Abstract
Lipid A structure is a critical determinant of the interaction between pathogens and the innate immune system. Previously, we demonstrated the presence of non- and monophosphorylated tetra-acylated lipid A structures in the outer membrane of Porphyromonas gingivalis, an agent of human periodontal disease. These modifications to lipid A structure lead to evasion and suppression of innate defenses mediated by Toll-like receptor 4 (TLR4) and cationic antimicrobial peptides. In this investigation, we examined the influence of growth temperature on P. gingivalis lipid A structure and recognition by TLR4 as an example of an environmental influence which is known to vary between healthy and diseased sites in the periodontium. We demonstrate that P. gingivalis grown at a normal body temperature produces mainly nonphosphorylated and monophosphorylated tetra-acylated lipid A structures, whereas bacteria grown at 39°C and 41°C intended to mimic increasing levels of inflammation, producing increasing proportions of monophosphorylated, penta-acylated lipid A. The temperature-dependent alteration in lipid A renders the bacterium significantly more potent for activating TLR4 and more susceptible to killing by β-defensins 2 and 3. This is the first report of a lipid A remodeling system linked to temperature shifts associated with a deregulated inflammatory response. Temperature elevation at sites of inflammation in the periodontium may be a significant environmental regulator of the lipid A modification systems of P. gingivalis, which will influence the interaction of this organism with the innate host defense.
Lipopolysaccharide (LPS) is the major constituent of the outer leaflet of the outer membrane of all Gram-negative bacteria, where it plays an important structural role as well as mediates interactions with the environment. It consists of three regions, as follows: lipid A, which is embedded in the outer membrane and is responsible for the proinflammatory endotoxin properties of the molecule, and the core oligosaccharide and O-polysaccharide side chain, which both extend from the surface of the cell and mediate external interactions. Lipid A is considered to be an archetypal microbe-associated molecular pattern (MAMP), a molecular motif found in a range of microbes which the innate immune system recognizes as nonself through an extensive repertoire of evolutionary conserved receptors. Recognition of lipid A by Toll-like receptor 4 (TLR4) facilitates a robust proinflammatory response by the host immune system that promotes clearance of the bacteria from protected tissues (24). Furthermore, phosphate groups on lipid A bestow an overall negative charge to the outer surface of Gram-negative bacteria, which promotes the selective membrane binding of host cationic antimicrobial peptides and, hence, bacterial killing at mucosal epithelial surfaces (3).
Lipid A is a β-1,6-linked d-glucosamine disaccharide phosphorylated in the 1 and 4′ positions and N- and/or O-acylated at positions 2, 3, 2′, and 3′ with various amounts of fatty acids and 3-hydroxy fatty acids (20). Modifications to this basic lipid A structure are frequently observed (19). Remodeling may involve removal of one of the 1 or 4′ phosphate groups through specific lipid A phosphatases or modification of these groups with amine-containing substituents, which reduces the overall net negative charge and thereby lowers the affinity of cationic antimicrobial peptides. Structural modification also occurs through removal of acyl chains from the glucosamine disaccharide by specific lipid A deacylases to produce penta-, tetra- and occasionally triacylated forms, such as those in Pseudomonas aeruginosa and Helicobacter pylori (10, 23).
We have previously examined the lipid A composition of Porphyromonas gingivalis, which is considered to be an important agent in human periodontal disease—a chronic inflammatory condition of the supporting tissues of the teeth. These studies have demonstrated a very large degree of flexibility in the structure of lipid A in this bacterium: in addition to di- and monophosphorylated and penta- and tetra-acylated moieties (16), a spectrum of nonphosphorylated lipid A species was also present (21). Such modifications influence the potency of the interaction of P. gingivalis lipid A with the innate host defense. For example, nonphosphorylated forms of lipid A are highly unusual and appear to form the basis for the ability of P. gingivalis to disguise itself from recognition by the TLR4 branch of the innate immune system (4).
Alterations to P. gingivalis lipid A structure are influenced by the concentration of environmental hemin, an essential micronutrient for the growth of this organism (1). Hemin enters the gingival crevice following vascular ulceration and release of blood into the gingival tissues, which is a characteristic feature of diseased sites. P. gingivalis grown under relatively low concentrations of hemin (1 μg/ml) produces a TLR4 agonist, albeit of lower potency than that of Escherichia coli lipid A, consisting of monophosphorylated penta-acylated lipid A, whereas in high hemin concentrations (10 μg/ml), an LPS antagonist predominates, comprising the monophosphorylated tetra-acylated species. Hence, under low-hemin conditions, the bacterium adopts an immune-evasive phenotype, while in a high-hemin environment, the bacterium is immunosuppressive.
