Abstract
Staphylococcus aureus clinical isolates are capable of producing at least two distinct types of biofilm mediated by the fibronectin-binding proteins (FnBPs) or the icaADBC-encoded polysaccharide intercellular adhesin (PIA). Deletion of the major autolysin gene atl reduced primary attachment rates and impaired FnBP-dependent biofilm production on hydrophilic polystyrene in 12 clinical methicillin-resistant S. aureus (MRSA) isolates but had no effect on PIA-dependent biofilm production by 9 methicillin-susceptible S. aureus (MSSA) isolates. In contrast, Atl was required for both FnBP- and PIA-mediated biofilm development on hydrophobic polystyrene. Here we investigated the role of Atl in biofilm production on hydrophilic polystyrene. The alternative sigma factor σB, which represses RNAIII expression and extracellular protease production, was required for FnBP- but not PIA-dependent biofilm development. Furthermore, mutation of the agr locus enhanced FnBP-dependent biofilm development, whereas a sarA mutation, which increases protease production, blocked FnBP-mediated biofilm development. Mutation of sigB in MRSA isolate BH1CC lowered primary attachment rates, in part via reduced atl transcription. Posttranslational activation or inhibition of Atl activity with phenylmethylsulfonyl fluoride and polyanethole sodium sulfonate or mutation of the Atl amidase active site interfered with lytic activity and biofilm development. Consistent with these observations, extracellular DNA was important for the early stages of Atl/FnBP-dependent biofilm development. Further analysis of atl regulation revealed that atlR encodes a transcriptional repressor of the major autolysin and that an atlR::Tcr mutation in BH1CC enhanced biofilm-forming capacity. These data reveal an essential role for the major autolysin in the early events of the FnBP-dependent S. aureus biofilm phenotype.
Staphylococcus aureus and S. epidermidis are among the most common hospital pathogens associated with a wide variety of infections, including those involving indwelling medical devices. Intensive care unit patients are particularly at risk of acquiring device-related infections, which involve biofilms. Such infections are particularly problematic because cells in the biofilm are significantly more resistant to host defenses and antimicrobial agents, resulting in significant morbidity and mortality. Differences in the biofilm mechanisms of S. epidermidis and S. aureus have been recognized (45). Production of the icaADBC-encoded polysaccharide intercellular adhesin (PIA) or polymeric N-acetylglucosamine (PNAG) is now a well-defined mechanism of biofilm development in both species. Carriage of the ica locus is strongly associated with biofilm-forming capacity in S. epidermidis isolates implicated in device-related infections compared to commensal strains (72). In contrast, the correlation between ica carriage and biofilm-forming capacity in S. aureus is more ambiguous, even though this locus is maintained, expressed, and regulated in almost all S. aureus isolates (48). Thus, the role of ica in S. aureus biofilm development is complex, particularly given that icaADBC-independent biofilm development has been described in this organism (4, 17, 47, 48) (and, more recently, in S. epidermidis [27]).
Predictably, icaADBC-independent biofilm development mechanisms involve cell surface components such as teichoic acids (58) and microbial surface components recognizing adhesive matrix molecules (18). Twenty-eight surface proteins have been identified in S. aureus (20), of which 21 are predicted to contain LPXTG motifs anchored to the cell wall by the srtA-encoded sortase (42). Deletion of srtA interferes with the normal display of LPXTG surface proteins and results in severe virulence defects (42). The LPXTG-containing surface proteins Bap (34), Aap/SasG (16, 57), SasC (60), and protein A (43) are known mediators of biofilm development.
We recently identified a new S. aureus biofilm phenotype mediated by the LPXTG-anchored fibronectin (Fn)-binding proteins (FnBPs) FnBPA and FnBPB (17, 46, 47). Using a collection of clinical isolates from Beaumont Hospital, Dublin, Ireland, our studies revealed that biofilm development in methicillin-resistant S. aureus (MRSA) strains was FnBP dependent and ica independent and was triggered by mild acid stress in medium supplemented with glucose (47, 48). Importantly, deletion of both fnb genes had no effect on primary attachment, suggesting that they are involved in biofilm accumulation or maturation (47). In contrast, PIA production played a more important role in methicillin-susceptible S. aureus (MSSA) biofilm development and was triggered by osmotic stress in medium supplemented with NaCl (47, 48). Mutations in fnbAB had no effect on biofilm production by MSSA strains expressing a PIA-dependent biofilm phenotype (47). A role for the FnBPs in citrate-induced S. aureus biofilm production was also reported in the laboratory strain RN6390 (61). More recently, by systematically mutating LPXTG protein genes, Vergara-Irigaray et al. (66) identified an MRSA isolate, designated 132, capable of switching between PIA- and FnBP-dependent biofilm production. Spontaneous or induced mutations in the agr global regulatory locus, which repress expression of the FnBPs, promoted biofilm production by strain 132 (66). Similarly, mutation of LexA, a repressor of fnbB expression (6), also increased FnBP-mediated biofilm production (66). Importantly, using a mouse foreign body infection model, an isogenic fnbAB mutant was significantly less able to colonize a subcutaneous implanted catheter than the wild type or icaADBC mutant strains (66), suggesting that the FnBP-dependent biofilm phenotype may be an important virulence determinant.
To begin to investigate the early events in the FnBP-dependent biofilm phenotype, we turned our attention to the major autolysin, which has previously been implicated in primary attachment to surfaces in S. epidermidis (25) and S. aureus (7). Atl is a wall-anchored 1,256-amino-acid bifunctional peptidoglycan hydrolase that plays an important role in daughter cell separation following cell division. Atl proprotein contains amidase (Ami) and glucosaminidase (GL) domains separated by three direct repeats, R1, R2, and R3. Proteolytic cleavage of pro-Atl via an uncharacterized mechanism generates a 62-kDa amidase with C-terminal R2 and R3 repeats (Ami-R1,2) and a 51-kDa glucosaminidase with an N-terminal R3 repeat (R3-GL) (25, 49, 62). The structure of the S. epidermidis Ami zinc-dependent metalloenzyme was recently solved, and amino acids involved in coordinating the zinc ion and catalytic activity were identified (73). The repeat domains of Atl target the Ami-R1,2 and R3-GL enzymes to the septal region of the cell surface, thus localizing peptidoglycan hydrolysis to the site of cell separation (2, 71). Schlag et al. (59) recently elucidated the mechanism underpinning localized peptidoglycan hydrolysis. Their study revealed that wall teichoic acid (WTA) prevents Atl binding to the cell wall and that Ami-R1,2 binding at the cross-wall region was increased, presumably due to a lower WTA concentration (59). This mechanism explains why the Atl-derived peptidoglycan hydrolases are primarily active at the site of cell separation and why rates of autolysis are higher in WTA mutants (22, 32, 38, 65). The repeat domains of Atl have also been shown to bind various host extracellular matrix proteins, including vitronectin and fibronectin (1, 24-26).
In this study, we constructed atl deletion mutants in clinical isolates of MRSA and MSSA and characterized the role of the global regulators SarA, Agr, and SigB in Atl-dependent, FnBP-mediated biofilm production on hydrophilic polystyrene. The relationship between autolytic activity, extracellular DNA (eDNA) release, and biofilm production was examined. Transcriptional regulation of atl by AtlR was examined and correlated with the biofilm phenotype. Our findings reveal an essential role for Atl in ica-independent S. aureus biofilm production.
MATERIALS AND METHODS
S. aureus strains.
The S. aureus strains and plasmids used this study are described in Table 1. Clinical S. aureus isolates from Beaumont Hospital, Dublin, have been described previously (47, 48). A previously characterized genetically and geographically diverse collection of 88 MRSA and 55 MSSA isolates (54, 55) was kindly provided by D. A. Robinson.
TABLE 1.
