Abstract
The microsporidia are a diverse phylum of obligate intracellular parasites that infect all major animal groups and have been recognized as emerging human pathogens for which few chemotherapeutic options currently exist. These organisms infect every tissue and organ system, causing significant pathology, especially in immune-compromised populations. The microsporidian spore employs a unique infection strategy in which its contents are delivered into a host cell via the polar tube, an organelle that lies coiled within the resting spore but erupts with a force sufficient to pierce the plasma membrane of its host cell. Using biochemical and molecular approaches, we have previously identified components of the polar tube and spore wall of the Encephalitozoonidae. In this study, we employed a shotgun proteomic strategy to identify novel structural components of these organelles in Encephalitozoon cuniculi. As a result, a new component of the E. cuniculi developing spore wall was identified. Surprisingly, using the same approach, a heretofore undescribed filamentous network within the lumen of the parasitophorous vacuole was discovered. This network was also present in the parasitophorous vacuole of Encephalitozoon hellem. Thus, in addition to further elucidating the molecular composition of seminal organelles and revealing novel diagnostic and therapeutic targets, proteomic analysis-driven approaches exploring the spore may also uncover unknown facets of microsporidian biology.
The microsporidia are a diverse phylum of obligate intracellular parasites containing over 1,200 species representing nearly 150 genera (14, 41). Fourteen species have been shown to infect humans (10). In recent decades, these unicellular parasites have garnered considerable attention as opportunistic gastrointestinal parasites of AIDS patients; however, the spectrum of disease is broad, encompassing every tissue type and most organ systems, as well as immunocompetent individuals (10, 12, 42). Due to the emerging nature and diversity of human-pathogenic species, highly effective and safe treatments are not yet available for all of the pathogenic microsporidia (9). This highlights the need for a better appreciation of the basic biology of the microsporidia. Such research efforts have naturally focused on the spore, the infectious stage of these organisms.
The microsporidian spore is a thick-walled, environmentally resistant cell, which upon germination is capable of extruding its internal polar filament and thereby inoculating its contents into a nearby host cell. The spore wall, polar tube, and other specialized organelles, such as the polaroplast and posterior vacuole, play critical roles in the infectious process of the microsporidia (42, 46). In a resting spore, the polar tube lies coiled within the cell; at this stage, it is solid and is thus referred to as a polar filament (28, 37). Upon an appropriate species-specific environmental stimulus (38), an increasing internal osmotic pressure develops and causes the polaroplast and posterior vacuole to swell and the polar filament to erupt from the anterior, thinnest point of the spore wall. During this process, the polar filament everts, becoming a hollow polar tube through which the sporoplasm passes. The proteinaceous, chitinous spore wall also plays a key role in the infectious process, as it must not only withstand environmental degradative stresses in the resting spore but also resist the increasing turgor pressure during germination until the moment of rupture (13).
Investigations of the molecular composition of both the polar tube and spore wall have been facilitated by the immunodominance of component proteins in natural infections and parasite lysate immunizations. Polyclonal and monoclonal antibodies isolated from sera of infected or immunized animals have led to the detection of polar tube proteins (PTPs) and spore wall proteins (SWPs) from human-infecting microsporidia of the family Encephalitozoonidae via immunohistochemical techniques (3, 29, 39). For example, a 51-kDa protein (SWP1) of the exospore of Encephalitozoon cuniculi was identified with the aid of a monoclonal antibody, and the gene was cloned by immunoscreening an expression library (5). Homologs of this protein have been identified in Encephalitozoon hellem (5) and Encephalitozoon intestinalis (18). Using proteomic techniques, two endospore-associated proteins, SWP3/EnP2 and EnP1, were discovered in E. cuniculi (32, 45). EnP1 was later localized to the exospore as well and found to mediate adherence to host cells (36). Similar techniques were used to localize a putative chitin deacetylase to the E. cuniculi plasma membrane-endospore interface, suggesting a role in microsporidian endospore development (6).
With the exception of the putative chitin deacetylase (6), SWPs and PTPs identified thus far have no conserved functional domains or homology to known proteins (5, 45) and only limited similarity to each other (discussed in reference 45). The first two Encephalitozoonidae spore wall proteins, SWP1 and SWP2 (5, 18), are both cysteine rich and show conservation in their N termini, but SWP3/EnP2 of E. cuniculi is dissimilar to these proteins (45), and the three PTPs of E. cuniculi differ in molecular weight and amino acid composition. While immunohistochemical data suggest structural roles for all of these proteins, little is known about how they accomplish their specific functions. In addition, it is possible that other, perhaps lower-abundance component proteins of the spore wall and polar tube await discovery.
Our laboratory has previously developed a differential solubility-based subcellular fractionation procedure for the isolation of polar tube and spore wall protein from microsporidian spores (22). This resulted in the identification of Encephalitozoon cuniculi PTP1 (21, 22, 24) and SWP3/EnP2 (32, 45) as well as PTPs of other Encephalitozoonidae (24) and Glugea americanus (25). In the current study, a shotgun proteomic strategy was employed to identify novel structural components from polar tube and spore wall fractions of Encephalitozoon cuniculi. As a result, novel components of the developing spore wall and a heretofore undescribed filamentous network within the parasitophorous vacuole were identified, the latter of which is also present in the sister species Encephalitozoon hellem.
(Portions of this work have been published as the Ph.D. thesis of Kaya Ghosh (15).)
MATERIALS AND METHODS
Culturing of microsporidia to maximize spore yield and ensure purity.
E. cuniculi- or E. hellem-infected rabbit kidney cell line RK13 (American Type Cell Culture Collection, Manassas, VA) cultures were maintained as previously described (22) with a reduced serum concentration of 7%. Spores were pelleted from culture supernatants by centrifugation at 2,500 × g and washed in Dulbecco's modified phosphate-buffered saline (DPBS). Host cell debris and nonspore material were solubilized by incubation with 1% sodium dodecyl sulfate (SDS) in DPBS at 37°C and removed by three DPBS washes and centrifugations.
Fractionation of spores.
Polar tube and spore wall proteins were fractionated from whole, purified E. cuniculi spores as previously described (21-24, 40). Briefly, culture-purified spores were resuspended in 1 ml DPBS in a 2.0-ml screw-cap plastic bead-beater tube (Sarstedt, Newton, NC) half filled with 400 to 600 μm acid-washed glass beads (Sigma) and mechanically disrupted by three 2-min pulses in a Mini-Beadbeater (Biospec Products, Bartlesville, OK). Spores were cooled on ice for 1 min between pulses. Spore homogenate was recovered from the glass beads by puncturing the bottom of the Beadbeater tube with a 23-gauge needle, placing the tube in a 10- by 13-mm glass tube, and centrifuging at 1,000 × g for 10 min. The clear buffer overlying the pellet was used to wash the glass beads, and the centrifugation procedure was repeated twice. The pellet of disrupted spores was transferred to a low-retention microcentrifuge tube (Fisher) and washed five times with 1% SDS/DPBS and three times with DPBS to remove SDS. The pellet was washed once with 9 M urea and incubated with 2% dithiothreitol (DTT) in 25 mM Tris (pH 7.4) with Complete protease inhibitor cocktail (Roche) for 2 h at room temperature. The supernatant containing solubilized polar tube protein was separated from the pellet consisting largely of insoluble spore wall material, and the pellet was washed three times in buffer by resuspension and centrifugation, and both fractions were stored at −80°C.
Proteomic analysis of spore lysate. (i) DTT-solubilized material.
