Abstract
As bacterial pathogens develop resistance against most currently used antibiotics, novel alternatives for treatment of microbial infectious diseases are urgently needed. Targeting bacterial virulence functions in order to disarm pathogens represents a promising alternative to classical antibiotic therapy. Type IV secretion systems, which are multiprotein complexes in the cell envelope that translocate effectors into host cells, are critical bacterial virulence factors in many pathogens and excellent targets for such “antivirulence” drugs. The VirB8 protein from the mammalian pathogen Brucella was chosen as a specific target, since it is an essential type IV secretion system component, it participates in multiple protein-protein interactions, and it is essential for the assembly of this translocation machinery. The bacterial two-hybrid system was adapted to assay VirB8 interactions, and a high-throughput screen identified specific small-molecule inhibitors. VirB8 interaction inhibitors also reduced the levels of VirB8 and of other VirB proteins, and many of them inhibited virB gene transcription in Brucella abortus 2308, suggesting that targeting of the secretion system has complex regulatory effects in vivo. One compound strongly inhibited the intracellular proliferation of B. abortus 2308 in a J774 macrophage infection model. The results presented here show that in vivo screens with the bacterial two-hybrid assay are suited to the identification of inhibitors of Brucella type IV secretion system function.
The increasing resistance to classical antibiotics necessitates the development of alternative therapeutic strategies against microbial infectious diseases (36, 47). Genomics-based approaches, which are aimed at identifying novel targets (29), have potential to yield new therapeutic approaches; it is nevertheless foreseeable that resistance will eventually develop against drugs that target vital cell functions. Alternative strategies comprise phage therapy, the stimulation of the host immune system, and the development of “antivirulence” drugs that specifically target bacterial virulence functions but not vital cell functions (4, 7, 16, 30). The rationale underlying the latter approach is that these molecules will disarm pathogens, permitting their elimination from the body by the immune system, and that the selection pressure for the development of resistance mutations will be reduced, as they do not target vital cellular functions. Recent years have seen significant advances in this area, especially in type III secretion (T3S) systems, where promising molecules were discovered (22, 34). Interestingly, many of the active molecules belong to the class of salicylidene acylhydrazides and have broad-spectrum activity against Yersinia, Chlamydia, Salmonella, and Shigella species (33, 37, 39, 46). These molecules were isolated using cell-based high-throughput screening (HTS) measuring T3S system functions in living cells, and their targets have not been unequivocally identified. In contrast, we have pursued a different approach based on a well-characterized target with known X-ray structure from the Brucella type IV secretion (T4S) system (45).
T4S systems are multiprotein complexes that translocate macromolecules, such as DNA, proteins, and DNA-protein complexes, across the cell envelope of Gram-negative bacteria (3, 5, 15). They are essential virulence factors of many important pathogens, such as Agrobacterium, Brucella, Helicobacter, and Legionella, and this makes them excellent model targets for the development of antivirulence drugs (7). They typically comprise more than 10 protein components that assemble into a transmembrane complex, and the best-studied system, from the plant pathogen Agrobacterium tumefaciens, with its 12 components (VirB1 to VirB11 and VirD4), serves as a reference model. X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy approaches have provided several individual T4S protein structures (VirB5, VirB8, VirB10, VirB9, VirB11, and VirD4) (8, 24, 26, 27, 45, 49, 50). Recent cryo-electron microscopy (cryo-EM) and X-ray crystallographic studies revealed that three of them, VirB7, VirB9, and VirB10, form a macromolecular complex that likely represents a portion of the core T4S system and translocation channel (13, 25). Two of the available protein structures, VirB8 and VirB11, are from Brucella species, which cause the most widespread zoonotic disease (more than 500,000 cases per year), with significant economic losses of livestock and morbidity in humans in South and Central America and in Mediterranean and Arabic countries (2, 10, 43, 51). In addition, Brucella is considered a potential bioterror threat (48), as it is easily transmitted by aerosols and it causes long-lasting severe infections that require treatment with two antibiotics, such as doxycycline and rifampin or streptomycin, over 4 to 6 weeks (2). In spite of the aggressive antibiotic therapies used in humans, relapses are frequent, and this may be due to the fact that Brucella is an intracellular pathogen that grows inside cells of the reticuloendothelial system (12). Antivirulence drugs that deprive the pathogen of its essential virulence factor, the T4S system, would constitute alternatives to or enhancements of current antibiotic treatment regimens. Previous screening efforts to isolate T4S inhibitors led to the isolation of molecules that impact Helicobacter pylori VirB11 ATPase activity and the T4S-mediated transfer of broad-host-range plasmids, respectively, but these molecules had limited potency and specificity (23, 28).
Here, we pursued an approach inspired by previous X-ray crystallographic studies and structure-function analyses suggesting that dimerization is important for VirB8 functionality (40, 45; D. Sivanesan and C. Baron, submitted for publication). VirB8 is a bitopic inner membrane protein that undergoes multiple interactions with other T4S components via its periplasmic C-terminal domain, and it is an essential assembly factor (6, 11, 31, 44). We designed a cell-based assay for the isolation of small molecular inhibitors of VirB8 interactions with the goal of isolating T4S inhibitors. Screening a small-molecule library identified several specific inhibitors of VirB8 interactions, and one of these molecules strongly reduced the intracellular proliferation of Brucella abortus 2308. Our results show that we have identified hit molecules (specific VirB8 interaction inhibitors) that may be suitable for development into leads for antivirulence drugs that disarm Brucella.
MATERIALS AND METHODS
Bacterial growth conditions.
Escherichia coli strain JM109 was used as the host for cloning, and strain BTH101 was used for bacterial two-hybrid (BTH) assays in LB or phosphate-buffered tryptone-glucose-yeast extract-phosphate (TGYEP) medium (32), as indicated. The B. abortus 2308 wild type and the virB deletion strain ADH4.2 were cultured on tryptic soy agar (TSA) (Difco/Becton-Dickinson, Sparks, MD) or in tryptic soy broth (TSB) at 37°C on a rotary shaker. To test expression of VirB proteins, B. abortus cultures were inoculated in TSB and incubated at 37°C with shaking at 200 rpm for 18 to 24 h. The bacteria were sedimented and resuspended in an equal volume of 1× modified minimal E medium, pH 5 (35, 41). The cultures were incubated in this virB expression-inducing medium at 37°C with shaking at 200 rpm for 4 to 5 h; the bacteria were sedimented and resuspended in 2× Laemmli sample buffer at a concentration of 1 × 1010 CFU/ml and heated at 100°C for 5 min, and the total protein from 1 × 108 CFU (0.01 ml) was loaded per well for separation by SDS-PAGE. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes by electroblotting and detected using primary polyclonal rabbit sera specific for VirB5 (1:1,000), VirB8 (1:5,000), VirB9 (1:5,000), VirB10 (1:5,000), VirB11 (1:5,000), VirB12 (1:50,000), Bcsp31 (1:5,000), and, as a secondary antibody, goat anti-rabbit IgG (1:5,000) conjugated to horseradish peroxidase (HRP). HRP activity was detected with a chemiluminescent substrate (Perkin-Elmer, Waltham, MA).