Strains
| Strain or plasmid | Relevant detailsa | Reference or source |
|---|---|---|
| Strains | ||
| S. aureus | ||
| 8325-4 | 8325 derivative cured of prophages, 11-bp deletion in rsbU | 28 |
| RN4220 | Restriction-deficient derivative of 8325-4 | 33 |
| ICA1 | ΔicaABDC::Tcr, isogenic 8325-4 mutant | 17 |
| DU5883 | fnbA::TcrfnbB::Emr, isogenic 8324-5 mutant | 21 |
| PC6911 | agr::Tcr, isogenic 8325-4 mutant | 44 |
| PC400 | sigB::Tcr, isogenic 8325-4 mutant | 12 |
| UAMS-240 | sarA::Tcr, isogenic RN6390 mutant | 8 |
| WA400 | ΔRNAIII::cat86, isogenic 8325-4 mutant | 5, 29 |
| JT1392 | Δatl::Cmr, isogenic RN4220 mutant | 63 |
| ATLR1 | atlR::Tcr, isogenic RN4220 mutant | This study |
| DAR34 | MRSA, SCCmec type II, MLST type 8, clonal complex 8 | 55 |
| DAR35 | MRSA, SCCmec type II, MLST type 8, clonal complex 8 | 55 |
| DAR69 | MRSA, SCCmec type II, MLST type 239, clonal complex 239 | 55 |
| DAR75 | MSSA, MLST type 5, clonal complex 5 | 55 |
| DAR87 | MSSA, MLST type 11, clonal complex 5 | 55 |
| DAR107 | MSSA, MLST type 8, clonal complex 8 | 55 |
| DAR119 | MRSA, SCCmec type IV, MLST type 8, clonal complex 8 | 55 |
| DAR124 | MRSA, SCCmec type III, MLST type 239, clonal complex 239 | 55 |
| DAR174 | MSSA, MLST type 8, clonal complex 8 | 55 |
| DAR177 | MSSA, MLST type 5, clonal complex 5 | 55 |
| DAR203 | MRSA, SCCmec type III, MLST type 239, clonal complex 239 | 55 |
| BH2 | MRSA, SCCmec type IV, clonal complex 22 | 48 |
| BH3 | MRSA, SCCmec type II, MLST type 8, clonal complex 8 | 48 |
| BH4 | MRSA, SCCmec type II, MLST type 8, clonal complex 8 | 48 |
| BH5 | MRSA, SCCmec type IV, clonal complex 22 | 48 |
| BH10 | MRSA, SCCmec type IV, MLST type 22, clonal complex 22 | 48 |
| BH1CC | MRSA, SCCmec type II, MLST type 8, clonal complex 8 | 48 |
| BH40 | MSSA, MLST type 1099, clonal complex 5 | 48 |
| BH48 | MSSA, MLST type 8, clonal complex 8 | 48 |
| BH49 | MSSA, MLST type 6, clonal complex 6 | 48 |
| BH51 | MSSA, MLST type 5, clonal complex 5 | 48 |
| E. coli | ||
| TOPO | recA1endA1 lac [F′ proAB lacIq Tn10 (Tetr)] | Invitrogen |
| Rosetta | F−ompThsdSB(rB− mB−) galdcm λ(DE3 [lacIlacUV5-T7 gene 1 ind1sam7nin5]) pLysSRARE (Cmr) | Invitrogen |
| Plasmids | ||
| pT181 | 4.45-kb S. aureus plasmid containing tetA(K) | 31 |
| pCR-Blunt II-Topo | PCR cloning vector, Kmr Zeor | Invitrogen |
| pBT2 | Temperature-sensitive E. coli-Staphylococcus shuttle vector; Apr (E. coli), Cmr (Staphylococcus) | 10 |
| pBlue-tet | pBluescript containing the tetA gene from pT181 on a 2,352-bp HindIII fragment. | This study |
| pLI50 | E. coli-Staphylococcus shuttle vector; Apr (E. coli), Cmr (Staphylococcus) | 36 |
| pSAatlR1 | 4,418-bp PCR product containing the atlR gene amplified from 8325-4 using primers SAatl1 and SAatl2 and cloned into plasmid pCR-Blunt II-TOPO | This study |
| pSAatlR2 | Deletion of 44-bp fragment containing an EcoRV site from pSAatlR2 by PstI-XhoI digestion, followed by T4 polymerase treatment and ligation | This study |
| pSAatlR3 | 2,236-bp SwaI-SmaI fragment containing the tetA gene from pBlue-tet cloned into EcoRV site located in the atlR gene of pSAatlR2 | This study |
| pSAatlR4 | 8,686-bp BamHI-SmaI fragment containing atlR::Tcr allele from pSAatlR3 cloned into pBT2 digested with BamHI-SmaI | This study |
| patlRTOPO | 967-bp PCR product containing the atlR gene amplified from 8325-4 using primers atlR-Cmp_for and atlR-Cmp_rev cloned into pCR-Blunt II-TOPO | This study |
| patlR | 985-bp EcoRI fragment containing the atlR gene from patlRTOPO cloned into the EcoRI site of pLI50 | This study |
| patlTOPO | 4,312-bp PCR product containing the atl gene amplified from BH1CC using primers SAatlcomp1_for and SAatlcomp1_rev cloned into pCR-Blunt II-TOPO | This study |
| pLI50-tet | 2,413-bp XbaI-HindIII fragment containing the tetA gene from pBlue-tet cloned into XbaI-HindIII-digested pLI50 | This study |
| patl | 4,330-bp EcoRI fragment containing the atl gene from patlTOPO cloned into the EcoRI site of pLI50-tet | This study |
| patlH265A | patl with an Ami H265A mutation | This study |
| pTATLR1 | 420-bp PCR product containing the atlR gene amplified from 8325-4 using primers SA_AtlR1 and SA_AtlR2 cloned into pCR-Blunt II-TOPO | This study |
| pMAL-c2X | Maltose-binding protein fusion vector | NEB |
| pMATLR1 | EcoRI fragment from pTATLR1 cloned into the EcoRI site downstream of malE in pMAL-c2x | This study |
SCCmec, staphylococcal cassette chromosome mec; MLST, multilocus sequence typing.
Media and growth conditions.
Escherichia coli strains were grown at 37°C on LB medium supplemented, when required, with ampicillin (Ap; 100 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (Cm; 30 μg/ml). Overnight Express medium (Novagen) was used for purification of protein from E. coli. S. aureus strains were grown at 30°C or 37°C on brain heart infusion (BHI; Oxoid) medium supplemented, when required, with chloramphenicol (10 to 20 μg/ml), erythromycin, and tetracycline (Tc; 2.5 μg/ml). BHI broth was supplemented where indicated with 1% glucose or 4% NaCl. BHI and BHI glucose media were also supplemented, where indicated, with phenylmethylsulfonyl fluoride (PMSF; 3 μM; Sigma) or polyanethole sodium sulfanate (PAS; 500 μg/ml; Sigma).
Genetic techniques.
Genomic DNA and plasmid DNA were prepared using Wizard genomic DNA and plasmid purification kits (Promega). Prior to DNA extraction, cells were pretreated with 5 to 10 μl of a 1-mg/ml solution of lysostaphin (Ambi Products, Lawrence, NY) in 100 μl 50 mM EDTA to facilitate subsequent lysis. Restriction and DNA-modifying enzymes (Roche and NEB) were used according to the manufacturers' instructions. All oligonucleotide primers used for PCR, reverse transcription-PCR (RT-PCR), and DNA sequencing were supplied by MWG Biotech, Germany (Table 2) .
TABLE 2.