Residual detergent was removed from the DTT-solubilized spore supernatant material via repeated extraction with ethyl acetate (EA) by the addition of 10 volumes of fresh water-saturated EA followed by vigorous vortexing and removal of the upper organic layer containing EA and SDS (47). This extraction procedure was repeated six times. Residual EA was evaporated for 20 min under N2 gas. This material was then digested in solution with bead-immobilized trypsin (Pierce, Rockford, IL), according to the manufacturer's directions, overnight at 37°C in an Eppendorf Thermomixer shaker (Eppendorf, Westbury, NY). The digest was quenched by the addition of trifluoroacetic acid (TFA) to 0.15%. This material was then analyzed by nanospray liquid chromatography coupled with tandem electrospray ion-trap mass spectrometry (nanoLC/MS-MS) on an LTQ linear ion trap mass spectrometer (Thermo Scientific, San Jose, CA) interfaced with a TriVersa NanoMate nanoelectrospray ion source (Advion BioSciences, Ithaca, NY). An Ultimate Plus nano-high-performance liquid chromatography (HPLC) system with a Famous autosampler (Dionex Corporation, Sunnyvale, CA) was coupled with the TriVersa NanoMate. Peptides were loaded on a C18 μ-Precolumn cartridge (5 μm, 100 Å, 300-μm by 5-mm inside diameter [i.d.]) from the autosampler with a 25-μl sample loop at a flow rate of 15 μl/min. After injection of 20 μl sample and washing for 20 min, the precolumn was switched in line with the analytical column, a C18 PepMap100 (3 μm, 100 Å, 75-μm by 150-mm i.d.; Dionex Corporation, Sunnyvale, CA). Mobile phase B (80% acetonitrile/water plus 0.1% formic acid) was increased from 2 to 55% over 70 min, held for 5 min, increased to 95% over 20 min, and held at 95% for 5 min. The flow rate used was 250 nanoliters/min, and mobile phase A consisted of 5% acetonitrile/water plus 0.1% formic acid. The 10 most intense ions, having a charge state of +2 to +4, determined from an initial survey scan from 300 to 1,800 m/z, were selected for MS-MS. MS-MS was performed using an isolation width of 3 m/z, a normalized collision energy of 35%, and a minimum signal intensity of 500 counts. The dynamic exclusion option is enabled. Once a certain ion is selected twice for MS-MS in 30 s, this ion is excluded from being selected again for MS-MS during the next period of 120 s. Data files were created from the raw LTQ mass spectrometer LC/MS-MS data. The created data files were then merged using the merge script tool from Matrix Science. The subsequent combined merge file was used to search the E_cuniculi 11 database using the following parameters: trypsin, 2 missed cleavages; variable modifications of carbamidomethylation (Cys), deamidation (Asn and Gln), and oxidation (Met); monoisotopic masses; peptide mass tolerance of 3.0 Da; product ion mass tolerance of 0.6 Da. Proteins were considered identified if they had at least one bold red peptide (the most logical assignment of a peptide to a protein; prevents duplicate homologous proteins from being reported) and an ion score cutoff of 40 or greater.
(ii) DTT-insoluble material.
In order to proteolytically digest the sample and subject it to nanoLC/MS-MS, it was first partially solubilized in 70% trifluoroacetic acid. The reducing agent Bond-Breaker TCEP [Tris(2-carboxyethyl)phosphine] (Pierce, Rockford, IL) was added to 50 μM, and the chemical cleavage agent cyanogen bromide was added to 20 mM. Oxygen was displaced from the reaction by the aeration of nitrogen gas into the solution with a gel-loading pipette tip, and the solution was incubated for 18 h at room temperature in the dark. The mixture was then evaporated overnight under nitrogen gas, and the pellet was resuspended in 100 mM NH4HCO3 (pH 8.5) in a sonicating water bath for 30 min at room temperature. Urea was added to 8 M, and the sample was sonicated for 15 min, after which point some material was still insoluble. This material was reserved, and the rest of the procedure was carried out on the solubilized material. Endoproteinase Lys-C (Roche Applied Science, Indianapolis, IN) was added according to the manufacturer's directions, and the reaction mixture was incubated overnight at 37°C. The urea concentration was then adjusted to 2 M by the addition of 8.5 M NH4HCO3, and bead-immobilized trypsin (Pierce, Rockford, IL) was added. The reaction was quenched, the material was analyzed by nanoLC/MS-MS, and data were used to search the E_cuniculi 11 database as described above.
Bioinformatic analysis of selected proteins. (i) In silico analysis.
Hypothetical genes for which peptides were detected were analyzed for similarity to all proteins in the complete NCBInr protein database using the BLAST algorithm (2). The molecular weight (MW), isoelectric point (pI), grand average hydropathicity (GRAVY), and number of transmembrane helices for each hypothetical protein were calculated or predicted using TarO version 2.0 (http://www.compbio.dundee.ac.uk/taro) (31) and XtalPred (http://ffas.burnham.org/XtalPred-cgi/xtal.pl) (35). Signal peptide and subcellular compartment predictions were made using the TargetP 1.1 server (http://www.cbs.dtu.dk/services/TargetP/) (11). Phosphorylation site predictions were made using the NetPhos 2.0 server (4).
(ii) Identification of homologs in E. hellem and E. intestinalis.
Raw Illumina reads from an ongoing E. hellem genome project were assembled using Velvet (48) and Consed (17) with parameters described previously (8). Predicted open reading frames (ORFs) on all preliminary scaffolds were searched for homologs of ECU10_1500 and ECU10_1070 using BLAST (2), and the sequence quality and coverage of putative matches were confirmed manually. In both cases, a single protein-coding gene with a high degree of sequence similarity was identified. The E. intestinalis genome (8) was also searched for homologs of these proteins using BLAST.
Cloning and expression of hypothetical E. cuniculi genes.
Mixed parasite/host genomic DNA was prepared from infected RK13 cell cultures using the Qiagen DNeasy blood and tissue kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Forward and reverse primers specific for each E. cuniculi hypothetical gene were designed with 5′ overhangs complementary to the pET-41 Ek-LIC vector (EMD Biosciences, Gibbstown, NJ) directional cloning site and used to PCR amplify hypothetical E. cuniculi genes. PCR was performed using 45 μl PCR SuperMix (Invitrogen, Carlsbad, CA), 27.5 ng purified RK13/parasite DNA, 200 nM each primer, 2.9 mM MgCl2 (final concentration), and primers for hypothetical genes ECU10_1500 (5′GACGACGACAAGATGGATGGATGGTCTGGC3′ [forward], 5′GAGGAGAAGCCCGGTTCAACTACGGCTCTCCTCCT3′ [reverse]) or ECU10_1070 (5′GACGACGACAAGATGACACAAGAAAGTACGATG3′ [forward], 5′GAGGAGAAGCCCGGTTCAGAACATCCCAAAGTC3′ [reverse]). DNA was initially denatured for 5 min at 94°C followed by five initial amplification cycles and 30 final amplification cycles (1-min denaturation at 94°C, 1-min annealing at 49.4/68.3°C (ECU10_1500) or 48.0/67.0°C (ECU10_1070), extension at 72°C for 1 min 45 s (ECU10_1500) or 54 s (ECU10_1070), and 7-min final extension at 72°C in an Eppendorf Mastercycler gradient thermal cycler (Eppendorf, Westbury, NY). PCR products were subjected to agarose gel electrophoresis and examined by ethidium bromide staining. Bands were excised from the gel using a clean scalpel under UV illumination. PCR products were purified from free primers, nucleotides, enzyme, buffer salts, and agarose using the silica membrane-based QIAquick gel extraction kit (Qiagen, Valencia, CA). Purified PCR products were resuspended in H2O and stored at −20°C.
PCR products were directionally cloned into the pET-41 EK-LIC vector as per the manufacturer (EMD Biosciences, Gibbstown, NJ). Briefly, vector-compatible overhangs on the PCR insert were generated by treating purified PCR product with T4 DNA polymerase at 22°C for 30 min, followed by inactivation of the enzyme at 75°C for 20 min. The insert was then annealed to the vector at 22°C for 5 min, and the ligation product was used to transform a chemically competent recA endA cloning strain of Escherichia coli (NovaBlue GigaSingles competent cells provided with the pET-41-EkLIC vector kit). For each hypothetical gene, five to 10 colonies were screened for transformants by PCR with vector-specific primers (forward primer, STagF [5′GAACGCCAGCACATGGAC3′]; reverse primer, pET41R [5′AGGCGCGCCAAGGCCTG3′]). Plasmids were isolated from two or three PCR-positive colonies using the silica membrane-based QIAprep spin miniprep kit (Qiagen, Valencia, CA) and were sequenced with the STagF primer by the Albert Einstein College of Medicine Sequencing Facility using a model 3730 DNA analyzer (Applied Biosystems, Foster City, CA).
Five to 10 ng of sequence-verified plasmid was used to transform a chemically competent protease-deficient strain of Escherichia coli harboring a plasmid encoding 7 tRNAs for rare codons in E. coli [genotype F− ompT hsdSB(rB− mB−) gal dcm pRARE2 (Camr), Rosetta 2 competent cells, EMD Biosciences, Gibbstown, NJ]. Two-milliliter Luria-Bertani broth (LB) cultures supplemented with Overnight Express autoinduction system 1 (EMD Biosciences, Gibbstown, NJ) in 14-ml round-bottom polypropylene tubes (BD Falcon 352018, Franklin Lakes, NJ) were inoculated with transformant colonies, and cultures were incubated at 37°C for 20 h with 250 rpm agitation. Bacterial cells were pelleted from culture and frozen at −80°C.
Expression of fusion protein was evaluated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) of bacterial lysates followed by Coomassie staining of gels and immunoblotting. Immunodetection of the vector-encoded glutathione S-transferase (GST) fusion tag was by primary antibody incubation with a 1:1,000 dilution of anti-GST hybridoma supernatant (gift of Peter Davies, Albert Einstein College of Medicine) followed by secondary antibody incubation with a 1:1,000 dilution of peroxidase-conjugated polyclonal goat anti-mouse IgG (H+L) (Thermo Scientific, Rockford, IL). Immunoblots were visualized by chemiluminescence with ECL Western blotting system (GE Healthcare Bio-Sciences, Piscataway, NJ).
