Abstract
The Rgg-like regulators, a family of transcription factors commonly found in many Gram-positive bacteria, play multiple roles, especially in the control of pathogen virulence. Here, we report an rgg homologue from a Chinese isolate, 05ZYH33, of Streptococcus suis serotype 2 (SS2). Deletion of the rgg gene in SS2 increased its adhesion to Hep-2 cells and hemolytic activity in vitro. Significantly, inactivation of the rgg gene attenuated SS2 virulence in an experimental piglet infection model. Using DNA microarrays and quantitative reverse transcription-PCR, we found that the Rgg regulator affects the transcriptional profile of 15.87% (n = 345) of all of the annotated chromosomal genes, including those involved in nonglucose carbohydrate metabolism, DNA recombination, protein biosynthesis, bacterial defense mechanisms, and others. It was experimentally verified that the deletion of rgg in SS2 reduced the utilization of nonglucose carbohydrates, such as lactose and maltose. In addition, the rgg gene was found to be associated with changes in the bacterial microscopic phenotype and growth curve. These data suggested that Rgg in SS2 is a global transcriptional regulator that plays an important role in promoting SS2 bacterial survival during pathogen-host interaction.
Streptococcus suis serotype 2 (SS2) is a leading swine pathogen that occasionally infects humans who engage in close contact with swine and pork-derived products (7, 14, 35, 38). Before our report of two epidemic outbreaks of human SS2 in China, it was thought that SS2 could only cause sporadic human cases (7, 38, 43, 49). In view of this epidemiological history of human SS2 infection, both outbreaks (37, 43, 49) and sporadic cases (9, 27) are now believed to take place in China. Comparative genomics analyses from three different groups (4, 15, 48) have suggested that virulent Chinese strains of SS2 feature a specific, 89-kb-long DNA fragment (dubbed 89K by our group [4]). Further genetic studies showed preliminary evidence that 89K may function as a pathogenicity island (22). Besides well-known virulence-associated factors, such as capsular polysaccharide (36) and suilysin (18), 17 additional virulence-associated components have been identified (9), including a two-component signal transduction system (TCSTS), SalK-SalR (22); an orphan regulator, CovR (31); lipoteichoic acid (11); and others.
Like many other pathogenic bacteria, SS2 is transmitted via the respiratory route and remains localized in the palatine tonsils but subsequently escapes from the immune system and disseminates via the blood circulation system, finally invading various host organs (1). In order to colonize and infect different regions within the host, SS2 requires a regulatory network that senses changing surroundings and responds to environmental signals. Such responses typically involve genome-wide changes in transcriptional activation or repression of specific genes through interactions among multiple regulators. In recent years, thorough research and analyses of global regulatory networks in Streptococcus pyogenes have been made (11); however, the corresponding knowledge regarding SS2 is relatively limited (8, 31). Rgg-like regulators, a family of transcriptional factors, are widely distributed in Gram-positive bacteria, including S. pyogenes (26), Streptococcus gordonii (41), Lactococcus lactis (34), and others. Rgg-like proteins have a conserved helix-turn-helix motif and several invariant residues in the amino terminus that are necessary for specific binding to the promoter regions of Rgg-regulated genes. Rgg protein plays various roles in many bacteria: (i) in S. gordonii, Rgg positively regulates extracellular glucosyltransferase expression (40); (ii) in L. lactis, the Rgg-like regulator GadR is required for glutamate-dependent acid tolerance (34); (iii) in Streptococcus mutans, the Rgg-like protein MutR controls the expression of the mutacin antibiotic (33); and (iv) in S. pyogenes, Rgg (also known as RopB) is necessary for the expression of secreted proteins such as SpeB (3, 21, 26). In fact, Rgg could affect the transcription of multiple genes throughout the genome by altering the expression of known and putative transcriptional regulators, thereby achieving bacterial adaptive responses to various environmental conditions (5).
In the present study, we characterized a functional member of the Rgg protein family of transcriptional regulators from a Chinese SS2 isolate, 05ZYH33. Our data clearly demonstrated that SS2 Rgg protein is a global transcriptional regulator that both contributes to the virulence of SS2 and controls nonglucose carbohydrate metabolism.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in the present study are listed in Table 1. The SS2 strains were grown at 37°C in Todd-Hewitt broth (THB; Difco Laboratories, Detroit, MI) liquid medium, on THB agar plates containing 5% (vol/vol) sheep blood (43), or in 10 ml of chemically defined medium (CDM) (5). The CDM was prepared without glucose, and maltose, lactose, fructose, or mannose was added separately to a final concentration of 1% (wt/vol). Escherichia coli DH5α was maintained in Luria-Bertani (LB) broth liquid medium or plated on LB agar at 37°C overnight with aeration. The antibiotic concentrations used for SS2 transformations were 100 μg/ml for spectinomycin (Spc; Sigma) and 5 μg/ml for chloromycetin (Cm; Sigma). The antibiotic concentrations used for plasmid selection in E. coli was 50 μg of ampicillin (Amp; Sigma)/ml.
TABLE 1.
Summary of bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics and/or functiona | Source or reference |
|---|---|---|
| Bacterial strains | ||
| 05ZYH33 | Virulent strain isolated from a dead patient with STSS | Lab collection |
| Δrgg mutant | Isogenic Δrgg deletion mutant of strain 05ZYH33; Spcr | This study |
| CΔrgg mutant | Complemented Δrgg mutant; Spcr Cmr | This study |
| 05HAS68 | Avirulent strain isolated from a healthy pig | Lab collection |
| E. coli DH5α | Cloning host for recombinant vector | Promega |
| Plasmids | ||
| pUC18 | Cloning vector; Ampr | Promega |
| pUC18-Spc | With an Spcr gene | This study |
| pUC::rgg | A recombinant vector with the background of pUC18, designed to knock out gene rgg; Ampr Spcr | This study |
| pSET1 | E. coli-S. suis shuttle vector; Cmr | 42 |
| pSET2 | E. coli-S. suis shuttle vector; Spcr | 42 |
| pSET1::rgg | pSET1 containing the intact rgg gene and its upstream promoter; Cmr Spcr | This study |
Ampr, ampicillin resistant; Cmr, chloromycetin resistant; Spcr, spectinomycin resistant.
Knockout of rgg and functional complementation.