Oligonucleotides
| Target | Primer name | Primer sequence (5′-3′) |
|---|---|---|
| atl | atl1 | AAATTGCTCTTGCGTTGTCA |
| atl2 | CAGTTGCCACTGCTCGAATA | |
| atlR | atlR-Cmp_for | GCGTGATCAAAAACCATAACG |
| atlR-Cmp_rev | TGTGTGCACAACGACACTAAA | |
| icaA | SAicaEON1 | TACCTTGCCTAACCCGTAC |
| SAicaEON2 | GTTGGCTCAATGGGGTCTAA | |
| fnbA | FnBPAfwd | CGCGGATCCGGTACAGATGTAACAAGTAAAG |
| FnBPArev | GACGCGTCGACTTAATTCGGACCATTTTTCTCATT | |
| spa | SAspa1 | TAATTGGTGCAACTGGGACA |
| SAspa2 | CACATTCAAAGCCCCACTTT | |
| atl | SAatlcomp1_for | GGGGGACTTTTGATCCT |
| SAatlcomp2_rev | CAGTTGCCACTGCTCGAATA | |
| atlR | SAatl1 | GCTGGAACATTGGCAAAAAT |
| SAatl2 | AAGCTGGTGCAGTTTCTGGT | |
| sarA | SAsarA1 | GCGTTGATTTGGGTAGTATGC |
| SAsarA2 | TCACCAAATTGCGCTAAACA | |
| agr | SAagr1 | GGGGCTCACGACCATACTTA |
| SAagr2 | CGGGGTAGGAAATTGTAGCA | |
| atlR | atlR_for | GTGAATAGAAGATTTGGACAACG |
| atlR_rev | TGTACGTGCTTGATCAGTCAAA | |
| atl | SAatlFOR | AAGCAGCTGAGACGACACAA |
| SAatlREV | TTGCTGTTTTTGGTTGGACA | |
| atl promoter | SAatlProm1 | CACGACAATAGCAAACACATAATTTAG |
| SAatlProm2 | GCAACATGAACATAGGATCAAAAGTCA | |
| atlR | SA_AtlR1 | ATGTATAAACAACTTGAAAAACTTATTACA |
| SA_AtlR2 | TTACAACGATATGCGTCGTTGATTTAACTT | |
| atl | SaH265A_5′ | GAAGGTATCGTAGTTGCTGATACAGCTAATGATCa |
| SaH265A_3′ | GATCATTAGCTGTATCAGCAACTACGATACCTTCa |
Underlining indicates nucleotide changes to generate an H265A amino acid substitution in active sites of the Atl amidase enzyme.
Construction of Δatl::Cmr mutants.
The Δatl::Cmr allele from strain JT1392 was transduced into clinical isolates using phage 80α as described previously (48). Candidate mutants were confirmed by PCR analysis using the SAatlFOR and SAatlREV primers (data not shown). The Δatl::Cmr mutants were complemented using the multicopy Staphylococcus-E. coli shuttle vector pLI50 (36), in which the Cmr gene was replaced with the tetA (Tcr) gene from pT181, which had first been cloned on a 2,352-bp HindIII fragment into pBluescript to generate pBlue-tet. The atl gene was amplified from MRSA isolate BH1CC on a 4,312-bp PCR product using primers SAatlcomp1_for and SAatlcomp1_rev and cloned into pCR-Blunt II-TOPO to generate patlTOPO, before being subcloned on an EcoRI fragment into pLI50-tet to create patl (Table 1).
Construction of an atlR::Tcr mutation.
A 4,418-bp fragment containing the atlR gene from strain 8325-4 was amplified by PCR with Phusion high-fidelity DNA polymerase (NEB) using primers SAatl1 and SAatl2 (Table 2) and cloned into pCR-Blunt II-TOPO to create pSAatlR1. To facilitate this, an EcoRV site was first removed from pSAatlR1 by digestion with PstI and XhoI, treatment with T4 polymerase, and religation to generate pSAatlR2. The Tcr gene from pBlue-tet was subcloned on a 2,236-bp SwaI-SmaI fragment into the now unique EcoRV site in the atlR gene of pSAatlR2 to create pSAatlR3. An 8,686-bp BamHI-SmaI fragment containing the atlR::Tcr allele from pSAatlR3 was then ligated into pBT2 digested with BamHI-SmaI to create pSAatlR4. Allele replacement of temperature-sensitive pSAatlR4 in RN4220 was achieved following repeated growth (three subcultures) at 42°C for 24 h without antibiotic selection and then selection of Tcr colonies on BHI agar plates. Tcr colonies were then screened for sensitivity to Cm to confirm plasmid loss, and PCR was used to verify the presence of the atlR::Tcr allele on the chromosome. Bacteriophage 80α was used to transduce the mutation into other S. aureus strains and clinical isolates.
Construction of AmiE H265A mutation.
The histidine at amino acid 265 in the AmiE domain of Atl was recently shown to be essential for the catalytic activity of the S. epidermidis enzyme (73). H265 was mutated to alanine using Phusion polymerase and primers SaH265_5′ and SaH265A_3′ (Table 2), which were essentially the same as those described by Zoll et al. (73) but adapted for the S. aureus atl gene sequence. Plasmid patlTOPO was used as the template. Successful mutagenesis of the DNA sequence encoding the H265 residue was associated with the loss of a BspHI restriction site, and candidate plasmids harboring the mutation were digested with this enzyme before being confirmed by sequencing. The atlH265A allele was subsequently subcloned from patlTOPO into pLI50-tet on an EcoRI fragment, before being electroporated into RN4220 and ultimately into the BHI1CC Δatl::Cmr mutant.
Biofilm assays.
Semiquantitative determinations of biofilm formation under static conditions using Nunclon tissue-culture-treated, hydrophilic 96-well polystyrene plates (Nunc, Denmark) or untreated, hydrophobic 96-well polystyrene plates (Sarstedt, Germany) were performed on the basis of the method of Christensen et al. (15), as described previously (47, 48). Where indicated, hydrophobic 96-well plates were precoated with various concentrations of human fibronectin (Sigma) for 24 h at 4°C before biofilm assays were performed.
Bacterial adherence to hydrocarbon assay.
Cultures were grown with shaking in BHI medium at 37°C to an A600 of 1.0 before they were washed twice in Dulbecco's phosphate-buffered saline (PBS; Oxoid) and resuspended in 4.8 ml 1× PBS (A600 = 0.5). Two hundred microliters of p-xylene (Sigma) was added to the cell suspension, and the mixture was vortexed vigorously for 1 min. After 20 min (to allow phase separation), the percentage of cells (determined spectrophotometrically at A600) retained in the lower, aqueous phase was measured. Each experiment was repeated three times.
Primary attachment assays.
Primary attachment assays were performed on the basis of the method of Lim et al. (37). Overnight cultures grown at 37°C in BHI glucose medium were diluted in the same medium to approximately 300 CFU/100 μl. One hundred microliters of the adjusted suspension was spread on Nunclon tissue-culture-treated, hydrophilic polystyrene petri dishes (Nunc). After incubation at 37°C for 30 min, the petri dishes were gently rinsed three times with 5 ml of sterile PBS (pH 7.5) and covered with 15 ml of molten 0.8% BHI agar cooled to 48°C. Where indicated, cells were pretreated with 1 mg/ml DNase I for 2 h at 37°C and resuspended in BHI glucose medium before the primary attachment assay was performed. Primary attachment was expressed as a percentage of the numbers of CFU remaining on the petri dishes after the cultures were washed compared to the initial numbers of CFU. Each experiment was repeated three times.
RNA purification and analysis.
Bacterial cells were collected and immediately stored at −20°C in RNAlater (Ambion) to ensure maintenance of RNA integrity prior to purification. Total RNA was subsequently isolated using a GenElute total RNA purification kit (Sigma), according to the manufacturer's instructions, after it was washed in 500 μl of 50 mM EDTA and a 5- to 10-min pretreatment of the cells with 5 to 10 μl of a 1-mg/ml solution of lysostaphin in 100 μl 50 mM EDTA. Purified RNA was eluted and stored in RNAsecure resuspension solution (Ambion), and the integrity of the RNA was confirmed by agarose gel electrophoresis. Residual DNA present in RNA preparations following purification was removed using DNA-free DNase treatment and removal reagents (Ambion). The concentration of RNA was determined using a NanoDrop spectrophotometer. Real time RT-PCR was performed on a LightCycler instrument using a SYBR green I RNA amplification kit (Roche), following the manufacturer's recommended protocol. For RT-PCRs, reverse transcription was performed at 61°C for 20 min, followed by a denaturation step at 95°C for 30 s and 35 amplification cycles of 95°C for 2 s, 50°C for 5 s, and 72°C for 8 s. Melting curve analysis was performed at 45°C to 95°C (temperature transition, 0.1°C per s) with stepwise fluorescence detection (42, 44). For LightCycler RT-PCR, RelQuant software (Roche) was used to measure the relative expression of target genes. The gyrB gene was used as the internal standard in all RT-PCR experiments. RT-PCR experiments were performed at least three times, and average data with standard deviations are presented.