Confirmation of identity of recombinant ECU10_1500 by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS).
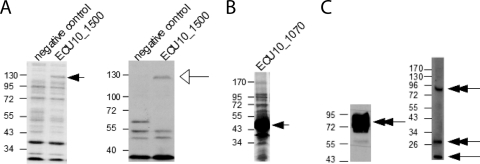
Bacterial protein lysate was subjected to SDS-PAGE and Coomassie blue staining. The overexpressed band which unexpectedly migrated at approximately 120 kDa (Fig. 1 A, black arrow) was excised with a clean scalpel, cut into 1 mm3 pieces, and placed into a low-retention 1.5-ml microcentrifuge tube (Fisher). A slice from a protein-free region of the gel was processed identically for use as a control. The gel pieces were destained three times in 300 μl of 50 mM NH4HCO3/50% acetonitrile for 35 to 40 min in a shaking 35°C water bath. Gel pieces were dehydrated by the addition of 25 mM acetonitrile (100 μl). The acetonitrile was removed by pipette, and the gel was dried further in a SpeedVac concentrator (Thermo Fisher Scientific, Waltham, MA) for 3 min. The gel pieces were incubated in 100 μl 10 mM dithiothreitol (DTT) in 25 mM NH4HCO3 for 30 min at 56°C and 15 min at room temperature to reduce the proteins. The DTT was removed, and alkylation was accomplished by the addition of 100 μl 55 mM iodoacetamide in 25 mM NH4HCO3 and incubation at 45°C in the dark at room temperature. The iodoacetamide was removed and 100 μl 25 mM NH4HCO3 was added to the tube of gel pieces, which was then vortexed, placed in a sonicating water bath, vortexed, and centrifuged. The NH4HCO3 was removed, and this vortex-sonication-centrifugation cycle was repeated two times with 1:1 50 mM NH4HCO3/acetonitrile. The solution was removed and dehydrated with acetonitrile as before, with an additional vortex-sonication-centrifugation step prior to complete dehydration in a SpeedVac. The gel pieces were then rehydrated by the addition of 30 μl 20 ng/μl sequencing-grade modified trypsin (Promega, Madison, WI) in 25 mM NH4HCO3 and placement of the microcentrifuge tubes on ice for 45 min. All but 2 μl of the trypsin solution was removed and replaced with 50 μl 25 mM NH4HCO3. Proteolytic digestion proceeded for 16 h at 37°C. The digest was halted by the addition of 1% trifluoroacetic acid to a final concentration of 0.1%.
FIG. 1.
(A and B) Heterologous expression of E. cuniculi genes in E. coli. Expression of recombinant GST-tagged ECU10_1500 (A) and ECU10_1070 (B) evaluated by SDS-PAGE (A, left, and B, black arrows) and immunoblot (A, right, white arrow) analyses. Bands are visibly overexpressed as detected by Coomassie staining (arrows). As expression of ECU10_1500 was not as robust as that of ECU10_1070, it was also detected by immunoblotting with anti-GST antibody (A, white arrow). The wells to the left in the Coomassie gel and immunoblot in panel A are negative controls (i.e., bacteria transformed with the same expression vector encoding an unrelated parasite gene). The GST antibody also reacts with GST and adjacent vector-encoded tags at ∼32 kDa and nonspecific bands from bacterial proteins at ∼50 kDa. (C) Murine antisera to recombinant, hypothetical E. cuniculi proteins detect cognate proteins in spore lysates. SDS-insoluble, DTT-solubilized spore fractions immunoblotted with mouse antisera to ECU10_1500 (left) or ECU10_1070 (right). Arrows indicate bands of approximate calculated sizes; double-headed arrows indicate bands of unexpected sizes. Although ECU10_1500 antiserum recognized a band of approximately 80 kDa rather than the 50-kDa size calculated from its ORF, the GST-tagged protein migrated aberrantly slowly in recombinant bacterial lysates as well (A) and was shown by mass spectrometry to correspond to the ECU10_1500 ORF (see Fig. S2 and Table S1 in the supplemental material). While nonspecific high-molecular-weight bands are recognized by antiserum to ECU10_1070, a low-molecular-weight band of the approximate expected size (11 kDa) is also detected.
The digest solution was bound to a preequilibriated C18 ZipTip (Millipore, Billerica, MA) according to the manufacturer's directions and eluted with α-cyano-4-hydroxycinnamic acid matrix (Sigma-Aldrich, St. Louis, MO) onto a 100-well MALDI-TOF target sample plate (Applied Biosystems, Foster City, CA). Mass spectrometric data were collected using an AB Voyager system DE-STR MALDI-TOF mass spectrometer (Applied Biosystems, Foster City, CA). The data were acquired in reflector mode with a laser intensity of 1,610 relative units. A mass range of 500 to 4,000 Da was examined, and 300 laser shots were averaged for each mass spectrum. The spectrum was internally calibrated on prominent autolysis peaks of trypsin (842.51 Da, monoisotopic; 1,045.56 Da, monoisotopic; 2,212.43 Da, average), and the α-cyano-4-hydroxycinnamic acid matrix trimer (568.14 Da, monoisotopic).
Monoisotopic m/z peaks from the acquired spectrum were manually identified and used to conduct a trypsin peptide mass fingerprint search of the E_cuniculi 11 (13 October 2004) database with Mascot (Matrix Science Inc., Boston, MA.). The following parameters were stipulated: carbamidomethyl (fixed), deamidated (variable), Gln → pyro-Glu, Glu → pyro-Glu (variable), oxidation (variable), peptide mass tolerance of ±0.5 Da, peptide charge state of +1, and 1 maximum missed cleavage.
Production of antisera to recombinant E. cuniculi hypothetical gene products.
Recombinant parasite proteins were purified from endogenous E. coli lysate proteins by acrylamide gel electrophoresis followed by visualization of protein bands by reverse zinc staining and destaining using the zinc stain and destain kit (Bio-Rad, Hercules, CA) and excision of the band with a clean scalpel. Excised gel bands were processed immediately for immunization or stored at −80°C.
Female 6- to 10-week-old BALB/c mice were immunized with either a slurry of crushed ECU10_1500 protein gel band in DPBS or with ECU10_1070 protein electroeluted from the gel band and mixed with Freund's complete adjuvant (Sigma-Aldrich, St. Louis, MO). Gel bands representing five lane widths of an 8- by 10-cm gel were crushed with a polypropylene Kontes pellet pestle in an accompanying microcentrifuge tube (Fisher, Pittsburgh, PA). DPBS was added to 1 ml, and the slurry was passed several times through an 18- and then a 23-gauge needle. Alternatively, protein was electroeluted from freshly excised (not frozen) gel bands by electrophoresis in 0.025% SDS in standard Western blotting Tris-glycine buffer for 400 to 500 V·h at 4°C in a D-Tube dialyzer (EMD Biosciences, Gibbstown, NJ). Eluate was dialyzed at 4°C against three 1-liter changes of DPBS diluted 1:1 with H2O, evaporated by vacuum centrifugation to 0.3 ml, mixed 1:2 with Freund's complete adjuvant, and emulsified for 1 min in a 1-ml syringe with a motor-driven pestle.
Groups of two or three mice were immunized with each protein. Each mouse was injected subcutaneously and intraperitoneally using a 23-gauge needle with a total of 0.2 to 0.3 ml acrylamide/PBS slurry or 0.2 ml adjuvant emulsion. Blood (0.2 ml per mouse) was collected by retroorbital bleeding 4 weeks postimmunization. Cells were pelleted from whole blood by centrifugation at 5,000 rpm for 15 min in a tabletop microcentrifuge, and the sera were stored at 4°C or −80°C. Mice were reimmunized (boosted) with acrylamide/PBS slurry or Freund's incomplete adjuvant, and blood was again collected 4 weeks after reimmunization and serum was isolated.
Immunoblots against crude bacterial lysates or fractionated, DTT-solubilized parasite lysates were used to evaluate serum specificity. Nitrocellulose membranes were blocked and antibodies diluted in 1% bovine serum albumin (BSA) plus 5% nonfat dry milk (NFDM). Mouse antisera to ECU10_1500 (diluted 1:500) or to ECU10_1070 (diluted 1:2,500) were used as primary antibody; the remainder of the immunoblotting procedure was performed as described above.
Immunolocalization of hypothetical proteins in situ. (i) Immunofluorescence microscopy.