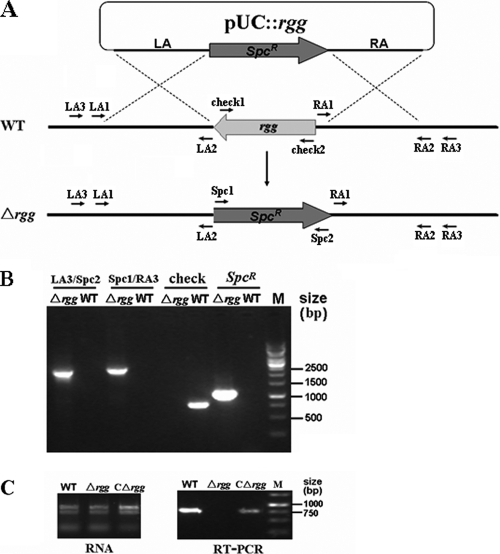
rgg deletion was performed as previously described (8, 22). Briefly, the Spc resistance (Spcr) gene cassette (amplified from pSET2) was inserted into a pUC18 vector (Promega) to create the recombinant plasmid pUC18-Spc. Two DNA fragments (LA and RA) flanking the rgg gene were cloned into pUC18-Spc to generate the knockout plasmid, pUC::rgg. The pUC::rgg plasmid was electroporated into 05ZYH33 competent cells (22). The acquired Spcr transformants were confirmed by using multiplex-PCR (Table 2). The fidelity of the double-crossover recombination was confirmed in the mutants by PCR using flanking primers (LA3/RA3) lying outside the homologous regions, followed by direct DNA sequencing. Reverse transcription-PCR (RT-PCR) detection was carried out to confirm the successful inactivation of the rgg gene in the deletion mutant, designated Δrgg.
TABLE 2.
Primers used for PCR amplification and detection
| PCR type and primer | Sequences (5′-3′)a | Restriction enzyme or size (bp) | Function or gene |
|---|---|---|---|
| General PCR | |||
| LA1 | AAGCTTTGGTAATTTGGTCGTTTAGAA | HindIII | Left arm of rgg |
| LA2 | CTGCAGTTTGTTGTCCTATATTTATAA | PstI | |
| RA1 | GGATCCCAGACCTCCTTTTGTATATAA | BamHI | Right arm of rgg |
| RA2 | GAATTCTCTAGAGCCTGCAGGTATAAT | EcoRI | |
| Spc1 | CTGCAGGTTTTCTAAAATCTGAT | PstI | Spcr gene |
| Spc2 | GGATCCGTTCGTGAATACATGTTATA | BamHI | |
| Check1 | CTATCGACCGCTCGTAAAAG | Internal region of rgg | |
| Check2 | TTTGGGAAAACGTTGAAATT | ||
| LA3 | ATCTCACCGCTTTTACTTGGTTTAG | For combined PCR detection | |
| RA3 | GAGGATTATCAGGGGAATTTTG | ||
| rggC-F | GGATCCGCTTGAAGACGCAAACAT | BamHI | rgg gene and its upstream promoter |
| rggC-R | GAATTCGCAACCACTACTCAATAA | EcoRI | |
| Real-time PCRb | |||
| 0155-F | GTGACCAAATGGTTCTTGAC | 261 | gapdh |
| 0155-R | ATTCAGTAGCAGCAGCTTTC | ||
| 0178-F | CAGAATCCGCTAAGTCAGTC | 252 | ef |
| 0178-R | TAAGGAATGCCTTGATACGA | ||
| 0475-F | GCATCCGACATTGAAGCCTA | 239 | srtF |
| 0475-R | AAATCAGAAAACAGACGGGC | ||
| 0565-F | CGGAATTGAATCTGAAAGAG | 262 | cps2B |
| 0565-R | CCAGCTAATAAACCAAGCAA | ||
| 0581-F | CCACGTTGTTAGCTTTACCA | 237 | neuA |
| 0581-R | ATCGAATCTTCCTTCGTCAT | ||
| 0903-F | TATGCGGAGTTTCAGACATT | 247 | Integrase |
| 0903-R | CAATAAGTCGTTCACCTGCT | ||
| 0906-F | TGGCTGACCTCAGTAAACAC | 264 | nisK |
| 0906-R | TGCTCTTTCAATGTCCCTAA | ||
| 0994-F | CTATCCTTTGGAGAGGAGGT | 253 | 05SSU0994 |
| 0994-R | TGGCAATGTAAATCATGCTA | ||
| 1492-F | CTGGAGATTGAGTTGGATGT | 261 | fbp |
| 1492-R | TTTCTGACGCTTATGGATTT | ||
| 1932-F | GTTGTAACGGGATCTAGTCG | 239 | Glucokinase regulatory protein |
| 1932-R | GCTAATTTCACGTCTTGGTT | ||
| 2134-F | ATCGCCTTTACAAACTACGA | 257 | malC |
| 2134-R | CATGGGAGCAAGAAGATAAC |
The underlined sequences are the restriction sites.
The sizes and genes/proteins refer to both primers (forward [F] and reverse [R]) in a given primer pair (e.g., 0155-F and 0155-R, etc.).
To create a construct for functional complementation, the DNA fragment covering the rgg coding region plus its 286-bp upstream promoter sequence was amplified from the chromosomal DNA of 05ZYH33 by PCR using the primers rggC-F and rggC-R. The resulting PCR product was cloned into an E. coli-S. suis shuttle vector, pSET1 (42), to yield plasmid pSET1::rgg. This plasmid was used for subsequent functional complementation via electroporation (8, 31).
Transmission electron microscopy.
To visualize the effect of the rgg gene on morphological changes in 05ZYH33, transmission electron microscopy was performed according to our previously reported method (31). The samples from agar-grown bacteria were fixed in 5% glutaraldehyde for 2 h, postfixed with 2% osmium tetroxide for 2 h, dehydrated in a graded series of acetone washes, and embedded in epoxy resin. Thin sections were post-stained with uranyl acetate and lead citrate and then examined with a JEM-1010 TEM (JEOL, Ltd., Tokyo, Japan) at an accelerating voltage of 80 kV (19, 31). Capsule thickness measurements were made on 25 randomly chosen cells from each strain.
Hemolysin activity assay.
Hemolysin activity was tested as previously described (18, 31) with slight modifications. In brief, sterile culture supernatant fluids were prepared from cultures of wild-type (WT) and Δrgg strains. Serial 2-fold dilutions (150 μl) of test samples were prepared in 96-well microplates with 10 mM phosphate-buffered saline (PBS) buffer (pH 7.4) as the diluent. Next, 150 μl of 2% human red blood cells was added. After incubation for 30 min at 37°C, the plates were centrifuged at 1,000 × g for 10 min, and 150 μl of supernatant was transferred to a new plate for spectrophotometric measurements at 540 nm. Hemolysin units (HU) were defined as the reciprocal of the highest dilution of supernatant inducing at least 50% lysis of erythrocytes.
Flow cytometry analysis.
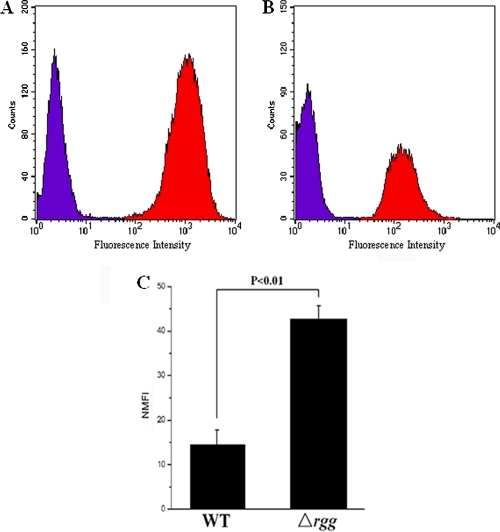
Bacteria were harvested, washed, and resuspended in PBS. Carboxyfluorescein diacetate succinimidyl ester (CFDA-SE) solution was added to the bacterial suspensions to a final concentration of 10 μM, and the cells were incubated with gentle rotation at 37°C for 20 min. Human laryngeal epithelial cell line Hep-2 (CCTCC GDC004) cells were cultured to monolayer at 37°C and 5% CO2 in RPMI 1640 medium (Gibco), detached with 0.05% trypsin, washed twice, and resuspended in PBS. Labeled bacteria and Hep-2 cells were coincubated (ratio 100:1) with gentle agitation at 37°C for 2 h and then fixed with 4% (wt/vol) paraformaldehyde for flow cytometry analysis (16, 24). Flow cytometry data are represented by histograms, which indicate the distribution of cells as follows: the x axis indicates the fluorescence intensity, and the y axis indicates the number of particles. By measuring the mean fluorescence intensity (MFI) values of labeled bacteria and Hep-2 cells with adherent bacteria, the normalized MFI (NMFI) values were calculated according to the method of Hytönen et al. (16).