Measurement of autolytic and bacteriolytic activity.
Triton X-100 (TRX-100)-induced autolysis assays were performed essentially as described by Mani et al. (39). Overnight cultures of S. aureus were subcultured in BHI medium and incubated at 37°C with shaking (200 rpm) to an A600 of 1.0. Cells were then pelleted and washed twice with ice-cold PBS and subsequently resuspended in PBS containing 0.02% Triton X-100. The suspensions were then incubated with shaking (200 rpm) at 37°C. A600 readings were taken at 0 min, at 10 min, and then at 30-min intervals. Triton X-100-induced autolysis was measured and is indicated as a percentage of the initial A600 (A600i).
Bacteriolytic activity in supernatants from growing cultures was assayed by measuring the rate of change in turbidity of the substrate (heat-killed RN4220 cells) as described previously (19). Nine milliliters of filter-sterilized culture supernatant was mixed with 1 ml heat-killed RN4220 cells suspended in PBS to an initial A600 of 0.5. The cell-supernatant mixture was then incubated at 30°C with shaking (200 rpm) for 4 h, and the rate of change in turbidity was measured at 30-min intervals. Bacteriolytic activity is expressed as percent lysis at time t, where lysis at time t is equal to [A600i − A600 at time t)/A600i]. Each experiment was repeated three times.
Construction of maltose-binding protein (MBP)-AtlR expression vector and purification of recombinant AtlR protein.
The atlR gene was amplified by PCR with Phusion high-fidelity DNA polymerase using primers SA_AtlR1 and SA_AtlR2 and ligated into plasmid pCR-Blunt II-TOPO to create pTATLR1. pTATLR1 was digested with EcoRI, and the atlR gene was subcloned into the EcoRI site downstream of the malE gene in pMal-c2x vector (NEB) to generate pMATLR1. pMATLR1 was transformed into E. coli strain Rosetta/pLysSRARE, and overnight cultures were grown in Overnight Express medium (Novagen) containing Ap at 50 μg/ml and Cm at 30 μg/ml. Cells were resuspended in 5 ml column buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT]) and sonicated on ice for 2 min (15-s bursts with 30-s intervals). Following refrigerated centrifugation at 14,000 rpm for 30 min, the soluble fraction was added to 50 ml column buffer and passed through an amylose resin column (NEB) at a flow rate of 1 ml/min. The column was washed and eluted with column buffer containing 10 mM maltose. Purified fractions were collected in 500-μl aliquots and analyzed on a 10% SDS-polyacrylamide gel. The concentration of purified protein was measured using a NanoDrop spectrophotometer, and the recombinant protein was stored at −80°C in 50-μl aliquots.
Electrophoretic mobility shift assay.
The atl promoter was amplified by PCR using Phusion high-fidelity DNA polymerase (NEB) and the biotinylated primers SAatlProm1 and SAatlProm2. The product was purified first from a 2% agarose gel and then from a nondenaturing 5% acrylamide gel. The DNA concentration of the free probe was determined using a NanoDrop spectrophotometer. Twenty-five nanograms of biotinylated probe was added to recombinant MBP-AtlR protein in a 20-μl binding reaction mixture containing 0.2 μg poly(dI-dC), 5% glycerol, 5 mM MgCl2, 10 mM Tris, 50 mM KCl, and 1 mM DTT at pH 7.5). The reaction mixture was incubated at room temperature for 20 min, loaded onto a 5% nondenaturing polyacrylamide gel, and electrophoresed at 100 V for 65 min. DNA was transferred onto a Biodyne-B nylon membrane (Pall) at 4°C in prechilled 0.5% TBE (Tris-borate-EDTA) buffer at 80 V for 60 min using an electroblotting apparatus (Bio-Rad) and cross-linked to the membrane under UV light for 10 min. The biotinylated probes were detected using a LightShift chemiluminescent electrophoretic mobility shift assay (EMSA) kit (Pierce Chemicals, Rockford, IL) and a FluorChem FC2 imaging system (Alpha Innotech).
Statistical analysis.
Two-tailed, two-sample equal-variance Student's t tests (Microsoft Excel 2007 software) were used to determine statistically significant differences in primary attachment, biofilm-forming capacity, and lytic activity.
RESULTS
Role of the major autolysin in biofilm phenotype of MRSA isolate BH1CC.
We recently reported a new role for the fibronectin-binding proteins FnBPA and FnBPB in S. aureus biofilm production (47). FnBP-dependent biofilm development is triggered by mild acid stress in BHI glucose medium, and our findings implicated the FnBPs in biofilm accumulation and not primary attachment to surfaces (47). To investigate the early events in FnBP-dependent biofilm production, we investigated the role of the major autolysin, Atl, in the MRSA clinical isolate BH1CC, which produces an FnBP-dependent biofilm. Atl has previously been implicated in primary attachment of S. epidermidis (25) and S. aureus (7) to surfaces. A Δatl::Cmr deletion mutation (63) was transduced from laboratory strain RN4220 into BH1CC and its isogenic fnbAB::Tcr double mutant using phage 80α. The BH1CC atl null mutant formed large macroscopic cell aggregates in broth cultures (data not shown); was more hydrophilic (P < 0.01), as determined by a bacterial adherence to hydrocarbon assay (data not shown); and exhibited significantly reduced (P < 0.0001) TRX-100-induced autolytic activity (Fig. 1A).
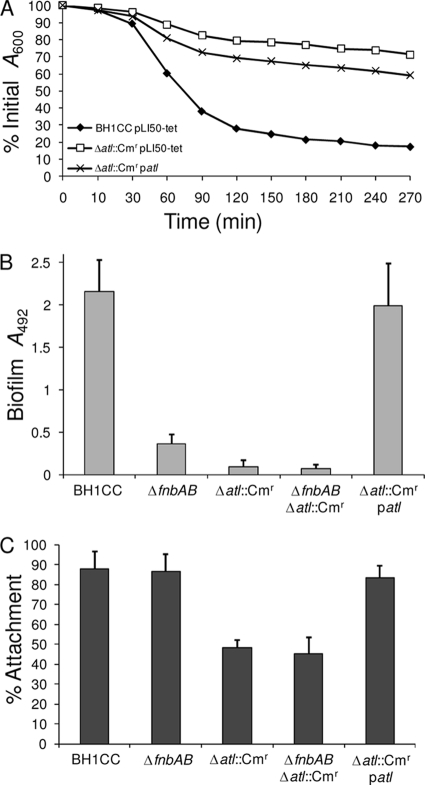
FIG. 1.
Phenotypic impact of Δatl::Cmr mutation in MRSA isolate BH1CC. (A) Triton X-100-induced autolysis of BH1CC/pLI50-tet, BH1CC Δatl::Cmr'/pLI50-tet, and BH1CC Δatl::Cmr/patl. Three independent experiments were performed, and results of a representative experiment are shown. (B) Biofilm phenotypes of BH1CC, BH1CC fnbAB::Tcr, BH1CC Δatl::Cmr, BH1CC fnbAB::Tcr Δatl::Cmr, and BH1CC Δatl::Cmr/patl grown for 24 h at 37°C in BHI glucose medium in hydrophilic 96-well polystyrene plates. Assays were repeated three times, and standard deviations are indicated. (C) Primary attachment phenotypes on hydrophilic, polystyrene petri dishes for BH1CC, BH1CC fnbAB::Tcr, BH1CC Δatl::Cmr, BH1CC fnbAB::Tcr Δatl::Cmr, and BH1CC Δatl::Cmr/patl grown to stationary phase in BHI glucose medium. Primary attachment is expressed as the percentage of the numbers of CFU attached to the petri dish after it was washed. Experiments were repeated three times, and standard deviations are indicated. *, P < 0.001.