Antisera to expressed recombinant parasite proteins were tested via an immunofluorescence assay (IFA) for reactivity against E. cuniculi-infected RK13 host cell culture. Antisera to recombinant hypothetical protein ECU10_1500 was also tested against E. hellem-infected RK13 host cell culture and evaluated by immunofluorescence and immunoelectron microscopy. Infected host cell cultures on four-well Permanox plastic chamber slides (Thermo Fisher Science, Rochester, NY) or poly-d-lysine plus laminin-coated glass coverslips (BD, Franklin Lakes, NJ) were fixed with 2% formaldehyde/DPBS for 30 min at room temperature, permeabilized for 20 min in 0.2% Triton X-100/DPBS, and blocked overnight in 3% BSA/0.2% Triton X-100/DPBS at 4°C. Subsequent steps, including antibody incubations and washes, were performed in 1% BSA/0.2% Triton X-100/DPBS. Fixed host cell cultures were incubated with 1:1,000 dilutions of mouse antisera to recombinant parasite proteins at 37°C for 1.5 h followed by a 1:500 dilution of goat anti-mouse IgG conjugated to Alexa Fluor 488 or 594 (Invitrogen, Carlsbad, CA) for 1 h at 37°C and 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) for 15 min at room temperature before the final wash. Coverslips were mounted with ProLong antifade mounting medium (Invitrogen, Carlsbad, CA) and cured overnight in the dark. Stained slides were viewed with an epifluorescence-equipped microscope (Zeiss AxioVert 200 M; Carl Zeiss, Goettingen, Germany). Images were captured with a Zeiss Axiocam monochrome digital camera (Carl Zeiss, Goettingen, Germany), and false color was assigned according to the known emission wavelength of each fluorophore. Where indicated, incremental 0.2-μm Z-stack images were captured from culture slides and deconvolved using the iterative maximum likelihood algorithm in the deconvolution module of Zeiss AxioVision software.
(ii) Correlative light-immunoelectron microscopy.
E. hellem was chosen for analysis by immunoelectron microscopy (immunoEM), as parasitophorous vacuoles were more frequently stained by ECU10_1500 antiserum than were those of E. cuniculi. Infected host cell cultures grown on treated glass coverslips were processed as they were for immunofluorescence, through incubation with mouse antiserum, inclusive, and incubated with a 1:50 dilution of Alexa Fluor 488 FluoroNanogold (Nanoprobes, Yaphank, NY) for 25 min at 37°C and 35 min at room temperature. Cultures were postfixed in 2% formaldehyde/DPBS for 3 min at room temperature and washed three times in DPBS and three times in H2O. The cultures were then enhanced by silver deposition for 15 or 25 min with the LI silver enhancement kit (Nanoprobes, Yaphank, NY) as per the manufacturer's directions and imaged using bright-field optics with a Nikon DMicrophot-FXA microscope. Images were captured using a Nikon FX-35DX digital camera controlled by Nikon Digital Sight DS-L1 software and visualization screen (Nikon Instruments, Melville, NY).
To process for immunoEM, the cultures were dehydrated in graded ethanols and embedded in situ in Epon resin. Glass coverslips were removed by treatment with hydrofluoric acid. Thin sections were cut with a microtome and mounted on Formvar-coated grids, stained with uranyl acetate, and observed and photographed with a JEOL 1200 EX transmission electron microscope in the Albert Einstein College of Medicine Analytical Imaging Facility.
Quantitative analysis of EM data.
The ultrastructure of the elongated tubular or filamentous network within the parasitophorous vacuole of E. hellem was quantitatively analyzed in order to discern its laminar composition. Using ImageJ software (1) to analyze a representative tract, the average gray value (0 = black, 255 = white) for each pixel along the 60-pixel-wide cross-section of the filamentous structure was plotted. As the area bounded by the dotted lines is 84 pixels in height, each plotted gray value is the mean of result from 84 measurements.
RESULTS
Proteins detected by LC/MS-MS of fractionated spores.
Our laboratory has previously developed a differential solubility-based subcellular fractionation procedure for the isolation of polar tube and spore wall protein from microsporidian spores (22) and has subjected the DTT-solubilized material to reversed-phase HPLC (21-23) and two-dimensional gel electrophoresis (45), resulting in the detection and purification of component proteins of these organelles. In an effort to identify additional component proteins, including low-abundance species, we subjected both the DTT-soluble and -insoluble fractions to LC/MS-MS. Proteins corresponding to peptides identified by LC/MS-MS in the DTT-soluble and DTT-insoluble fractions are listed in Tables 1 and 2, respectively. As expected, peptides corresponding to PTP1, -2, and -3 and to proteins of the spore wall, including SWP1 and -3 and EnP1, were detected from these fractions (14.7%, 49.0%, 6.4%, 5.1%, 9.0%, and 7.6% sequence coverage [95% confidence interval], respectively). A number of species commonly regarded as “contaminants” in high-sensitivity proteomic experiments (e.g., translation elongation factors, ribosomal proteins, etc.) were also detected, presumably due to their high abundance. In addition, peptides corresponding to 11 and six hypothetical proteins in the DTT-soluble and -insoluble fractions were detected, respectively (Tables 1 and 2). Results from this proteomic analysis led to the selection of ECU10_1500 and ECU10_1070 (22.6% and 13.9% sequence coverage [95% confidence interval], respectively) for the production of murine antisera for immunolocalization in situ.
TABLE 1.
Proteins identified by LC/MS-MS-detected peptides of fractionated E. cuniculi spore lysate, DTT-soluble fraction
| Functional category | Swiss-Prot accession no. | Description |
|---|---|---|
| Characterized polar tube proteins (PTPs) | O76942 | PTP_ENCCU, major polar tube protein precursor (major PTP) |
| Q8SRT0 | PTP2 | |
| Q8SU21 | PTP3 | |
| Characterized spore wall proteins (SWPs) | Q9XZV1 | SWP1_ENCCU, SWP1 precursor |
| Q8SWL3 | EnP1 | |
| Q8SWI4 | SWP3 | |
| Q8SU65 | Putative chitin deacetylase | |
| Other annotated proteins | Q8SVQ9 | Zinc finger protein |
| Q8SQL7 | Thioltransferase (glutaredoxin) | |
| Q8SSL7 | Phosphoacetyl-glucosamine mutase | |
| Q8SSI6 | RAN-specific GTPase-activating protein | |
| Q8SRE1 | Peptidyl-prolyl cis-trans isomerase (PPIase) (rotamase) | |
| Q8SSA1 | Nucleolar protein of the GAR family | |
| Q8SSE8 | Heat shock protein HSP90 homolog | |
| Q8SQY5 | Tryptophanal tRNA synthetase | |
| Q8SS29 | Translation elongation factor 1 alpha | |
| Q8SR93 | General transcription factor | |
| Q8SQM2 | 40S ribosomal protein S28 | |
| Q8SQX3 | 40S ribosomal protein S7 | |
| Hypothetical proteins | Q8STR5 | Hypothetical protein ECU09_1010 |
| Q8SUB5 | Hypothetical protein ECU10_1500 | |
| Q8SUD4 | Hypothetical protein ECU10_1070 | |
| Q8SUM7 | Hypothetical protein ECU08_1390 | |
| Q8SUP6 | Hypothetical protein ECU08_1020 | |
| Q8SUS9 | Hypothetical protein ECU08_0420 | |
| Q8SVC2 | Hypothetical protein ECU06_0650 | |
| Q8SW99 | Hypothetical protein ECU02_1330 | |
| Q8SWJ8 | Hypothetical protein ECU01_1070 | |
| Q8SWP0 | Hypothetical protein ECU01_0440 | |
| Q8SV33 | Hypothetical protein ECU07_0400 |
TABLE 2.
Proteins identified by LC/MS-MS-detected peptides of fractionated E. cuniculi spore lysate, DTT-insoluble fraction
| Functional category | Swiss-Prot accession no. | Description |
|---|---|---|
| Characterized polar tube proteins (PTPs) | O76942 | PTP_ENCCU, major polar tube protein precursor (major PTP) |
| Q8SRT0 | PTP2 | |
| Q8SU21 | PTP3 | |
| Characterized spore wall proteins (SWPs) | Q9XZV1 | SWP1_ENCCU, spore wall protein 1 precursor |
| Q8SWL3 | EnP1 | |
| Q8SWI4 | SWP3 | |
| Other annotated proteins | Q8SVQ9 | Zinc finger protein |
| Q8SRL6 | Required for nuclear division (spindle pole body duplication) | |
| Q8SQS2 | NIFS-like protein (cysteine desulfurase) involved in Fe-S cluster synthase | |
| Q8SRZ2 | Vacuolar ATP synthase subunit E (fragment) | |
| Q8SWI8 | Similarity to DNA helicase | |
| Q8SVC0 | Protein kinase domain | |
| Q8SS77 | Histone H3 | |
| Q8SQP4 | Histone H4 | |
| Q8SS29 | Translation elongation factor 1 alpha | |
| Q8SQT7 | Translation elongation factor 2 | |
| Q8SS53 | 40S ribosomal protein S11 | |
| Q8SR65 | 40S ribosomal protein S23 | |
| Q8SRN2 | 40S ribosomal protein S26 | |
| Q8SS34 | 40S ribosomal protein S27 | |
| Q8SRQ3 | 60S ribosomal protein L5 | |
| Q8SUW1 | Similarity to ribosomal protein L5 | |
| Q8SSM6 | 60S ribosomal protein L8 | |
| Q8SR18 | 60S ribosomal protein L44 (L42 in yeast) | |
| Hypothetical proteins | Q8SUK1 | Hypothetical protein ECU08_1730 |
| Q8SUK4 | Hypothetical protein ECU08_1700 | |
| Q8SUQ8 | Hypothetical protein ECU08_0810 | |
| Q8SUV9 | Hypothetical protein ECU07_1550 | |
| Q8SV27 | Hypothetical protein ECU07_0500 | |
| Q8SV33 | Hypothetical protein ECU07_0400 |
In silico analysis of ECU10_1500 and ECU10_1070.