Experimental infection of piglets.
To probe the possible role of the rgg gene in SS2 virulence, 3-week-old specific-pathogen-free (SPF)-piglets (6 pigs/group) were challenged with the Δrgg mutant and compared to those challenged by either the WT strain or the complemented strain CΔrgg. An avirulent strain, 05HAS68, was utilized as a negative control. All of the SPF piglets were inoculated intravenously at a dose of 108 CFU per piglet (verified by plating, optical density at 600 nm [OD600] of 0.90 for both strains). The infected piglets were monitored for clinical signs and survival times. All animal experiments were performed in a biosafety level 3 facility and approved by the local ethical committee. After the experiments, the sacrificed piglets were autopsied, and sections were prepared from the specimens for light microscopy observation, as reported previously (43).
RNA isolation and real-time RT-PCR.
SS2 strains were grown to post-exponential phase (OD600 = 0.90) in the absence of antibiotics. Total RNA was isolated by using an SV total RNA isolation system (Promega) according to the manufacturer's instructions. The absence of contaminating DNA was confirmed by PCR-based assays (8, 22). The qualified RNA was then used for cDNA synthesis, as well as further microarray analysis.
A subset of genes was selected to validate the microarray data with SYBR green detection as described previously (31). The primers (Table 2) were designed according to the published genomic sequence of 05ZYH33 (4). SYBR green detection was used in the quantitative RT-PCR (8, 31). Each reaction was conducted in triplicate two-step multiplex RT-PCRs with SYBR premix ExTaq (TaKaRa) on an Opticon 2 (MJ Research). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal reference gene. The relative expression level was measured by using the 2−ΔΔCT method (23).
DNA microarray-based analysis.
DNA microarray creation and analyses were performed as previously described (8, 22, 31). The microarray data were then normalized by quantile normalization and logarithmically transformed before further analysis. Statistical analysis was then carried out, i.e., using the Student t test and the generation of P values, on these data. Genes with changes >2-fold were regarded as candidate targets (17).
Statistical analysis.
All assays were performed in triplicate at least three times. Statistical analysis was performed by using a Student t test. Differences were considered statistically significant when the calculated P value was <0.05.
Microarray data accession number.
The microarray data presented in here have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE21940.
RESULTS
Characterization of the rgg gene in 05ZYH33.
The rgg gene on the chromosome of 05ZYH33 was predicted to encode a 287-amino-acid protein product. This regulator was suggested to exhibit 28.2, 26.2, 20.1, 39.7, and 31.5% identity to multiple Rgg-like regulators of Streptococcus agalactiae NEM316, S. pneumoniae R6, S. pyogenes M1, S. mutans UA159, and S. gordonii Challis substrain CH1, respectively. Bioinformatics analysis showed that this Rgg protein possesses a putative DNA-binding helix-turn-helix motif at its N terminus and contains three invariant residues: glycine (G5), arginine (R12), and tryptophan (W150), all of which are critical for fully active Rgg-like regulators (25).
Microbiological characterization of the Δrgg mutant.
An isogenic rgg mutant of SS2 strain 05ZYH33 was constructed through homologous recombination (Fig. 1 A). One putative mutant was found from more than 100 Spcr transformants. The mutation was confirmed by combined PCR detection (Fig. 1B), RT-PCR (Fig. 1C), and direct DNA sequencing (data not shown).
FIG. 1.
Construction and characterization of the isogenic Δrgg mutant. (A) Diagram of gene rgg knockout via double-crossover recombination. pUC::rgg is the plasmid used for the gene rgg knockout. LA and RA indicate the left and right arms of rgg. Two pairs of specific primers (Check1/Check2 and Spc1/Spc2) were used for PCR detection of the existence of gene rgg and the Spcr gene in the genome of SS2. The flanking primers (LA3/RA3) locating outside LA and RA were designed to confirm double-crossover recombination in mutant by combined PCR detection. WT, wild-type strain of SS2, 05ZYH33. (B) Multiple-PCR analyses of the Δrgg mutant. (C) RT-PCR detection of rgg gene transcripts. A pair of primers, Check1/Check2, was used here.
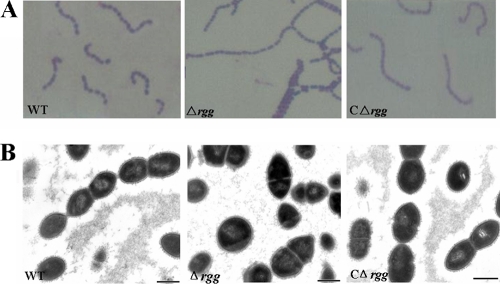
The effects of rgg deletion on the basic biological properties of SS2 were investigated in terms of morphology, hemolytic activity, and growth in vitro. Morphological examination by Gram staining showed that inactivation of rgg resulted in much longer chain length, as well as abnormal morphology. These qualities were significantly different from those of the WT strain (Fig. 2 A). Transmission electron microscopy-based observations showed obvious differences in microscopic phenotype between the Δrgg and WT strains under normal culture conditions. The Δrgg mutant grew as chains of individual cells that lost the typical shape characteristic of S. suis, showing an altered cell diameter and irregular margins (Fig. 2B). The microscopic phenotype of S. suis was restored after complementation. Further, measurement of capsule thickness showed no obvious differences between 05ZYH33 (range, 34.7 to 56.3 nm) and the Δrgg mutant strain (range, 36.3 to 59.6 nm).
FIG. 2.
Microbiological characterization of WT, Δrgg, and CΔrgg strains. (A) Gram-stained images of bacteria under light microscopy (magnification, ×1,000). (B) Transmission electron micrographs of bacteria. Bars, 500 nm. Bacteria were cultured in THB.
