different virulence potentials. We examined 69 V. vulnificus biotype 1 strains that were genotyped by several methods and evaluated them for virulence in a subcutaneously inoculated iron dextran-treated mouse model. By examining the relationships between skin infection, systemic liver infection, and presumptive death (a decrease in body temperature), we determined that liver infection is predicated on severe skin infection and that death requires significant liver infection. Although most strains caused severe skin infection, not every strain caused systemic infection and death. Strains with polymorphisms at multiple loci (rrn, vcg, housekeeping genes, and repetitive DNA) designated profile 2 were more likely to cause lethal systemic infection with more severe indicators of virulence than were profile 1 strains with different polymorphisms at these loci. However, some profile 1 strains were lethal and some profile 2 strains did not cause systemic infection. Therefore, current genotyping schemes cannot strictly predict the virulence of V. vulnificus strains and further investigation is needed to identify virulence genes as markers of virulence.
Vibrio vulnificus is a halophilic bacterium that is naturally present in estuarine waters and contaminates oysters and other shellfish. It is an opportunistic pathogen of humans that causes primary septicemia and wound infection in susceptible hosts and is the leading cause of reported seafood-related deaths due to infection in the United States (for a review, see reference 17). V. vulnificus strains are divided into three biotypes, with human disease caused predominantly by biotype 1 and disease of eels caused predominantly by biotype 2. Biotype 3 was recently isolated from wound infections in Israel and represents an emerging form of this species (3). Several factors have been definitively shown to contribute to virulence of V. vulnificus, including capsular polysaccharide (60), the ability to acquire iron (38, 45, 61), type IV pili (46), flagella (37, 47), RTX toxin (30, 35, 39), and others (17, 27). To date, no single factor has been identified that can distinguish between naturally occurring virulent and less virulent isolates of V. vulnificus.
In susceptible humans, V. vulnificus causes a rapid, fulminating disease process resulting in extensive tissue damage (reviewed in reference 17). Primary septicemia is characterized by high fever, chills, hypotension, and septic shock. In addition, secondary cutaneous skin lesions often form in the extremities marked by bullae containing hemorrhagic fluid (6). Mortality rates for susceptible individuals who develop fulminating primary septicemia are greater than 50% (20-22). Wound infections caused by V. vulnificus are generally associated with the handling of shellfish or exposure of existing wounds to contaminated seawater. Patients exhibiting wound infections often experience painful swelling and redness in the infected area. In extreme cases, skin infections can lead to severe cellulitis, necrotizing fasciitis, and myositis requiring surgical debridement of infected tissue or amputation of the limb (2, 6, 22); sepsis can also follow skin infection. Therapeutic intervention is often difficult, since death from sepsis can occur in less than 24 h after contact with bacteria.
The predisposing conditions for infection by V. vulnificus include, but are not limited to, liver disease, hemochromatosis, diabetes, and immune compromise (9, 20, 32, 33). However, the number of individuals experiencing severe infection with V. vulnificus each year compared to the number of predisposed people who consume raw oysters is relatively small. A single oyster can harbor up to 1,000 different V. vulnificus strains (8). A critical question remains whether or not all V. vulnificus isolates have the same potential to cause human disease or if only a subset has full virulence potential. If the latter were the situation, then devising a test to identify oysters that are contaminated with more virulent isolates could be used to selectively manage harvested oysters.
Several studies have examined DNA sequence polymorphisms in different loci to genotype V. vulnificus strains. Real-time PCR of the 16S rRNA (rrn) polymorphic variants identified three major types: A, B, and hybrid AB ribotypes (11, 44, 57). The rrn B type was predominantly associated with clinically isolated strains, and the A and AB types were associated with environmentally derived strains. DNA polymorphism at the vcg locus, detected by a simple PCR method, identified vcgC versus vcgE genotype strains predominantly associated with clinical or environmental isolation, respectively (48). vcg typing generally matched rrn typing (11, 16, 49). Bisharat et al. (4, 5) used multilocus sequence typing (MLST) of housekeeping genes to type 159 V. vulnificus strains and constructed an online database for V. vulnificus MLST (http://pubmlst.org/vvulnificus/). V. vulnificus strains generally segregated into two clusters, with cluster 1 composed mainly of environmental isolates and cluster 2 composed primarily of clinical isolates (5). Cohen et al. (14) subsequently examined six housekeeping genes in 67 strains by MLST and developed a different nomenclature (lineage 1 and lineage 2) with designations that were the opposite of those of Bisharat and coworkers (5). Almost all lineage 1 strains (similar to MLST cluster 2) were of clinical origin, while lineage 2 (similar to MLST cluster 1) comprised most environmental isolates. Furthermore, most lineage 1 strains possessed a genomic island that was absent from most lineage 2 strains examined in that study. Repetitive extragenic palindromic DNA PCR (rep-PCR), which is based on polymorphisms at numerous loci (56), was applied to the analysis of 68 strains that were divided into seven groups (11). Most clinical isolates were in three groups, and most environmental isolates were in one group. The same study examined polymorphisms in intergenic sequences at the group 1 capsule wzb locus by PCR and identified three groups: group 1 generally correlated with clinical derivation, group 2 generally corresponded to environmental isolation, and group 0 failed to generate an amplicon and hence was untypeable and was composed exclusively of environmental strains. All of these studies related the genotype of a strain to the source of its isolation, implying that strains associated with clinical isolation would be of increased virulence. Bogard and Oliver (7) showed that the vcg genotype correlates with a virulence attribute, serum resistance, with vcgC genotype strains being more serum resistant. However, the relationship of genotype to virulence in animal models remained to be determined.
Our laboratory recently participated in a multicenter study that examined 25 clinical and 25 oyster-derived V. vulnificus strains for predictive markers of virulence, as well as frequencies of virulent strains among these populations, based on a mouse model of disease (15). That study showed that the vast majority of both clinical and oyster strains could cause skin infection in a subcutaneously (s.c.) inoculated iron dextran-treated mouse model. Clinical strains were significantly more adept at causing systemic liver infection and decreased body temperature, which is used as a surrogate marker for death. However, that study did not identify genetic markers for virulence of strains. We also used this mouse model to examine the virulence of small sets of clinical and environmental V. vulnificus strains and determined that some of the environmental strains either grew more slowly in vivo or were killed more effectively by the host (51). However, neither of these studies provided a comprehensive analysis of genotype and animal virulence.
In the present study, we performed a detailed analysis of the virulence properties of 33 clinical isolates and 36 environmental isolates of V. vulnificus biotype 1 in our s.c. inoculated iron dextran-treated mouse model. Most strains caused severe skin infection, which was a necessary prerequisite for systemic liver infection. Liver infection was a prerequisite step for death, which was determined by the surrogate criterion of a decrease in body temperature below 33°C. Even in the presence of severe skin infection, only a subset of strains had the potential to cause liver infection and death. Higher virulence potential, indicated by liver infection, a body temperature decrease, or death, was highly correlated with, but not exclusive to, the genotype that was common to clinical isolates (rrn type B, vcgC, and MLST group 2). Because genotypic classification did not correlate 100% with virulence, the relationship was one of propensity and was not predictive of virulence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Sixty-nine opaque, encapsulated V. vulnificus biotype 1 isolates were from clinical (n = 33) and environmental (n = 36) sources (Table 1). Forty-nine strains of V. vulnificus biotype 1 were obtained from a study in collaboration with the Food and Drug Administration and the Interstate Shellfish Sanitation Conference (15). V. vulnificus biotype 1 strains added to the collection included nine environmental strains from the Tampa Bay region of the Gulf of Mexico and one clinical strain from the Florida Department of Public Health, kindly provided by Valerie Harwood; previously described clinical strains MO6-24/0 (25, 51, 60) and ATCC 23907 (25); MLT365, MLT367, MLT403, VV1009, 2400112, and LL728, previously examined by our group for virulence (51, 52); and genomically sequenced strains CMCP6 (29) and YJ016 (13). All strains were confirmed as V. vulnificus by species-specific vvhA PCR and DNA probe as previously described (10, 59).
TABLE 1.