In the current work, we aimed to examine the influence of growth temperature on lipid A modification in P. gingivalis as an example of a critical environmental parameter which varies at sites of increasing inflammation. Diseased sites in periodontal tissues display all the hallmarks of inflammation, and temperature differentials between sublingual and subgingival sites of up to +4°C have been recorded in periodontal patients (9, 11, 17). We also assessed the functional consequences of temperature-dependent alterations to P. gingivalis lipid A structure and demonstrate that surprisingly minor changes in growth temperature of 2 to 4°C have significant effects on the interaction of this bacterium with the host defense.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
Porphyromonas gingivalis W50 was grown in a 2-liter-capacity chemostat (Applikon Biotechnology Ltd., Worcestershire, United Kingdom) at a dilution rate (D) of 0.1 h−1, corresponding to a mean generation time of 6.9 h, as described previously (18). Briefly, the pH of the culture was maintained at 7.0 (±0.1) by the automatic addition of 1 M NaOH and 0.5 M HCl, and the temperature was controlled at 37°C (±0.1°C). The medium used was BHI (brain heart infusion) broth (Oxoid, Basingstoke, United Kingdom) supplemented with 5 mg/liter of hemin (Sigma, Poole, United Kingdom) to achieve excess hemin levels. P. gingivalis W50 was grown to late logarithmic phase in an anaerobic batch culture, and 100 ml of the culture was used to inoculate the chemostat. In subsequent experiments, chemostat cultures were started at 37°C prior to increasing the temperature to either a constant 39°C (±0.1°C) or 41°C (±0.1°C). At each temperature, the chemostat was allowed to achieve a steady state (approximately 4 days; equivalent to 13 to 14 mean generation times) after inoculation, and samples were removed during each steady-state period for analysis over 6 days. Daily measurements of the optical density at 540 nm and dry weight and viable counts of steady-state cultures were performed.
Isolation of LPS and lipid A.
LPS was isolated from freeze-dried whole bacterial cells (10 mg) using TRIzol (Invitrogen) as described previously by Yi and Hackett (26), and lipid A was purified from this material by mild acid hydrolysis in 10 mM sodium acetate (pH 4.5) containing 1% sodium dodecyl sulfate (SDS). For SDS-urea-polyacrylamide gel electrophoresis (PAGE) experiments, LPS was prepared using an extraction kit from Intron Biotechnology (South Korea). LPS preparations (10 to 20 μg) from P. gingivalis W50 grown in a batch culture (37°C) and in continuous cultures were subjected to SDS-urea-PAGE in polyacrylamide slab gels at 10°C using procedures described by Inzana and Apicella (14); gels were then silver stained as previously described. The structure of lipid A from P. gingivalis W50 grown at different temperatures was subsequently determined by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS).
Structural analysis of lipid A by MALDI-TOF MS.
MALDI-TOF MS was performed using a Kratos Axima curved field reflection instrument (Kratos Analytical Ltd., Manchester, United Kingdom) fitted with a nitrogen laser operating at 337 nm using pulsed extraction in negative linear mode. Lipid A was analyzed using norharmane (9H-pyrido[3,4]indole) as a matrix in methanol at a concentration of 10 mg/ml. A total of 0.5 μl of lipid A solution in water (100 pmol/μl), together with 0.5 μl of matrix solution, was applied to the MALDI plate and allowed to dry in air. The instrument was calibrated using the peptides des-Arg1-bradykinin (mass, 904.0), angiotensin 1 (mass, 1,296.5), and neurotensin (mass, 1,672.3), and average masses were used throughout.
Sensitivity of P. gingivalis cells to HDPs.
Host defense peptides (HDPs) were synthesized by Fmoc (9-fluorenylmethoxy carbonyl) chemistry and purified by HPLC. Human β-defensin 2 (HBD2, GIGNPVTCLKSGAICHPVFCPRRYKGIGYCGLPGTKCCLL, with disulfide bonds C-1-C-5, C-2-C-4, and C-3-C-6) and β-defensin 3 (HBD3, GIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKK, with disulfide bonds C-1-C-5, C-2-C-4, and C-3-C-6) were supplied by Peptides International. Peptides (>99% pure) were dissolved in 0.01% (vol/vol) acetic acid at a concentration of 500 μg/ml and stored in salinized vials at −80°C until needed.
To determine killing by HDPs, P. gingivalis cells were removed from the chemostat and immediately placed in an anaerobic workstation (Don Whitley Scientific, Otley, United Kingdom) under an atmosphere of 10% CO2, 10% H2, and 80% N2 at 37°C. They were washed twice in reduced transport fluid (RTF) and resuspended in RTF to an optical density at 540 nm corresponding to 1 × 106 CFU/ml. Doubling dilutions of peptides were made in RTF in wells of a polypropylene microtiter plate, and 100 μl bacterial suspension was added, giving a final volume of 200 μl and final peptide concentrations ranging from 3.12 μg/ml to 50 μg/ml. After incubation for 30 min, aliquots from each well were diluted in RTF, and viable counts were determined by inoculation onto Columbia blood agar plates.