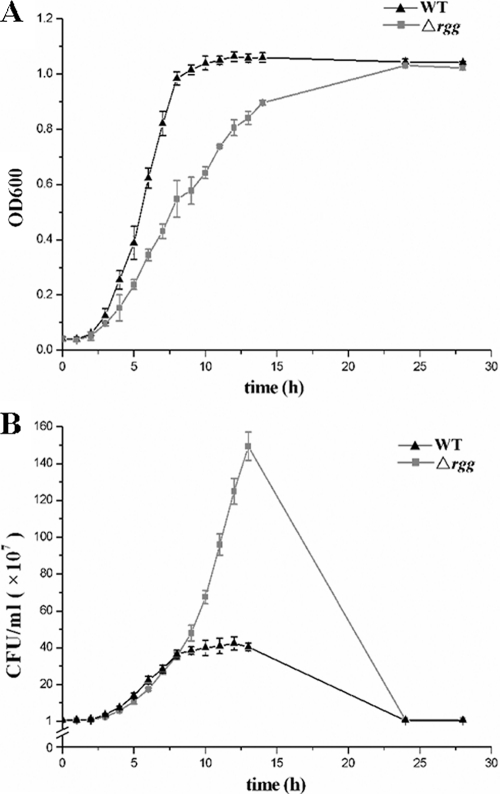
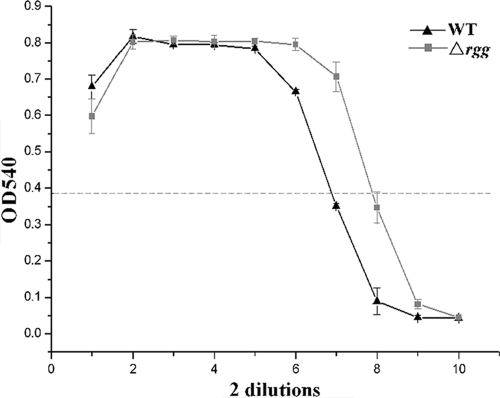
The growth kinetics of the Δrgg strain were compared to those of the WT strain by measuring OD600 values, and we found that the growth of the Δrgg strain was 40% slower in the first 8 to 9 h than the WT strain (Fig. 3 A). However, when we determined the growth rates of Δrgg and WT cultures by counting the CFU, no obvious differences in the CFU counts were observed during the initial 8 to 9 h of growth. At this time, the WT strain had already reached the post-exponential-growth phase, but the Δrgg strain continued to reproduce until the culture reached an OD600 similar to that of the WT strain (Fig. 3B). We noted that the OD600 of the Δrgg culture appeared to be lower than that of the WT culture, indicating that the rgg deletion changed the light-scattering properties in this SS2 strain. Nevertheless, the growth kinetics of the complemented strain CΔrgg returned to normal (data not shown). In addition, hemolytic activity was significantly higher in culture supernatant fluids obtained from the Δrgg mutant strain than in those obtained from the WT strain (Fig. 4), suggesting that Rgg inactivation could enhance hemolytic activity in SS2.
FIG. 3.
Growth characteristics of the WT and Δrgg mutant. (A) The cell density is measured spectrometrically at 600 nm, and the data were collected at the indicated times. (B) Separate aliquots of the bacterial suspensions were serially diluted and plated to determine CFU numbers per milliliter.
FIG. 4.
Titration of hemolytic activities of the WT and Δrgg mutant. The horizontal line means the highest dilution of supernatant inducing at least 50% lysis of erythrocytes.
Adhesion of Δrgg to epithelial cells.
The bacteria were found to be effectively labeled with CFDA-SE, as shown in Fig. 5. There is an ∼500-fold increase in the MFI of labeled bacteria compared to unlabeled bacteria (Fig. 5A). There is a >50-fold increase in the MFI of cells incubated with bacteria compared to Hep-2 cells alone (Fig. 5B). The incubation of the cells with unlabeled bacteria did not affect the fluorescence of the cells (data not shown). This result indicated that the flow cytometric approach is suitable for assaying the adhesion of SS2 to Hep-2 cells. Fluorescently labeled WT and Δrgg mutant strains were allowed to coculture with Hep-2 cells, and the fluorescence of the cells was then measured. The MFI of the cells showed that the binding of the Δrgg mutant to Hep-2 cells is increased ∼3-fold compared to the WT strain (Fig. 5C). This observation suggested that the Rgg regulator might have a role in the adherence of SS2 to human epithelial cells.
FIG. 5.
Evaluation of adhesion ability of Δrgg mutant by using flow cytometry. (A) Histogram of fluorescence intensity of unlabeled bacteria (purple peak) and CFDA-SE-labeled bacteria (red peak). (B) Histogram of fluorescence intensity of Hep-2 cells alone (purple peak) and Hep-2 cells incubated with labeled bacteria (red peak). (C) Adhesion of Δrgg mutant to Hep-2 cells compared to the WT strain. The normalized mean fluorescence intensities (NMFI) of Hep-2 cells after incubation with the bacteria are shown as columns with standard errors.
Experimental infection of piglets.
Experimental infection of piglets was carried out to address the relationship between the rgg gene and SS2 virulence. All SPF piglets inoculated intravenously with the WT strain (positive control) developed serious clinical symptoms, such as high fever, swollen joints, shivering, and central nervous system failure within 24 h, and none of them survived. In contrast, the six piglets infected with an avirulent strain, 05HAS68 (negative control), survived without any obvious symptoms during the entire monitoring period. The six piglets challenged with the Δrgg strain exhibited markedly lower morbidity and mortality rates, compared to those infected with the WT strain. Five piglets in this group exhibited joint swelling and high fever, but only two of them died (on days 10 and 14 postinfection), and the others survived the 20-day observation period. All of the piglets infected with the complemented strain CΔrgg died within 2 to 3 days, similar to the median survival time observed in the WT group. Therefore, we concluded that the rgg gene plays a role in the pathogenicity of Chinese SS2 isolates.
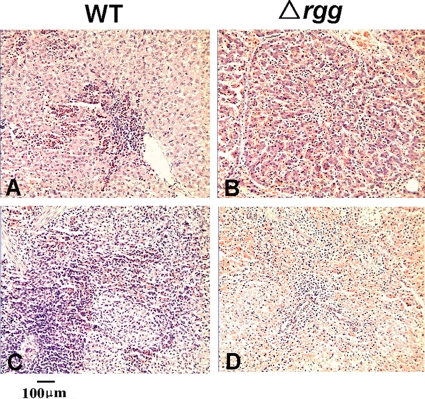
Pathological examination was conducted by light microscopy. The WT-infected group exhibited pathological changes in liver, spleen, and kidney tissues. Although the hepatic lobule structure and tandem arrangement of the hepatic plates were relatively normal, hepatic cell degeneration and necrosis were seen in the areas surrounding the lobule. Massive infiltration of neutrophils was observed in the convergent zone (Fig. 6 A). The structure of the spleen was also affected; areas of both red and white pulp were clearly visible, with some reduction in the white pulp. There were epithelioid nodules within the spleen parenchyma, and some of the nodules showed fusion (Fig. 6C). For the mutant-infected group, similar pathological alterations occurred in these tissues, but to a lesser degree. There were no obvious pathological lesions except for mild hyperemia in hepatic tissue (Fig. 6B). White pulp was increased significantly, and no epithelioid nodules akin to those described above were found in splenic tissue (Fig. 6D). Joint lesions were observed in piglets in both groups. The postmortem examination showed that the piglets exhibiting joint swelling and lameness had various degrees of serous or purulent arthritis. No positive histological sign of meningitis was apparent (data not shown), although bacteria were recovered from the brains of all of the sick piglets. Together, these results suggested that Rgg inactivation had led to changes in the virulence phenotype.