Strains examined in this study and summary of genotyping and virulence data
| Strain | Sourcea | rrn type | vcg genotype | rep-PCR | Capsule PCR | Lineage | MLST | Virulence group | No. with skin infection/totalb | Skin CFU count/g (log10)c | No. with liver infection/totalb | Liver CFU count/g (log10)c | No. dead/totalb | Temp (°C)c |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2400112 | C | B | C | 3 | 4 | 5/5 | 7.7 ± 1.2 | 5/5 | 5.0 ± 1.9 | 3/5 | 32.5 ± 1.7 | |||
| 106-2A | E | B | C | NGd | 2 | 3 | 4/5 | 6.9 ± 1.8 | 0/5 | 2.2 ± 0.1 | 0/5 | 37.8 ± 0.4 | ||
| 246-0058 Cape Canaveral | C | A | E | 7 | 0 | 3 | 5/5 | 6.8 ± 2.2 | 0/5 | 2.5 ± 0.0 | 0/5 | 36.1 ± 1.1 | ||
| 98-640 DP-E9 | E | A | E | 4 | 2 | 2 | 1 | 2 | 3/5 | 6.0 ± 2.8 | 0/5 | 2.5 ± 0.0 | 0/5 | 35.9 ± 0.7 |
| 98-641 DP-G8 | E | AB | E | 4 | 2 | 1 | 2 | 4/5 | 6.5 ± 2.5 | 0/5 | 2.5 ± 0.0 | 0/5 | 34.5 ± 0.6 | |
| 98-783 DP-A1 | E | A | E | 4 | 1 | 1 | 2 | 3/4 | 6.3 ± 2.2 | 0/4 | 2.5 ± 0.0 | 1/4 | 34.5 ± 1.3 | |
| 99-505 DP-C8 | E | AB | E | 4 | 2 | 1 | 2 | 5/5 | 6.2 ± 2.1 | 2/5 | 3.2 ± 0.9 | 1/5 | 34.6 ± 1.4 | |
| 99-509 DP-A6 | E | A | E | 4 | 1 | 2 | 4 | 8/10 | 6.7 ± 2.3 | 7/10 | 4.8 ± 2.4 | 4/10 | 33.5 ± 2.7 | |
| 99-520 DP-B8 | E | AB | E | 4 | 2 | 1 | 3 | 5/5 | 7.9 ± 0.6 | 0/5 | 2.5 ± 0.0 | 0/5 | 36.3 ± 0.4 | |
| 99-537 DP-G7 | E | A | E | 4 | 2 | 1 | 3 | 5/5 | 8.2 ± 0.3 | 2/5 | 3.0 ± 0.7 | 0/5 | 37.2 ± 1.2 | |
| 99-540 DP-B6 | E | A | E | 4 | 2 | 2 | 1 | 3 | 5/5 | 7.7 ± 0.4 | 0/5 | 2.5 ± 0.0 | 0/5 | 36.3 ± 0.6 |
| 99-578 DP-B1 | E | B | C | 3 | 1 | 2 | 4 | 5/5 | 7.9 ± 0.2 | 4/5 | 4.9 ± 1.4 | 4/5 | 32.3 ± 1.9 | |
| 99-581 DP-C7 | E | A | E | 6 | 2 | 1 | 3 | 5/5 | 7.2 ± 0.6 | 2/5 | 3.0 ± 0.6 | 0/5 | 36.8 ± 0.3 | |
| 99-584 DP-B12 | E | A | E | 4 | 2 | 1 | 2 | 4/10 | 4.6 ± 2.0 | 0/10 | 2.5 ± 0.0 | 0/10 | 35.2 ± 1.2 | |
| 99-609 DP-A4 | E | A | E | 4 | 2 | 2 | 1 | 3 | 5/5 | 7.9 ± 0.4 | 3/5 | 2.8 ± 0.3 | 1/5 | 35.1 ± 1.8 |
| 99-622 DP-E4 | E | A | E | 4 | 2 | 1 | 4 | 5/5 | 7.7 ± 0.3 | 2/5 | 3.6 ± 1.6 | 0/5 | 34.1 ± 1.1 | |
| 99-623 DP-F5 | E | AB | E | 4 | 2 | 1 | 2 | 5/5 | 5.4 ± 2.0 | 0/5 | 2.5 ± 0.1 | 0/5 | 35.5 ± 0.8 | |
| 99-624 DP-C10 | E | A | E | 4 | 2 | 1 | 3 | 4/4 | 6.8 ± 2.5 | 1/4 | 3.2 ± 1.3 | 1/4 | 35.7 ± 2.1 | |
| 99-625 DP-D8 | E | AB | E | 4 | 2 | 1 | 4 | 5/5 | 8.1 ± 0.2 | 3/5 | 4.0 ± 1.6 | 2/5 | 34.1 ± 3.3 | |
| 99-645 DP-C4 | E | A | E | 5 | 2 | 1 | 4 | 5/5 | 8.3 ± 0.4 | 4/5 | 4.7 ± 1.9 | 2/5 | 34.1 ± 2.0 | |
| 99-736 DP-C7 | E | AB | E | 4 | 2 | 1 | 2 | 7/10 | 6.1 ± 2.5 | 3/10 | 3.2 ± 1.1 | 2/10 | 34.3 ± 2.3 | |
| 99-738 DP-B5 | E | A | E | 4 | 2 | 1 | 5 | 5/5 | 8.3 ± 0.3 | 5/5 | 5.4 ± 1.0 | 2/5 | 33.5 ± 1.3 | |
| 99-742 DP-A9 | E | AB | E | 4 | 2 | 1 | 4 | 5/5 | 6.5 ± 2.0 | 3/5 | 4.4 ± 1.7 | 3/5 | 33.7 ± 1.7 | |
| 99-743 DP-B6 | E | B | E | 3 | 1 | 1 | 3 | 4/5 | 7.3 ± 2.5 | 0/5 | 2.5 ± 0.0 | 0/5 | 35.1 ± 1.6 | |
| 99-779 DP-D2 | E | A | E | 6 | 0 | 2 | 1 | 3 | 5/5 | 6.8 ± 2.5 | 1/5 | 3.4 ± 0.5 | 1/5 | 34.4 ± 2.4 |
| 99-780 DP-E1 | E | A | E | 5 | 1 | 1 | 3 | 5/5 | 7.2 ± 1.6 | 1/5 | 2.7 ± 0.5 | 0/5 | 35.4 ± 1.9 | |
| 99-796 DP-E7 | E | AB | E | 4 | 2 | 1 | 4 | 5/5 | 8.4 ± 0.3 | 5/5 | 4.5 ± 1.4 | 0/5 | 35.2 ± 2.2 | |
| ATCC 29307 | C | B | C | 4 | 5/5 | 8.1 ± 0.0 | 3/5 | 5.0 ± 2.6 | 0/5 | 34.6 ± 1.5 | ||||
| ATL-9572 | C | A | E | 3 | 2 | 1 | 1 | 0/5 | 3.0 ± 0.0 | 0/5 | 2.5 ± 0.0 | 0/5 | 35.4 ± 1.0 | |
| ATL-9580 | C | B | C | 3 | 1 | 2 | 5 | 5/5 | 8.3 ± 0.2 | 5/5 | 5.8 ± 0.0 | 5/5 | 29.6 ± 0.0 | |
| ATL-9824 | C | B | C | 4 | 1 | 2 | 2 | 3/5 | 6.1 ± 2.9 | 2/5 | 3.4 ± 1.2 | 1/5 | 34.7 ± 2.0 | |
| CDC 9003-97 (FLA 9509) | C | B | C | 9 | 1 | 2 | 4 | 5/5 | 8.3 ± 0.1 | 5/5 | 5.1 ± 0.5 | 5/5 | 29.7 ± 1.0 | |
| CDC 9005-97 (DAL 7-9087) | C | A | E | 3 | 2 | 1 | 1 | 3 | 9/9 | 7.0 ± 1.4 | 3/9 | 2.6 ± 0.3 | 0/10 | 36.0 ± 0.6 |
| CDC 9030-95 (ORL 1506) FLDHRS-383C | C | AB | E | 4 | 2 | 2 | 1 | 5 | 10/10 | 7.8 ± 0.3 | 10/10 | 5.7 ± 1.1 | 10/10 | 29.2 ± 1.4 |
| CDC 9031-96 (ATL 6-1306) | C | AB | E | 4 | 2 | 1 | 3 | 5/5 | 6.9 ± 1.9 | 3/10 | 2.7 ± 0.6 | 2/10 | 35.3 ± 2.6 | |
| CDC 9032-95 (ORL-8074) FLDHRS-407C | C | B | C | 3 | 1 | 2 | 4 | 5/5 | 7.9 ± 0.5 | 5/5 | 5.4 ± 0.1 | 5/5 | 29.9 ± 0.5 | |
| CDC 9038-96 (LOS 7318) | C | B | C | 2 | 1 | 1 | 2 | 2 | 5/5 | 6.1 ± 1.7 | 0/5 | 2.5 ± 0.0 | 0/5 | 34.9 ± 0.8 |
| CDC 9053-96 (FLA 8869) | C | B | C | 9 | 1 | 2 | 2 | 4/5 | 4.9 ± 2.1 | 1/5 | 2.8 ± 0.7 | 0/5 | 35.5 ± 0.6 | |
| CDC 9060-96 (DAL 7-9002) | C | B | C | 4 | 2 | 2 | 5 | 5/5 | 7.6 ± 0.5 | 5/5 | 6.3 ± 1.0 | 5/5 | 29.6 ± 1.2 | |
| CDC 9062-96 (LOS 7343) | C | B | C | 9 | 1 | 1 | 2 | 4 | 10/10 | 8.1 ± 0.9 | 6/10 | 3.5 ± 1.4 | 4/10 | 33.4 ± 3.6 |
| CDC 9067-96 (DAL 7-9000) | C | B | C | 9 | 1 | 2 | 4 | 5/5 | 7.9 ± 0.3 | 5/5 | 5.3 ± 1.1 | 4/5 | 31.8 ± 1.1 | |
| CDC 9070-96 (DAL 79040) | C | B | C | 4 | 1 | 2 | 2 | 3/5 | 5.9 ± 2.7 | 3/5 | 3.7 ± 1.2 | 2/5 | 34.0 ± 2.5 | |
| CDC 9074-96 (ATL 71491) | C | B | C | 9 | 1 | 2 | 5 | 5/5 | 8.2 ± 0.3 | 5/5 | 6.5 ± 1.4 | 5/5 | 27.9 ± 1.9 | |
| CDC 9075-96 (ATL 71503) | C | AB | E | 4 | 2 | 1 | 2 | 4/5 | 6.0 ± 2.5 | 2/5 | 3.7 ± 1.2 | 1/5 | 34.9 ± 3.3 | |
| CDC 9076-96 (ATL 71504) | C | B | C | 9 | 1 | 2 | 5 | 5/5 | 8.5 ± 0.1 | 5/5 | 5.4 ± 1.4 | 0/5 | 35.1 ± 2.1 | |
| CDC 9149-95 (NSV 5829) | C | AB | E | 4 | 2 | 1 | 4 | 5/5 | 8.0 ± 0.3 | 5/5 | 4.9 ± 1.5 | 3/5 | 33.0 ± 2.5 | |
| CDC 9340-95 (ORL 8324) FLDHRS-721C | C | B | C | 1 | 1 | 2 | 4 | 5/5 | 8.0 ± 0.3 | 4/5 | 4.2 ± 1.3 | 3/5 | 32.9 ± 3.2 | |
| CDC 9342-95 (LOS 6966) | C | B | C | 4 | 2 | 2 | 2 | 3/5 | 5.9 ± 2.6 | 0/5 | 2.5 ± 0.1 | 0/5 | 35.8 ± 1.1 | |
| CDC 9345-95 (DAL 6-5000) | C | B | C | NG | 2 | 2 | 1 | 5/5 | 4.0 ± 1.3 | 1/5 | 2.6 ± 0.1 | 0/10 | 36.0 ± 0.5 | |
| CDC 9348-95 (NSV 5830) | C | AB | E | 4 | 2 | 1 | 5 | 5/5 | 8.1 ± 0.0 | 5/5 | 6.7 ± 0.2 | 4/5 | 31.1 ± 1.1 | |
| CDC 9349-95 (NSV 5736) | C | B | C | 3 | 1 | 2 | 5 | 5/5 | 8.3 ± 0.5 | 5/5 | 7.3 ± 0.9 | 4/5 | 31.6 ± 0.0 | |
| CDC 9352-94 (ATL 9823) | C | B | C | 1 | 1 | 2 | 4 | 5/5 | 8.4 ± 0.5 | 3/5 | 3.3 ± 0.8 | 0/5 | 37.3 ± 1.0 | |
| CMCP6 | C | B | C | NG | 1 | 1 | 2 | 4 | 5/5 | 7.2 ± 0.3 | 5/5 | 4.3 ± 1.2 | 4/5 | 31.2 ± 2.0 |
| FDOH 6325 | C | A | E | 7 | 0 | 4 | 7/10 | 6.5 ± 2.4 | 7/10 | 4.2 ± 1.2 | 6/10 | 31.3 ± 4.3 | ||
| LL728 | C | B | C | NG | 4 | 4/5 | 7.5 ± 2.1 | 4/5 | 4.8 ± 1.3 | 4/5 | 33.2 ± 1.9 | |||
| MO6-24/O | C | B | C | 9 | 1 | 1 | 2 | 4 | 5/5 | 7.9 ± 0.1 | 5/5 | 5.2 ± 0.7 | 5/5 | 29.0 ± 2.1 |
| MLT365 | E | B | C | 8 | 1 | 1/5 | 3.7 ± 1.7 | 1/5 | 2.5 ± 0.1 | |||||
| MLT367 | E | B | C | 8 | 1 | 2/6 | 3.5 ± 1.1 | 0/6 | 2.5 ± 0.0 | |||||
| MLT403 | E | B | C | NG | 1 | 1/5 | 3.5 ± 1.1 | 0/5 | 2.5 ± 0.0 | |||||
| S1-10 | E | B | C | 9 | 1 | 2 | 2/5 | 5.1 ± 2.9 | 1/5 | 2.9 ± 1.1 | 0/5 | 37.5 ± 1.7 | ||
| S1-13 | E | A | E | 6 | 1 | 4 | 5/5 | 8.1 ± 0.2 | 4/5 | 4.7 ± 1.9 | 4/5 | 32.5 ± 3.8 | ||
| S1-8 | E | A | E | 4 | 0 | 2 | 0/5 | 4.5 ± 0.0 | 0/5 | 2.5 ± 0.0 | 0/5 | 37.4 ± 0.5 | ||
| S2-10 | E | A | E | 3 | 0 | 4 | 5/5 | 8.3 ± 0.4 | 2/5 | 3.4 ± 1.7 | 2/5 | 35.8 ± 3.9 | ||
| S2-20 | E | B | C | 3 | 1 | 4 | 4/4 | 8.2 ± 0.3 | 4/4 | 4.8 ± 0.7 | 4/4 | 32.6 ± 3.4 | ||
| S2-22 | E | B | C | 9 | 1 | 3 | 4/5 | 6.8 ± 1.9 | 1/5 | 2.5 ± 0.7 | 0/5 | 38.3 ± 0.3 | ||
| S3-16 | E | B | C | 3 | 0 | 1 | 1/5 | 3.2 ± 0.0 | 0/5 | 2.4 ± 0.2 | 0/5 | 36.8 ± 0.6 | ||
| S3-5 | E | A | E | 4 | 0 | 4 | 5/5 | 7.5 ± 1.3 | 4/5 | 4.7 ± 1.1 | 2/5 | 34.5 ± 2.1 | ||
| VV1009 | C | B | C | NG | 4 | 4/5 | 6.9 ± 2.3 | 4/5 | 4.6 ± 1.3 | 3/5 | 34.3 ± 3.1 | |||
| YJ016 | C | B | C | 2 | 2 | 1 | 2 | 5 | 5/5 | 7.7 ± 0.3 | 8/8 | 5.7 ± 0.2 | 8/8 | 29.1 ± 3.6 |
C, clinical; E, environmental.