Plasmid construction and bacterial two-hybrid assay.
Plasmids for bacterial two-hybrid assays were constructed using standard procedures, and assays were conducted as described previously (44).
Growth of cultures and reporter enzyme assay.
The quantification of the functional complementation mediated by interaction between two proteins was obtained by measuring β-galactosidase activities in liquid cultures. Cultures of the cya-deficient strain BTH101 (32) containing the appropriate vectors were cultivated by shaking (200 rpm) in 2 ml LB or TGYEP medium supplemented with the appropriate amount of antibiotics and 1 mM isopropyl-β-d-thiogalactopyranoside to induce expression of the T18 and T25 fusion proteins. The β-galactosidase activities were measured from 20 μl culture incubated with 80 μl Z buffer (0.06 M Na2HPO4·7H2O, 0.04 M NaH2PO4·H2O, 0.01 M KCl, 0.001 M MgSO4) containing 8 mg/ml 2-nitrophenyl-β-d-galactopyranoside, 0.01% SDS, and 50 mM β-mercaptoethanol (modified from reference 17). The reaction mixtures were incubated for 0.5 h at room temperature, and the reactions were stopped with 100 μl 1 M Na2CO3. The end products were measured at 420 nm and 550 nm with a SpectramaxPlus384 Microplate Spectrophotometer Reader (Molecular Devices) plate reader. Specific activity was calculated as follows: Miller units = [OD420 − (1.75 × OD550)]/[t × OD600 × (volume in ml)] × 1,000, where OD600 is the optical density at 600 nm after 12 h of incubation and t is the time needed for color formation.
Assay optimization and validation for high-throughput screening use.
For high-throughput screening, BTH101 cultures containing the appropriate vectors were grown under semianaerobic conditions in 96-well plates in TGYEP-medium (1% casein, 0.5% yeast, 1.2% glucose, 0.1 M potassium phosphate buffer, pH 6.5; modified from reference 9).
The Z′ factor for the assay was calculated to assess the variability of the method and the difference between positive and negative controls {Z′ factor = 1 − [3 × (σp + σn)]/|μp − μn|, where σ is the standard deviation of positive (p) and negative (n) controls and μ is the mean of positive (p) and negative (n) controls} (52). Briefly, 96-well plates were divided into two parts; the positive control, BTH101 carrying plasmids pUT18CB8 and pKT25B8, was placed in 48 wells, and the negative control, BTH101 carrying plasmids pKT25B8 and pUTC18, was placed in the remaining 48 wells, followed by incubation without shaking for 12 h at 37°C; the β-galactosidase activities were determined; and the Z′ factor was calculated.
High-throughput screening.
Plasmids pUT18CB8 and pKT25B8 (positive control) and pKT25B8 and pUTC18 (negative control) were freshly transformed into strain BTH101 and used to inoculate the overnight cultures for screening in TGYEP medium (100 μg/ml ampicillin and 50 μg/ml kanamycin). The 96-well plates were prefilled in the wells of columns 2 to 11 with 3 μl of 1 mM the different compound solutions to be tested dissolved in dimethyl sulfoxide (DMSO) (15 μM final concentration); the wells of columns 1 and 12 were filled with 3 μl of pure DMSO and were used for the negative and positive controls without compounds. The loading of the samples was conducted with a Biomek FX liquid handler at the McMaster University HTS laboratory from premade compound-containing plates, and all assays were performed in duplicate.
TGYEP medium supplemented with 1 mM isopropyl-β-d-thiogalactopyranoside and antibiotics as described above was inoculated with the appropriate overnight cultures to an OD600 of 0.05 and immediately added in a volume of 0.2 ml to the predetermined wells of 96-well plates with the compounds to give a 15 μM final concentration or with 3 μl of DMSO for positive and negative controls. OD readings of the 96-well plates at 420, 550, and 600 nm were subsequently collected in the SpectramaxPlus384 Microplate Spectrophotometer Reader (Molecular Devices). The plates were incubated standing at 37°C for 12 h, and 20 μl of the cultures was subsequently transferred to two new empty 96-well plates, followed by determination of the β-galactosidase activity, as described above, in duplicate.
The data analysis was performed in Microsoft Excel. The wells containing the positive and negative controls were used to determine the relative activities per plate and to calculate the activity values for each well containing the compounds. The Z factor (52) for each plate was calculated to assess the variability of the assay per plate. The Miller unit data obtained from each well were calculated as an absolute percentage of the residual activity. The residual activities of the plate and of the replica plate were plotted against each other, and the hit zone was estimated as the total average minus the standard deviation.
Quantitative RT-PCR analysis.
Wild-type B. abortus 2308 was grown as described above to express the virB genes. After a 5-h incubation, RNA was isolated using the Ribo-Pure Bacteria kit from Ambion. Briefly, the bacteria were resuspended in 350 μl RNAwiz. The suspension was then added to 0.5 ml zirconium beads and shaken on a vortex for 45 min to break open the bacteria. After centrifugation, the supernatant was transferred to a new tube, and 0.2 volume of chloroform was added. This suspension was mixed very well, and after a 10-min incubation, the two phases were separated by centrifugation. The aqueous phase was transferred to a new tube and 0.5 volume of 100% ethanol was added. This solution was put onto a column, which was washed with 700 μl wash solution 1 and two times with 500 μl wash solution 2/3 by centrifugation. The RNA was eluted two times in 25 μl water. Before cDNA synthesis, the RNA was first treated twice with DNase and then cleaned up using Qiagen's RNeasy MinElute cleanup kit. For the quantitative reverse transcription-PCR (qRT-PCR), cDNA was synthesized from 500 ng of total RNA, and gene expression was determined with primers for virB1 (RTvirB-F, 5′-TGCCATTCCTTGTCCTCGC-3′; RTvirB-R, 5′-CCCACCAACGACGCCTATTG-3′), virB6 (RTvirB6-F, 5′-TATGATTGCCACGACTGCTGTC-3′; RTvirB6-R, 5′-ATGACGAGTTGCGAAAACGG-3′), virB11 (RTvirB11F, 5′-CAGGCAAGACCACACTGATGAAAG-3′; RTvirB11R, 5′-CGCTTCGCTCGGATAAAACAG-3′), vjbR (RTvjbR-F, 5′-TCTTTTCATCGCCGTATCCG-3′; RTvjbR-R, 5′-GGTCTTCGTCCTCCACCTCG-3), bcsp31 (RTbcsp31-F, 5′-ATGATGGCAAGGGCAAGGTG-3′; RTbcsp31-R, 5′-CTGCGACCGATTTGATGTTTG-3′), and gyrA (RTgyrA-F, 5′-TGATGCCCTCGTGCGTATG-3′; RTgyrA-R, 5′-TTCCGTGACCTTTTCCAGACG-3′). The fold change over untreated B. abortus 2308 was calculated.
Macrophage cytotoxicity.