Deletion of atl in isolate BH1CC significantly impaired (P < 0.0001) biofilm formation on hydrophilic, tissue-culture-treated polystyrene in BHI glucose medium (Fig. 1B). Furthermore, the Δatl::Cmr mutation resulted in a further, significant (P < 0.001) reduction in biofilm production in the BH1CC fnbAB::Tcr double mutant (Fig. 1B). The atl null mutant biofilm defect correlated with significantly decreased (P < 0.0001) levels of primary attachment to hydrophilic polystyrene (Fig. 1C). Introduction of patl into BH1CC Δatl::Cmr partially complemented TRX-100-induced autolysis over a 4-hour time period (Fig. 1A) and almost fully complemented autolytic activity over a 24-hour time period (data not shown). In addition, patl fully complemented both biofilm-forming capacity (P < 0.001) (Fig. 1B) and primary attachment (Fig. 1C) in the BH1CC Δatl::Cmr mutant. These data reveal that biofilm development is significantly reduced but not abolished in the fnbAB mutant, whereas impaired primary attachment in the atl mutant completely abolishes biofilm formation in BH1CC.
Role of atl in biofilm phenotype of MRSA and MSSA clinical isolates.
To further evaluate the role of Atl in the S. aureus biofilm phenotype, phage 80α transduction was used to transduce the Δatl::Cmr allele into a range of MRSA and MSSA strains. PCR was used to verify atl null mutants. Using previously characterized clinical and laboratory strains (47, 48), three MRSA strains which form fnbAB-dependent, icaADBC-independent biofilms and four MSSA strains which produce icaADBC-dependent, FnBP-independent biofilms were initially examined. The atl deletion significantly reduced (P < 0.0001) FnBP-dependent biofilm development on hydrophilic polystyrene in all three MRSA isolates grown in BHI glucose medium (Fig. 2A). None of these isolates make a significant biofilm in BHI or BHI NaCl medium (data not shown). In contrast, the atl deletion had no impact on NaCl-induced, PIA-dependent biofilm production in the four MSSA isolates tested (P > 0.05) (Fig. 2B). Mutation of atl abolished glucose-induced biofilm production in an additional eight MRSA clinical isolates (Fig. 2C) but had no significant effect on NaCl-induced biofilm development in five MSSA clinical isolates (Fig. 2D).
FIG. 2.
Contribution of the atl, fnbAB, and icaADBC loci to biofilm regulation in clinical isolates of S. aureus. (A) Biofilm phenotypes of MRSA isolates BH3, BH4, and BH10 and their isogenic ΔicaADBC::Tcr, fnbAB::Tcr, Δatl::Cmr, and fnbAB::Tcr Δatl::Cmr mutants grown for 24 h at 37°C in BHI glucose medium in hydrophilic 96-well polystyrene plates. WT, wild type. (B) Biofilm phenotypes of MSSA isolates BH48, BH49, BH51, and 8325-4 and their isogenic ΔicaADBC::Tcr, fnbAB::Tcr, and Δatl::Cmr mutants grown for 24 h at 37°C in BHI NaCl medium in hydrophilic 96-well polystyrene plates. (C) Biofilm phenotypes of MRSA isolates DAR34, DAR35, DAR69, DAR119, DAR124, DAR203, BH2, and BH5 and their isogenic Δatl::Cmr mutants grown for 24 h at 37°C in BHI glucose medium in hydrophilic 96-well polystyrene plates. (D) Biofilm phenotypes of MSSA isolates DAR75, DAR87, DAR107, DAR174, and DAR177 and their isogenic Δatl::Cmr mutants grown for 24 h at 37°C in BHI NaCl medium in hydrophilic 96-well polystyrene plates. Biofilm assays were repeated at least three times, and standard deviations are indicated.
The absence of a role for Atl in PIA-mediated biofilm formation on hydrophilic polystyrene prompted us to examine biofilm production on hydrophobic polystyrene. Interestingly, biofilm production by strain 8325-4 on hydrophobic polystyrene was induced in BHI glucose medium (P < 0.0001) but not in BHI NaCl medium (Fig. 3A). Furthermore, glucose-induced biofilm production by 8325-4 on hydrophobic polystyrene was impaired by the Δatl::Cmr and ΔicaADBC::Tcr mutations (P < 0.0001) (Fig. 3A). In contrast, the biofilm phenotypes of BH1CC, BH1CC Δatl::Cmr, and BH1CC ΔicaADBC::Tcr observed on hydrophobic polystyrene (Fig. 3A) were similar to those observed on hydrophilic polystyrene. These data reveal an essential role for Atl in PIA-mediated biofilm production on hydrophobic polystyrene but indicate that the major autolysin is dispensable for PIA-mediated biofilm production on hydrophilic polystyrene. In contrast, Atl was required for FnBP-promoted biofilm production on both hydrophilic and hydrophobic polystyrene.
FIG. 3.
(A) Biofilm phenotypes of strains 8325-4, 8325-4 Δatl::Cmr, 8325-4 ΔicaADBC::Tcr, BH1CC, BH1CC Δatl::Cmr, and BH1CC ΔicaADBC::Tcr grown for 24 h at 37°C in BHI, BHI NaCl, and BHI glucose media in hydrophobic 96-well polystyrene plates. (B) Biofilm phenotypes of strains BH1CC, BH1CC Δatl::Cmr, and BH1CC ΔfnbAB::Tcr grown for 24 h at 37°C in BHI glucose medium in 96-well polystyrene plates coated with increasing concentrations of human fibronectin. Biofilm assays were repeated at least three times, and standard deviations are indicated.
To assess the physiological relevance of our findings, we investigated the role of Atl and the FnBPs in biofilm production on polystyrene plates coated with increasing concentrations of human Fn. Biofilm production by isolate BH1CC was not significantly different on surfaces coated with different concentrations of Fn (Fig. 3B). Similarly, the BH1CC fnbAB::Tcr mutant remained biofilm negative on Fn-coated polystyrene (Fig. 3B). However, on surfaces coated with Fn concentrations of ≥2.5 μg/ml, BH1CC Δatl::Cmr produced significantly more biofilm than it did on uncoated polystyrene (P < 0.0001) (Fig. 3B). Nevertheless, biofilm production by BH1CC Δatl::Cmr remained significantly less than that by the wild type even on Fn-coated polystyrene (P < 0.0001) (Fig. 3B). Thus, these data indicate that the role of Atl in primary attachment and early biofilm formation on naked polystyrene can be partially overcome on Fn-coated surfaces, which presumably facilitates some FnBP-mediated attachment and biofilm accumulation. In the absence of the FnBPs, it appears that no significant attachment or biofilm accumulation is possible.
Regulation of Atl/FnBP-mediated biofilm phenotype.
To further investigate Atl/FnBP-mediated biofilm production, we examined the roles of the global regulators Agr, SarA, and SigB, which are centrally involved in the control of virulence and biofilm. Agr negatively regulates biofilm production by encoding the surfactant-acting δ toxin (67), repressing protein A (43), and upregulating extracellular protease production (9). SarA and SigB regulate the agr locus (8, 14, 35) and also control biofilm production (3, 11, 35, 48, 64). A sigB::Tcr allele (12) transduced into isolate BH1CC using phage 80α resulted in a 6-fold reduction in biofilm production in BHI glucose medium (P < 0.0001), which was complemented by the sigB operon on plasmid pLI50 (psigB) (Fig. 4A). The sigB mutation affected biofilm production at the level of primary attachment (P < 0.0001) (Fig. 4B), and real-time PCR analysis using RNA extracted from cultures grown at 37°C in BHI glucose medium to an A600 of 4 (pH 6) revealed an approximately 4-fold reduction in atl transcription in the sigB mutant compared to that in BH1CC (P < 0.0001) (Fig. 4C). Multicopy expression of atl in the sigB mutant also increased biofilm production (P < 0.0001) (Fig. 4D). Because SigB represses RNAIII expression and extracellular protease production (9), we examined the impact of a sigB RNAIII double mutation on biofilm production in BH1CC. Biofilm-forming capacity was restored in the sigB RNAIII double mutant (Fig. 4D), suggesting that σB influences Atl-dependent biofilm production by increasing extracellular protease production and at the level of primary attachment via reduced atl transcription.