Number of mice with detectable skin infection, liver infection, or a body temperature of less than 33°C over the total number of mice inoculated.
Mean ± standard deviation.
NG, not grouped. No other strains exhibited greater than 85% homology.
Bacteria were grown in Luria-Bertani broth containing 0.85% (wt/vol) NaCl (LB-N) or on LB-N plates containing 1.5% (wt/vol) agar. Unless otherwise noted, all reagents and media were purchased from Difco (Franklin Lakes, NJ). Strains were stored at −70°C in LB-N with 35% (vol/vol) glycerol. For mouse infections, a static overnight starter culture of the bacteria was grown at room temperature in LB-N broth in a culture tube. On the day of infection, the starter culture was diluted 1:20 into fresh LB-N broth and shaken at 37°C until the optical density at 600 nm reached approximately 0.4, which was previously determined to be 1.5 × 108 CFU/ml. Bacteria were diluted in phosphate-buffered saline (PBS) to a concentration appropriate for use. Total CFU counts in the inoculum were confirmed by dilution and plating.
Infection of mice.
The iron dextran-treated mouse model was described previously (51, 52). Briefly, 7- to 10-week old female ICR mice (Harlan Sprague-Dawley, Indianapolis, IN) were housed under specific-pathogen-free conditions. All experiments were approved by the University of Florida Institutional Animal Care and Use Committee. Groups of five mice were injected intraperitoneally (i.p.) with 250 μg of iron dextran (Sigma Aldrich) per g of body weight 1 to 2 h prior to infection. Mice were injected s.c. with approximately 1,000 CFU suspended in 0.1 ml of PBS in the lower back and were monitored for lethargy, scruffiness, and a decrease in body temperature. Body temperature was measured rectally using a mouse rectal probe (AD Instruments, Colorado Springs, CO) attached to a Fluke Thermocouple (Cole-Parmer) and recorded immediately prior to euthanasia. Mice were euthanized by carbon dioxide asphyxiation when their body temperatures fell below 33°C, when their gross appearance suggested severe distress, or at 20 h postinfection. Following euthanasia, the skin was peeled back from head to tail, revealing the s.c. injection site with or without a lesion. The injection site and musculature were photographed. Three indicators were used to determine virulence in the mouse model: the CFU count/g in s.c. skin lesions, the CFU count/g in liver tissue, and body temperature. For statistical analysis, if a mouse died before its temperature could be taken, it was assigned the value of the surviving infected mouse within the group with the most severe infection. Mice without s.c. skin lesions were assigned minimum detectable values for statistical analysis of skin lesion and liver CFU counts/g.
Genotyping of V. vulnificus strains.
Molecular typing of polymorphic DNA sequences was performed as described previously. vcg genotype was determined by PCR using the primers of Rosche et al. (48) to classify strains as being of the vcgC or the vcgE genotype. The capsule genotype (0, 1, or 2) was determined by PCR using primers of Chatzidaki-Livanis et al. (12). The 16S rRNA (rrn) type was determined using the real-time PCR method and primers of Chatzidaki-Livanis et al. (11) to detect polymorphisms in the 16S rRNA sequence yielding rrn types A, B, and AB (11, 44, 57). rep-PCR was performed exactly as described by Chatzidaki-Livanis et al. (11). Although we did not perform MLST analysis, we used MLST data of Bisharat et al. (5), Cohen et al. (14), and the V. vulnificus MLST website (http://pubmlst.org/vvulnificus/) (26) for the strains examined in our study.
Statistical analysis.
Skin lesion and liver CFU counts were log10 transformed for statistical analysis. SAS version 9.2 (SAS, Cary, NC) was used for all statistical analyses. Two-sided tests were used for all hypothesis testing. The level of significance was set at 0.05 for all tests of hypotheses. The relationships among quantitative indicators of virulence (skin and liver CFU counts/g and body temperature) were tested using Spearman correlational analysis. We used multiple-regression analysis to simultaneously test the relationship of skin and liver CFU counts/g to body temperature.
The CLUSTER procedure in SAS was used to examine the relationships of strains, as determined by their skin and liver CFU counts. The objective in cluster analysis is to group like observations together when the underlying structure is unknown. A distance measure, which determines the similarity of elements, is calculated, and data points with the smallest distances between them are grouped together. The data with the next smallest distances are added to each group, etc., until all observations end up together in one large group organized by subcategories of homogeneity. This procedure is useful to detect clusters with practical meaning.
Differences in skin lesion and liver CFU counts and body temperature between sets of V. vulnificus strains were tested using analysis of variance (ANOVA) (when there were three or more levels of genotype), followed by Tukey multiple-comparison testing or two-sample Student t tests with unequal group variances assumed (when there were two levels of genotype). Chi-square analysis was used to compare the incidences of detectable skin infection, detectable liver infection, and death (a body temperature of less than 33°C) between groups of strains. Chi-square analysis (and Fisher's exact testing in the case of data sparseness) was used to test the relationship between the source of isolation and the genotype.
RESULTS
High levels of skin infection are necessary, but not sufficient, to cause systemic infection and death in s.c. inoculated mice.
Using the s.c. inoculated iron-dextran-treated mouse model, we previously determined that almost all strains of V. vulnificus biotype 1, regardless of clinical or oyster isolation, were capable of causing severe skin infection (15). However, clinical isolates produced significantly higher CFU counts in livers and significantly lower body temperatures (a surrogate marker for death) at the time of euthanasia. These results suggested that the clinical strains were preselected for the ability to cause systemic disease and that some variation in the virulence potential of V. vulnificus strains in the wild might exist, i.e., that there is a subset of strains with the potential to cause systemic disease and a subset that lacks this potential. The present study was undertaken to examine how molecular typing of V. vulnificus strains relates to virulence in our mouse model. We also added 8 clinical and 12 environmental (oysters or seawater) V. vulnificus strains to the group that was previously examined in the mouse model (15), bringing the totals to 33 clinical strains and 36 environmental strains (Table 1).
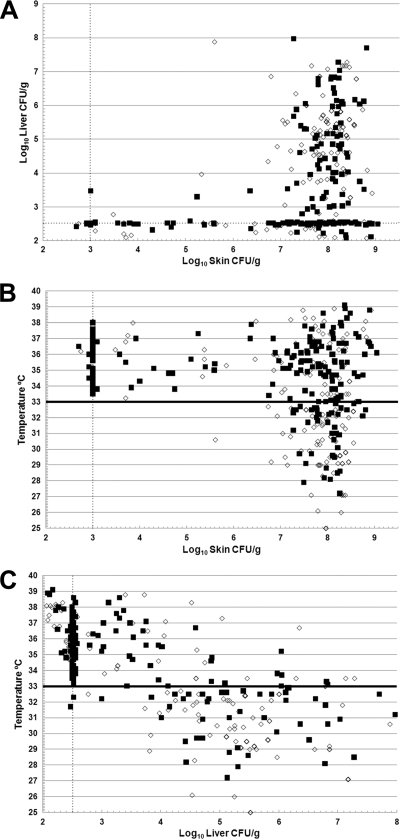
The three indicators of virulence (the skin CFU count/g, the liver CFU count/g, and a decrease in body temperature) were examined to determine if relationships exist among them that could explain the disease process in more detail using pairwise linear regression analysis between the measures of virulence for every mouse in this study (n = 390). There was a positive correlation between the skin lesion CFU count/g and the liver CFU count/g (Fig. 1A) (Spearman correlation = 0.48, P < 10−6). As the number of bacteria in skin lesions increased, the number of bacteria in the liver increased. However, there was a threshold effect, with a minimum level of skin infection required for systemic liver infection. With only a few exceptions (5 of 194 mice with detectable liver infections), liver infection was undetectable until the skin infection was greater than 106.5 CFU/g. We used a decrease in body temperature from 37°C to less than 33°C as a surrogate marker for death (51). There were highly significant correlations between the skin lesion CFU count/g and the liver CFU count/g with decreased body temperature (Fig. 1B and C). For the skin CFU count and body temperature, the Spearman correlation coefficient was −0.24 (P = 4.3 × 10−6, n = 374). For the liver CFU count and body temperature, the Spearman correlation coefficient was −0.73 (P < 10−6, n = 374). The results of multiple-regression analysis showed that after adjusting for the liver CFU count, the effects of the skin CFU count were not significant (P = 0.71). In contrast, after adjusting for the skin CFU count, the liver CFU count was highly predictive of the body temperature (P < 10−6). For every 10-fold increase in the liver CFU count, the body temperature dropped by 1.5°C, and for every 10-fold increase in the liver CFU count, the odds of death increased 5-fold. Furthermore, with only one exception, the body temperature did not drop below 33°C until the skin CFU count reached greater than 106.7 CFU/g skin lesion. However, even with skin infections above this level, only 132 (46%) of 290 mice were classified as dead. Therefore, during the first 20 h of infection, it was possible for mice to have a severe skin infection but not lethal systemic disease. In contrast, as soon as liver infections rose above the minimum detectable level of 102.6 CFU/g, body temperatures began to drop below 33°C, and as liver infections increased, body temperatures further decreased. In fact, when the liver infection level was greater than 104.1 CFU/g, only 25 (17%) of 147 mice had body temperatures above that at which they were considered moribund. It should be noted that several of these surviving mice likely would have died with further decreasing body temperatures had they not been euthanized for humane purposes, given their physical appearance. Therefore, severe skin infection is a necessary prerequisite for liver infection, which is a prerequisite for death. However, severe skin infection does not necessarily lead to systemic infection.