The percentage of J774 death was determined by measuring lactate dehydrogenase (LDH) release using the CytoTox 96 nonradioactive cytotoxicity assay kit (Promega, Madison, WI) following instructions provided by the manufacturer.
Tissue culture and Brucella infection assays.
The mouse macrophage-like cell line J774A.1 was cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Rockville, MD) supplemented with 10% heat-inactivated fetal bovine serum and 1% nonessential amino acids (DMEMsup), and macrophage infection assays (gentamicin protection) were conducted as described previously (20, 21). All experiments were performed independently in triplicate at least three times, and the standard error for each time point was calculated.
Protein biochemical work.
VirB8 and the dimer site variant VirB8M102R were overproduced and purified as described previously (40). Analytical ultracentrifugation to quantify the effects of inhibitors on dimerization was carried out in a Beckman Coulter XL-A analytical ultracentrifuge with three concentrations of VirB8 (A280 = 0.1, 0.3, and 0.5) in a total volume of 120 μl at 4°C in the presence of a stoichiometric (1:1) ratio of inhibitor to protein concentration (2.6, 7.9, and 13.2 μM) at three different rotor speeds (20,000, 24,000, and 28,000 rpm). All experiments were conducted in 1% DMSO, which was used to dissolve the inhibitor. The data were analyzed as described above using Origin 6.0 (Microcal) and the Sedenterp program.
RESULTS
Adaptation of the bacterial two-hybrid assay and high-throughput screening for the isolation of inhibitors.
Analysis of the X-ray structures of the periplasmic domains of Brucella suis and A. tumefaciens C58 VirB8 proteins suggested that they form dimers, and structure-function analyses showed that the predicted dimer interfaces are very important for T4S system function in these pathogens (44; Sivanesan and Baron, submitted) (note that VirB8 proteins are identical in all Brucella species). Based on these results, we concluded that dimer formation may be a suitable target for small-molecule inhibitors of T4S, and we established a BTH system-based assay for the dimerization of Brucella VirB8 in the periplasm (44). The BTH assay makes use of adenylate cyclase (AC), which is not active when expressed as two fragments (T18 and T25) in the cytoplasm of Escherichia coli strain BTH101 lacking the cyaA gene encoding AC. Enzyme activity is restored if these fragments are brought into close proximity by fusion to interacting proteins, such as full-length VirB8 fused to the C termini of T18 and T25, which places the C terminus of VirB8 into the periplasm. The cyclic AMP (cAMP) signal cascade subsequently activates the lacZ promoter, and the β-galactosidase activity can be quantified. Compared to the yeast two-hybrid system used by others to measure VirB8 interactions (18), this assay places the C terminus of the protein in the periplasm of a Gram-negative cell, and this localization reflects the natural environment of the target VirB8. We reasoned that inhibitors identified using this approach would have a higher likelihood to be effective in bacteria than those identified using alternative in vivo or in vitro approaches. The BTH assay also monitors the dimerization of VirB10 and the formation of VirB8-VirB10 heterodimers, which served as controls for inhibitor specificity during the course of the study (44).
As a first step, the assay was adapted for HTS. To this end, we modified the medium from the LB originally used to TGYEP medium, which better supported growth and assay reproducibility under semianaerobic conditions in the 96-well microtiter plates used for screening. We next determined the Z′ factor, a statistical measure to assess the suitability of assays for the HTS format (52). To this end, we performed 1,920 assays (960 positive and 960 negative controls) in 20 microtiter plates, and the Z′ factor was 0.59, indicative of acceptable signal-to-noise and signal-to-background ratios in the compound and control wells, respectively. We next screened the Canadian Compound Collection of 29,567 compounds (http://www.ccbn-rcbc.ca) in duplicate, and the results of the β-galactosidase assays are shown in Fig. 1. When the cutoff was set to 50% (2 standard deviations below the positive-control mean), 345 molecules were identified as primary hits. Control assays were conducted next in order to determine which of these molecules were specific for VirB8 interactions.
FIG. 1.
High-throughput screening of the Canadian Compound Collection for inhibitors of VirB8 dimerization. E. coli strain BTH101 expressing VirB8 as a fusion to the T18 and T25 domains of adenylate cyclase was cultivated in 96-well microtiter plates in TGYEP medium, and the β-galactosidase activities reflecting this protein-protein interaction were determined in the absence and in the presence of 29,567 molecules from the Canadian Compound Collection. Shown is a plot of the two screening replicates reported as percent residual activity relative to the average of the high controls. Hits were identified as molecules reducing the activity to less than 50% in both replicates (shown in gray), a statistical cutoff 2 standard deviations below the high control mean.
Identification of VirB8 interaction-specific inhibitors.
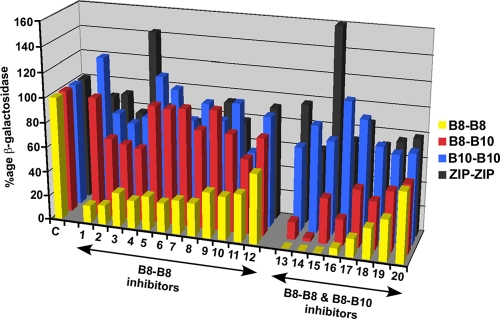
The BTH assay is based on the restoration of interactions between the T18 and T25 domains of AC in the cytoplasm, enabling cAMP formation and signal transduction to activate the lacZ gene, followed by β-galactosidase-mediated conversion of a chromogenic substrate for readout. This signal cascade contains multiple possible targets for small-molecule inhibitors, and in order to minimize false positives, we assessed the abilities of the compounds to inhibit the VirB8-VirB8 interaction, as well as VirB8-VirB10, VirB10-VirB10, and interactions between two leucine zippers. Of the 345 primary hits, 145 inhibited cell growth, 102 did not show reproducible inhibition, and 15 inhibited all four assays; all these molecules were excluded from further analysis. Twenty-eight molecules inhibited both VirB8-VirB8 and VirB8-VirB10 assays, and 20 inhibited only the VirB8-VirB8 assay; detailed inhibition data for these 48 molecules are presented in Table S1 in the supplemental material. The effects of the 20 most potent molecules, named B8I-1 to B8I-20, which were analyzed further as explained below, are shown in Fig. 2. While the inhibition of VirB8 dimerization was our primary target, small-molecule binding to VirB8 may impact its conformation and thereby also impact the VirB8-VirB10 interaction, as VirB10 is believed to be closely associated with VirB8 in the T4S apparatus (18, 25). We therefore considered the molecules that inhibited both VirB8 interactions potentially interesting. In contrast, we excluded molecules with impacts on VirB10 dimerization and the leucine zipper assay from further analysis, as they likely target components of the cAMP signal transduction pathway. Most of the compounds comprise aromatic structures, and they belong to different chemical classes, suggesting that the inhibitory potential is not linked to a specific family of molecules (Fig. 3; the nomenclature is explained in Table 1).
FIG. 2.