TLR4 activation assays.
HEK293 cells were obtained from American Type Culture Collection (Manassas, VA). HEK293 cells were plated in 96-well plates at a density of 4 × 104 cells per well and transfected the following day by a standard calcium phosphate precipitation method as described previously (5) with the following plasmids: a reporter plasmid bearing a firefly luciferase coding sequence under the control of a NF-κB-inducible basal TATA box-containing promoter (pNF-κB-TA-Luc) (Clontech), a plasmid bearing the Renilla luciferase coding sequence under the control of a constitutive β-actin promoter (pβ-actin Renilla Luc) as an internal standard for variation in transfection efficiency (13), a modified pDisplay vector encoding human TLR4 (phTLR4) under the control of a cytomegalovirus (CMV) promoter, and a pEF-BOS vector encoding human MD-2 (phMD-2) under the control of the elongation factor 1α (EF-1α) promoter. In some experiments, an expression plasmid encoding human membrane CD14 (phmCD14) was included in the transfection mixture (12). After overnight growth of the transfected HEK293 cells to allow expression of the TLR receptor complexes, the test wells were stimulated in triplicate for 4 h at 37°C with the indicated doses and sources of LPS in Dulbecco modified Eagle medium (DMEM) containing 10% human serum. Following stimulation, the HEK293 cells were rinsed with phosphate-buffered saline and then lysed with 50 μl of passive lysis buffer (Promega, Madison, WI). Luciferase activity was measured using the dual-luciferase assay reporter system (Promega, Madison, WI). Data are expressed as relative NF-κB-dependent luciferase activity levels, which represent the mean inductions ± standard deviations (SD). Data were analyzed by two-tailed, unpaired Student's t tests (GraphPad Prism), and P of <0.05 was considered indicative of statistical significance.
RESULTS
Growth of P. gingivalis at different temperatures.
As reported previously (18), P. gingivalis grew stably at all three temperatures tested, although optimal growth was shown at 37°C. The value of the chemostat is that it imposes a constant, fixed growth rate on the culture. Therefore, the cells were grown at the same constant mean generation time of 6.9 h at all three temperatures. The cell yields were lower at the higher temperatures in terms of dry weight (21% and 36% at 39°C and 41°C versus 37°C, respectively), but the differences were relatively modest in terms of viable counts (7% and 13%, respectively).
Structural analyses of lipid A.
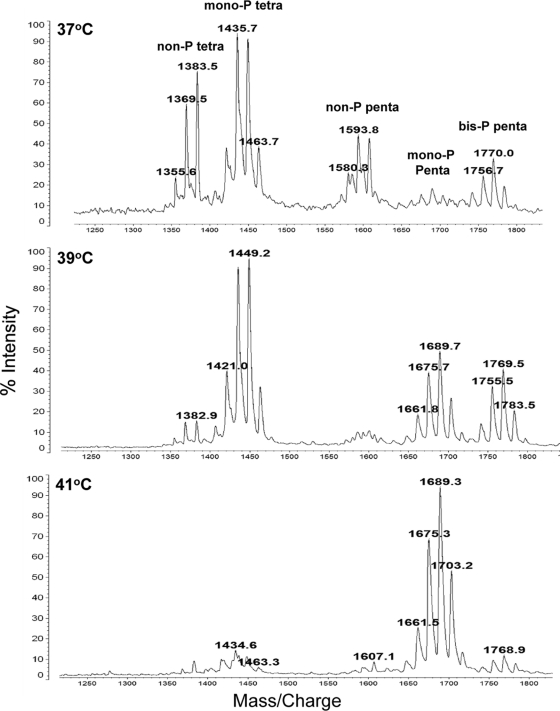
In agreement with previous data, MALDI-TOF MS analysis of lipid A from the LPS of P. gingivalis W50 showed the presence of five main clusters of peaks at approximately m/z 1,376, m/z 1,449, m/z 1,593, m/z 1,693, and m/z 1,772 corresponding to nonphosphorylated tetra-acylated, monophosphorylated tetra-acylated, nonphosphorylated penta-acylated, monophosphorylated penta-acylated, and bisphosphorylated penta-acylated species, respectively (21). However, the relative proportions of these species varied dramatically in response to growth at different temperatures (Fig. 1). For example, the clusters at m/z 1,370 to m/z 1,384 and m/z 1,580 to m/z 1,606, corresponding to nonphosphorylated tetra-acylated and nonphosphorylated penta-acylated species, respectively, were present in much larger amounts at 37°C and were either present in smaller amounts or nondetectable when P. gingivalis was grown at 39°C and 41°C. Similarly, the proportion of monophosphorylated tetra-acylated species (cluster at m/z 1,435 to m/z 1,464) was considerably reduced in the lipid A of cells grown at 41°C compared to that of cells grown at 37°C and 39°C. The signals for the cluster of peaks at m/z 1,657 to m/z 1,690, corresponding to monophosphorylated penta-acylated species, were absent in the cells grown in continuous culture at 37°C, whereas they were present in increasing amounts in lipid A of cells grown at 39°C and 41°C. The higher m/z values of this cluster with increasing growth temperature probably reflect the incorporation of slightly longer-chain fatty acids.