FIG. 1.
Relationships among the virulence attributes of all of the mice in the s.c. inoculated iron dextran-treated mouse model. The skin CFU count/g, the liver CFU count/g, and the body temperature at euthanasia were examined relative to each other. (A) Liver infection versus skin infection. (B) Body temperature versus skin infection. (C) Body temperature versus liver infection. Symbols: ▪, mice infected with vcgE genotype V. vulnificus; ⋄, mice infected with vcgC genotype V. vulnificus. The line at 33°C in panels B and C shows the body temperature cutoff for death in the model. The dotted lines for the skin and liver CFU counts show the minimum detectable values.
These data were also examined based on strain means, and the same conclusions were drawn with regard to skin versus liver infection and liver infection versus death. Only 2 of the 69 strains failed to cause a detectable skin infection in any mice exposed to an inoculum of 1,000 CFU. Seventeen strains failed to cause a detectable liver infection in any mice, and 35 strains failed to cause a liver infection in half or more of the inoculated mice. Of the 45 strains that caused a mean skin infection level of greater than 106.7 CFU/g, 32 caused a drop in body temperature to a mean of less than 33°C. Of the 35 strains that caused liver infections in greater than half of the inoculated mice, all but 4 killed some of the mice. Based on these relationships and as we reported previously (15), encapsulated V. vulnificus biotype 1 strains have a nearly universal ability to cause skin infection in s.c. inoculated iron dextran-treated mice, but the ability to cause a systemic liver infection that leads to death is a separable characteristic.
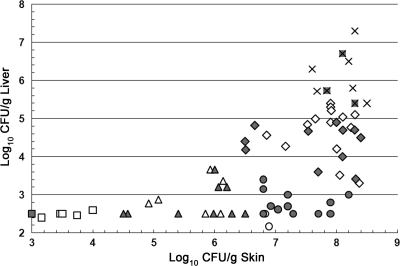
Because of the complex pattern relating the skin and liver CFU counts among strains in this study, we performed a cluster analysis to examine the relationships of strains relative to the skin and liver CFU counts. A cluster distance of 1.0 generated five cluster groups of strains with interpretable characteristics (Fig. 2). Group 1 strains exhibited very low levels of skin infection with undetectable liver infection. These are the most attenuated of the strains. Group 2 strains had low skin infection levels with undetectable to low liver infection levels. Group 3 strains exhibited high levels of skin infection but undetectable to low levels of liver infection. Group 4 made up the largest set of strains and exhibited high levels of skin infection with moderate to high levels of liver infection. The nine strains in group 5 exhibited very high skin and liver infection levels and were the most virulent of the strains examined.
FIG. 2.
Grouping of V. vulnificus strains based on the relationship between skin and liver CFU counts. The CLUSTER program of SAS was used to examine the skin and liver CFU counts of V. vulnificus strains. Strains with a cluster distance of less than 1.0 were considered a group. Group 1, □, very low level in skin, undetectable in liver; group 2, Δ, low level in skin and undetectable to low level in liver; group 3, ○, high level in skin and undetectable to low level in liver; group 4, ⋄, high level in skin and moderate to high level in liver; group 5, ×, very high level in skin and very high level in liver. vcgC genotype strain symbols are white; vcgE genotype strain symbols are gray.
The source of a strain correlates with but does not predict virulence.
We investigated the relationship of the clinical versus environmental source of isolation of a V. vulnificus strain to its virulence potential. Clinical strains caused significantly higher skin CFU counts, on average, than did environmental strains after s.c. inoculation of iron dextran-treated mice (P = 9.4 × 10−4, t test). However, it should be noted that both sets of strains caused high levels of skin infection (mean CFU counts/g of 107.2 and 106.5, respectively). As we reported previously (15), clinical strains also caused significantly higher liver CFU counts and lower body temperatures than did environmental strains (P < 10−6, t tests). The clinical strains caused detectable liver infections in 65% of the mice and death in 49% of the mice, compared to 34% and 20% for the environmental strains, with highly significant differences (P < 10−6, χ2 test). However, 9 of 36 environmental strains exhibited high virulence in terms of causing greater than 104 CFU/g liver, and 13 of 33 clinical strains failed to cause this level of liver infection. Therefore, the relationships between the source of a strain and its virulence are not absolute in either direction, and the source of a strain does not predict its potential virulence. The lethality of the infections by the clinical strains for the patients was known for 24 of the strains, so we asked if differences in virulence attributes exist between the strains that caused lethal versus nonlethal infections. For all three measures of virulence, there were no significant differences between the strains that killed the patients and those that did not. Given all of the variables involved in determining the cause of death in a given case (e.g., the general health of the patient, the quality of health care, access to health care), this is not a surprising result.
Genotype of V. vulnificus strains is correlated with but does not predict the ability to cause systemic infection and death.
We genotyped our strain collection based on rrn type, vcg, rep-PCR, and capsule locus as described above. We used the MLST data from Bisharat et al. (5) and the online V. vulnificus MLST database (http://pubmlst.org/vvulnificus/), which included 50 of our 69 strains, and 12 of our strains were included in MLST data from Cohen et al. (14). The 69 strains were examined for relationships among these genotyping schemes and virulence potential in two ways. First, the mean CFU count/g skin, CFU count/g liver, and body temperature were compared for each mouse among genotypes. Second, the incidences of detectable skin infection, detectable liver infection, and death were also compared among genotypes.
rrn type.
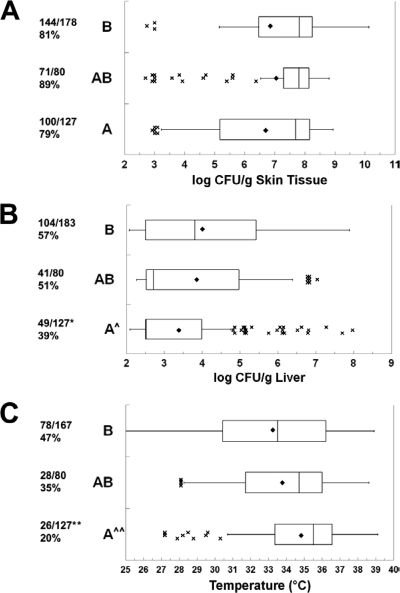
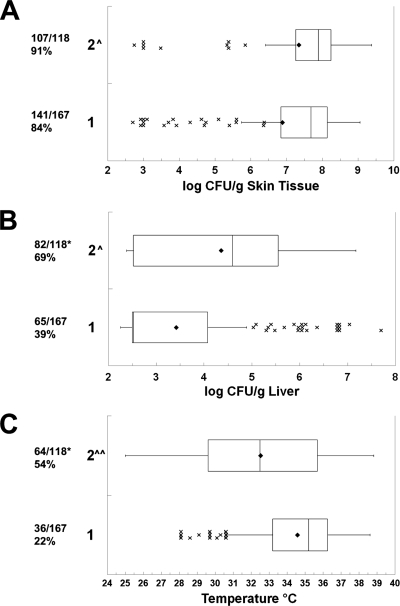
Based on rrn sequences, there were 22 A, 34 B, and 13 AB V. vulnificus strains in our collection, as determined using PCR with primers from Chatzidaki-Livanis et al. (11). There were highly significant differences among the rrn types for the clinical versus environmental sources of the strains (P = 4.8 × 10−4, χ2 test). The numbers of clinical/environmental strains of the A, AB, and B types were 4/18, 5/8, and 24/10, respectively. The B strains were significantly more likely to be from a clinical source than were the A and AB strains (P = 1.3 × 10−4 and P = 0.042, respectively, χ2 test). This distribution of rrn genotypes is similar to that reported by others (11, 44, 57). Because of the large sample sizes, the asymmetric distribution of data points, and frequent outlier data, the magnitudes of skin and liver infections and body temperature are presented as box plots. As shown in Fig. 3, there were no significant differences in the abilities of the three rrn types to cause skin infection in terms of incidence or magnitude. The asymmetric distribution of skin infection toward higher numbers of CFU/g for all three types can be seen by the means being lower than the medians, the third quartile of data spanning a smaller range of CFU counts/g than the second quartile of data, and the outlier data at the low end of the infection scale. This is consistent with the data presented in Fig. 1 showing a skewing of skin infection toward the higher end of the spectrum. The type A strains caused a significantly lower incidence of detectable systemic infection (P = 0.0016) and significantly lower levels of liver infection (P = 0.0014) than did type B strains. As opposed to the skin CFU data, the liver CFU data were skewed toward lower CFU counts/g for types A and AB (note the extremely narrow second quartiles). The outlier data for types A and AB demonstrate that the low levels of liver infection were not uniform for all of the strains of these types. The incidence of death was significantly different among the rrn types (P = 1.9 × 10−5, χ2 test), with the incidence of death for A strains being significantly lower than that for the AB and B rrn types (P = 3.2 × 10−6 and P = 0.020, respectively). This is explained by the type A strains causing a significantly higher body temperature at the time of euthanasia than the type B and AB strains (P = 4.4 × 10−5 and P = 0.039, respectively). There was not a significant difference in body temperature between type B and type AB strains. In summary, the rrn type B and AB strains were more proficient at causing systemic infection and death than were the type A strains. The similarity in type B and AB strains is interesting given the fact that the type AB strains are genetically more related to the type A strains by most of the other genotyping methods (see below). Although these results suggest an increased virulence potential for the type B strains, consistent with their tendency to be of clinical origin, it should be noted that many type B strains were avirulent by all measures. Conversely, some type A strains exhibited very high virulence. The former observation could be explained by loss of virulence factors upon subculture, but the latter observation indicates that the rrn genotype does not absolutely predict virulence.
FIG. 3.
Relationship between 16S rrn ribotype and virulence. Box plots show the relationships between rrn type and (A) skin infection, (B) liver infection, and (C) body temperature at euthanasia. The rrn genotypes are shown on the left. The fractions are the numbers of mice with detectable skin infections (A), detectable liver infections (B), or body temperatures of less than 33°C (surrogate for death) (C) over the total number of mice of that genotype for which data are available. The percent positive is shown below the fraction. The boxes show the second and third quartiles of data with the median in between. Symbols: ♦, mean; ×, outlier data (greater than 1.5 times the range of the second and third quartiles from the median, designated by the whiskers); ^, rrn type A significantly lower than type B (ANOVA); *, rrn type A significantly lower than type B (χ2); ^^, rrn type A significantly lower than types B and AB (ANOVA); **, rrn type A significantly lower than types B and AB (χ2). P values are in the text.
vcg gene polymorphism.