Controls of the specificity of VirB8 interaction inhibitors. E. coli strain BTH101 expressing fusions of VirB8 or VirB10 or of interacting leucine zippers (ZIP) to the T18 and T25 domains of adenylate cyclase was cultivated in 96-well microtiter plates in TGYEP medium, and the β-galactosidase activities reflecting the protein-protein interactions were determined in the absence (C [control]) and presence of 20 inhibitors that specifically inhibited VirB8-VirB8 or VirB8-VirB8 and VirB8-VirB10, but not VirB10-VirB10 and ZIP-ZIP interactions (the identities of the molecules are given in Table 1). Averages from 8 replicates are shown, and the standard deviations are not shown for clarity.
FIG. 3.
Chemical structures of the 20 VirB8 interaction inhibitors chosen for further analysis after specificity tests.
TABLE 1.
Compounds identified in the study, with commercial supplier and impacts on VirB8-VirB8 and VirB18-VirB10 interaction determined by the BTH assay
| Compound name | Supplier |
Inhibition of interaction ofa: |
||
|---|---|---|---|---|
| Code | Name | VirB8-VirB8 | VirB8-VirB10 | |
| B8I-1 | 5175083 | Chembridge | + | − |
| B8I-2 | 5367227 | Chembridge | + | + |
| B8I-3 | 150035 | Sigma | + | + |
| B8I-4 | BTB01692 | Maybridge | + | + |
| B8I-5 | SPB05636 | Maybridge | + | + |
| B8I-6 | 5141291 | Chembridge | + | − |
| B8I-7 | 5309022 | Chembridge | + | − |
| B8I-8 | 5105343 | Chembridge | + | − |
| B8I-9 | 5104971 | Chembridge | + | − |
| B8I-10 | 5352775 | Chembridge | + | + |
| B8I-11 | 5238271 | Chembridge | + | − |
| B8I-12 | 5380602 | Chembridge | + | + |
| B8I-13 | 5102966 | Chembridge | + | − |
| B8I-14 | 5119025 | Chembridge | + | − |
| B8I-15 | 5107400 | Chembridge | + | − |
| B8I-16 | 5108183 | Chembridge | + | + |
| B8I-17 | 5146442 | Chembridge | + | − |
| B8I-18 | XBX00150 | Maybridge | + | − |
| B8I-19 | SEW05753 | Maybridge | + | − |
| B8I-20 | GK02122 | Maybridge | + | + |
+, inhibition; −, no inhibition.
Assays of toxicity for Brucella and mammalian cells.
We next wanted to determine whether the VirB8 interaction inhibitors impact the T4S complex in its natural environment in Brucella and during infection assays. To exclude molecules that inhibit bacterial growth or the viability of mammalian cells, we tested the 19 (molecule B8I-5 was no longer available from the supplier) most potent inhibitors of VirB8 interactions first for cytotoxicity against mammalian cells using a lactate dehydrogenase release assay. This approach revealed that 4 compounds were highly toxic at concentrations as low as 12.5 μM, and they were excluded from further analysis (Table 2). Second, we tested the impact of the VirB8 inhibitors on the growth of B. abortus 2308 in rich TSB and mammalian cell cultivation medium (DMEM), which is used for infections. With the exception of compound B8I-3, which reduced bacterial growth only in DMEM, none of the compounds inhibited growth (Table 3). The 15 molecules that were not toxic for mammalian cells were then tested for their impacts on the T4S system in Brucella and on its intracellular replication in J774 macrophages.
TABLE 2.
Toxicities of VirB8 interaction inhibitors against mammalian cellsa
| Compound | Concn (μM) | % Cytotoxicity |
|---|---|---|
| DMSO | NA | 0 |
| B8I-1 | 50 | 96 |
| 25 | 93 | |
| 12.5 | 2 | |
| 5 | 0 | |
| B8I-2 | 50 | 9 |
| 25 | 0 | |
| 12.5 | 0 | |
| 5 | 0 | |
| B8I-3 | 50 | 20 |
| 25 | 0 | |
| 12.5 | 0 | |
| 5 | 0 | |
| B8I-4 | 50 | 25 |
| 25 | 19 | |
| 12.5 | 11 | |
| 5 | 0 | |
| B8I-6 | 50 | 17 |
| 25 | 10 | |
| 12.5 | 13 | |
| 5 | 14 | |
| B8I-7 | 50 | 100 |
| 25 | 100 | |
| 12.5 | 100 | |
| 5 | 100 | |
| B8I-8 | 50 | 13 |
| 25 | 17 | |
| 12.5 | 14 | |
| 5 | 12 | |
| B8I-9 | 50 | 7 |
| 25 | 6 | |
| 12.5 | 8 | |
| 5 | 8 | |
| B8I-10 | 50 | 12 |
| 25 | 10 | |
| 12.5 | 14 | |
| 5 | 15 | |
| B8I-11 | 50 | 53 |
| 25 | 36 | |
| 12.5 | 16 | |
| 5 | 13 | |
| B8I-12 | 50 | 9 |
| 25 | 8 | |
| 12.5 | 8 | |
| 5 | 6 | |
| B8I-13 | 50 | 100 |
| 25 | 100 | |
| 12.5 | 100 | |
| 5 | 27 | |
| B8I-14 | 50 | 4 |
| 25 | 0 | |
| 12.5 | 0 | |
| 5 | 0 | |
| B8I-15 | 50 | 50 |
| 25 | 16 | |
| 12.5 | 0 | |
| 5 | 0 | |
| B8I-16 | 50 | 0 |
| 25 | 1 | |
| 12.5 | 0 | |
| 5 | 0 | |
| B8I-17 | 50 | 16 |
| 25 | 8 | |
| 12.5 | 4 | |
| 5 | 3 | |
| B8I-18 | 50 | 53 |
| 25 | 50 | |
| 12.5 | 48 | |
| 5 | 37 | |
| B8I-19 | 50 | 42 |
| 25 | 45 | |
| 12.5 | 49 | |
| 5 | 0 | |
| B8I-20 | 50 | 10 |
| 25 | 17 | |
| 12.5 | 0 | |
| 5 | 0 |
Determined using the cytotox96 kit (Promega). The averages of results from parallel experiments in three wells are shown. The DMSO used to dissolve the compounds was not toxic, and values were set to 0%. NA, not applicable.
TABLE 3.
Toxicities of VirB8 interaction inhibitors at concentrations not toxic for mammalian cells against B. abortus 2308
| Compound | Concn (μM) | Toxicitya |
|
|---|---|---|---|
| TSB | DMEM | ||
| DMSO | NA | 1.00 | 1.00 |
| B8I-1 | 12.5 | 0.99 | 1.00 |
| B8I-2 | 12.5 | 1.00 | 1.01 |
| B8I-3 | 12.5 | 1.00 | 0.84 |
| B8I-3 | 5 | 1.00 | 0.96 |
| B8I-4 | 5 | 1.00 | 0.99 |
| B8I-6 | 25 | 0.99 | 0.99 |
| B8I-7 | 5 | 1.00 | 0.99 |
| B8I-8 | 12.5 | 1.00 | 0.99 |
| B8I-9 | 50 | 1.00 | 0.98 |
| B8I-10 | 50 | 1.00 | 0.99 |
| B8I-11 | 12.5 | 0.98 | 1.00 |
| B8I-12 | 50 | 1.01 | 1.00 |
| B8I-14 | 12.5 | 1.00 | 0.99 |
| B8I-15 | 12.5 | 0.99 | 1.01 |
| B8I-16 | 12.5 | 1.02 | 1.04 |
| B8I-17 | 12.5 | 1.00 | 1.01 |
| B8I-19 | 5 | 0.99 | 0.98 |
| B8I-20 | 12.5 | 0.99 | 0.99 |
B. abortus 2308 was grown in rich medium (TSB) or in DMEM. The ratios of surviving bacteria grown in the presence of VirB8 interaction inhibitors to bacteria grown in their absence are shown (averages of duplicate assays). NA, not applicable.