FIG. 1.
Variation in lipid A structure determined by MALDI-TOF MS from P. gingivalis cells grown in continuous culture at 37°C, 39°C, and 41°C. Bacteria grown at 37°C produce mainly tetra-acylated, nonphosphorylated lipid A and tetra-acylated, monophosphorylated lipid A bearing a 1-phosphate. However, when the growth temperature is maintained at 41°C, a penta-acylated, monophosphorylated lipid A predominates concomitant with reduction in both the nonphosphorylated and monophosphorylated, tetra-acylated lipid A structures.
Activation of TLR4 by LPS.
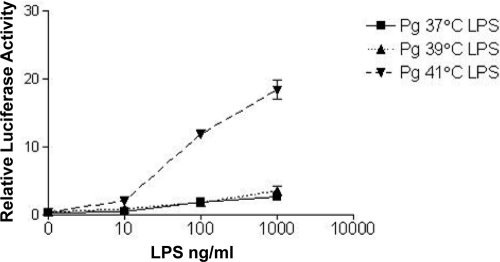
We next determined the effect of different lipid A structures produced at different growth temperatures on TLR4 activation by examining the abilities of LPS preparations to stimulate NF-κB activation in HEK 293 cells expressing recombinant human TLR4 and MD-2 (Fig. 2). LPS from cells grown at 41°C was a significantly more potent TLR4 agonist than LPS prepared from cells grown at either 37°C or 39°C. These data support the lipid A structural information, which demonstrated a temperature-dependent shift of lipid A from inert, nonphosphorylated and antagonizing, monophosphorylated, tetra-acylated structures at 37°C to the higher-potency, TLR4 agonist, monophosphorylated, penta-acylated structure at 41°C.
FIG. 2.
Activation of TLR4 by LPS derived from P. gingivalis grown in continuous culture at 37°C, 39°C, and 41°C. HEK293 cells transfected with human TLR4 (hTLR4), hMD-2, firefly luciferase, and Renilla luciferase were treated with LPS from cells grown at different temperatures. TLR4 activation is represented by the ratio of NF-κB-dependent firefly luciferase activity to β-actin promoter-dependent Renilla luciferase activity. LPS at 41°C is significantly more potent than LPS at 37°C and 39°C at stimulating TLR4-dependent NF-κB activation. P = <0.0001 for 100 ng LPS at 41°C versus 37°C; P = <0.0001 for 1,000 ng LPS at 41°C versus 37°C. Pg, P. gingivalis.
Activation of TLR4 by intact P. gingivalis cells.
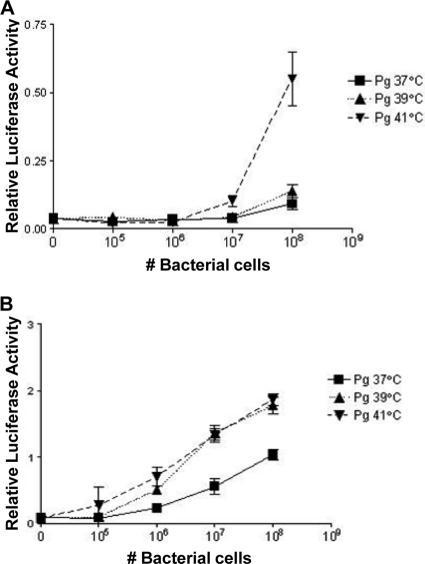
The results derived from experiments showing the relative abilities of LPS isolated from P. gingivalis grown at 37°C or 41°C to stimulate innate immune responses in human cells via TLR4 suggests that the temperature-dependent control of lipid A influences the ability of intact P. gingivalis cells to interact with the host innate immune system. To investigate the ability of intact P. gingivalis grown at either 37°C or 41°C to stimulate LPS-dependent innate immune responses, we employed the HEK-TLR4 assay (5). The results of these experiments demonstrate that bacteria grown at 41°C are significantly more potent in activating TLR4 compared to bacteria grown at 37°C at titers ranging from 107 to 108 bacteria (Fig. 3A). In addition, the differential activation between bacteria grown at either 41°C or 37°C was observed at titers as low as 106 bacteria when membrane CD14 (mCD14) is present in the signaling complex (Fig. 3B). mCD14 increases the sensitivity of TLR4 to LPS compared to soluble CD14 (sCD14), and the saturating dose is reached at relatively lower concentrations of agonist (6). It is possible that the bacterial cells grown at 39°C contain sufficient amounts of agonist lipid A to be more efficiently stimulatory in the presence of mCD14, while the 41°C sample has already saturated the TLR4 response in the presence of mCD14.