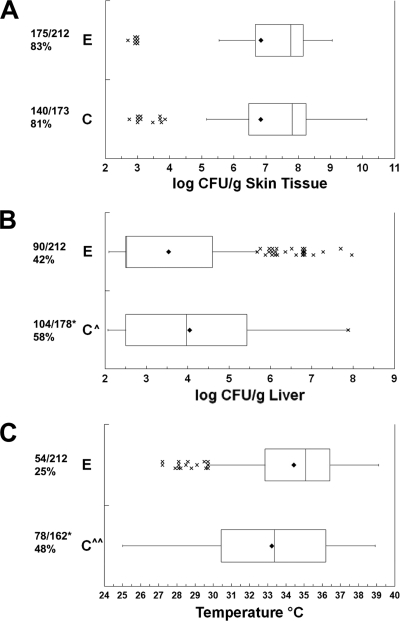
We used the PCR primers of Rosche et al. (48) to examine the vcg polymorphism of our 69 strains. As had been noted when this genotyping scheme was developed (48), vcgC strains were predominantly isolated from clinical cases rather than environmental sources (24 and 9 strains, respectively), whereas vcgE strains were predominantly from environmental sources rather than clinical sources (27 and 9 strains, respectively) (P = 7.4 × 10−5, χ2 test). The incidence and magnitude of skin infections were not significantly different between the vcgC and vcgE genotypes (Fig. 4). However, the vcgC genotype-infected mice had a significantly higher incidence of systemic infection (P = 0.0017, χ2 test) and higher numbers of CFU/g liver (P = 0.0013) than those infected with vcgE genotype strains. vcgC genotype strain-infected mice were significantly more likely to be classified as dead than were vcgE genotype-infected mice (P = 5.4 × 10−6, χ2 test), and the mean body temperatures of euthanized mice were significantly lower after vcgC genotype strain infection than after vcgE genotype strain infection (P = 0.0001). In summary, most vcgC genotype strains were more adept than vcgE genotype strains at causing systemic infection and death in mice; however, 20 of 36 vcgE genotype strains were capable of causing lethal systemic infections in some mice (note the outlier data in Fig. 4B and C), with 6 of these strains killing more than half of the inoculated mice.
FIG. 4.
Relationship between vcg type and virulence. Box plots show the relationships between vcg type and (A) skin infection, (B) liver infection, and (C) body temperature at euthanasia. The vcg genotypes are shown on the left. For details, see the legend to Fig. 3. Symbols: ^, vcg type C significantly higher than vcg type E (ANOVA); ^, vcg type C significantly lower than vcg type E (ANOVA); *, vcg type C significantly higher than vcg type E (χ2).
MLST.
Fifty of the strains in our collection have been typed by MLST (5) (http://pubmlst.org/vvulnificus/), with 29 in cluster 1 and 21 in cluster 2. The isolates in the MLST clusters were from highly significantly different sources, with MLST cluster 1 consisting mainly of environmental isolates (22 of 29 strains) and cluster 2 consisting mainly of clinical isolates (20 of 21 strains) (P < 10−6, χ2 test). The incidence of skin infection was not significantly different between the cluster 1 and cluster 2 genotype isolates (P = 0.12, χ2 test) (Fig. 5). Although the levels of skin infection were high for both MLST clusters (means of 106.9 and 107.3 CFU/g for clusters 1 and 2, respectively), cluster 2 strains caused significantly higher skin CFU counts than did cluster 1 strains (P = 0.037). The incidence of detectable systemic infection was nearly twice as high for cluster 2 as for cluster 1 (P < 10−6, χ2 test), and MLST cluster 2 strains caused significantly higher liver infection levels than MLST cluster 1 strains (P < 10−6). Similarly, the incidence of death was more than twice as high for cluster 2 as for cluster 1 (P < 10−6, χ2 test) and cluster 2 strains caused significantly lower body temperatures than did cluster 1 strains (P < 10−6). Despite these highly significant associations with virulence, 20 of the 29 cluster 1 strains still caused detectable systemic infection and 15 of these strains caused the death of at least one inoculated mouse. Conversely, six cluster 2 strains failed to kill any mice and two failed to cause detectable liver infection. Therefore, there is a correlative relationship between MLST and virulence but it is not absolute.
FIG. 5.
Relationship between MLST and virulence. Box plots show the relationships between MLST and (A) skin infection, (B) liver infection, and (C) body temperature at euthanasia. The MLST genotypes are shown on the left. For details, see the legend to Fig. 3. Symbols: ^, MLST 2 significantly higher than MLST 1 (ANOVA); ^^, MLST 2 significantly lower than MLST 1 (ANOVA); *, MLST 2 significantly higher than MLST 1 (χ2).
Twelve of our 69 strains were described by Cohen et al. (14) in their study detailing two principal lineages of V. vulnificus based on MLST, although a different set of genes was examined than in the MLST analysis described immediately above (5). Lineage 1 was associated with clinical strains, while lineage 2 was associated with environmental isolates. There were no significant differences in any of the measures of virulence compared between lineage 1 and 2 strains in our collection (data not shown), most likely due to the small sample size of strains in our collection that had been genetically characterized by Cohen et al. (14).
rep-PCR.
Chatzidaki-Livanis et al. (11) used repetitive DNA sequence PCR to classify 68 V. vulnificus strains into seven groups. Because rep-PCR data analysis is dynamic and based on the overall relatedness of all of the strains in the database at a given time, group designations can change as more strains are examined. The addition of strains to the previously described rep-PCR database for the present study caused the number of groups, defined as having greater than 85% homology, to increase to nine, and some of the rep-PCR designations changed from the previous report (Table 1). Furthermore, six strains were not grouped (i.e., they were not 85% homologous to any other strains) and one strain was not examined by rep-PCR. The largest group was group 4, with 29 strains, followed by groups 3 and 9, with 11 and 9 strains, respectively. All of the other groups had three or fewer strains. Groups 1, 2, and 7 consisted of only clinical isolates, whereas groups 5, 6, and 8 consisted of only environmental isolates. Group 3 was about evenly split, with six clinical and five environmental strains. Group 4 was predominantly environmental, with 20 of 29 strains being of environmental origin. Group 9 was predominantly clinical, with seven of nine strains being of clinical isolation. The groups were significantly different with regard to the source of isolation (P = 4.8 × 10−3, Fisher's exact test). The relationship between rep-PCR and other genetic typing systems is discussed below and is very complex.
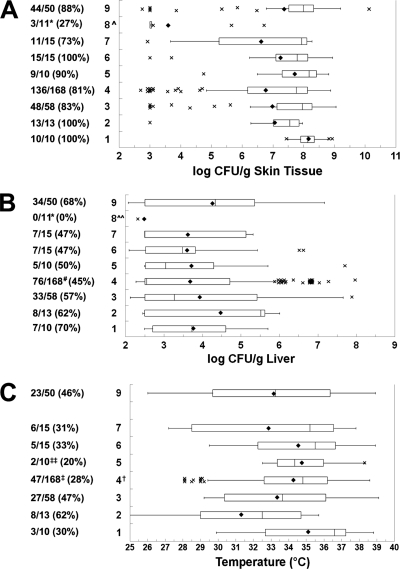
With the following exception, there were no significant differences in the skin CFU counts/g among the rep-PCR groups (Fig. 6). Interestingly, the two group 8 strains had significantly lower incidences of detectable skin infection (1.4 × 10−5 ≤ P ≤ 0.020) and produced significantly lower skin CFU counts/g than every other group (3.2 × 10−6 ≤ P ≤ 0.0027). Groups 1, 2, 3, and 9 caused liver infections in >50% of the inoculated mice, while groups 4, 5, 6, 7, and 8 caused liver infections in ≤50% of the inoculated mice (P = 4.1 × 10−3, χ2 test). Although the mean liver infection levels by rep-PCR group ranged from 102.5 CFU/g (undetectable) to 104.5 CFU/g, only two significant differences in liver CFU counts were noted between the groups; that of group 8 was significantly lower than that of groups 9 and 2 (P = 0.018 and 0.047, respectively). It is likely that some lack of significance in the liver CFU count was due to the small numbers of strains in some groups and assigning the minimum detectable level to negative samples. The rep-PCR groups differed significantly in incidence of death (P = 0.049, Fisher's exact test). Groups 1, 4, and 5 killed less than one-third of the inoculated mice, while the other groups were more lethal. Group 4 produced a significantly lower incidence of death than groups 2, 3, and 9 (0.009 ≤ P ≤ 0.017), and group 5 produced a significantly lower incidence than group 2 (P = 0.046). The mean body temperatures ranged from 35.1°C (group 1, slightly low) to 31.3°C (group 2, moribund). The mean body temperature of group 4 was significantly higher than that of group 2 (P = 0.019). Note that group 8, which was the least virulent rep-PCR group, did not have body temperature data available, although its low level of liver infection would predict a normal body temperature. In summary, as would be expected from dividing the data set for virulence criteria into nine groups by rep-PCR, it became more difficult to detect significant differences among the groups. However, rep-PCR group 8 was consistently among the least virulent, followed by rep-PCR group 4.
FIG. 6.
Relationship between rep-PCR type and virulence. Box plots show the relationships between rep-PCR group and (A) skin infection, (B) liver infection, and (C) body temperature at euthanasia. The rep-PCR genotypes are shown on the left. For details, see the legend to Fig. 3. Symbols: ^ and *, group 8 significantly lower than all other groups (ANOVA and χ2, respectively); ^^, group 8 significantly lower than groups 2 and 9 (ANOVA); #, group 4 significantly lower than group 9 (χ2); †, group 4 significantly higher than group 2 (ANOVA); ‡, group 4 significantly lower than groups 2, 3, and 9 (χ2); ‡‡, group 5 significantly lower than group 2 (χ2).
Capsule locus polymorphism.