VirB8 interaction inhibitors reduce VirB protein levels and virB gene transcription.
The levels of many VirB proteins are reduced in a virB8 gene deletion mutant of B. abortus 2308 (20), and this may be due to the destabilization of these proteins when the complex does not assemble properly in the absence of VirB8. The inhibition of VirB8 dimerization may similarly impact T4S complex stability, and to assess this possibility, B. abortus 2308 was cultivated in virulence gene-inducing minimal medium in the presence of VirB8 interaction inhibitors, and VirB protein levels were analyzed by Western blotting. Cultivation in the presence of 10 of the inhibitors strongly reduced the amounts of VirB5, VirB9, VirB10, and VirB11 (Fig. 4), which is similar to observations made in the virB8 gene deletion strain (20), and there was no effect on the levels of a control protein that is not involved in T4S (Bcsp31). Interestingly, these molecules also reduced the amounts of VirB8, suggesting that the inhibition of dimerization may destabilize the protein or that they have additional effects, e.g., on virB operon expression. To assess the latter possibility, we measured the impacts of these molecules on virB operon transcription using a strain carrying a chromosomal virB-lacZ fusion. We observed a reduction of transcription to levels of 30% to 60% of the DMSO control in the case of nine molecules (B8I-2, B8I-3, B8I-8, B8I-9, B8I-11, B8I-12, B8I-14, B8I-16, and B8I-17), which in some cases was as pronounced as that by a C12-homoserine lactone (HSL) (30% of control), a known inhibitor of virB operon transcription (reference 19 and data not shown).
FIG. 4.
Impacts of VirB8 interaction inhibitors on VirB protein levels in B. abortus 2308. Brucella was cultivated in a virulence gene-inducing minimal medium in the presence of small molecules, as indicated. C, DMSO control; HSL, C12-homoserine lactone; and VirB8 interaction inhibitors (B8I- plus numbers above lanes). B. abortus cells were lysed, followed by SDS-PAGE and Western blotting with specific antisera as indicated.
To gain more detailed insights into the effects on transcription, we compared the effects of C12-HSL and of the most potent molecule, B8I-2, on virB gene transcription by quantitative RT-PCR. After cultivation of B. abortus 2308 in the presence of C12-HSL (5 μM) and B8I-2 (10 μM), RNA was isolated and analyzed by quantitative RT-PCR. This analysis revealed that C12-HSL and B8I-2 have strongly negative effects on virB gene levels as measured with probes specific for virB1, virB6, and virB11 (Fig. 5). Gyrase mRNA levels that did not change were taken as negative standards, and similarly, the levels of mRNA encoding the control protein Bcsp31 did not change significantly. Interestingly, we noticed that both C12-HSL and B8I-2 modestly reduced the levels of vjbR mRNA, albeit not as strongly as virB mRNA levels, suggesting that quorum-sensing regulation may be involved in the effects of B8I-2 on virB transcription. The above results show that many of the VirB8 dimerization inhibitors have strong negative effects on the T4S system in Brucella, and we next tested whether they impact bacterial virulence in an infection model.
FIG. 5.
Impacts of B8I-2 and C12-HSL on transcription in B. abortus 2308. Brucella was cultivated in virulence gene-inducing minimal medium in the presence of small molecules as indicated. qRT-PCR was performed using primers for virB1, virB6, virB11, vjbR, gyrA, and bcsp31. The fold change was calculated over untreated Brucella. B8I-2, 10 μM; C12-HSL, 5 μM.
VirB8 interaction inhibitor B8I-2 strongly reduces intracellular survival of Brucella in macrophages.
To determine their T4S-dependent capacity for intracellular survival, B. abortus 2308 was cocultivated with macrophages for 20 min. After this cocultivation, extracellular bacteria were washed away with phosphate-buffered saline (PBS) and growth medium containing gentamicin, and the compounds were added to each well. Survival was determined after 48 h of infection by lysis of the macrophages and plating to determine the number of intracellular bacteria. The inhibitors were added, together with gentamicin, at concentrations of 12.5 and 5 μM, respectively. In the absence of inhibitors, we observed a 5-log-unit difference between wild-type bacteria and either a T4S mutant or the wild type treated with the virB gene transcription inhibitor C12-homoserine lactone (5 μM) (Fig. 6 A). Whereas most inhibitors did not significantly impact intracellular replication, molecule B8I-2 had a very significant negative effect (Table 4) on growth, as measured after 48 h. Tests with a larger number of concentrations revealed a 4-log-unit reduction of Brucella proliferation at concentrations of 8 and 12.5 μM, respectively (Fig. 6A and Table 4) (P = 0.00001 for 5 μM; P = 0.000003 for 12.5 μM). A time course study showed that the addition of B8I-2 as late as 2 h after infection inhibited intracellular replication measured after 48 h, but the strongest effects were observed when it was added during infection, suggesting that the molecule impacts an early event of the infection process (Fig. 6B). To gain more detailed insights into the timing of inhibitor action, we measured the kinetics of intracellular replication 1, 5, 24, and 48 h after infection, showing that B8I-2 inhibits replication between 5 and 24 h (Fig. 6C and Table 5). Microscopic observations and assays of lactate dehydrogenase release into the culture supernatant showed that the presence of the inhibitor does not increase cell lysis in infected cells (Table 5). The time course of infection in the presence of B8I-2 is very similar to that of virB gene deletion mutants (20, 21), suggesting that the effect is due to direct effects on the secretion system. Interestingly, B8I-2 is a salicylidene acylhydrazide (Fig. 6D) and therefore similar to members of a class of compounds that were shown to be active against a wide variety of T3S systems in different bacteria (34).
FIG. 6.
Effects of inhibitor B8I-2 on the intracellular replication of B. abortus 2308. (A) Replication of the Brucella wild type (wt) and virB deletion mutant (ΔvirB) inside J774 macrophages and effects when C12-homoserine lactone (5 μM) and B8I-2 at the concentrations shown were added at the end of the cocultivation period. (B) Replication of the Brucella wild type and virB deletion mutant inside J774 macrophages and effects when B8I-2 (10 μM) was added at the end of the cocultivation period, after the wash with fresh medium, or at the times shown after the elimination of extracellular bacteria with gentamicin. (C) Time course of effects of B8I-2 on intracellular survival and replication. B8I-2 was added at 10 μM at the beginning of the assay and maintained throughout the 48-h period. The data shown are compiled from two independent assays, each performed in triplicate. (D) Structure of B8I-2, a salicylidene acylhydrazide. The error bars represent the standard deviations; the asterisks indicate significance at a P value of <0.05.