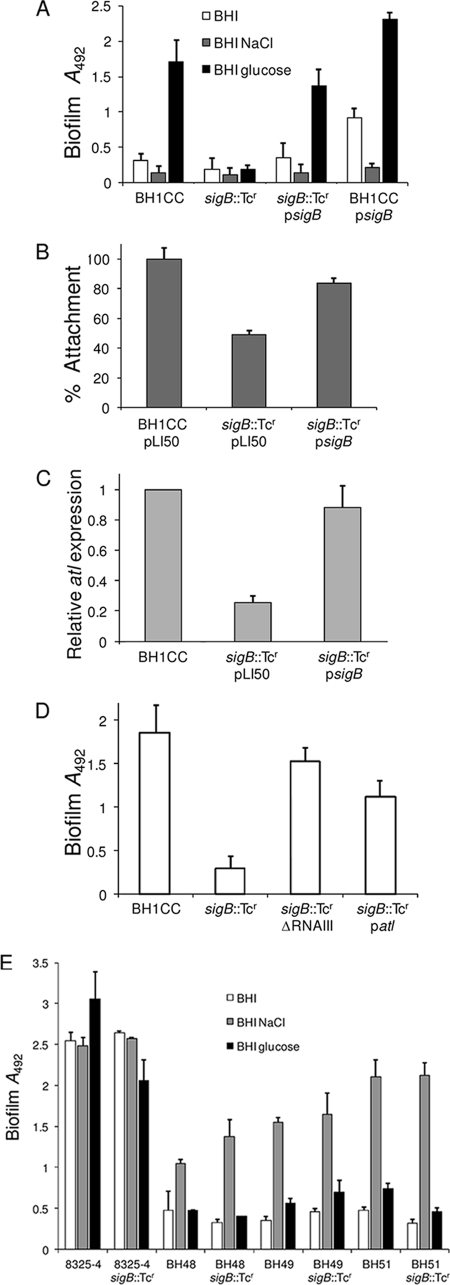
FIG. 4.
Contribution of σB to S. aureus biofilm regulation. (A) Impact of a sigB::Tcr mutation and carriage of multicopy psigB (sigB) on biofilm development in MRSA isolate BH1CC grown for 24 h at 37°C in BHI, BHI NaCl, and BHI glucose media in hydrophilic 96-well polystyrene plates. (B) Primary attachment of BH1CC/pLI50, BH1CC sigB::Tcr, and BH1CC sigB::Tcr/psigB cells grown in BHI glucose medium to hydrophilic polystyrene petri dishes. Primary attachment is expressed as the percentage of the numbers of CFU attached to the petri dish after it was washed relative to the initial numbers of CFU. Experiments were repeated three times, and standard deviations are indicated. (C) Comparison of relative atl transcription by real time RT-PCR in BH1CC/pLI50, BH1CC sigB::Tcr, and BH1CC sigB::Tcr/psigB. Total RNA was extracted from cultures grown to an A600 of 4 (pH 6) at 37°C in BHI glucose medium. (D) Biofilm phenotypes of BH1CC, BH1CC sigB::Tcr, BH1CC sigB::Tcr ΔRNAIII::Cmr, and BH1CC sigB::Tcr/patl grown for 24 h at 37°C in BHI glucose medium in hydrophilic 96-well polystyrene plates. (E) Biofilm phenotypes of MSSA isolates 8325-4, BH48, BH49, and BH51 and their isogenic sigB::Tcr mutants grown for 24 h at 37°C in BHI, BHI NaCl, and BHI glucose media in hydrophilic 96-well polystyrene plates. Biofilm assays were repeated at least three times, and standard deviations are indicated.
The sigB::Tcr mutation also resulted in a 3.5-fold reduction in glucose-induced biofilm production in MRSA isolates BH4 and BH10 (data not shown) (P < 0.0001). In contrast, the sigB mutation had no significant effect on PIA-dependent biofilm production by 8325-4 or MSSA clinical isolates BH48, BH49, and BH51 (Fig. 4E). These data revealed a significant difference in the role of σB in regulating FnBP- and PIA-promoted S. aureus biofilm development and are in keeping with σB acting as a negative regulator of the agr locus (35), which promotes extracellular protease production (9). Consistent with this, introduction of an agr::Tcr mutation into isolate BH1CC resulted in a significant 2-fold increase in early BH1CC biofilm production after 6 h of growth (P < 0.001) (Fig. 5A). Similarly, a BH1CC atl agr double mutant also displayed significantly impaired biofilm-forming capacity (P < 0.0001) (Fig. 5A), indicating that reduced protease production in the Agr mutant cannot promote biofilm in the absence of Atl. Unlike σB, the global regulator SarA is a positive regulator of the Agr locus (13), but deletion of sarA is also known to increase extracellular protease production (3), suggesting that the role of sarA in biofilm regulation is complex. Nevertheless, mutation of sarA completely impaired early BH1CC biofilm production (Fig. 5A). The mutations in atl and sarA also impaired biofilm production after 24 h of growth (Fig. 5B), whereas the impact of the agr mutation was not significant after this period of growth (Fig. 5B).
FIG. 5.
Contribution of the sarA, agr, and atl loci to early biofilm (6 h) (A) and mature biofilm (24 h) (B) production by isolate BH1CC grown at 37°C in BHI glucose medium in hydrophilic 96-well polystyrene plates. Biofilm assays were repeated at least three times, and standard deviations are indicated.
Relationship between lytic activity and biofilm production in BH1CC.
To further investigate the possible role of extracellular proteases in isolate BH1CC bacteriolytic activity and biofilm production, we used the serine protease inhibitor PMSF, which has previously been shown to increase the release of extracellular autolytic enzymes (19). Deletion of atl in BH1CC was accompanied by an approximately 9-fold decrease in extracellular bacteriolytic activity compared to that of BH1CC (data not shown), suggesting that bacteriolytic activity was predominantly the result of Atl activity (Fig. 6A). Bacteriolytic activity and biofilm production were similar (P > 0.05) in BHI medium culture supernatants with or without PMSF (Fig. 6A and B). Consistent with the findings of a previous study (53), bacteriolytic activity in BH1CC BHI glucose medium cultures was 10-fold lower than that in BHI medium cultures (Fig. 6A). However, addition of PMSF to BHI glucose medium restored lytic activity to wild-type levels (Fig. 6A). Furthermore, the restoration of bacteriolytic activity in BHI glucose medium cultures by PMSF correlated with the complete inhibition of BH1CC biofilm production (P < 0.0001) (Fig. 6B). These data indicate that reduced bacteriolytic activity in BHI glucose medium contributes to the BH1CC biofilm phenotype.
FIG. 6.

Relationship between autolytic activity and biofilm production in isolate BH1CC. (A) Bacteriolytic activity of culture supernatants from BH1CC grown in BHI and BHI glucose media in the absence and presence of 3 μM PMSF. Bacteriolytic activity was monitored by measuring the decrease in turbidity of the supernatant mixed with heat-killed cells, as described in the Materials and Methods. (B) Biofilm production by BH1CC grown in the absence and presence of 3 μM PMSF in BHI and BHI glucose media at 37°C in hydrophilic 96-well polystyrene plates. (C) Triton X-100-induced autolysis of BH1CC in the absence and presence of 500 μg/ml PAS. BH1CC Δatl::Cmr was included as a control. The strains were grown to an A600 of 1.0 in BHI medium at 37°C, before being washed in PBS and resuspended in 0.02% Triton X-100. The A600 was monitored, and autolysis was expressed as a percentage of the initial A600. The results are representative of those from at least three independent experiments. (D) Biofilm production by BH1CC grown in the absence and presence of 500 μg/ml PAS in BHI and BHI glucose media at 37°C in hydrophilic 96-well polystyrene plates. Biofilm assays were repeated at least three times, and standard deviations are indicated.
In contrast to PMSF, PAS inhibits lytic activity without affecting viability (69, 70). Indeed, PAS inhibited Triton X-100-induced autolytic activity in isolate BH1CC significantly more than mutation of atl (Fig. 6C). The BH1CC biofilm-forming capacity was also completely abolished by PAS in both BHI medium and BHI glucose medium (P < 0.0001) (Fig. 6D). Similarly, PAS completely inhibited Atl/FnBP-dependent biofilm formation in 12 MRSA clinical isolates (DAR27, BH3, BH4, DAR34, DAR35, DAR119, BH2, BH5, BH10, DAR69, DAR124, and DAR203) (data not shown).