FIG. 6.
Light microscopy observation of the autopsy specimens from the SS2-infected piglets. Sectioned liver (A and B) and spleen (C and D) tissues were sampled from piglets infected by the WT or Δrgg mutant.
DNA microarray analysis.
To obtain a comprehensive view of the extent to which rgg impacts gene expression in SS2, global gene transcription profiles were determined by DNA microarray analysis for the Δrgg mutant and WT strains grown to post-exponential phase. The effectiveness of the microarray data was confirmed by real-time quantitative RT-PCR. Ten genes demonstrating various expression levels by microarray analysis were chosen for RT-PCR analysis. There were strong positive correlations (r2 = 0.93; Fig. 7) between the results obtained with the two methods.
FIG. 7.
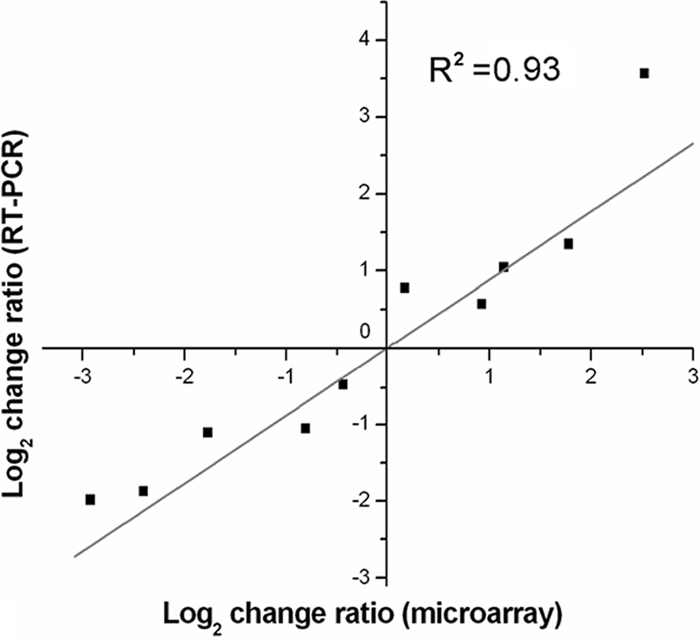
Correlation between DNA microarray data and real-time PCR results. The fold changes in transcript levels of 10 selected genes determined by both methods were log2 transformed, and the values were plotted against each other to evaluate their correlation. The genes analyzed are listed in the Table 2.
Inactivation of rgg alters the global gene transcription profile.
Globally, 15.87% of the genes represented on the microarray (n = 345) were differentially transcribed in the Δrgg strain compared to the WT strain. Among these, 195 genes were upregulated, and 150 genes were downregulated (see Table S1 in the supplemental material). These genes were involved in diverse cellular activities, including physiological metabolism, information storage and processing, cell division, and prophage integration (Table 3). Meanwhile, within each functional category, the tendency for genes to be up- or downregulated was easily discerned. Many transcripts (n = 72) encoding proteins associated with information storage and processing were the main part of upregulated genes in the Δrgg strain (Table 3). However, downregulated transcripts principally focused on genes related to metabolism (n = 63), especially glycometabolism. These data provide genome-wide insights into the regulation of Rgg in SS2.
TABLE 3.
Summary of genes differentially expressed in an Δrgg mutant during post-exponential growth as demonstrated by DNA microarray analysis
| Functional classification | No. of genesa |
|
|---|---|---|
| Upregulation | Downregulation | |
| 1. Metabolism | (37) | (63) |
| 1.1 Carbohydrate | 6 | 38 |
| 1.2 Fatty acid | 3 | 3 |
| 1.3 Nucleotide | 6 | 10 |
| 1.4 Amino acid | 16 | 8 |
| 1.5 Cofactor/vitamin | 2 | 2 |
| 1.6 Energy production | 4 | 2 |
| 2. Information storage and processing | (72) | (32) |
| 2.1 Regulators | 15 | 14 |
| 2.2 Protein biosynthesis and modification | 27 | 12 |
| 2.3 DNA replication/repair | 13 | 6 |
| 2.4 Chaperones | 2 | |
| 2.5 Defense mechanisms | 15 | |
| 3. Transport system | 7 | 7 |
| 4. Phage integrase family | 9 | |
| 5. Other | (70) | (48) |
| 5.1 Cellular division | 3 | 2 |
| 5.2 Cell wall biogenesis | 7 | 2 |
| 5.3 Membrane proteins | 9 | 5 |
| 5.4 Cytosolic proteins | 4 | 2 |
| 5.5 Uncharacterized or hypothetical protein | 47 | 37 |
| Total | 195 | 150 |
Total numbers for each classification are indicated in parentheses.
The Δrgg mutant showed changes in transcription for 29 genes encoding known or putative transcriptional regulators, with increased transcription for 15 genes and decreased transcription for 14 genes. These regulators are predicted to control the expression of genes involved in various cellular activities, including carbohydrate metabolism, lipid metabolism, and DNA replication. This indicated that at least some of the gene expression changes related to growth and metabolic processes in the mutant strain were probably caused by indirect effects through other regulatory circuits. Among these were three TCSTSs—05SSU0883/05SSU0884, 05SSU0906/05SSU0907, and 05SSU0943/05SSU0944—the latter two of which, located in 89K, were approximately 5-and 2-fold more abundant, respectively, in the Δrgg strain. TCSTSs are widespread among bacteria and play important roles in gene expression regulation to provoke a variety of responses to environmental stimuli. Thus, these observations imply that Rgg is part of the regulatory network in SS2 and may influence the activity of other regulatory systems to exert rigorous global regulation.
Microarray analysis showed that 26.6% (n = 21) of a total of 79 genes located in 89K exhibited altered transcript abundance, which obviously exceeded the proportion of regulated genes in the whole genome (15.87%). Moreover, 20 of 21 regulated genes in 89K were upregulated. Interestingly, it seemed that the deletion of rgg also had serious effects on the expression of genes putatively involved in DNA recombination events in SS2. In total, nine genes encoding phage integrase or recombinase in the Δrgg strain were upregulated, with a maximum increase of >9-fold (05SSU0953), but none were downregulated. Furthermore, we found that several genes involved in defense mechanisms were markedly altered by Rgg inactivation, of which a total of 15 gene transcripts were more abundant in the Δrgg strain. These defense mechanisms included the ABC-type multidrug transport system, the antimicrobial peptide transport system, a putative bacteriocin operon, and the restriction/modification system, which offers some protection against bacteriophage attack. It was also found that transcripts of several cell division genes, including divIVA (05SSU0487), ftsE (05SSU1411), and ftsI (05SSU1742), which play an important role in the bacterial division machinery, were differently expressed in the Δrgg mutant.
Deletion of rgg influences protein biosynthesis and modification.