Homologs of ECU10_1500 and ECU10_1070 were detected in the E. intestinalis (data not shown) and E. hellem genomes. The ECU10_1500 open reading frame is 1,371 bp long, encoding a 50.1-kDa protein of 456 amino acids; its 1,413-bp-long homolog in E. hellem encodes a 51.4-kDa protein of 470 amino acids. An amino acid alignment of these two proteins is shown in Fig. S1 in the supplemental material. At the time of this writing, neither of these proteins exhibited significant similarity by BLAST analysis to any GenBank proteins or possessed putative conserved domains, except for a putative calcium-binding domain in ECU10_1500. At the amino acid level, the E. cuniculi ECU10_1500 is 51% identical to its E. hellem homolog, and each is predicted to be slightly acidic (pI of 4.39 and 4.67, respectively) and moderately hydrophilic (GRAVY scores of −0.87 and −0.81, respectively) and to lack transmembrane domains. The ECU10_1070 open reading frame and its homolog in E. hellem are 306 bp, encoding 11.2- and 11.5-kDa proteins (respectively) of 101 amino acids (an alignment is shown in Fig. S1 in the supplemental material). At the amino acid level, these two proteins are 71% identical and are each predicted to be slightly acidic (pIs of 4.08 and 4.29, respectively) and moderately hydrophilic (GRAVY scores of −0.25 and −0.45, respectively) and to lack transmembrane domains. No signal peptides were predicted for either of these proteins, which rather than definitively precluding their secretion, more likely reflects the limited utility of existing signal peptide prediction algorithms for divergent protists such as the microsporidia. Forty-three and 41 phosphorylation sites (P > 0.7) were predicted for ECU10_1500 and its E. hellem homolog, respectively. Two phosphorylation sites were predicted for ECU10_1500, and one was predicted for its E. hellem homolog (P > 0.7).
Construction and expression of E. cuniculi GST fusion proteins.
Gene-specific primers for E. cuniculi hypothetical genes ECU10_1500 and ECU10_1070 were designed with vector-compatible overhangs for cloning into a prokaryotic expression vector. The recombinant GST-tagged proteins were expressed in E. coli and detected by SDS-PAGE (Fig. 1A [left] and B) or immunoblotting (Fig. 1A, right). Because the recombinant ECU10_1500 GST-tagged protein did not migrate in SDS-PAGE according to its predicted size of 82 kDa (Fig. 1A, band indicated by black arrow in Coomassie-stained gel and white arrow in immunoblot), it was subjected to in-gel digestion and MALDI-TOF mass spectrometry/peptide mass fingerprinting. Ten manually identified monoisotopic peaks were shown to match tryptic peptides of hypothetical protein ECU10_1500 (see Table S1 in the supplemental material) at P values of <0.05, corresponding to 29% sequence coverage for ECU10_1500 (comparable to the average coverage of 30% of gel-excised GST fusion proteins in a study by Korf et al. [27]). Some of the assigned peptides and associated mass spectrum are indicated in Fig. S2 in the supplemental material. The ion score was below the significance threshold for assignment to any other E. cuniculi protein. Each fusion protein was cut out of a preparative SDS-PAGE gel, and three mice were immunized with the crushed gel band of ECU10_1500 fusion protein. As mice immunized similarly with ECU10_1070 fusion protein failed to generate reactive antisera (data not shown), this protein was electroeluted from the excised acrylamide gel band and emulsified with Freund's complete adjuvant prior to immunization.
Evaluation of murine antisera specificity by immunoblot.
Antisera from immunized mice were collected and demonstrated by immunoblotting to react with bands of the approximate expected size of the native protein in parasite lysates (Fig. 1C), taking into account the previously observed aberrantly slow migration of ECU10_1500 and, additionally, the possibility of posttranslational modifications (phosphorylation, glycosylation, etc.) indicated by in silico analysis. While unexpectedly high-molecular-weight bands were also detected by antiserum to ECU10_1070, this is not uncommon for polyclonal murine antisera when examining lysates of microsporidia.
Expression of ECU10_1070 in E. cuniculi in infected host cell culture.
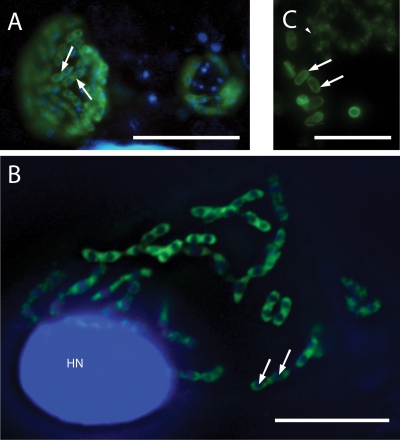
Antiserum to ECU10_1070 fusion protein stains the periphery of parasites that appear to be undergoing cytokinesis (Fig. 2 A, arrows); no staining of cell culture or parasite is observed with anti-GST antibody (45). The antiserum does not stain the periphery of mature, released spores as antiserum to SWP1 does (Fig. 2C, arrows), although the latter also stains the periphery of developing forms (Fig. 3 C, arrowhead). The occurrence of nuclei in pairs but separated by the deposition of ECU10_1070 protein is evident in the deconvolved image (Fig. 2B, arrows).
FIG. 2.
Immunofluorescence localization of ECU10_1070 and comparison to SWP1. Nuclei were counterstained with DAPI (blue). HN, host nucleus. (A) The protein encoded by ECU10_1070 is localized to a thick layer at the periphery of developing parasite stages floating in the lumen of the parasitophorous vacuole (arrows). Bar, 20 μm. (B) The deposition of ECU10_1070 protein at the periphery of parasites floating in the lumen of the parasitophorous vacuole and its expression by dividing parasites, indicated by the occurrence of nuclei in pairs (arrows), suggest that this protein is developmentally regulated and expressed specifically by sporonts. Bar, 20 μm. (C) In contrast to ECU10_1070, spore wall protein 1 (SWP1) is deposited at the periphery of mature spores (arrows) but is less abundant at the periphery of developing forms (arrowhead). Bar, 10 μm.
FIG. 3.
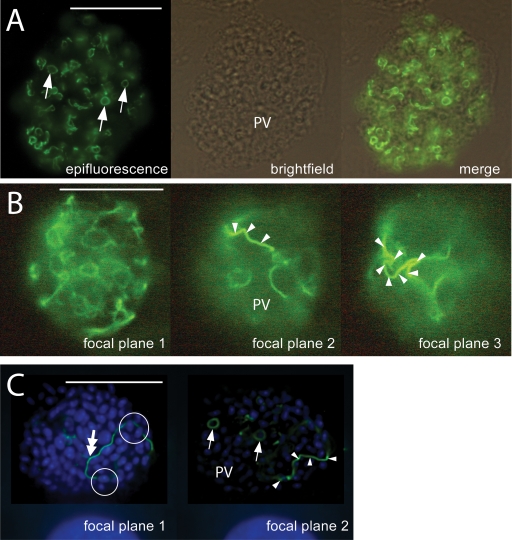
Immunofluorescence localization of ECU10_1500. (A) Appearance of ECU10_1500 in E. cuniculi. This protein localizes to novel structures of filamentous appearance (arrows) in the lumen of the parasitophorous vacuole (PV) which sometimes appear as small coils. Bar, 20 μm. (B) Immunofluorescence localization of ECU10_1500 in E. cuniculi: three focal planes of the same parasitophorous vacuole. The structures formed by the ECU10_1500 protein sometimes extend much of the length of the PV as long filamentous structures (arrowheads) in the lumen of the PV. Bar, 20 μm. (C) Immunofluorescence localization of ECU10_1500 in E. hellem: deconvolved Z stack. The antiserum to ECU10_1500 cross-reacts with E. hellem PV, staining structures with appearances similar to those in E. cuniculi. Nuclei were counterstained with DAPI. The filamentous structures stained by antiserum to this protein form small circles (single-headed arrows) and longer, extended forms (arrowheads) which appear to pass around rather than through parasites (circles) within the lumen of the PV. The filamentous structures can be seen to branch (double-headed arrow). Bar, 20 μm.