Impact of mutating the active site of the amidase enzyme on Atl-dependent biofilm production.
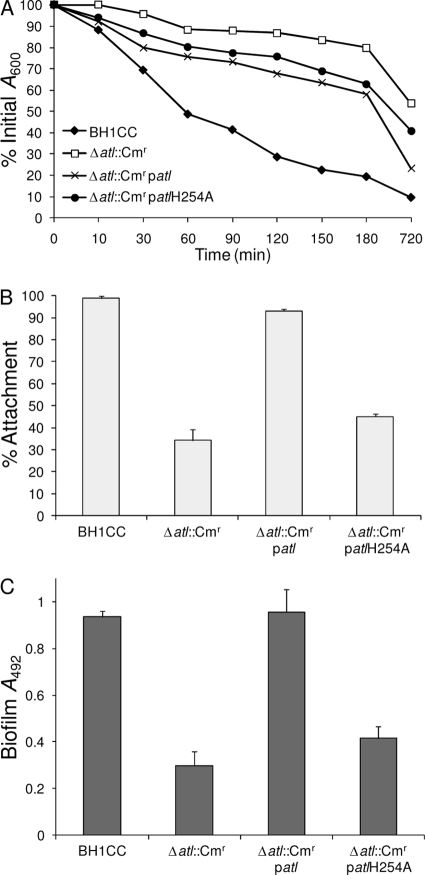
To more precisely investigate the relationship between Atl lytic activity and biofilm production, we used site-directed mutagenesis to mutate the H265 residue to alanine in the active site of the Ami enzyme from Atl. This histidine residue is involved in coordination of the zinc ion required for Ami catalytic activity, and its mutation to alanine has recently been shown to abolish Ami-mediated lytic activity without affecting expression of the protein or substrate binding (73). The plasmid carrying the H265A Atl allele, patlH265A, was capable of only partially complementing autolytic activity in the BH1CC Δatl::Cmr mutant compared to the capability of the wild-type atl plasmid (P < 0.005) (Fig. 7A), a finding consistent with the loss of amidase but not glucosaminidase activity encoded by the H265A Atl allele. The patlH265A plasmid also failed to complement primary attachment (Fig. 7B) and biofilm production (Fig. 7C) in the Δatl::Cmr mutant. Taken together, these data reveal that both posttranslational activation of Atl activity with PMSF and inhibition of Atl activity with PAS or by amino acid substitution interfered with Atl-promoted biofilm production. Apparently, inhibition or excessive activation of Atl-mediated lytic activity inhibits BH1CC biofilm production, suggesting that posttranslational control of Atl activity is a crucial determinant of biofilm production.
FIG. 7.
Impact of amidase active-site mutation on Atl-dependent autolytic activity, primary attachment, and biofilm production. (A) Triton X-100-induced autolysis of strains BH1CC, BH1CC Δatl::Cmr, BH1CC Δatl::Cmr/patl, and BH1CC Δatl::Cmr/patlH254A. Three independent experiments were performed, and the results of a representative experiment are shown. (B) Primary attachment of BH1CC, BH1CC Δatl::Cmr, BH1CC Δatl::Cmr/patl, and BH1CC Δatl::Cmr/patlH254A cells grown in BHI glucose medium to hydrophilic polystyrene. Primary attachment is expressed as the percent attachment after the cultures were washed. Experiments were repeated three times, and standard deviations are indicated. (C) Biofilm formation by BH1CC, BH1CC Δatl::Cmr, BH1CC Δatl::Cmr/patl, and BH1CC Δatl::Cmr/patlH254A grown for 24 h in BHI glucose medium in hydrophilic 96-well polystyrene plates. The results are the averages of three independent experiments, and standard deviations are indicated.
Role of extracellular DNA in BH1CC biofilm phenotype.
The importance of autolytic activity for Atl-dependent isolate BH1CC biofilm formation suggested that eDNA may play an adhesive role in this phenotype. We have previously reported that proteinase K significantly dispersed mature BH1CC biofilms, whereas treatment with sodium metaperiodate had no significant effect (47, 48). In contrast, PIA-mediated MSSA biofilms were dispersed with periodate but not proteinase K (47, 48). Pretreatment of stationary-phase BH1CC cells with DNase I reduced rates of primary attachment to levels similar to those for the BH1CC Δatl::Cmr mutant (Fig. 8A), revealing an important role for eDNA in primary attachment. DNase I added at time zero to BHI glucose medium cultures also significantly decreased BH1CC biofilm formation after 6 h (data not shown) and 24 h of growth (P < 0.0001) but had no effect on biofilm production by strain 8325-4, which produces a PIA-dependent biofilm (Fig. 8B). DNase I also impaired biofilm development by MRSA isolates from clonal complex 5 (CC5), CC8, CC22, and sequence type 239 (ST239) (P < 0.0001) (data not shown). In contrast, addition of DNase I had no effect on mature BH1CC biofilms grown in BHI glucose medium for 24 h (Fig. 8C). These data reveal a crucial role for eDNA in primary attachment and the early stages of Atl-dependent, FnBP-mediated MRSA biofilm formation (and are consistent with the proposed role of the FnBPs in biofilm maturation).
FIG. 8.
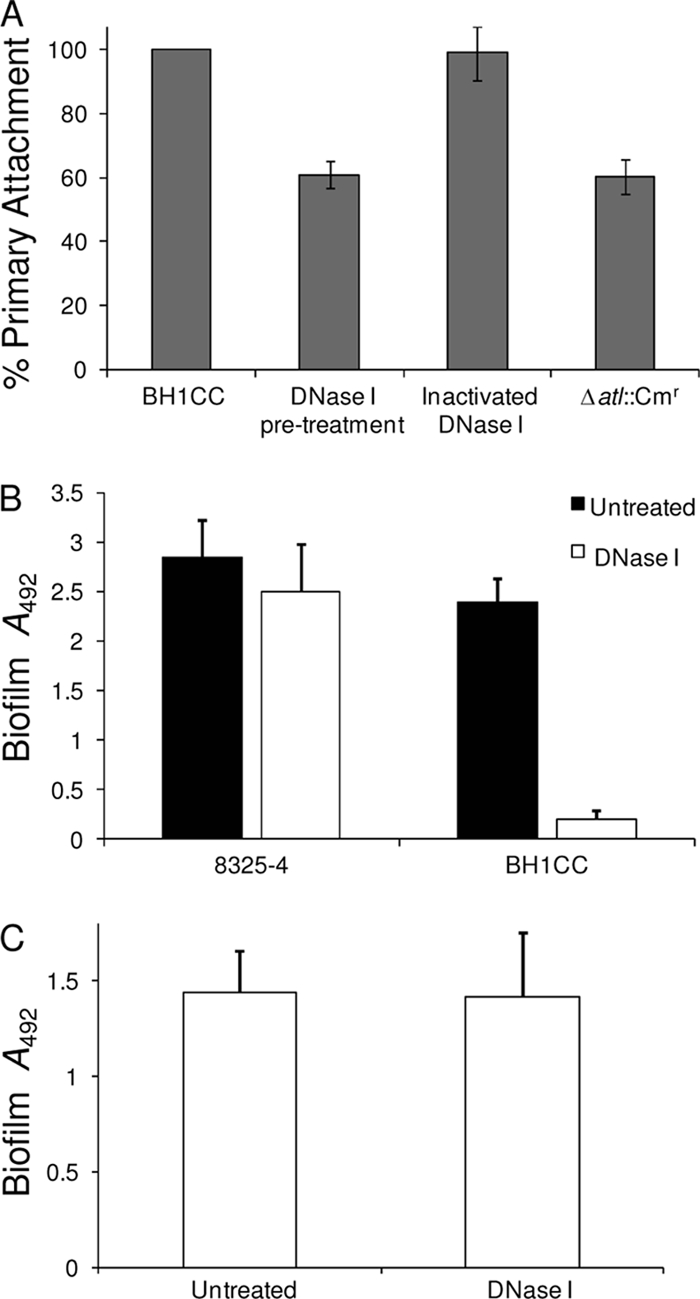
(A) Comparison of primary attachment to hydrophilic polystyrene by untreated BH1CC cells grown overnight in BHI glucose medium with cells pretreated for 2 h with 1 mg/ml DNase I or heat-inactivated DNase I (control). BH1CC Δatl::Cmr was included as a control. Primary attachment is expressed as the percent attachment after the cultures were washed. Experiments were repeated three times, and standard deviations are indicated. (B) Biofilm formation by isolates 8325-4 and BH1CC grown for 24 h in BHI glucose medium in hydrophilic 96-well polystyrene plates in the absence and presence of 0.5 mg/ml DNase I. The results are representative of those from three independent experiments, and standard deviations are indicated. (C) Susceptibility of 24-h BH1CC biofilms to dispersal by DNase I (0.5 mg/ml).