The genes encoding 5 of the 19 putative aminoacyl-tRNA synthetases (AARS) in the SS2 genome—tyrosyl-tRNA synthetase (05SSU0117), methionyl-tRNA synthetase (05SSU1245 and 05SSU1246), threonyl-tRNA synthetase (05SSU1366), cysteinyl-tRNA synthetase (05SSU1924), and glutamyl-tRNA synthetase (05SSU2027)—were more abundant in the Δrgg mutant. Meanwhile, transcription of the gene encoding peptidyl-tRNA hydrolase (05SSU0007) was also ∼3-fold more abundant in the Δrgg mutant. Furthermore, the differentially transcribed genes involved in amino acid metabolism were mostly upregulated, implying that rgg deletion may improve the availability of amino acids and the rate of protein synthesis to a certain extent. In addition, transcripts associated with protein processing and transport systems were more abundant in the Δrgg mutant. For example, the expression of groES/groEL, a classical molecular chaperone that plays an important role in the proper folding and assembly of macromolecular proteins, was increased, as was transcription of the srtF gene (05SSU0475), a member of the sortase family previously identified in a pilus gene cluster (10, 47). Upregulation of these transcripts and many genes encoding membrane surface proteins (i.e., the Sao protein) and cell wall components could potentially enhance adhesion of the Δrgg mutant to Hep-2 cells.
SS2 also has a tmRNA-mediated tagging and proteolysis system for prematurely terminated polypeptides, including a stable tmRNA encoded by ssrA (29), a tmRNA-binding protein encoded by the smpB gene (required for the tmRNA to enter the ribosome [20]), the ATP-dependent serine protease ClpP responsible for degrading tmRNA-tagged polypeptides (13), and some regulatory ATPase subunits encoded by clpX and clpL for ClpP activity (12). This system is induced by nutritional stress and functions to degrade amino acid starvation-induced aberrant proteins during the growth of S. pyogenes (39). Transcriptome analysis of the Δrgg strain revealed that the expression of the smpB (05SSU1390) and clpL (05SSU0389 and 05SSU0390) genes were all approximately half as abundant as those in the WT strain. This observation suggested that Rgg probably enhances the function of the proteolysis system for eliminating harmful abnormal proteins generated by nutritional deficiencies in the host environment.
Rgg-associated expression profile of genes involved in metabolism.
Transcripts encoding enzymes associated with amino acid and lipid metabolism were also influenced by Rgg inactivation. As mentioned above, the genes related to amino acid metabolism were mainly increased, with some exceptions. For example, the transcript abundance for genes encoding enzymes associated with arginine metabolism (arcBC, 05SSU0626, and 05SSU0627) was decreased compared to the parent strain. Apparently, carbohydrate metabolism-related genes were mainly inhibited by Rgg inactivation; 38 such genes were involved in the utilization of nonglucose carbohydrates. For example, products of genes in the tagatose 6-phosphate pathway gene cluster (lacABC, 05SSU1041 to 05SSU1043), which is involved in digesting lactose efficiently, were less abundant in Δrgg mutants than in WT bacteria. The same was true for genes involved in the fructose and mannose (05SSU0449 to 05SSU0454) or maltose (05SSU2131 to 05SSU2137, 05SSU0392 to 05SSU0394) metabolism and transport system. Similarly, transcript levels for the glg operon (05SSU1013 to 05SSU1016), which is responsible for the reciprocal transformation of glycogen and glucose, and the amyX gene (05SSU2138), which encodes pullulanase, an enzyme that hydrolyzes starch into glucose and maltose, were all reduced in the Δrgg mutant.
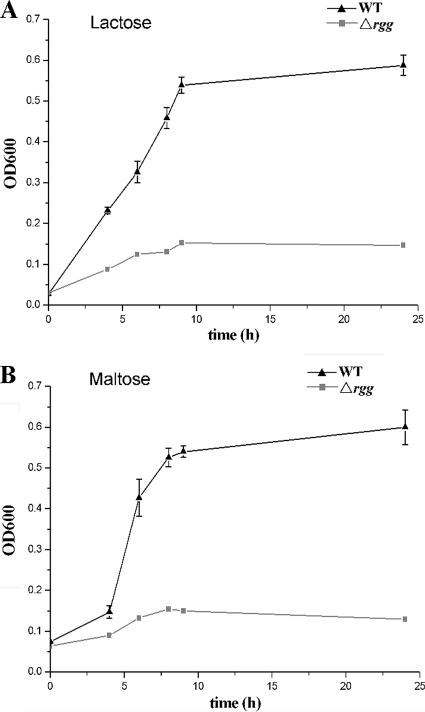
To probe the physiological role of Rgg-associated genes in the metabolism of nonglucose carbohydrates, we used CDM containing these sugars as the primary carbon source. After 24 h of incubation in medium containing 1% lactose, the growth yield of the WT strain reached an OD600 of 0.5. In contrast, the Δrgg mutant did not grow well in this medium (OD600 < 0.15). Similar results were confirmed in medium containing maltose (Fig. 8). However, there was no obvious difference in the growth of the WT and mutant strains when maintained in medium with fructose or mannose as the primary carbon source. This could be explained by the probable existence of separate fructose/mannose transport systems in SS2.
FIG. 8.
Growth of WT and Δrgg mutant strains on different carbon sources. Chemically defined media were used in which either 1% maltose (A) or 1% lactose (B) was supplemented as the primary carbon source.
DISCUSSION
Since pathogenic bacteria often encounter a series of different niches in the host during the various stages of the diseases they cause, it is a great advantage to use global regulatory networks to control the expression of different virulence factors in response to changing environmental cues throughout the infection process (21). In many bacteria, the Rgg regulator has been found, and it has been suggested to control a variety of genes and phenotypes. Here, we identified a member of the Rgg-like transcriptional regulators from a Chinese isolate of SS2. To assess the impact of Rgg on SS2 infections, we initiated our studies by constructing a strain bearing a knockout of the rgg gene and compared the expression profiles of this strain with those of its WT parent during post-exponential growth by using microarray technology. Microarray analysis revealed that Rgg directly or indirectly altered the transcription of 345 genes dispersed throughout the SS2 genome. These changes affected a variety of cellular functions, including nonglucose carbohydrate metabolism, DNA replication/repair, protein biosynthesis, and amino acid metabolism, etc.
Rgg inactivation affected the bacterial phenotype of SS2 in obvious ways, including microscopic phenotype, growth kinetics, and optical density. This phenotype has not been described in the literature of functional research on Rgg-like regulators in other streptococci. Subsequent microarray-based results provided a possible explanation for this phenotype: the transcript of the divIVA gene, which is required for correct cell division and chromosome segregation in most Gram-positive bacteria (46), was half as abundant in Δrgg. In some other streptococcus species, e.g., S. pneumoniae, and in Synechococcus elongatus, disruption of divIVA resulted in cells having a reduced separation frequency and undergoing aberrant polar division, leading to the formation of anucleate minicells and chains of unseparated cells two or three times longer than those of the WT cells (6, 28). These descriptions are very similar to the microscopic phenotype of our Δrgg mutants, suggesting that Rgg may influence the cell division and propagation of SS2 through regulation of divIVA gene expression. Transcripts of genes encoding the FtsE and FtsI proteins, which are responsible for the assembly of constitutive protein and the synthesis of peptidoglycan, respectively, during cell division (45), were also more abundant in the Δrgg mutant. It was reported previously that decreased expression of ftsI resulted in overexpression of divIVA and clear changes in the bacterial morphology of Corynebacterium glutamicum (44). Therefore, it is reasonable to speculate that transcriptional alteration of these genes, which play critical roles in the maintenance of proper cell division and shape, should be responsible for the morphological changes seen in the Δrgg mutants, although their concrete functions are not yet determined by experiments.