TABLE 4.
Impacts of VirB8 interaction inhibitors on intracellular replication in macrophagesa
| Compound | Concn (μM) | Avg replication | SD |
|---|---|---|---|
| DMSO | NA | 4.98 | 0.20 |
| C12-HSL | 5 | 0.17 | 0.41 |
| B8I-1 | 12.5 | 5.18 | 0.31 |
| B8I-1 | 5 | 5.36 | 0.20 |
| B8I-2 | 12.5 | 1.17 | 1.61 |
| B8I-2 | 8 | 1.17 | 1.83 |
| B8I-2 | 5 | 3.85 | 0.23 |
| B8I-2 | 4 | 4.09 | 0.37 |
| B8I-2 | 1 | 4.84 | 0.26 |
| B8I-3 | 12.5 | 5.56 | 0.25 |
| B8I-3 | 5 | 5.44 | 0.31 |
| B8I-4 | 5 | 4.61 | 0.39 |
| B8I-6 | 12.5 | 4.46 | 0.40 |
| B8I-6 | 5 | 4.39 | 0.44 |
| B8I-8 | 12.5 | 3.99 | 0.60 |
| B8I-8 | 5 | 4.33 | 0.57 |
| B8I-9 | 12.5 | 4.74 | 0.53 |
| B8I-9 | 5 | 4.69 | 0.46 |
| B8I-10 | 12.5 | 4.87 | 0.44 |
| B8I-10 | 5 | 4.71 | 0.51 |
| B8I-11 | 12.5 | 3.99 | 0.67 |
| B8I-11 | 5 | 4.48 | 0.73 |
| B8I-12 | 12.5 | 4.23 | 0.85 |
| B8I-12 | 5 | 4.13 | 0.41 |
| B8I-14 | 12.5 | 4.73 | 0.60 |
| B8I-14 | 5 | 4.33 | 0.83 |
| B8I-15 | 12.5 | 5.15 | 0.33 |
| B8I-15 | 5 | 5.18 | 0.26 |
| B8I-16 | 12.5 | 5.13 | 0.05 |
| B8I-16 | 5 | 5.16 | 0.07 |
| B8I-17 | 12.5 | 5.13 | 0.20 |
| B8I-17 | 5 | 5.18 | 0.24 |
| B8I-20 | 12.5 | 5.53 | 0.27 |
Intracellular growth of B. abortus 2308 after 48 h was quantified by lysis of macrophages, dilution, and plating (replication is shown on a logarithmic scale). Averages from three replicates with three wells per replicate are shown. NA, not applicable.
TABLE 5.
Time course showing the impacts of inhibitor B8I-2 at 10 μM on intracellular replication in macrophages and on cell survival
| Time (h) | Presence of B8I-2 (10 μM)b | Avg replicationa | SD | % Cytotoxicity |
|---|---|---|---|---|
| 1 | − | 4.60 | 0.11 | 0 |
| 1 | + | 4.54 | 0.11 | 0 |
| 5 | − | 4.38 | 0.09 | 0 |
| 5 | + | 4.63 | 0.15 | 0 |
| 24 | − | 3.56 | 0.19 | 0 |
| 24 | + | 1.24 | 0.69 | 0 |
| 48 | − | 5.31 | 0.20 | 0 |
| 48 | + | 1.91 | 1.78 | 0 |
Intracellular growth of B. abortus 2308 was quantified by lysis of macrophages, dilution, and plating (replication is shown on a logarithmic scale). The toxicity of B8I-2 was assessed using the cytotox96 kit (Promega). Averages from two replicates with three wells per replicate are shown.
+, present; −, absent.
To assess whether potent VirB8 interaction inhibitors have similar broad-spectrum activity against other T4S systems, we tested the impact of the qualitatively most potent VirB8 interaction inhibitors, B8I-1 to B8I-5, on the T4S system-mediated conjugative transfer of plasmids RP4 and pKM101 between E. coli cells. None of the molecules impacted the conjugative transfer at concentrations as high as 50 μM, showing that, unlike some T3S system inhibitors, they do not have broad-spectrum activity against T4S (results not shown).
The VirB8 interaction inhibitor B8I-2 inhibits dimerization in vitro.
Since the experiments described above raise some questions about the mechanism of inhibitor action, we verified whether binding of inhibitor B8I-2 impacts the dimerization of VirB8, the primary target in the BTH assay. VirB8 and the dimer site variant VirB8M102R were overproduced and purified, and their dimerization was analyzed by analytical ultracentrifugation as in previous work (40). The apparent dissociation constant (Kd) was calculated to be 144 μM for wild-type VirB8, whereas the dimer site variant VirB8M102R was a monomer (Fig. 7), which is very similar to the data we previously reported (40). In contrast, when the experiment with VirB8 was carried out in the presence of stoichiometric amounts of B8I-2 (1:1 ratio), the protein was also present as a monomer (Fig. 7). These results are consistent with the effect of B8I-2 in the BTH assay and suggest that it inhibits the dimerization of VirB8.
FIG. 7.
Sedimentation equilibrium analysis of the dimerization of VirB8. Analytical ultracentrifugation to analyze the self-association of wild-type VirB8 alone (WT) or in the presence of B8I-2 (WT + B8I-2) and of the dimer site variant VirB8M102R (M102R). The lower graphs show a representative fit of the experimental data to a monomer/dimer model in the case of VirB8 and to a single-species (i.e., monomer) model in the case of VirB8 plus B8I-2 and M102R. The upper graphs show the residuals of the fit. The representative fits shown here were taken from the data obtained for proteins at an A280 of 0.5 and rotor speeds of 20,000, 24,000, and 28,000 rpm in a Beckman Coulter XL-A analytical ultracentrifuge.
DISCUSSION
Here, we present the results of a structure-inspired HTS approach aimed at identification of T4S inhibitors targeting the dimerization of one of its key components, VirB8. The amino acids at the dimer interface were shown to be important for virulence in B. abortus 2308 (40), and more recent work in the A. tumefaciens systems further substantiated the importance of this region of the protein for T4S (Sivanesan and Baron, submitted). In contrast to other groups that identified T3S inhibitors using cell-based screens, we used a structurally well-characterized target, and this allowed us to identify compounds that interfere with an interaction required for its function. This approach with a well-characterized target may facilitate future development of the small molecules through iterations of chemical synthesis, followed by functional assays and structural analysis of VirB8-inhibitor complexes.