Expression of ECU10_1500 in E. cuniculi in infected host cell culture.
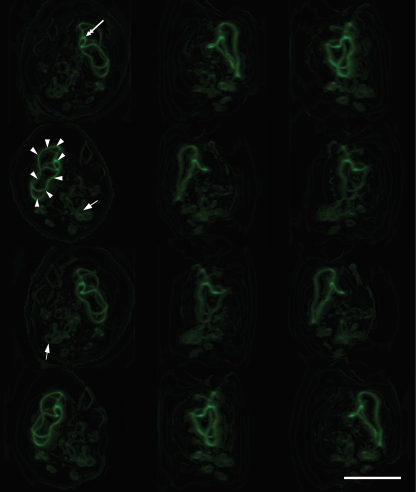
Antiserum to ECU10_1500 stains structures of filamentous appearance in the lumen of the parasitophorous vacuole of E. cuniculi (Fig. 3A and B) and E. hellem (Fig. 3C and 4). These structures appear as small coils (Fig. 3A and C, arrows) or longer filaments (Fig. 3B and C and 4, arrowheads), sometimes within the same vacuole. In deconvolved images, these filamentous structures seem to pass around rather than through developing parasites (Fig. 3C, circles). A montage of a rotated three-dimensional (3-D) reconstruction of a deconvolved stack of images (Fig. 4) suggests that the longer filaments seen in epifluorescence micrographs (Fig. 3B and C and 4, arrowheads) may actually be part of a larger coil (Fig. 4, arrowheads) and that the protein forms a branching network (branch points [Fig. 3C and 4, double-headed arrows]) within the lumen of the parasitophorous vacuole.
FIG. 4.
Immunofluorescence localization of ECU10_1500 in E. hellem. A montage of a rotated 3-D reconstruction of a deconvolved Z stack of images suggests that the longer filaments seen by epifluorescence microscopy may actually be part of a larger coil (arrowheads) and that the protein forms a branching network (branch point, double-headed arrow) within the lumen of the parasitophorous vacuole (PV). It is also clear that within a PV that contains a large network, smaller coils may also be seen (single-headed arrows). Bar, 20 μm.
Correlative light/immunoelectron microscopy of ECU10_1500.
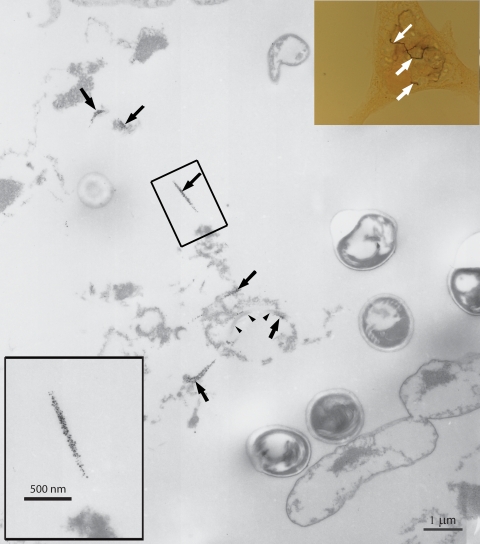
In E. hellem-infected host cell cultures stained with antiserum to ECU10_1500, silver enhancement of gold-conjugated secondary antibody for 25 min produced a brown precipitate visible by bright-field microscopy, revealing structures of filamentous appearance consistent in appearance with those observed by immunofluorescence microscopy (Fig. 5, inset in upper right corner, white arrows). In samples silver enhanced for 15 min, this precipitate was visible by transmission electron microscopy as irregularly shaped structures 4 to 25 nm in diameter (Fig. 5, inset in bottom left corner; Fig. 5 and 6, black arrows). The silver precipitate was deposited along long, narrow tracts (Fig. 5 and 6, black arrows) in the lumen of the parasitophorous vacuole, consistent with the immunostaining pattern of ECU10_1500 antiserum by light microscopy (Fig. 3 and 4). The precipitate appears to be deposited upon structures of filamentous or tubular appearance (Fig. 5, black arrowheads, and 6, first and second insets), one of which appears to branch (Fig. 6, first inset, white arrowhead) as observed by immunofluorescence microscopy (Fig. 3C and 4, white double-headed arrows). The diameter of this filamentous or tubular structure varies along its length and between filaments, ranging approximately from 80 to 140 nm. Ultrastructurally, in longitudinal section it has a slightly electron-dense core (Fig. 6, second inset, white double-headed arrow) surrounded by an electron-lucent middle layer (Fig. 6, second inset, black double-headed arrow) and a very fine, slightly electron-dense outer layer (Fig. 6, second inset, black double arrowhead). The silver precipitate is not deposited upon cross-sections or longitudinal sections of the polar filament within spores or developing stages of the parasite (Fig. 6, black arrowheads).
FIG. 5.
Immunolocalization of ECU10_1500 in E. hellem-infected host culture by silver-enhanced correlative light microscopy/immunoEM. A brown precipitate visible by bright-field microscopy formed on structures of filamentous appearance (inset, upper right corner, white arrows) consistent in appearance with those observed by immunofluorescence. This precipitate was visible as small (4- to 25-nm) electron-dense dots (inset, lower left corner) on structures of filamentous or tubular appearance (arrowheads).
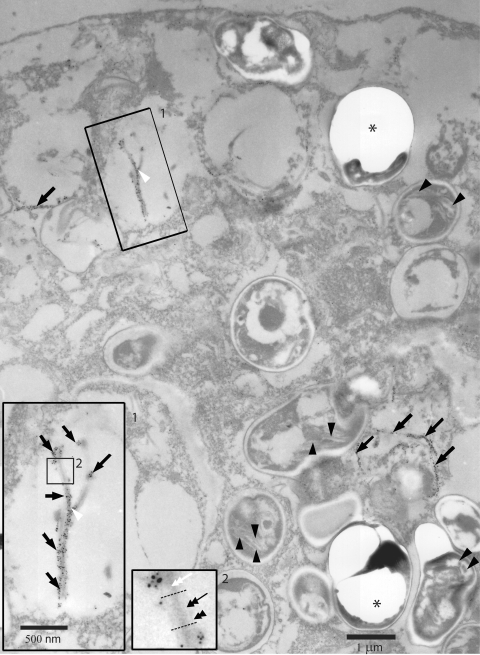
FIG. 6.
Immunolocalization of ECU10_1500 in E. hellem-infected host culture by silver-enhanced correlative light microscopy/immunoEM. ECU10_1500 is detected along long tubular or filamentous structures (black arrows) of ∼0.1-μm diameter in the parasitophorous vacuole and not along sections of the developing polar tube inside parasites (black arrowheads). These structures appear to branch, consistent with findings by light microscopy (white arrowhead, first inset). At higher magnifications (first and second insets), the structure is of trilaminate appearance with an inner, slightly electron-dense core (white double-headed arrow), an electron-lucent intermediate layer (black double-headed arrow), and a very fine, slightly electron-dense outer layer (black double-arrowhead). Asterisks indicate spores or sporogonic stages dislodged during tissue processing for EM.
Multilaminar ultrastructure of filaments comprising the intravacuolar network.
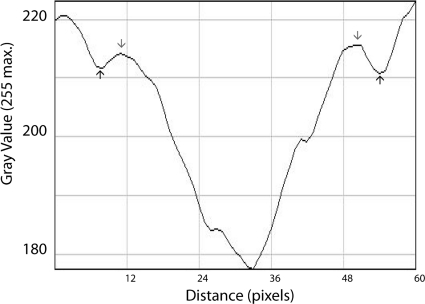
A plot of the gray intensity along a cross-section of the filament is approximately bilaterally symmetric and comprises two peaks (Fig. 7, gray arrows) and three valleys (Fig. 7, black arrows and the large central valley of the lowest gray value). The small valleys (black arrows) correspond to the fine gray outer border of the filament (Fig. 6, second inset, black double arrowhead); the peaks correspond to the intermediate electron-lucent layer (Fig. 6, second inset, black double-headed arrow); and the deep central valley corresponds to the moderately electron-dense core (Fig. 6, second inset, white double-headed arrow). The varied electron density along the cross-section of the filament and the symmetry of the pixel intensity plot (Fig. 7) suggest a filamentous structure of annular or trilaminate composition.
FIG. 7.
Quantitative analysis of the ultrastructure of intravacuolar filaments. A representative tract (the area bounded by the dotted lines in Fig. 6, second inset) was analyzed. This area is 84 pixels in height; each plotted gray value is the mean of 84 measurements. The trilaminate composition of the immunostained filaments within the parasitophorous vacuole is evident as peaks and valleys of pixel intensity (gray values: 0 = black, 255 = white): the black arrows correspond to the fine gray outer layer of the filament; the gray arrows correspond to the intermediate electron-lucent layer; and the central valley corresponds to the moderately electron-dense core of the filament.