The deletion of rgg in SS2 also resulted in the alteration of virulence phenotypes. The mutant exhibited remarkable phenotypic changes, such as hyperadhesion to epithelial cells and increased hemolytic activity. However, we observed that the Δrgg strain had significantly lower lethality than the WT strain in our piglet infections. Pathological examination of the two groups of sacrificed piglets revealed that many organs suffered less damage in the mutant-infected group. Therefore, we can conclude that inactivation of Rgg in SS2 attenuates its pathogenicity in the piglet infection model. In S. pyogenes, Rgg affects virulence through the regulation of extracellular product expression, as well as interaction with several regulatory systems, including Mga, CovRS, and FasBCAX (21). The present results did not indicate that Rgg regulates the secretory virulence-associated factor EF or the orphan response regulator CovR in SS2. Moreover, CovR inactivation did not affect transcription of the rgg gene (31). In addition, we found that the capsule thickness of the Δrgg mutant was similar to that of the WT strain. Therefore, the most likely reason for attenuation of virulence is that Rgg inactivation influences growth and metabolism in the mutant strain.
The inactivation of Rgg in SS2 altered the transcription of several putative phage-associated genes, including 21 genes located in 89K and 9 genes encoding integrase, which were almost entirely transcriptionally upregulated. This finding was consistent with previous research on rgg gene inactivation in S. pyogenes strain NZ131 (5). One possible explanation for the phenomenon, proposed by Dmitriev et al. (5), is that rgg seemed to be a vestige of a prophage-encoded regulatory gene. In SS2 strain 05ZYH33, bioinformatics analysis showed that the rgg gene has a GC content of 36.1%, similar to that of 89K (36.8%), and much lower than the average genomic GC content of 41.1%. These results seemed to corroborate our previous suspicion that 89K was a large integrated alien fragment in the SS2 genome. Currently, research on the function of integrase in SS2 and the integration-excision reaction of 89K is an ongoing process.
Also, we found that Rgg inactivation repressed transcript abundance for enzymes involved in the metabolism of arginine. In S. pyogenes, each enzyme of the arginine deiminase system was expressed in both the exponential and the stationary phases of growth in Δrgg mutants, although these enzymes were synthesized only in the stationary phase of growth in WT strains (2). As previously reported, Rgg inactivation in S. pyogenes caused transcription of the arginine deiminase operon (arcABC) to be greatly increased during exponential growth and decreased during post-exponential growth (5). In the present study, we focused on comparing SS2 wild-type and Δrgg mutant transcript levels in the post-exponential phases of growth. Therefore, our findings of transcriptional downregulation of enzymes associated with arginine catabolism in the Δrgg strain were concordant with those of previous studies in S. pyogenes.
In addition, the transcriptome analyses revealed that the SS2 Rgg regulator could repress expression of several genes associated with various bacterial defense mechanisms. For bacteria, these defense mechanisms, including multidrug transport and antimicrobial peptide biosynthesis, are very important in that they enable them to survive in ecological niches occupied by diverse commensal microorganisms or exposed to antibiotic treatment. Furthermore, restriction/modification enzymes could digest foreign DNA and protect the bacterial chromosome DNA against phage attacks (30). However, as a pathogenic bacterium, SS2 could invade into normally sterile sites of the host or penetrate into infected cells and enter into an intracellular persistence state to escape antibiotic therapy. In these niches, expression of the defense mechanisms discussed above would appear wasteful for SS2 under nutritional stress conditions and therefore might be inhibited by an elaborate regulatory network. Since it has been shown in many studies that multidrug efflux systems are regulated by environmental signals and global regulators (32), our microarray data suggest that Rgg in SS2 may play a role in the negative regulation of these types of defense systems.
In many host tissues, SS2 would confront environments that supply sufficient sugars, including glycogen, starch, galactose, fructose, and maltose, other than glucose. DNA microarray analysis and experimental evidence revealed that Rgg could increase transcription of genes associated with metabolism of a variety of nonglucose carbohydrates. The Rgg regulator may therefore contribute to the adaptation of SS2 to nonglucose carbohydrate sources and distinct nutritional conditions during pathogenesis. Accordingly, it may have potential as a drug target in the development of therapeutics against severe SS2 infections.
During the infection process, SS2 may encounter nutritional deficiencies or be bathed in growth-suppressing substances within the host, for instance, under the intracellular persistence state or in the phagosome. Under such conditions, in order to survive, the bacteria are forced to enter the stationary growth phase and transform their metabolic mode (21). On the basis of the present experimental and microarray analysis results, we suggest that the Rgg regulator in SS2 plays a critical role in global regulatory networks to activate transcription of genes involved in the metabolism of nonglucose carbohydrates. At the same time, it appears to decrease unnecessary expression of bacterial surface proteins responsible for adherence or internalization, thus facilitating the survival of SS2 under nutritional stress conditions within the host.
Supplementary Material
Acknowledgments
We thank Daisuke Takamatsu at the National Institute of Animal Health of Japan for providing the generous gifts of the E. coli-S. suis shuttle vectors pSET1 and pSET2.
This study was supported by grants from the Key Program of National Natural Science Foundation of China (grant 30730081); the National Basic Research Program (grant 973) of China (2006CB504400); National Scientific and Technical Supporting Program (2006BAD06A01); the General Program of National Natural Science Foundation of China (grants 30972638, 81071317, and 36071848); the National High-Tech Research and Development Project 863 (2006AA0Z455); the Natural Science Foundation of Jiangsu Province, China (BK2010025, BK2010113, BK2010114, BK2008066, and BK2009042); the Military Tackle Key Problems Project in Science and Technology (06G039); the Foundation of Innovation of Medical Science and Technology (07Z045); and the 122 Project of Talent Cultivating in Health Professions.