The HTS approach identified several hundred potential inhibitors, and follow-up controls eliminated most compounds, with only 48 acting as specific inhibitors of VirB8 interactions. This reduction in the number of molecules is typical for screening projects, which tend to discover a large number of primary hits but eliminate 90% or more of the molecules in follow-up controls. Our choice of a cell-based screen using an assay in Gram-negative bacteria was aimed at isolating molecules that do not inhibit vital cell functions in bacteria, and it was therefore not surprising that VirB8 interaction inhibitors were generally not toxic for Brucella. However, only one of the molecules (B8I-2) had a significant impact on the intracellular growth of B. abortus 2308 in macrophages, suggesting that the other molecules may not be sufficiently potent or that they may not be able to penetrate into mammalian cells. We did not measure penetration into mammalian cells in the context of this work, but the analysis of the negative impact of VirB8 interaction inhibitors on VirB protein levels and virB operon transcription suggests that many of them reach their target in B. abortus. A notable exception was molecule B8I-1, which inhibited the VirB8 interaction in the BTH assay. In contrast to B8I-2 and many other molecules, B8I-1 did not reduce VirB protein levels in B. abortus, suggesting that it does not penetrate into the cells. Alternatively, the inhibitor may be able to bind to VirB8 alone as in the BTH assay, but not in the context of an assembled T4SS in which VirB8 interacts with other proteins. This explanation may apply equally to other molecules that are active in the BTH assay but not effective in Brucella. It will be interesting in future to modify this molecule and others by chemical synthesis in order improve their potency as inhibitors and their ability to penetrate into cells.
The molecule B8I-2 is the most promising inhibitor resulting from these studies, and several features underline this conclusion. First, it inhibited intracellular growth of B. abortus 2308 in J774 macrophages at relatively low μM concentrations, which compares very advantageously with the secretion system inhibitors isolated by others, which require high μM concentrations for biological effects. Second, B8I-2 is a salicylidene acylhydrazide and is structurally similar to a class of molecules that have broad-spectrum activity against T3S systems (34). This similarity may be fortuitous, as benzylidene acylhydrazides are common building blocks in chemical libraries generated for HTS experiments, and T3S systems do not contain VirB8 homologs that could serve as targets. Also, the most active molecules isolated here did not have broad-spectrum activity against other T4S systems, arguing for a specific target interaction. However, the T3S target could have structural similarities to VirB8, and it will therefore be interesting to test the effect of B8I-2 on the T3S and, reciprocally, to test the effect of T3S inhibitors on VirB8 interactions and T4S system function. Third, the observation that B8I-2 and several other VirB8 interaction inhibitors reduce virB operon transcription in B. abortus 2308 is intriguing, since similar observations were reported in the case of salicylidene acylhydrazide T3S inhibitors (33, 38). However, in contrast to the active T3S inhibitors, the 20 molecules that are VirB8 interaction inhibitors in the BTH assay described here belong to different chemical classes, and they were all identified based on their specific activity against one common target, VirB8. Therefore, it is very unlikely that they all inhibit different alternative targets with the common result of reduced transcription of the virB operon in B. abortus 2308. A more likely explanation for the effect on virB gene transcription is that the inhibition of dimerization inhibits VirB8 interactions (VirB8-VirB8 and in many cases also VirB8-VirB10), and as a consequence, other T4SS components do not assemble properly and are destabilized. The accumulation of misfolded and improperly assembled proteins may induce a feedback loop, e.g., similar to the envelope stress response system, such as CpxA/CpxR, and this response may reduce virB gene transcription. A CpxA/CpxR system regulates Dot/Icm components and T4S substrates in Legionella (1), but until now, little was known with regard to the implications of such systems for T4S-related processes in Brucella. The fact that C12-HSL and B8I-2 have qualitatively similar negative effects on virB gene transcription and that they modestly reduce levels of the vjbR mRNA suggests that the quorum-sensing regulon may be involved. In future studies, it would be interesting to conduct comparative transcriptome analysis to assess whether VirB8 interaction inhibitors and HSLs drive similar responses. The regulation of virB operon transcription is complex, and nutrient deprivation and quorum sensing are the best-characterized effectors (42). The work described here may lead to the discovery of another regulatory circuit and thereby advance understanding of virulence gene regulation in Brucella.
In conclusion, using a target-based HTS in a cell-based assay, we have isolated a series of potent hit molecules that may have potential as leads for the development of antivirulence drugs. The development of biochemical assays to measure target interaction and structural studies (analysis of VirB8 inhibitor complexes) are the next steps required on the path to drug development. Given the abundance of structural information on secretion systems, similar approaches to identify inhibitors of type III or other secretion systems should discover hit molecules that are suitable for leads and drug development in future. In addition to their potential as drugs, secretion system inhibitors could prove to be valuable tools for investigating the mechanisms of secretion systems or the time course of T4S activity during infection (14).
Supplementary Material
Acknowledgments
We thank the coworkers of the McMaster HTS laboratory, especially Jan Blanchard and Cecilia Murphy, for assistance with high-throughput screening and consultation. We thank Nadeem Siddiqui for assistance with AUC experiments that were conducted at the Biophysical platform at the Institute for Research in Immunology and Cancer (IRIC), Université de Montréal.
Work in the laboratory of C.B. is supported by grants from the Canadian Institutes of Health Research (CIHR grants XNO-83099 and MOP-84239), the Bristol Myers-Squibb Chair Hans Selye and the Canada Foundation for Innovation (CFI), the Ontario Innovation Trust (OIT), and the FRSQ (Fonds de la Recherche en Santé du Québec). Work in R.T.'s laboratory is supported by PHS grant AI050553 from the NIH.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 20 December 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Altman, E., and G. Segal. 2008. The response regulator CpxR directly regulates expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J. Bacteriol. 190:1985-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ariza, J., et al. 2007. Perspectives for the treatment of brucellosis in the 21st century: the Ioannina recommendations. PLoS Med. 4:e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backert, S., and T. F. Meyer. 2006. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9:207-217. [DOI] [PubMed] [Google Scholar]
- 4.Baron, C. 2010. Antivirulence drugs to target bacterial secretion systems. Curr. Opin. Microbiol. 13:100-105. [DOI] [PubMed] [Google Scholar]
- 5.Baron, C. 2005. From bioremediation to biowarfare: on the impact and mechanism of type IV secretion systems. FEMS Microbiol. Lett. 253:163-170. [DOI] [PubMed] [Google Scholar]
- 6.Baron, C. 2006. VirB8: a conserved type IV secretion system assembly factor and drug target. Biochem. Cell Biol. 84:890-899. [DOI] [PubMed] [Google Scholar]
- 7.Baron, C., and B. Coombes. 2007. Targeting bacterial secretion systems: benefits of disarmament in the microcosm. Infect. Disord. Drug Targets 7:19-27. [DOI] [PubMed] [Google Scholar]