FIG. 3.
Activation of TLR4 by whole cells of P. gingivalis grown in continuous culture at 37°C, 39°C, and 41°C. HEK293 reporter cells transfected with hTLR4 and hMD-2 (A) or hTLR4, hMD-2, and hmCD14 (B) were treated with whole cells of P. gingivalis grown at different temperatures. (A) Bacteria grown at 41°C were significantly more potent at activating TLR4 than bacteria grown at 37°C, with titers ranging from 107 to 108 bacteria. (B) In addition, the differential activation between 41°C bacteria and 37°C bacteria was observed at titers as low as 106 bacteria when mCD14 was present in the signaling complex. P = 0.0049 for 106 bacteria grown at 41°C versus 37°C; P = 0.0046 for 107 bacteria grown at 41°C versus 37°C; P = 0.0011 for 108 bacteria grown at 41°C versus 37°C.
Synthesis of TLR4 antagonist at different temperatures.
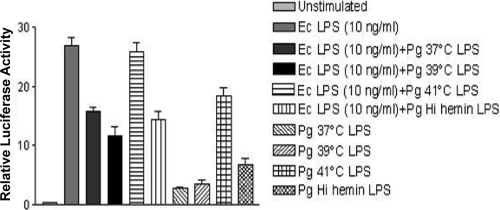
We next examined the ability of the different P. gingivalis LPS preparations to function as TLR4 antagonists since P. gingivalis LPS containing monophosphorylated, tetra-acylated lipid A has been shown to antagonize Escherichia coli LPS-dependent activation of TLR4 (5) (Fig. 4). The lipid A structures depicted in Fig. 1 for the 37°C- and 41°C-grown P. gingivalis suggest that the monophosphorylated tetra-acylated lipid A structures perform a dominant antagonist role at TLR4 in the 37°C LPS preparations and that the monophosphorylated, penta-acylated lipid A structures perform a dominant agonist role at TLR4 in the 41°C LPS preparations. To test this hypothesis, P. gingivalis LPS was mixed with E. coli LPS to assess its ability to function as a TLR4 antagonist. P. gingivalis LPS generated in a high-hemin-containing batch culture was included as a positive control for TLR4 antagonist LPS (5). As we had seen previously, LPS from P. gingivalis cells grown at 41°C was a significantly more potent agonist at TLR4 than LPS from cells grown at lower temperatures (Fig. 3). However, we also observed significant antagonist activity toward E. coli-dependent activation of TLR4 by LPS from 37°C- and 39°C-grown, cells which was not apparent in the LPS from 41°C-grown P. gingivalis. Hence, growth at a raised temperature downregulates the production of TLR4 antagonist activity in P. gingivalis LPS, which is consistent with the loss of monophosphorylated tetra-acylated lipid A under the higher-temperature growth conditions.
FIG. 4.
TLR4 antagonist activity in LPS derived from P. gingivalis grown at 37°C, 39°C, and 41°C. LPS from P. gingivalis grown at 37°C, 39°C, or 41°C was mixed with E. coli LPS to test their relative abilities to function as TLR4 antagonists. P. gingivalis LPS generated in high hemin at 37°C was included as a positive control for antagonist LPS activity. LPS at 37°C has potent antagonist activity against E. coli LPS in contrast to the 41°C LPS preparation. Ec, E. coli.
Sensitivity of P. gingivalis cells to host defense peptides.