Editor: A. Camilli
Footnotes
Published ahead of print on 13 December 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Charland, N., V. Nizet, and C. E. Rubens. 2000. Streptococcus suis serotype 2 interactions with human brain microvascular endothelial cells. Infect. Immun. 68:637-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaussee, M. A., E. A. Callegari, and M. S. Chaussee. 2004. Rgg regulates growth phase-dependent expression of proteins associated with secondary metabolism and stress in Streptococcus pyogenes. J. Bacteriol. 186:7091-7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaussee, M. S., D. Ajdic, and J. J. Ferretti. 1999. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect. Immun. 67:1715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, C., et al. 2007. A glimpse of streptococcal toxic shock syndrome from comparative genomics of Streptococcus suis 2 Chinese isolates. PLoS One 2:e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dmitriev, A. V., et al. 2006. The Rgg regulator of Streptococcus pyogenes influences utilization of nonglucose carbohydrates, prophage induction, and expression of the NAD-glycohydrolase virulence operon. J. Bacteriol. 188:7230-7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fadda, D., et al. 2003. Characterization of divIVA and other genes located in the chromosomal region downstream of the dcw cluster in Streptococcus pneumoniae. J. Bacteriol. 185:6209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng, Y., H. Zhang, Y. Ma, and G. F. Gao. 2010. Uncovering newly emerging variants of Streptococcus suis, an important zoonotic agent. Trends Microbiol. 18:124-131. [DOI] [PubMed] [Google Scholar]
- 8.Feng, Y., et al. 2008. Functional definition and global regulation of Zur, a zinc uptake regulator in a Streptococcus suis serotype 2 strain causing streptococcal toxic shock syndrome. J. Bacteriol. 190:7567-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng, Y., et al. 2009. Recurrence of human Streptococcus suis infections in 2007: three cases of meningitis and implications that heterogeneous S. suis 2 circulates in China. Zoonoses Public Health 56:506-514. [DOI] [PubMed] [Google Scholar]
- 10.Fittipaldi, N., et al. 2010. Mutations in the gene encoding the ancillary pilin subunit of the Streptococcus suis srtF cluster result in pili formed by the major subunit only. PLoS One 5:e8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fittipaldi, N., et al. 2008. d-Alanylation of lipoteichoic acid contributes to the virulence of Streptococcus suis. Infect. Immun. 76:3587-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frees, D., and H. Ingmer. 1999. ClpP participates in the degradation of misfolded protein in Lactococcus lactis. Mol. Microbiol. 31:79-87. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman, S., E. Roche, Y. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottschalk, M., M. Segura, and J. Xu. 2007. Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim. Health Res. Rev. 8:29-45. [DOI] [PubMed] [Google Scholar]
- 15.Holden, M. T., et al. 2009. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One. 4:e6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hytönen, J., S. Haataja, and J. Finne. 2006. Use of flow cytometry for the adhesion analysis of Streptococcus pyogenes mutant strains to epithelial cells: investigation of the possible role of surface pullulanase and cysteine protease, and the transcriptional regulator Rgg. BMC Microbiol. 6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ichikawa, J. K., et al. 2000. Interaction of pseudomonas aeruginosa with epithelial cells: identification of differentially regulated genes by expression microarray analysis of human cDNAs. Proc. Natl. Acad. Sci. U. S. A. 97:9659-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs, A. A., P. L. Loeffen, A. J. van den Berg, and P. K. Storm. 1994. Identification, purification, and characterization of a thiol-activated hemolysin (suilysin) of Streptococcus suis. Infect. Immun. 62:1742-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacques, M., M. Gottschalk, B. Foiry, and R. Higgins. 1990. Ultrastructural study of surface components of Streptococcus suis. J. Bacteriol. 172:2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karzai, A. W., M. M. Susskind, and R. T. Sauer. 1999. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J. 18:3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 22.Li, M., et al. 2008. SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS One 3:e2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 24.Logan, R. P., et al. 1998. A novel flow cytometric assay for quantitating adherence of Helicobacter pylori to gastric epithelial cells. J. Immunol. Methods 213:19-30. [DOI] [PubMed] [Google Scholar]
- 25.Loughman, J. A., and M. G. Caparon. 2007. Contribution of invariant residues to the function of Rgg family transcription regulators. J. Bacteriol. 189:650-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma, Y., Y. Feng, D. Liu, and G. F. Gao. 2009. Avian influenza virus, Streptococcus suis serotype 2, severe acute respiratory syndrome-coronavirus and beyond: molecular epidemiology, ecology, and the situation in China. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:2725-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyagishima, S. Y., C. P. Wolk, and K. W. Osteryoung. 2005. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol. Microbiol. 56:126-143. [DOI] [PubMed] [Google Scholar]
- 29.Muto, A., C. Ushida, and H. Himeno. 1998. A bacterial RNA that functions as both a tRNA and an mRNA. Trends Biochem. Sci. 23:25-29. [DOI] [PubMed] [Google Scholar]
- 30.O'Sullivan, O., et al. 2009. Comparative genomics of lactic acid bacteria reveals a niche-specific gene set. BMC Microbiol. 9:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan, X., et al. 2009. The orphan response regulator CovR: a globally negative modulator of virulence in Streptococcus suis serotype 2. J. Bacteriol. 191:2601-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piddock, L. J. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi, F., P. Chen, and P. W. Caufield. 1999. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl. Environ. Microbiol. 65:652-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanders, J. W., et al. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27:299-310. [DOI] [PubMed] [Google Scholar]
- 35.Segura, M. 2009. Streptococcus suis: an emerging human threat. J. Infect. Dis. 199:4-6. [DOI] [PubMed] [Google Scholar]
- 36.Smith, H. E., et al. 1999. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 67:1750-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sriskandan, S., and J. D. Slater. 2006. Invasive disease and toxic shock due to zoonotic Streptococcus suis: an emerging infection in the East? PLoS Med. 3:e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staats, J. J., I. Feder, O. Okwumabua, and M. M. Chengappa. 1997. Streptococcus suis: past and present. Vet. Res. Commun. 21:381-407. [DOI] [PubMed] [Google Scholar]
- 39.Steiner, K., and H. Malke. 2001. relA-Independent amino acid starvation response network of Streptococcus pyogenes. J. Bacteriol. 183:7354-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sulavik, M. C., and D. B. Clewell. 1996. Rgg is a positive transcriptional regulator of the Streptococcus gordonii gtfG gene. J. Bacteriol. 178:5826-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sulavik, M. C., G. Tardif, and D. B. Clewell. 1992. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J. Bacteriol. 174:3577-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takamatsu, D., M. Osaki, and T. Sekizaki. 2001. Construction and characterization of Streptococcus suis-Escherichia coli shuttle cloning vectors. Plasmid 45:101-113. [DOI] [PubMed] [Google Scholar]
- 43.Tang, J., et al. 2006. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 3:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valbuena, N., et al. 2006. Morphological changes and proteome response of Corynebacterium glutamicum to a partial depletion of FtsI. Microbiology 152:2491-2503. [DOI] [PubMed] [Google Scholar]
- 45.Vicente, M., A. I. Rico, R. Martínez-Arteaga, and J. Mingorance. 2006. Septum enlightenment: assembly of bacterial division proteins. J. Bacteriol. 188:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vicente, M., and M. García-Ovalle. 2007. Making a point: the role of DivIVA in streptococcal polar anatomy. J. Bacteriol. 189:1185-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, C., et al. 2009. The involvement of sortase A in high virulence of STSS-causing Streptococcus suis serotype 2. Arch. Microbiol. 191:23-33. [DOI] [PubMed] [Google Scholar]
- 48.Ye, C., et al. 2009. Clinical, experimental, and genomic differences between intermediately pathogenic, highly pathogenic, and epidemic Streptococcus suis. J. Infect. Dis. 199:97-107. [DOI] [PubMed] [Google Scholar]
- 49.Yu, H., et al. 2006. Human Streptococcus suis outbreak, Sichuan, China. Emerg. Infect. Dis. 12:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.