- 8.Bayliss, R., et al. 2007. NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system. Proc. Natl. Acad. Sci. U. S. A. 104:1673-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Begg, Y. A., J. N. Whyte, and B. A. Haddock. 1977. The identification of mutants of Escherichia coli deficient in formate dehydrogenase and nitrate reductase activities using dye indicator plates. FEMS Microbiol. Lett. 2:47-50. [Google Scholar]
- 10.Boschiroli, M. L., V. Foulongne, and D. O'Callaghan. 2001. Brucellosis: a worldwide zoonosis. Curr. Opin. Microbiol. 4:58-64. [DOI] [PubMed] [Google Scholar]
- 11.Cascales, E., and P. J. Christie. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Celli, J., and J. P. Gorvel. 2004. Organelle robbery: Brucella interactions with the endoplasmic reticulum. Curr. Opin. Microbiol. 7:93-97. [DOI] [PubMed] [Google Scholar]
- 13.Chandran, V., et al. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 29:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charpentier, X., et al. 2009. Chemical genetics reveals bacterial and host cell functions critical for type IV effector translocation by Legionella pneumophila. PLoS Pathog. 5:e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:415-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clatworthy, A. E., E. Pierson, and D. T. Hung. 2007. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3:541-548. [DOI] [PubMed] [Google Scholar]
- 17.Cowie, A., et al. 2006. An integrated approach to functional genomics: construction of a novel reporter gene fusion library for Sinorhizobium meliloti. Appl. Environ. Microbiol. 72:7156-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das, A., and Y.-H. Xie. 2000. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9 and VirB10 interact with one another. J. Bacteriol. 182:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delrue, R. M., et al. 2005. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell. Microbiol. 7:1151-1161. [DOI] [PubMed] [Google Scholar]
- 20.den Hartigh, A. B., H. G. Rolan, M. F. de Jong, and R. M. Tsolis. 2008. VirB3 to VirB6 and VirB8 to VirB11, but not VirB7, are essential for mediating persistence of Brucella in the reticuloendothelial system. J. Bacteriol. 190:4427-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.den Hartigh, A. B., et al. 2004. Differential requirements for VirB1 and VirB2 during Brucella abortus infection. Infect. Immun. 72:5143-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felise, H. B., et al. 2008. An inhibitor of gram-negative bacterial virulence protein secretion. Cell Host Microbe 4:325-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez-Lopez, R., et al. 2005. Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology 151:3517-3526. [DOI] [PubMed] [Google Scholar]
- 24.Fronzes, R., P. J. Christie, and G. Waksman. 2009. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7:703-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fronzes, R., et al. 2009. Structure of a type IV secretion system core complex. Science 323:266-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomis-Rüth, F. X., et al. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637-641. [DOI] [PubMed] [Google Scholar]
- 27.Hare, S., R. Bayliss, C. Baron, and G. Waksman. 2006. A large domain swap in the VirB11 ATPase of Brucella suis leaves the hexameric assembly intact. J. Mol. Biol. 360:56-66. [DOI] [PubMed] [Google Scholar]
- 28.Hilleringmann, M., et al. 2006. Inhibitors of Helicobacter pylori ATPase Cagalpha block CagA transport and cag virulence. Microbiology 152:2919-2930. [DOI] [PubMed] [Google Scholar]
- 29.Hughes, D. 2003. Exploiting genomics, genetics and chemistry to combat antibiotic resistance. Nat. Rev. Genet. 4:432-441. [DOI] [PubMed] [Google Scholar]
- 30.Hung, D. T., E. A. Shakhnovich, E. Pierson, and J. J. Mekalanos. 2005. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science 310:670-674. [DOI] [PubMed] [Google Scholar]
- 31.Judd, P. K., R. B. Kumar, and A. Das. 2005. Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc. Natl. Acad. Sci. U. S. A. 102:11498-11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karimova, G., Nathalie Dautin, and Daniel Ladant. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kauppi, A. M., R. Nordfelth, H. Uvell, H. Wolf-Watz, and M. Elofsson. 2003. Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem. Biol. 10:241-249. [DOI] [PubMed] [Google Scholar]
- 34.Keyser, P., M. Elofsson, S. Rosell, and H. Wolf-Watz. 2008. Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram-negative bacteria. J. Intern. Med. 264:17-29. [DOI] [PubMed] [Google Scholar]
- 35.Kulakov, Y. K., P. G. Guigue-Talet, M. R. Ramuz, and D. O'Callaghan. 1997. Response of Brucella suis 1330 and B. canis RM6/66 to growth at acid pH and induction of an adaptive acid tolerance response. Res. Microbiol. 148:145-151. [DOI] [PubMed] [Google Scholar]
- 36.Levy, S. B., and B. Marshall. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10:S122-S129. [DOI] [PubMed] [Google Scholar]
- 37.Muschiol, S., et al. 2006. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U. S. A. 103:14566-14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Negrea, A., et al. 2007. Salicylidene acylhydrazides that affect type III protein secretion in Salmonella enterica serovar typhimurium. Antimicrob. Agents Chemother. 51:2867-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nordfelth, R., A. M. Kauppi, H. A. Norberg, H. Wolf-Watz, and M. Elofsson. 2005. Small-molecule inhibitors specifically targeting type III secretion. Infect. Immun. 73:3104-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paschos, A., et al. 2006. Dimerization and interactions of Brucella suis VirB8 with VirB4 and VirB10 are required for its biological activity. Proc. Natl. Acad. Sci. U. S. A. 103:7252-7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patey, G., Z. Qi, G. Bourg, C. Baron, and D. O'Callaghan. 2006. Swapping of periplasmic domains between Brucella suis VirB8 and pSB102 VirB8 homologue allows heterologous complementation. Infect. Immun. 74:4945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rambow-Larsen, A. A., E. M. Petersen, C. R. Gourley, and G. A. Splitter. 2009. Brucella regulators: self-control in a hostile environment. Trends Microbiol. 17:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seleem, M. N., S. M. Boyle, and N. Sriranganathan. 2010. Brucellosis: a re-emerging zoonosis. Vet. Microbiol. 140:392-398. [DOI] [PubMed] [Google Scholar]
- 44.Sivanesan, D., M. A. Hancock, A. M. Villamil Giraldo, and C. Baron. 2010. Quantitative analysis of VirB8-VirB9-VirB10 interactions provides a dynamic model of type IV secretion system core complex assembly. Biochemistry 49:4483-4493. [DOI] [PubMed] [Google Scholar]
- 45.Terradot, L., et al. 2005. Crystal structures of the periplasmic domains of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 102:4596-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veenendaal, A. K., C. Sundin, and A. J. Blocker. 2009. Small-molecule type III secretion system inhibitors block assembly of the Shigella type III secreton. J. Bacteriol. 191:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walsh, C. 2003. Where will new antibiotics come from? Nat. Rev. Microbiol. 1:65-70. [DOI] [PubMed] [Google Scholar]
- 48.Yagupsky, P., and E. J. Baron. 2005. Laboratory exposures to brucellae and implications for bioterrorism. Emerg. Infect. Dis. 11:1180-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeo, H.-J., Q. Yuan, M. R. Beck, C. Baron, and G. Waksman. 2003. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc. Natl. Acad. Sci. U. S. A. 100:15947-15962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeo, H. J., S. N. Savvides, A. B. Herr, E. Lanka, and G. Waksman. 2000. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell 6:1461-1472. [DOI] [PubMed] [Google Scholar]
- 51.Young, E. J. 1995. An overview of human brucellosis. Clin. Infect. Dis. 21:283-289. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, J. H., T. D. Chung, and K. R. Oldenburg. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67-73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.