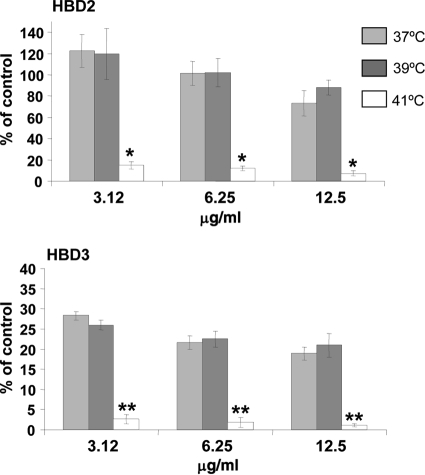
Resistance to antimicrobial cationic peptides represents another important aspect of the survival of bacterial pathogens at mucosal surfaces. A number of studies have demonstrated that modifications to lipid A, including dephosphorylation and decoration with aminoarabinose or phosphoethanolamine, provide a potent mechanism for bacteria to resist killing by cationic antimicrobial peptides. Recently, it was demonstrated that P. gingivalis resists killing to polymyxin B via production of tetra-acylated, nonphosphorylated lipid A, whereas mutant bacteria deficient in production of nonphosphorylated lipid A are highly susceptible to this antimicrobial (4). The lipid A structures depicted in Fig. 1 indicate that nonphosphorylated lipid A represents a significant proportion of total lipid A species in bacteria grown at 37°C compared to that of total lipid A species in bacteria grown at 41°C, suggesting that bacteria grown at 37°C will be more resistant to antimicrobial cationic peptide killing than bacteria grown at 41°C. Therefore, the ability of β-defensins, which are an important aspect of innate immunity in the oral mucosal epithelium (7), to kill P. gingivalis grown at different temperatures was assessed (Fig. 5). These experiments confirmed that bacteria grown at 41°C are more susceptible to β-defensins 2 and 3 than bacteria grown at 37°C, providing novel evidence that reduction of nonphosphorylated lipid A as a result of increased growth temperature also reduces the ability of this bacterium to evade host defenses.
FIG. 5.
Susceptibility to killing of P. gingivalis cells grown at different temperatures by human β-defensins. Bacterial cells grown at 37°C, 39°C, and 41°C were incubated with human β-defensin 2 or 3 at different concentrations for 30 min, and cell viability was then determined by colony counts. Bacteria grown at 41°C are significantly more susceptible to β-defensins 2 and 3 than bacteria grown at 37°C. *, P < 0.01 (37°C versus 39°C); **, P < 0.001 (37°C versus 39°C).
DISCUSSION
In agreement with previous studies, lipid A from P. gingivalis grown at 37°C showed the presence of four main clusters of peaks corresponding to nonphosphorylated tetra-acylated (m/z 1,376), monophosphorylated tetra-acylated (m/z 1,449), nonphosphorylated penta-acylated (m/z 1,593), and diphosphorylated penta-acylated (m/z 1,772) species. However, the relative proportion of these species changed significantly in response to growth at different temperatures. Lipid A from cells grown at 39°C contains much lower levels of the nonphosphorylated species and correspondingly higher levels of the monophosphorylated penta-acylated form at m/z 1,689, suggesting that just a 2°C shift in growth temperature leads to loss of phosphatase action at either the 1 or 4′ positions. The reduced phosphatase activity is maintained at 41°C and is accompanied by the loss of deacylase activity, such that lipid A from cells grown at 41°C contains primarily the monophosphorylated and diphosphorylated penta-acylated forms. Hence, in P. gingivalis, an increase in growth temperature over the narrow range of 37 to 41°C, which is relevant to the changes recorded in the host during an inflammatory response (11), leads to progressively fewer modifications to lipid A.
These structural changes lead to corresponding changes in the biological activity of lipid A. LPS from cells grown at 41°C was a significantly more potent TLR4 agonist than LPS prepared from cells grown at either 37°C or 39°C, reflecting the strong agonist activity of mono- and diphosphorylated penta-acylated lipid A (Fig. 2). We also examined the functional antagonism of TLR4 activation by E. coli LPS (Fig. 4). We observed significant antagonist activity toward E. coli-dependent activation of TLR4 by LPS from 37°C- and 39°C-grown cells, but this was not apparent in the LPS from 41°C-grown P. gingivalis. Hence, there are significant differences in the functional TLR4 activation of lipid A from P. gingivalis grown at different temperatures. The increased potency of TLR 4 activation by lipid A from P. gingivalis grown at increasing temperatures was mirrored by the activity of bacterial whole cells. Growth at different temperatures has been shown to influence a number of cell surface components of P. gingivalis. The expression of P. gingivalis fimbriae (25), the outer membrane proteins RagA and RagB (2), hemagglutinins, and proteases (18) are all reduced in cells grown at temperatures over 37°C. Hence, we are unable to rule out the possibility that other temperature-dependent alterations to the bacterial cell surface may also influence TLR4 activation. However, the close correspondence we report in this study between the degree of activation of TLR4 by whole cells and that by purified LPS from cultures at different growth temperatures suggests that temperature-dependent lipid A modification may have a significant influence on the inflammatory phenotype of the intact bacterium.