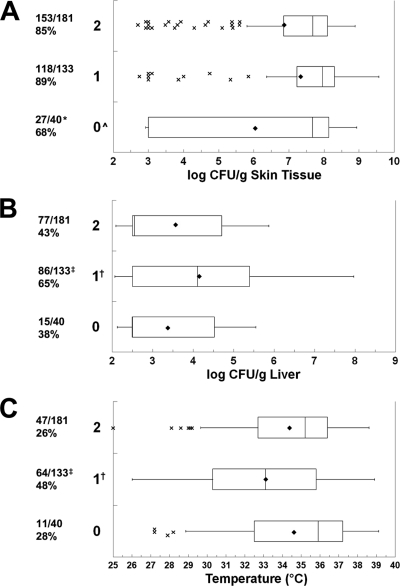
We used the PCR method of Chatzidaki-Livanis et al. (12) to classify three V. vulnificus alleles that are associated with intergenic, noncoding repetitive DNA regions that flank the wzb capsule locus: types 1 and 2 yielded distinct bands, and type 0 was not typeable. Unlike with the other genotyping systems, there was not a significant difference among the capsule allele types for clinical versus environmental sources (P = 0.076, χ2 test), with all types consisting of mixtures of clinical and environmental strains. The numbers of clinical/environmental strains of the 0, 1, and 2 types were 2/5, 16/9, and 11/19, respectively. Given the fact that most of the V. vulnificus strains examined to date can cause skin infection in s.c. inoculated iron dextran-treated mice at an inoculum of 1,000 CFU, it was very interesting that capsule genotyping enabled significant discrimination of skin infection levels between the alleles. The incidences of skin infections were significantly different based on capsule allele (P = 2.0 × 10−3, χ2 test), with allele 0 significantly lower than alleles 1 and 2 (P = 0.0014 and P = 0.016, respectively) (Fig. 7). The magnitude of skin infection for allele 0 was significantly lower than for alleles 1 and 2 (P = 0.0006 and P = 0.037, respectively). There were highly significant differences in the incidences of detectable liver infections among the alleles (P = 1.3 × 10−4, χ2 test). Allele 1 caused detectable liver infection significantly more frequently than did alleles 0 and 2 (P = 0.0022 and P = 0.00011, respectively). In terms of magnitude of liver infection, that for allele 1 was significantly greater than those for alleles 2 and 0 (P = 0.0023 and P = 0.013, respectively). Allele 1 strains were significantly more likely to cause death than were allele 0 and 2 strains (P = 0.021 and P = 5.0 × 10−5, χ2 test), and allele 1 strains produced significantly lower body temperatures than did allele 0 and 2 strains (P = 0.019 and P = 0.0020, respectively). In summary, capsule allele 1 strains were the most adept at causing systemic infection and death and nontypeable allele 0 strains were the most attenuated among all of those examined in this study, especially in terms of skin infection, which is a very rare characteristic. It should be noted that the nontypeable strains form opaque colonies and hence are likely encapsulated. Incidences of liver infection and death were similar for allele 2 strains and the attenuated allele 0 strains, even though allele 2 strains were significantly better at causing skin infection. Hence, allele 2 strains represent those that exhibit a block in the ability to cause systemic infection in the presence of significant skin infection.
FIG. 7.
Relationship between capsule PCR allele and virulence. Box plots show the relationships between capsule PCR type and (A) skin infection, (B) liver infection, and (C) body temperature at euthanasia. The capsule PCR genotypes are shown on the left. For details, see the legend to Fig. 3. Symbols: ^ and *, capsule PCR type 0 significantly lower than types 1 and 2 (ANOVA and χ2, respectively); † and ‡, capsule PCR type 1 significantly different from types 0 and 2 (ANOVA and χ2, respectively).
Relationships among genotyping systems.
We characterized nearly all of our 69 biotype 1 strains in terms of vcg genotype, rrn type, rep-PCR, and capsule PCR. MLST and lineage data were available for 50 and 12 strains, respectively. As noted above, there were highly significant correlations among the vcg genotype, MLST, and rrn type (P < 10−6, χ2 test). Every vcgC genotype strain was in MLST cluster 2, and every vcgE genotype strain was in MLST cluster 1. Every rrn type A and AB strain was of the vcgE genotype and in MLST cluster 1, while every rrn type B strain examined, with one exception, was of the vcgC genotype and in MLST cluster 2. rrn typing assays were repeated on this lone exception, and the results were consistent. As would be expected for two classification systems based on MLST data, the MLST and lineage results for the 12 strains for which data were available were in very good agreement, with MLST cluster 1 corresponding to lineage 2 and MLST cluster 2 corresponding to lineage 1. There was one exception; strain CDC 9005-97 was in MLST cluster 1 and belonged to lineage 1. We believe that the single exception to this relationship was reported erroneously in the lineage study (14) because the strain was rrn typed differently between our studies as well (rrn type A in our study versus type B in the study of Cohen et al. [14]). If this is so, then the MLST and lineage classifications are the same. These results are in agreement with proposed population structures in which V. vulnificus biotype 1 consists of two primary lineages: vcgC genotype/MLST cluster 2/rrn type B/lineage 1 and vcgE genotype/MLST cluster 1/rrn types A and AB/lineage 2 (49).
Nonrandom, but not absolute, relationships were observed between the capsule allele and genotype, the MLST cluster, and the rrn type (P = 3.8 × 10−6, P = 3.1 × 10−6, and P < 10−6, respectively). Capsule allele 0 strains were predominantly of the vcgE genotype (6/7) and rrn type A (6/7). Only one capsule allele 0 strain was typed by MLST, and it was in MLST cluster 1. Capsule allele 1 strains were predominantly of the vcgC genotype (20/25), in MLST cluster 2 (17/20), and of rrn type B (21/25). Capsule allele 2 strains were predominantly of the vcgE genotype (25/30), in MLST cluster 1 (25/29), and of either rrn type A (12/30) or AB (13/30). Hence, the capsule allele roughly corresponds to the model of two different overall genetic groups of biotype 1 V. vulnificus but there are numerous exceptions.
Because of the numerous groups based upon rep-PCR, the relationships with other genotyping systems are much more complex. Both rep-PCR group 1 strains were of the vcgC genotype, of rrn type B, in MLST cluster 2, and of capsule type 1. Both rep-PCR group 2 strains were of the vcgC genotype, of rrn type B, and in MLST cluster 2, with one strain being of capsule type 1 and the other being of type 2. The 11 strains of rep-PCR group 3 were divided among all elements of the vcg genotype, rrn type, MLST, and capsule type. rep-PCR group 3 was the most heterogeneous of the nine groups. The 29 strains of rep-PCR group 4 exhibited interesting clustering. Four group 4 strains were of the vcgC genotype, of rrn type B, in MLST cluster 2, and evenly split between capsule types 1 and 2. Twelve group 4 strains were of the vcgE genotype, of rrn type A, in MLST cluster 1 (three strains were not typed by MLST), and predominantly of capsule type 2 (with two of type 0 and two of type 1). The final 13 rep-PCR group 4 strains were uniformly of the vcgE genotype, of rrn type AB, in MLST cluster 1, and of capsule type 2. This set of strains contained all of the rrn type AB strains. rep-PCR groups 5 and 6 were of the vcgE genotype, of rrn type A, and in MLST cluster 1 (one strain was not typed). These were split among capsule types 0, 1, and 2. Similarly, the two strains of rep-PCR group 7 were of the vcgE genotype, of rrn type A, and in MLST cluster 1 but were both of capsule type 0. The two rep-PCR group 8 strains were of the vcgC genotype and of rrn type B, with no other typing data available. Finally, the nine strains in rep-PCR group 9 formed a homogeneous group that was of the vcgC genotype, of rrn type B, in MLST cluster 2 (two strains not typed), and of capsule type 1. These data indicate that among the larger rep-PCR groups, group 9 is the most homogeneous, group 3 is the most heterogeneous, and group 4 is split into three subgroups based on rrn typing.
DISCUSSION
A perplexing issue in understanding V. vulnificus disease is why so many people consume V. vulnificus-contaminated seafood, many of whom have underlying predispositions, yet so few become sick and die. Furthermore, many people experience wound infection but do not progress to sepsis (20, 31). Genetics and predisposing physiological or immunological conditions of the human host and elements of encounter (e.g., inoculum size) are important in determining what type of disease, if any, will ensue from the encounter. However, there is documented variation in virulence potential among V. vulnificus strains (1, 15, 43, 52-54, 58). Therefore, there has been a significant amount of work devoted to identifying phenotypic and genotypic markers that could predict the virulence potential of different V. vulnificus strains, and certain genotypes are found significantly more frequently in clinical or environmental strains than would be predicted by chance. However, such relationships are not proof of the virulence potential of a given strain. We therefore undertook the present study to examine our mouse model of V. vulnificus infection in detail using a large set of diverse V. vulnificus biotype 1 strains and to examine the relationships between genotypes and virulence potential.
Progression of infection in s.c. inoculated mice.
The vast majority of the strains (80%) caused severe skin infections when administered as a relatively small inoculum of 1,000 CFU, including strains with the genotypes that were mostly associated with environmental isolates. However, liver infection required a minimum level of skin infection of approximately 106.5 CFU/g (Fig. 1). Therefore, V. vulnificus appears to require a strong base of infection of the skin before breaking through to the rest of the body in this mouse model. Given the extreme ease with which V. vulnificus kills iron dextran-treated mice after i.p. inoculation (50% lethal dose, ∼10 CFU), it is surprising that such a high level of skin infection is required for systemic infection to ensue in the s.c. inoculation model. Perhaps an anatomical or host defense barrier prevents systemic spread from the skin until a critical mass of vibrios accumulates that is able to overwhelm this barrier. Injecting mice i.p. likely bypasses this anatomical or defense barrier. In this regard, Bogard and Oliver (7) reported that vcgC-type strains are more resistant to human serum than are vcgE-type strains, which may explain our observations of a greater potential for causing systemic infection. Hence, complement could be one such defense barrier to systemic infection. Finally, the requirement for a high cell density in skin tissues as a prerequisite step for systemic infection suggests that quorum sensing could be involved. V. vulnificus expresses both AI-1 and AI-2 autoinducers (28, 55), and a LuxR homologue, SmcR, involved in quorum sensing signal transduction has been studied for effects on gene expression and virulence (24, 36, 41, 42, 50). Interestingly, a V. vulnificus luxS (a gene encoding AI-2 production) mutant showed attenuated virulence in i.p. inoculated mice (28). In contrast, an smcR mutant did not show attenuated virulence in i.p. inoculated mice (50). To determine if quorum sensing is involved with the cell density-dependent systemic infection in our s.c. inoculation model, luxS and smcR mutants should be examined. It is also possible that density-dependent virulence is based on the accumulation of extracellular virulence factors or damage to host tissues rather than quorum sensor-mediated changes in gene expression.
The ability to cause death (a body temperature of less than 33°C) in our model was strongly linked to liver infection (Fig. 1). Even when their skin contained 1010 CFU/g V. vulnificus, the mice did not die unless significant liver infection occurred. This relationship may explain one component of human disease. The mortality rate for sepsis is greater than 50%, even with administration of antibiotics; however, the mortality rate for wound infection and necrotizing fasciitis is significantly lower (approximately 25%) (20, 21, 31, 34). Just as is the case for mice in our model, it is possible for human patients to have severe skin infections without systemic involvement leading to death. Therefore, it is critical to identify those V. vulnificus strains that have the potential to cause systemic disease and to understand the virulence attributes enabling systemic disease. In that regard, the usefulness and limitations of our s.c. inoculation model are worth noting. Most important, because inoculation is s.c., our model cannot probe the earliest stages of infection following the ingestion of contaminated food. The nature of the gastroenteritis caused by ingestion of V. vulnificus is very poorly documented, so analysis of this gastroenteritis should rely on oral inoculation models. One virulence factor involved in invasion beyond the intestine after oral inoculation is the RtxA1 toxin, as determined using oral inoculation models with mice (30). In contrast, RtxA1 has only a minor role in our s.c. inoculation model (J. L. Joseph et al., unpublished data). However, for severe, life-threatening sepsis after either ingestion or wound infection, our model should relate very well to human disease. Unfortunately, without having a set of clinical isolates that caused wound infection without subsequent systemic infection, it is impossible to directly relate virulence results obtained in our model to human disease.