DISCUSSION
In this study, the differential solubility of the microsporidian polar tube and spore wall was harnessed to identify via mass spectrometry and in situ immunolocalization additional components of the polar tube and spore wall of E. cuniculi. As a result, we have demonstrated a novel component of the developing spore wall. While additional components of the polar tube were not identified, we have demonstrated a heretofore undescribed intravacuolar network of proteinaceous, branching filaments from the DTT-soluble subcellular fraction. Although this fraction is presumed on the basis of negative-stain transmission electron microscopy (TEM) images (22) to consist chiefly of polar tube material, the copurification of ECU10_1500 is likely due to its having a similar SDS/DTT insolubility/solubility profile, as well as to its apparent abundance. Similarly, while previously identified SWPs (SWP1, EnP1, snd SWP3) and PTPs (PTP1, -2, and -3) were detected as expected in the DTT-insoluble or -soluble fraction, respectively, each was also detected in both fractions, probably also due to their high abundance or, in the case of the PTPs, incomplete solubilization. The appearance of SWP3 in the DTT-soluble fraction has previously been noted (46).
The 11-kDa ECU10_1070-encoded protein is localized to a thick layer at the periphery of developing parasite stages floating in the lumen of the parasitophorous vacuole (Fig. 2A [arrows] and B). The occurrence of nuclei in pairs (Fig. 2B, arrows) but separated by the deposition of ECU10_1070 protein indicates that these parasites are undergoing cell division. The location of the parasites and the nuclear configuration suggest that this protein is specifically expressed by sporonts, the stage at which the commitment to spore formation begins (7). The protein does not appear to be expressed in sporoblasts, the cells which undergo metamorphosis to form spores, as the final rounds of cell division occur in sporonts, and no parasites with isolated nuclei are stained. Nor does this antiserum stain the wall of mature spores, as does similarly prepared antiserum to SWP1 (Fig. 2C, arrows); note that SWP1 is also found at the periphery of developing forms (Fig. 2C, arrowhead). Thus, it is conceivable that ECU10_1070 forms an early scaffold in sporogony for the deposition of SWP1 (5), SWP3/EnP2 (32, 45), or EnP1 (32), the three proteins which have been shown to be components of the mature spore wall. It is a small (11-kDa) protein with no predicted transmembrane or putative conserved domains and, with the exception of a presumptive homolog in E. intestinalis (8; also data not shown), has no significant similarity to any proteins in the National Center for Biotechnology Information (NCBI) database except for a low-score match (BLAST score 51) to a component of the large ribosomal subunit of Enterocytozoon bieneusi, which is likely of no functional significance, especially given immunolocalization data.
The 51-kDa ECU10_1500 protein is localized to a heretofore undescribed branching network in the lumen of the parasitophorous vacuole of E. cuniculi (Fig. 3A and B). Antiserum to this protein cross-reacts with its presumable homolog in E. hellem, staining apparently identical structures (Fig. 3C and 4). By light and electron microscopy, this network appears to be composed of filamentous structures roughly 100 nm in diameter (Fig. 6, first and second insets). Within one parasitophorous vacuole, multiple structures of different sizes may exist, with some appearing as small, unbranching coils and others as larger, branching networks that extend through much of the parasitophorous vacuole (Fig. 3C and 4). While the diameter and multilaminate ultrastructure of the filament are somewhat reminiscent of those of the polar tube (Fig. 6, second insets), antiserum to ECU10_1500 does not stain polar filaments internal to parasites or polar tubes extruded from spores (Fig. 6, black arrowheads), and the network formed by this protein is found strictly in the lumen of the parasitophorous vacuole. Moreover, ECU10_1500 does not exhibit sequence similarity with PTPs of any microsporidia.
Because ECU10_1500 exhibits no significant similarity to any gene in the NCBI database and possesses no conserved functional domains other than a calcium-binding motif, it is difficult to speculate on the role of the filamentous network formed by this protein. Only one comparable protist-derived intravacuolar network, the so-called tubulovesicular network of Toxoplasma gondii (33, 34), presents itself in the literature. T. gondii is a ubiquitous, obligate intracellular parasite of the phylum Apicomplexa that also causes serious morbidity in immunosuppressed populations (19). Within 5 min of invading a host cell or being phagocytosed by a macrophage, this parasite begins elaborating a network of nanotubules within the lumen of the nascent parasitophorous vacuole (33). However, the ultrastructural appearance of this network is dissimilar to that formed by ECU10_1500: in cross-section, it is of membranous vesicular appearance (33) but is associated with several proteins derived from electron-dense granules which are thought to be involved in its formation (30); the tubules are hollow and somewhat smaller in diameter on average (60 to 90 nm) than the ECU10_1500 filaments (34); the network fills the Toxoplasma parasitophorous vacuole more profusely (33, 34) than has been observed in Encephalitozoon, and tubules of the network are continuous with the membrane of the parasitophorous vacuole (PVM) (34). Based on these observations, the authors of these studies have theorized that this network may serve to maximize the surface area for nutrient acquisition from the host into the parasitophorous vacuole. However, the appearance and discontinuity with the PVM of the intravacuolar network described herein do not support such speculation in the case of Encephalitozoon. Currently, delineation of protein function in the microsporidia depends on bioinformatic analysis and classical molecular biological and biochemical experiments that do not require genetic manipulation of the parasite. In view of this limitation, the antiserum to ECU10_1500 may be useful for immunoprecipitation of interacting proteins from E. cuniculi (and other closely related microsporidia), which would allow their subsequent identification by proteomic techniques. Although ECU10_1500 exhibits only 51% sequence identity to its homolog in E. hellem, discernment of conserved motifs within an alignment of presumptive homologs from additional organisms may provide clues as to functionality. The prediction of over 40 phosphorylation sites (P > 0.7) in the protein encoded by ECU10_1500 and its E. hellem homolog suggests the possibility of extensive posttranslational regulation. Differential detection of phosphorylated forms at distinct time points postinfection or in distinct host states, for example, may also serve to illuminate the role of the ECU10_1500-encoded protein and the unique intravacuolar network it comprises.
Due to the emerging nature of these pathogens, proteomic analysis-driven investigations of the microsporidia have been sparse, but the sequencing of the E. cuniculi genome by Katinka and colleagues (20) and of other microsporidian genomes has inaugurated a new era in microsporidiology wherein these approaches are coming into focus. Utilization of these genome data has enabled the compilation of a list of highly abundant proteins in E. cuniculi (6) and the identification of new spore wall proteins of Nosema bombycis, a silkworm-pathogenic species (43, 44). Elucidation of the components of the polar tube and spore wall has already practically impacted clinical diagnostic settings, where antibodies directed against these organelles have utility as diagnostic reagents (see reference 16). In addition, the apparent immunodominance of component proteins has aided efforts to understand the immune response to these pathogens (reviewed in reference 26). Finally, as the remarkable elasticity of the polar tube and tensile strength of the spore wall are critical to later stages of the germination process which occur downstream of osmotic swelling, a better appreciation of the molecular composition of these and functionally related organelles within the spore is likely to enhance our understanding of the germination process and general biology of the microsporidia. It is hoped that such efforts may ultimately uncover novel drug targets for these diverse and difficult-to-treat pathogens (42).
Supplementary Material
Acknowledgments
This work was supported by NIH grant 5R01AI031788-19 from the National Institute of Allergy and Infectious Diseases. Ongoing sequencing of the E. hellem genome is supported by Canadian Institutes for Health Research grant MOP-42517. P.K. is a Fellow of the Canadian Institute for Advanced Research and Senior Scholar of the Michael Smith Foundation for Health Research.
We thank Frank Macaluso of the Analytical Imaging Facility of the Albert Einstein College of Medicine Cancer Center Shared Resource (NIH grant 2P30CA013330) for technical assistance in electron microscopy sample processing and access to the JEOL 1200 electron microscope.
Editor: J. H. Adams
Footnotes
Published ahead of print on 10 January 2011.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Altschul, S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckers, P. J., G. J. Derks, T. Gool, F. J. Rietveld, and R. W. Sauerwein. 1996. Encephalocytozoon intestinalis-specific monoclonal antibodies for laboratory diagnosis of microsporidiosis. J. Clin. Microbiol. 34:282-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blom, N., S. Gammeltoft, and S. Brunak. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351-1362. [DOI] [PubMed] [Google Scholar]
- 5.Bohne, W., D. J. Ferguson, K. Kohler, and U. Gross. 2000. Developmental expression of a tandemly repeated, glycine- and serine-rich spore wall protein in the microsporidian pathogen Encephalitozoon cuniculi. Infect. Immun. 68:2268-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosson, D., L. Kuhn, G. Prensier, C. P. Vivares, and C. Texier. 2005. The putative chitin deacetylase of Encephalitozoon cuniculi: a surface protein implicated in microsporidian spore-wall formation. FEMS Microbiol. Lett. 247:81-90. [DOI] [PubMed] [Google Scholar]
- 7.Cali, A., and P. Takvorian. 1999. Developmental morphology and life cycles of the microsporidia, p. 85-128. In M. Wittner and L. M. Weiss (ed.), The microsporidia and microsporidiosis. American Society for Microbiology, Washington, DC.