We previously showed that P. gingivalis is inherently resistant to antimicrobial peptides (8) and that the resistance is independent of its proteolytic activity, which is capable of degrading these cationic peptides. Given that resistance may be linked to the decreased negative charge of an outer membrane consisting of nonphosphorylated lipid A, we examined how increasing temperature would affect the susceptibility of P. gingivalis cells to human β-defensins (HBDs). P. gingivalis grown at 41°C was highly sensitive to both HBD2 and HBD3 in contrast to cells grown at the two lower temperatures (Fig. 5). Hence, preservation of phosphorylated lipid A in 41°C cells is correlated with sensitivity to killing by antimicrobial peptides. These data also correlate well with our previous observation that strains of P. gingivalis deficient in lipid A phosphatase activity both fail to produce nonphosphorylated lipid A and are highly sensitive to cationic antimicrobial peptide killing (4). Overall, these data are consistent with the hypothesis that under the stress conditions of growth at inflammatory temperatures of >39°C, P. gingivalis no longer remodels the structure of lipid A that is observed routinely at 37°C. As a result, the immunoevasive and immunosuppressive activities of lipid A (both monophosphorylated and nonphosphorylated tetra-acylated lipid A), which is characteristic of cells grown at 37°C, are progressively lost as the temperature increases, leading to the generation of a cell surface (monophosphorylated, penta-acylated lipid A) which is a potent activator of TLR4 and which has an increased susceptibility to killing by the innate defense at 41°C. It is unlikely that a temperature of 41°C is routinely attained in the periodontium even at sites of a highly deregulated inflammatory response. However, the data in this report demonstrate the principle that relatively minor shifts in temperature can act as important environmental regulators of lipid A modification in P. gingivalis. Moreover, as it is well recognized that other environmental parameters are also capable of influencing the process of lipid A remodeling, it is plausible that combinations of these factors, including growth temperatures at less extreme levels than 41°C, will work in concert to influence the lipid A structure and function in this bacterium. Regulation of lipid A remodeling has previously been shown to be linked to changes in growth temperature in other pathogens. For example, lipid A from Yersinia pestis, grown at temperatures found in its mammalian host, comprises primarily the tetra-acylated species, whereas the hexa-acylated form predominates in lipid A from cells grown at 21°C, representative of growth in the flea midgut, due to upregulation of the acyl-transferase genes lpxP and msbB at this lower temperature (15, 22). The formation of the tetra-acylated lipid A at 37°C may be beneficial to the bacterium because of the low immunogenicity of this form, whereas the hexa-acylated form may be required for stability of the outer membrane at the lower temperatures of the insect vector. Similarly, the deacylase PagL in Pseudomonas aeruginosa, which is responsible for deacylation at the 3 position of lipid A, is also temperature regulated, being inactive at 15°C but fully active at 37°C. As a result, a hexa-acylated species predominates at the lower environmental temperature, whereas a penta-acylated form is abundant at the temperature encountered by this bacterium in a warm blooded host (10). In both of these examples, lipid A remodeling is associated with very marked temperature differences, where there are liable to be significant issues in relation to maintenance of membrane fluidity over temperature extremes spanning up to 20°C. In contrast, the striking differences in lipid A structure which we have observed in P. gingivalis when cultured between 37°C and 41°C comprise the first report of a lipid A remodeling system linked to the temperature shifts associated with a deregulated inflammatory response.
The underlying mechanisms for some of the lipid A modifications in P. gingivalis and the influence of hemin on these changes have recently been proposed following examination of P. gingivalis mutants defective in putative 1 and 4′ lipid A phosphatase genes—PG1773 and PG1587, respectively (4). These studies suggest that the remodeling of lipid A occurs via a series of sequential reactions that require the initial removal of the 4′ phosphate from penta-acylated lipid A precursors by PG1587 in order to produce both the nonphosphorylated lipid A and the tetra-acylated lipid A 1-phosphate species, presumably by supplying the optimal substrates for subsequent dephosphorylation and deacylation reactions. Furthermore, lipid A 1-phosphatase activity at PG1773 is suppressed under high-hemin-excess conditions. Future studies will examine the interplay between environmental hemin and growth temperature: low hemin at a low growth temperature and high-hemin-excess conditions at a high temperature may be regarded as the two environmental extremes experienced by this bacterium in the transition from health to disease.
Acknowledgments
This investigation was supported by the Medical Research Council (United Kingdom) grant G0501478642 and the Research Advisory Board of the Barts and The London Charity grant 643 RAB06/PJ/14.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 10 January 2011.