Relationships between genotypes.
As had been reported by others (11, 49), we found that V. vulnificus biotype 1 strains can be classified into two profiles. Profile 1 contains strains generally isolated from the environment (e.g., oysters and water) and is characterized by MLST cluster 1, vcgE, rrn types A and AB, and lineage 2. Profile 2 contains primarily clinical isolates and is characterized by MLST cluster 2, vcgC, rrn type B, and lineage 1. We propose this profile nomenclature to follow the MLST cluster numbering of Bisharat et al. (5), since they were the first to publish definitive detailed divisions based on the most widely accepted methodology, MLST. There were few, if any, exceptions to these relationships among genotypes in our strains. Typing of strains by PCR amplicon of the capsule locus followed this pattern closely. Capsule allele 1 strains were predominantly in profile 2, and capsule type 2 strains were predominantly in profile 1. Capsule allele 0, which is the result of a negative PCR and hence not necessarily a uniformly defined group, was primarily in profile 1. The relationship between rep-PCR genotype and profile is more complex and difficult to determine. This is due to the fact that we observed nine genotypes based on similarity in banding patterns at the 85% level, and several rep-PCR groups were very small (six of the rep-PCR groups were composed of only two or three strains). Because of the proposed horizontal gene transfer between V. vulnificus strains (5, 19), there will undoubtedly be strains with mixed characteristics between these profiles. An example of such an apparent crossover is V. vulnificus 99-743 DP-B6, which is of the vcgE genotype but of rrn type B.
Relationship between genotype and virulence.
As has been predicted based on correlations of genotypes with clinical versus environmental isolation of strains (14, 44, 48), our data presented herein demonstrate that these genetic profiles do, in fact, correlate with virulence potential in a mammalian model of disease. There are very few encapsulated V. vulnificus biotype 1 strains that cannot cause skin infection in this model when inoculated at 1,000 CFU, and increasing the inoculum by as little as 10-fold can enable such strains to cause a skin infection that is indistinguishable from that caused by the more virulent strains (51, 52). Profile 1 strains, primarily isolated from the environment, are less adept at causing systemic infection and death. Profile 2 strains have the greatest likelihood of causing systemic infection and death. Because the vcg genotype can be assessed in a single duplex PCR, as opposed to multiple DNA sequence analyses for MLST or complex PCR/restriction digestions for ribotyping, vcg genotyping is the most straightforward approach to genotyping a V. vulnificus strain into these profiles. However, one cannot assume that vcgE strains are less virulent and vcgC strains are highly virulent. In fact, three of the nine most virulent strains in our collection (group 5 in Fig. 2) are in profile 1 (vcgE). Therefore, profile 1 strains should not be labeled “avirulent.” Furthermore, when six vcgC strains isolated from the environment were examined for virulence, only two of them exhibited virulence typical of profile 2 strains and five of the six strains in the most attenuated group (group 1 in Fig. 2) are classified as profile 2. It therefore appears that the majority of vcgC genotype strains in the environment that have not been preselected for virulence by having been derived from patient samples may not be highly virulent. The vast majority of the vcgC genotype strains in our collection are of clinical origin and hence have been preselected for high virulence. Therefore, a more comprehensive analysis of virulence of vcgC genotype strains from the environment should be performed. In a recent analysis of 33 V. vulnificus biotype 1 strains isolated from aquacultured fish or pond water in Bangladesh, 100% of the strains were profile 2 (40). Preliminary examination of these strains also shows a range of virulence potentials in this mouse model (P. A. Gulig et al., unpublished data).
A confounding issue with all but one of these genotyping methods is the genetic basis of the typing system. MLST analysis is based on DNA sequence polymorphisms in housekeeping genes that should not be under intensive selective pressure for change, especially with regard to virulence. 16S rRNA (rrn) typing and 16S-23S intergenic sequence typing are based on DNA sequence polymorphisms within or between rRNA genes. The vcg polymorphism is based on a single locus of unknown function (48). rep-PCR is based on the amplification of repetitive DNA sequences, the functions of which are unknown, that are shared among numerous and diverse bacteria (56). The only genotyping method used in this study that is based on a confirmed virulence attribute is PCR amplification of the wzb capsule locus, and the DNA targets were in an intergenic region. We noted that capsule type 2, as opposed to the other types, was attenuated for systemic infection despite high levels of skin infection. At first, this result could suggest that the type 2 strains possess a capsular antigen that is less fit for enabling systemic infection. However, it should also be noted that most capsule type 2 strains were also profile 1, which exhibits the same virulence phenotype. In contrast, capsule type 0 strains exhibited remarkably low levels of skin infection and hence were defective at even the earliest stages of disease. Capsule type 1 strains were predominantly profile 2, which was most adept at causing systemic infection. Finally, if capsule type 2 was strictly linked to the production of a nonsystemic capsular antigen, then type 2 strains should not be able to cause systemic infection. However, genomically sequenced strain YJ016 is of capsule type 2 and is fully virulent, so this capsule type alone is not an attenuating trait. Most likely, the capsule PCR type is a marker for the overall genetic background of V. vulnificus strains, in which the true differential virulence genes are embedded.
Because these various loci are likely under different selective pressures for either change or homeostasis, because they are scattered throughout the genome of V. vulnificus, and because mechanisms exist for horizontal gene transfer among V. vulnificus strains (19), it is not surprising that none of these genotyping schemes completely separates strains into groups of uniform virulence. Furthermore, the genes that actually cause virulence in V. vulnificus strains are mostly unknown. Our ongoing studies are aimed at identifying these virulence attributes that enable systemic infection and death.
Genomic sequencing of various V. vulnificus strains should lead to the identification of relevant virulence genes. However, the only two published genomic sequences of V. vulnificus are from clinical strains with similar genotypes in profile 2: MLST cluster 2, vcgC, and rrn type B. YJ016 is in rep-PCR group 2 with capsule allele 2, while CMCP6 was not typed by rep-PCR (i.e., there was not another strain in our collection with 85% similarity in its banding pattern) and possessed capsule allele 1. We recently obtained first-pass SOLiD genomic DNA sequences of two environmental V. vulnificus strains, 99-520 DP-B8 and 99-738 DP-B5, that are in MLST cluster 1, vcgE, rep-PCR 4, capsule type 2; clinical strain MO6-24/0, which is in MLST cluster 2, of rrn type B, vcgC, capsule allele 1, and rep-PCR group 9; and biotype 2 strain ATCC 33149 (18). Profile 1 strain 99-520 DP-B8 is defective at causing systemic infection and death (group 3; Fig. 2 and Table 1), whereas profile 1 strain 99-738 DP-B5 is one of the most virulent strains in our collection (group 5; Fig. 2 and Table 1). We identified 80 genes that were common to the profile 2 strains (CMCP6, YJ016, and MO6-24/0) but were missing from the profile 1/biotype 1 strains and from the biotype 2 strain. Candidate virulence genes, which await experimental analysis, included several GGDEF proteins and a Flp pilus-coding region. Additionally, we identified 61 genes that were common to highly virulent profile 1 strain 99-738 DP-B5 and all three profile 2 strains but were missing from the other more typical profile 1 strain and the biotype 2 strain. These genes are interesting because they could explain the unusually high virulence of 99-738 DP-B5 and include genomic island XII identified by Cohen et al. (14), sialic acid catabolism, and mannitol catabolism. Jeong et al. (23) recently determined that the nanA gene involved with sialic acid catabolism is essential for virulence in V. vulnificus, and mannitol fermentation has been linked to the clinically associated genotypes (16). Because most of the systems commonly used to genotype V. vulnificus are not based on virulence genes and because the profiles do not enable prediction of virulence potential, it is crucial that the virulence genes that underlie the pathogenicity of V. vulnificus be identified. Identification of these virulence genes would not only improve the understanding of V. vulnificus pathogenesis but could lead to improved molecular screens to predict the virulence of V. vulnificus strains associated with seafood.
Acknowledgments
This work was supported by NIH grant R01 056056 and USDA grant CREES 2007-01955.
We thank Jorge Girón and Max Teplitski for critical review of the manuscript.
Editor: S. M. Payne
Footnotes
Published ahead of print on 3 January 2011.