- 8.Corradi, N., J. F. Pombert, L. Farinelli, E. S. Didier, and P. J. Keeling. 2010. The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nat. Commun. doi: 10.1038/ncomms1082. [DOI] [PMC free article] [PubMed]
- 9.Costa, S. F., and L. M. Weiss. 2000. Drug treatment of microsporidiosis. Drug Resist. Updat. 3:384-399. [DOI] [PubMed] [Google Scholar]
- 10.Didier, E. S., and L. M. Weiss. 2006. Microsporidiosis: current status. Curr. Opin. Infect. Dis. 19:485-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emanuelsson, O., H. Nielsen, S. Brunak, and G. von Heijne. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300:1005-1016. [DOI] [PubMed] [Google Scholar]
- 12.Franzen, C., and A. Muller. 2001. Microsporidiosis: human diseases and diagnosis. Microbes Infect. 3:389-400. [DOI] [PubMed] [Google Scholar]
- 13.Frixione, E., L. Ruiz, J. Cerbon, and A. H. Undeen. 1997. Germination of Nosema algerae (Microspora) spores: conditional inhibition by D2O, ethanol and Hg2+ suggests dependence of water influxupon membrane hydration and specific transmembrane pathways. J. Eukaryot. Microbiol. 44:109-116. [DOI] [PubMed] [Google Scholar]
- 14.Garcia, L. S. 2002. Laboratory identification of the microsporidia. J. Clin. Microbiol. 40:1892-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh, K. 2009. Ph.D. thesis. Identification of proteins of the infectious apparatus of Encephalitozoon cuniculi. Rutgers University, Newark, NJ.
- 16.Ghosh, K., and L. M. Weiss. 2009. Molecular diagnostic tests for microsporidia. Interdiscip. Perspect. Infect. Dis. 2009:926521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon, D. 2003. Viewing and editing assembled sequences using Consed. Curr. Protoc. Bioinformatics 11:11.2. [DOI] [PubMed] [Google Scholar]
- 18.Hayman, J. R., S. F. Hayes, J. Amon, and T. E. Nash. 2001. Developmental expression of two spore wall proteins during maturation of the microsporidian Encephalitozoon intestinalis. Infect. Immun. 69:7057-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill, D., and J. P. Dubey. 2002. Toxoplasma gondii: transmission, diagnosis and prevention. Clin. Microbiol. Infect. 8:634-640. [DOI] [PubMed] [Google Scholar]
- 20.Katinka, M. D., et al. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414:450-453. [DOI] [PubMed] [Google Scholar]
- 21.Keohane, E., et al. 1994. The identification and characterization of a polar tube reactive monoclonal antibody. J. Eukaryot. Microbiol. 41:48S. [PubMed] [Google Scholar]
- 22.Keohane, E. M., et al. 1996. Purification and characterization of human microsporidian polar tube proteins. J. Eukaryot. Microbiol. 43:100S. [DOI] [PubMed] [Google Scholar]
- 23.Keohane, E. M., et al. 1998. The molecular characterization of the major polar tube protein gene from Encephalitozoon hellem, a microsporidian parasite of humans. Mol. Biochem. Parasitol. 94:227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keohane, E. M., and L. M. Weiss. 1999. The structure, function, and composition of the microsporidian polar tube, p. 196-224. In M. Wittner and L. M. Weiss (ed.), The microsporidia and microsporidiosis. American Society for Microbiology, Washington, DC.
- 25.Keohane, E. M., et al. 1996. Purification and characterization of a microsporidian polar tube protein. Mol. Biochem. Parasitol. 79:255-259. [DOI] [PubMed] [Google Scholar]
- 26.Khan, I. A., M. Moretto, and L. M. Weiss. 2001. Immune response to Encephalitozoon cuniculi infection. Microbes Infect. 3:401-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korf, U., et al. 2005. Large-scale protein expression for proteome research. Proteomics 5:3571-3580. [DOI] [PubMed] [Google Scholar]
- 28.Lom, J., and J. Vavra. 1963. The mode of sporoplasm extrusion in microsporidian spores. Acta Protozool. 1:81-89. [Google Scholar]
- 29.Lujan, H. D., et al. 1998. Detection of microsporidia spore-specific antigens by monoclonal antibodies. Hybridoma 17:237-243. [DOI] [PubMed] [Google Scholar]
- 30.Mercier, C., et al. 2002. Biogenesis of nanotubular network in Toxoplasma parasitophorous vacuole induced by parasite proteins. Mol. Biol. Cell 13:2397-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overton, I. M., et al. 2008. TarO: a target optimisation system for structural biology. Nucleic Acids Res. 36:W190-W196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peuvel-Fanget, I., et al. 2006. EnP1 and EnP2, two proteins associated with the Encephalitozoon cuniculi endospore, the chitin-rich inner layer of the microsporidian spore wall. Int. J. Parasitol. 36:309-318. [DOI] [PubMed] [Google Scholar]
- 33.Sibley, L. D., J. L. Krahenbuhl, G. M. Adams, and E. Weidner. 1986. Toxoplasma modifies macrophage phagosomes by secretion of a vesicular network rich in surface proteins. J. Cell Biol. 103:867-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sibley, L. D., I. R. Niesman, S. F. Parmley, and M. F. Cesbron-Delauw. 1995. Regulated secretion of multi-lamellar vesicles leads to formation of a tubulo-vesicular network in host-cell vacuoles occupied by Toxoplasma gondii. J. Cell Sci. 108(Pt. 4):1669-1677. [DOI] [PubMed] [Google Scholar]
- 35.Slabinski, L., et al. 2007. XtalPred: a Web server for prediction of protein crystallizability. Bioinformatics 23:3403-3405. [DOI] [PubMed] [Google Scholar]
- 36.Southern, T. R., C. E. Jolly, M. E. Lester, and J. R. Hayman. 2007. EnP1, a microsporidian spore wall protein that enables spores to adhere to and infect host cells in vitro. Eukaryot. Cell 6:1354-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takvorian, P. M., and A. Cali. 1986. The ultrastructure of spores (Protozoa: Microsporidia) from Lophius americanus, the angler fish. J. Protozool. 33:570-575. [DOI] [PubMed] [Google Scholar]
- 38.Undeen, A. H., and N. D. Epsky. 1990. In vitro and vivo germination of Nosema locustae (Microspora: Nosematidae) spores. J. Invertebr. Pathol. 56:371-379. [Google Scholar]
- 39.Visvesvara, G. S., et al. 1994. Polyclonal and monoclonal antibody and PCR-amplified small-subunit rRNA identification of a microsporidian, Encephalitozoon hellem, isolated from an AIDS patient with disseminated infection. J. Clin. Microbiol. 32:2760-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weidner, E. 1976. The microsporidian spore invasion tube. The ultrastructure, isolation, and characterization of the protein comprising the tube. J. Cell Biol. 71:23-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss, L. M. 2001. Microsporidia: emerging pathogenic protists. Acta Trop. 78:89-102. [DOI] [PubMed] [Google Scholar]
- 42.Wittner, M., and L. M. Weiss. 1999. The microsporidia and microsporidiosis. American Society for Microbiology, Washington, DC.
- 43.Wu, Z., et al. 2008. Proteomic analysis of spore wall proteins and identification of two spore wall proteins from Nosema bombycis (Microsporidia). Proteomics 8:2447-2461. [DOI] [PubMed] [Google Scholar]
- 44.Wu, Z., Y. Li, G. Pan, Z. Zhou, and Z. Xiang. 2009. SWP25, a novel protein associated with the Nosema bombycis endospore. J. Eukaryot. Microbiol. 56:113-118. [DOI] [PubMed] [Google Scholar]
- 45.Xu, Y., et al. 2006. Identification of a new spore wall protein from Encephalitozoon cuniculi. Infect. Immun. 74:239-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu, Y., and L. M. Weiss. 2005. The microsporidian polar tube: a highly specialised invasion organelle. Int. J. Parasitol. 35:941-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeung, Y. G., E. Nieves, R. H. Angeletti, and E. R. Stanley. 2008. Removal of detergents from protein digests for mass spectrometry analysis. Anal. Biochem. 382:135-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zerbino, D. R., and E. Birney. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.