REFERENCES
- 1.Al-Qutub, M. N., et al. 2006. Hemin-dependent modulation of the lipid A structure of Porphyromonas gingivalis lipopolysaccharide. Infect. Immun. 74:4474-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonass, W. A., et al. 2000. Identification of ragAB as a temperature-regulated operon of Porphyromonas gingivalis W50 using differential display of randomly primed RNA. Infect. Immun. 68:4012-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brogden, K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238-250. [DOI] [PubMed] [Google Scholar]
- 4.Coats, S. R., et al. 2009. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4′-phosphatase activities. Cell. Microbiol. 11:1587-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coats, S. R., T. T. Pham, B. W. Bainbridge, R. A. Reife, and R. P. Darveau. 2005. MD-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J. Immunol. 175:4490-4498. [DOI] [PubMed] [Google Scholar]
- 6.Darveau, R. P., et al. 2004. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect. Immun. 72:5041-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devine, D. A. 2003. Antimicrobial peptides in defence of the oral and respiratory tracts. Mol. Immunol. 40:431-443. [DOI] [PubMed] [Google Scholar]
- 8.Devine, D. A., P. D. Marsh, R. S. Percival, M. Rangarajan, and M. A. Curtis. 1999. Modulation of antibacterial peptide activity by products of Porphyromonas gingivalis and Prevotella spp. Microbiology 145(Pt. 4):965-971. [DOI] [PubMed] [Google Scholar]
- 9.Fedi, P. F., Jr., and W. J. Killoy. 1992. Temperature differences at periodontal sites in health and disease. J. Periodontol. 63:24-27. [DOI] [PubMed] [Google Scholar]
- 10.Geurtsen, J., L. Steeghs, J. T. Hove, P. van der Ley, and J. Tommassen. 2005. Dissemination of lipid A deacylases (pagL) among gram-negative bacteria: identification of active-site histidine and serine residues. J. Biol. Chem. 280:8248-8259. [DOI] [PubMed] [Google Scholar]
- 11.Haffajee, A. D., S. S. Socransky, and J. M. Goodson. 1992. Subgingival temperature (I). Relation to baseline clinical parameters. J. Clin. Periodontol. 19:401-408. [DOI] [PubMed] [Google Scholar]
- 12.Hajjar, A. M., R. K. Ernst, J. H. Tsai, C. B. Wilson, and S. I. Miller. 2002. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat. Immunol. 3:354-359. [DOI] [PubMed] [Google Scholar]
- 13.Hajjar, A. M., et al. 2001. Cutting edge: functional interactions between Toll-like receptor (TLR) 2 and TLR1 or TLR6 in response to phenol-soluble modulin. J. Immunol. 166:15-19. [DOI] [PubMed] [Google Scholar]
- 14.Inzana, T. J., and M. A. Apicella. 1999. Use of a bilayer stacking gel to improve resolution of lipopolysaccharides and lipooligosaccharides in polyacrylamide gels. Electrophoresis 20:462-465. [DOI] [PubMed] [Google Scholar]
- 15.Kawahara, K., H. Tsukano, H. Watanabe, B. Lindner, and M. Matsuura. 2002. Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect. Immun. 70:4092-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumada, H., Y. Haishima, T. Umemoto, and K. Tanamoto. 1995. Structural study on the free lipid A isolated from lipopolysaccharide of Porphyromonas gingivalis. J. Bacteriol. 177:2098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niederman, R., C. Naleway, B. Y. Lu, Y. Buyle-Bodin, and P. Robinson. 1995. Subgingival temperature as a gingival inflammatory indicator. J. Clin. Periodontol. 22:804-809. [DOI] [PubMed] [Google Scholar]
- 18.Percival, R. S., et al. 1999. Effect of temperature on growth, hemagglutination, and protease activity of Porphyromonas gingivalis. Infect. Immun. 67:1917-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raetz, C. R., C. M. Reynolds, M. S. Trent, and R. E. Bishop. 2007. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76:295-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rangarajan, M., et al. 2008. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J. Bacteriol. 190:2920-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rebeil, R., et al. 2006. Characterization of late acyltransferase genes of Yersinia pestis and their role in temperature-dependent lipid A variation. J. Bacteriol. 188:1381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stead, C. M., A. Beasley, R. J. Cotter, and M. S. Trent. 2008. Deciphering the unusual acylation pattern of Helicobacter pylori lipid A. J. Bacteriol. 190:7012-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trent, M. S., C. M. Stead, A. X. Tran, and J. V. Hankins. 2006. Diversity of endotoxin and its impact on pathogenesis. J. Endotoxin. Res. 12:205-223. [DOI] [PubMed] [Google Scholar]
- 25.Xie, H., S. Cai, and R. J. Lamont. 1997. Environmental regulation of fimbrial gene expression in Porphyromonas gingivalis. Infect. Immun. 65:2265-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi, E. C., and M. Hackett. 2000. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst 125:651-656. [DOI] [PubMed] [Google Scholar]