REFERENCES
- 1.Amaro, C., E. G. Biosca, B. Fouz, A. E. Toranzo, and E. Garay. 1994. Role of iron, capsule, and toxins in the pathogenicity of Vibrio vulnificus biotype 2 for mice. Infect. Immun. 62:759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckman, E. N., G. L. Leonard, L. E. Castillo, C. F. Genre, and G. A. Pankey. 1981. Histopathology of marine vibrio wound infections. Am. J. Clin. Pathol. 76:765-772. [DOI] [PubMed] [Google Scholar]
- 3.Bisharat, N., et al. 1999. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354:1421-1424. [DOI] [PubMed] [Google Scholar]
- 4.Bisharat, N., et al. 2005. Hybrid Vibrio vulnificus. Emerg. Infect. Dis. 11:30-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bisharat, N., et al. 2007. The evolution of genetic structure in the marine pathogen, Vibrio vulnificus. Infect. Genet. Evol. 7:685-693. [DOI] [PubMed] [Google Scholar]
- 6.Blake, P. A., M. H. Merson, R. E. Weaver, D. G. Hollis, and P. C. Heublein. 1979. Disease caused by a marine vibrio. Clinical characteristics and epidemiology. N. Engl. J. Med. 300:1-5. [DOI] [PubMed] [Google Scholar]
- 7.Bogard, R. W., and J. D. Oliver. 2007. Role of iron in human serum resistance of the clinical and environmental Vibrio vulnificus genotypes. Appl. Environ. Microbiol. 73:7501-7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchrieser, C., V. V. Gangar, R. L. Murphree, M. L. Tamplin, and C. W. Kaspar. 1995. Multiple Vibrio vulnificus strains in oysters as demonstrated by clamped homogeneous electric field gel electrophoresis. Appl. Environ. Microbiol. 61:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bullen, J. J., P. B. Spalding, C. G. Ward, and J. M. Gutteridge. 1991. Hemochromatosis, iron and septicemia caused by Vibrio vulnificus. Arch. Intern. Med. 151:1606-1609. [PubMed] [Google Scholar]
- 10.Campbell, M. S., and A. C. Wright. 2003. Real-time PCR analysis of Vibrio vulnificus from oysters. Appl. Environ. Microbiol. 69:7137-7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatzidaki-Livanis, M., M. A. Hubbard, K. Gordon, V. J. Harwood, and A. C. Wright. 2006. Genetic distinctions among clinical and environmental strains of Vibrio vulnificus. Appl. Environ. Microbiol. 72:6136-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatzidaki-Livanis, M., M. K. Jones, and A. C. Wright. 2006. Genetic variation in the Vibrio vulnificus group 1 capsular polysaccharide operon. J. Bacteriol. 188:1987-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, C. Y., et al. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen, A. L., J. D. Oliver, A. DePaola, E. J. Feil, and E. F. Boyd. 2007. Emergence of a virulent clade of Vibrio vulnificus and correlation with the presence of a 33-kilobase genomic island. Appl. Environ. Microbiol. 73:5553-5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DePaola, A., et al. 2003. Analysis of Vibrio vulnificus from market oysters and septicemia cases for virulence markers. Appl. Environ. Microbiol. 69:4006-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drake, S. L., B. Whitney, J. F. Levine, A. DePaola, and L. A. Jaykus. 2010. Correlation of mannitol fermentation with virulence-associated genotypic characteristics in Vibrio vulnificus isolates from oysters and water samples in the Gulf of Mexico. Foodborne Pathog. Dis. 7:97-101. [DOI] [PubMed] [Google Scholar]
- 17.Gulig, P. A., K. L. Bourdage, and A. M. Starks. 2005. Molecular pathogenesis of Vibrio vulnificus. J. Microbiol. 43:118-131. [PubMed] [Google Scholar]
- 18.Gulig, P. A., et al. 2010. SOLiD sequencing of four Vibrio vulnificus genomes enables comparative genomic analysis and identification of candidate clade-specific virulence genes. BMC Genomics 11:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulig, P. A., M. S. Tucker, P. C. Thiaville, J. L. Joseph, and R. N. Brown. 2009. USER Friendly cloning coupled with chitin-based natural transformation enables rapid mutagenesis of Vibrio vulnificus. Appl. Environ. Microbiol. 75:4936-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hlady, W. G., and K. C. Klontz. 1996. The epidemiology of Vibrio infections in Florida, 1981-1993. J. Infect. Dis. 173:1176-1183. [DOI] [PubMed] [Google Scholar]
- 21.Hlady, W. G., R. C. Mullen, and R. S. Hopkin. 1993. Vibrio vulnificus infections from raw oysters: leading cause of reported deaths from foodborne illness in Florida. J. Fla. Med. Assoc. 80:536-538. [PubMed] [Google Scholar]
- 22.Hoffmann, T. J., B. Nelson, R. Darouiche, and T. Rosen. 1988. Vibrio vulnificus septicemia. Arch. Intern. Med. 148:1825-1827. [PubMed] [Google Scholar]
- 23.Jeong, H. G., et al. 2009. The capability of catabolic utilization of N-acetylneuraminic acid, a sialic acid, is essential for Vibrio vulnificus pathogenesis. Infect. Immun. 77:3209-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong, H. S., M. H. Lee, K. H. Lee, S. J. Park, and S. H. Choi. 2003. SmcR and cyclic AMP receptor protein coactivate Vibrio vulnificus vvpE encoding elastase through the RpoS-dependent promoter in a synergistic manner. J. Biol. Chem. 278:45072-45081. [DOI] [PubMed] [Google Scholar]
- 25.Jeong, K. C., et al. 2000. Construction and phenotypic evaluation of a Vibrio vulnificus vvpE mutant for elastolytic protease. Infect. Immun. 68:5096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, M. K., and J. D. Oliver. 2009. Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77:1723-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, S. Y., et al. 2003. Regulation of Vibrio vulnificus virulence by the LuxS quorum-sensing system. Mol. Microbiol. 48:1647-1664. [DOI] [PubMed] [Google Scholar]
- 29.Kim, Y. R., et al. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71:5461-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, Y. R., et al. 2008. Vibrio vulnificus RTX toxin kills host cells only after contact of the bacteria with host cells. Cell. Microbiol. 10:848-862. [DOI] [PubMed] [Google Scholar]
- 31.Klontz, K. C., et al. 1988. Syndromes of Vibrio vulnificus infections. Clinical and epidemiologic features in Florida cases, 1981-1987. Ann. Intern. Med. 109:318-323. [DOI] [PubMed] [Google Scholar]
- 32.Kraffert, C. A., and D. J. Hogan. 1992. Vibrio vulnificus infection and iron overload. J. Am. Acad. Dermatol. 26:140. [DOI] [PubMed] [Google Scholar]
- 33.Kumamoto, K. S., and D. J. Vukich. 1998. Clinical infections of Vibrio vulnificus: a case report and review of the literature. J. Emerg. Med. 16:61-66. [DOI] [PubMed] [Google Scholar]
- 34.Kuo, Y. L., S. J. Shieh, H. Y. Chiu, and J. W. Lee. 2007. Necrotizing fasciitis caused by Vibrio vulnificus: epidemiology, clinical findings, treatment and prevention. Eur. J. Clin. Microbiol. Infect. Dis. 26:785-792. [DOI] [PubMed] [Google Scholar]
- 35.Lee, J. H., et al. 2007. Identification and characterization of the Vibrio vulnificus rtxA essential for cytotoxicity in vitro and virulence in mice. J. Microbiol. 45:146-152. [PubMed] [Google Scholar]
- 36.Lee, J. H., et al. 2007. Identification and functional analysis of Vibrio vulnificus SmcR, a novel global regulator. J. Microbiol. Biotechnol. 17:325-334. [PubMed] [Google Scholar]
- 37.Lee, J. H., et al. 2004. Role of flagellum and motility in pathogenesis of Vibrio vulnificus. Infect. Immun. 72:4905-4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Litwin, C. M., T. W. Rayback, and J. Skinner. 1996. Role of catechol siderophore synthesis in Vibrio vulnificus virulence. Infect. Immun. 64:2834-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, M., A. F. Alice, H. Naka, and J. H. Crosa. 2007. The HlyU protein is a positive regulator of rtxA1, a gene responsible for cytotoxicity and virulence in the human pathogen Vibrio vulnificus. Infect. Immun. 75:3282-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahmud, Z. H., et al. 2010. Genetic characterization of Vibrio vulnificus strains from tilapia aquaculture in Bangladesh. Appl. Environ. Microbiol. 76:4890-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDougald, D., S. A. Rice, and S. Kjelleberg. 2000. The marine pathogen Vibrio vulnificus encodes a putative homologue of the Vibrio harveyi regulatory gene, luxR: a genetic and phylogenetic comparison. Gene 248:213-221. [DOI] [PubMed] [Google Scholar]
- 42.McDougald, D., S. A. Rice, and S. Kjelleberg. 2001. SmcR-dependent regulation of adaptive phenotypes in Vibrio vulnificus. J. Bacteriol. 183:758-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moreno, M. L., and M. Landgraf. 1998. Virulence factors and pathogenicity of Vibrio vulnificus strains isolated from seafood. J. Appl. Microbiol. 84:747-751. [DOI] [PubMed] [Google Scholar]
- 44.Nilsson, W. B., R. N. Paranjpye, A. DePaola, and M. S. Strom. 2003. Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. J. Clin. Microbiol. 41:442-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okujo, N., T. Akiyama, S. Miyoshi, S. Shinoda, and S. Yamamoto. 1996. Involvement of vulnibactin and exocellular protease in utilization of transferrin- and lactoferrin-bound iron by Vibrio vulnificus. Microbiol. Immunol. 40:595-598. [DOI] [PubMed] [Google Scholar]
- 46.Paranjpye, R. N., and M. S. Strom. 2005. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect. Immun. 73:1411-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ran Kim, Y., and J. Haeng Rhee. 2003. Flagellar basal body flg operon as a virulence determinant of Vibrio vulnificus. Biochem. Biophys. Res. Commun. 304:405-410. [DOI] [PubMed] [Google Scholar]
- 48.Rosche, T. M., Y. Yano, and J. D. Oliver. 2005. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol. Immunol. 49:381-389. [DOI] [PubMed] [Google Scholar]
- 49.Sanjuán, E., B. Fouz, J. D. Oliver, and C. Amaro. 2009. Evaluation of genotypic and phenotypic methods to distinguish clinical from environmental Vibrio vulnificus strains. Appl. Environ. Microbiol. 75:1604-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao, C. P., and L. I. Hor. 2001. Regulation of metalloprotease gene expression in Vibrio vulnificus by a Vibrio harveyi LuxR homologue. J. Bacteriol. 183:1369-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Starks, A. M., K. L. Bourdage, P. C. Thiaville, and P. A. Gulig. 2006. Use of a marker plasmid to examine growth and death of Vibrio vulnificus in infected mice. Mol. Microbiol. 61:310-323. [DOI] [PubMed] [Google Scholar]
- 52.Starks, A. M., et al. 2000. Pathogenesis of infection by clinical and environmental strains of Vibrio vulnificus in iron dextran-treated mice. Infect. Immun. 68:5785-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stelma, G. N., Jr., A. L. Reyes, J. T. Peeler, C. H. Johnson, and P. L. Spaulding. 1992. Virulence characteristics of clinical and environmental isolates of Vibrio vulnificus. Appl. Environ. Microbiol. 58:2776-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tison, D. L., and M. T. Kelly. 1986. Virulence of Vibrio vulnificus strains from marine environments. Appl. Environ. Microbiol. 51:1004-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valiente, E., et al. 2009. Vibrio vulnificus produces quorum sensing signals of the AHL-class. FEMS Microbiol. Ecol. 69:16-26. [DOI] [PubMed] [Google Scholar]
- 56.Versalovic, J., and J. R. Lupski. 2002. Molecular detection and genotyping of pathogens: more accurate and rapid answers. Trends Microbiol. 10:S15-S21. [DOI] [PubMed] [Google Scholar]
- 57.Vickery, M. C., W. B. Nilsson, M. S. Strom, J. L. Nordstrom, and A. DePaola. 2007. A real-time PCR assay for the rapid determination of 16S rRNA genotype in Vibrio vulnificus. J. Microbiol. Methods 68:376-384. [DOI] [PubMed] [Google Scholar]
- 58.Wong, H. C., S. H. Liu, and M. Y. Chen. 2005. Virulence and stress susceptibility of clinical and environmental strains of Vibrio vulnificus isolated from samples from Taiwan and the United States. J. Food Prot. 68:2533-2540. [DOI] [PubMed] [Google Scholar]
- 59.Wright, A. C., et al. 1996. Distribution of Vibrio vulnificus in the Chesapeake Bay. Appl. Environ. Microbiol. 62:717-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright, A. C., L. M. Simpson, J. D. Oliver, and J. G. Morris, Jr. 1990. Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect. Immun. 58:1769-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zakaria-Meehan, Z., G. Massad, L. M. Simpson, J. C. Travis, and J. D. Oliver. 1988. Ability of Vibrio vulnificus to obtain iron from hemoglobin-haptoglobin complexes. Infect. Immun. 56:275-277. [DOI] [PMC free article] [PubMed] [Google Scholar]