Transcriptional regulation of atl-dependent biofilm production.
To further investigate the role of atl transcriptional regulation in the biofilm phenotype, we turned our attention to the 417-bp gene downstream of atl (Fig. 9A). This gene (SA0904 in S. aureus N315) encodes a predicted 139-amino-acid MarR-type transcriptional regulator. We designated this gene atlR and used allele replacement to construct an atlR::Tcr mutant. Biofilm production by BH1CC, BH1CC atlR::Tcr, and the complemented atlR mutant (BH1CC atlR::Tcr/patlR) in BHI glucose medium was compared at 37°C and 30°C. These two temperatures were chosen because we have previously observed that biofilm production by BH1CC was repressed at 30°C compared to that at 37°C (46). The atlR::Tcr mutation had no significant effect (P < 0.05) on biofilm production at 37°C (Fig. 9B) but resulted in a 7-fold induction (P < 0.0001) at 30°C (Fig. 9B). Multicopy atl also activated BH1CC biofilm production at 30°C, whereas multicopy atlR significantly reduced (P < 0.0001) glucose-induced biofilm production at 37°C (Fig. 9B). Real-time PCR revealed a 5-fold induction in atl expression in BH1CC atlR::Tcr compared to that in BH1CC and the complemented mutant grown to an A600 of 2 in BHI glucose medium at 30°C (Fig. 9C). Electrophoretic mobility shift assays revealed a specific interaction between recombinant AtlR protein and an atl promoter probe (Fig. 9D). Taken together, these experiments revealed that AtlR acts as a repressor of atl transcription and biofilm production in BH1CC.
FIG. 9.
Contribution of AtlR to biofilm regulation in isolate BH1CC. (A) Chromosomal organization of the atlR and atl genes and location of the EcoRV site in atlR used in the construction of the atlR::Tcr allele. (B) Impact of an atlR::Tcr mutation and carriage of multicopy patlR (atlR) or patl (atl) on biofilm development in BH1CC grown for 24 h in BHI glucose medium at 37°C or 30°C in hydrophilic 96-well polystyrene plates. (C) Comparison of relative atl transcription by real time RT-PCR in BH1CC/pLI50, BH1CC atlR::Tcr/pLI50, and BH1CC atlR::Tcr/patlR at 30°C or 37°C. Total RNA was extracted from cultures grown to an A600 of 2 in BHI glucose medium. (D) Recombinant AtlR binding to the atl promoter. Increasing concentrations (0.015 to 0.5 μg) of recombinant AtlR protein were added to a biotinylated oligonucleotide patl probe. The protein-DNA interactions were competed with 100× specific cold competitor DNA or 100× nonspecific cold competitor DNA.
DISCUSSION
The study of biofilm production mechanisms independent of the icaADBC-encoded exopolysaccharide PIA has emerged as an important theme in staphylococcal biofilm research. We previously demonstrated a lack of correlation between activation of icaADBC expression and glucose-induced biofilm development in four MRSA clinical isolates and revealed the existence of a glucose/mild acid-induced icaADBC-independent biofilm phenotype in these MRSA strains (17, 47, 48). This ica-independent biofilm phenotype was impaired by mutations in the fnbAB genes (47). Consistent with our findings, Vergara-Irigaray et al. (66) recently described an MRSA isolate, strain 132, capable of switching between PIA-mediated biofilm production and FnBP-mediated biofilm production in medium supplemented with NaCl or glucose, respectively. Using a subcutaneous catheter colonization mouse model, an FnBP mutant was significantly less virulent than the wild type or a PIA mutant. A second research group also reported that the FnBPs are required for citrate-induced, ica-independent biofilm production in S. aureus (61). Thus, in addition to their known involvement in binding to host extracellular matrix components such as fibronectin, fibrinogen, and elastin (21, 41, 56, 68), the ability of the FnBPs to promote biofilm production appears to be a common and significant virulence determinant in clinical S. aureus isolates.
Our previous studies revealed that the fnbAB double mutation did not affect primary attachment, suggesting that FnBPA and FnBPB are involved in biofilm maturation (47). To investigate ica-independent primary attachment and early biofilm development, we turned our attention to the major autolysin, Atl, which has previously been implicated in S. epidermidis (25) and S. aureus SA113 (7) primary attachment. Mutation of atl in a number of clinical MRSA isolates which produce FnBP-dependent biofilms significantly impaired glucose-induced biofilm production and primary attachment to hydrophobic and hydrophilic polystyrene. Although the atl mutation also impaired PIA-dependent biofilm development by the laboratory MSSA strain 8325-4 on hydrophobic polystyrene, it did not affect biofilm production by 8325-4 on hydrophilic polystyrene. Thus, expression of PIA alone is sufficient for S. aureus biofilm production on hydrophilic polystyrene, whereas Atl is required for FnBP-promoted biofilm production on both hydrophilic and hydrophobic polystyrene. The role of Atl in PIA-mediated biofilm production on hydrophobic polystyrene is the focus of a separate study.
Atl/FnBP-mediated biofilm production on hydrophilic polystyrene was influenced by the global regulators agr, sarA, and sigB. Mutation of sigB impaired biofilm production in isolate BH1CC. Interestingly, this biofilm defect correlated with reduced levels of both primary attachment and atl transcription, suggesting that σB controls biofilm production, at least in part, by regulating atl expression. Furthermore, consistent with the findings of recent studies implicating σB in repression of RNAIII expression and extracellular protease production (9, 35, 40), we were able to rescue the biofilm defect of the BH1CC sigB mutant by introducing a second RNAIII deletion mutation. Deletion of the agr locus, which also represses the FnBPs (66), enhanced early biofilm development in BH1CC. Thus, σB is apparently required for ica-independent, surface protein-mediated biofilm production in S. aureus. In contrast, the role of σB in PIA-dependent biofilm production is more ambiguous. Valle et al. (64) reported that although σB does play a role in icaADBC transcriptional regulation, it was not required for PIA and biofilm production. Cerca et al. (11) also reported that σB can positively regulate icaADBC expression but demonstrated that σB was required for biofilm production in two separate S. aureus strains. Our findings revealed that σB was not required for PIA-dependent biofilm production on hydrophilic polystyrene by 8325-4 and three independent MSSA isolates. Mutation of sarA, which also leads to upregulation of extracellular proteases (3), impaired Atl-dependent biofilm production in BH1CC. Given that SarA is a positive regulator of the Agr locus (13), its role in controlling extracellular protease and biofilm production is clearly different from that of σB, which negatively regulates agr transcriptional activity (35). Nevertheless, these data indicate that regulatory mutations affecting extracellular protease activity strongly influence Atl/FnBP-dependent biofilm production.
The atlR gene, which is located immediately adjacent to atl on the chromosome, was found to encode a negative regulator of atl transcription. Inactivation of atlR in isolate BH1CC activated biofilm formation at 30°C but not 37°C. We previously reported that biofilm production by BH1CC was reduced at 30°C (46). Multicopy expression of the atl gene also increased biofilm production at 30°C, whereas overexpression of atlR reduced biofilm formation at 37°C. These biofilm phenotypes correlated well with the results of real-time RT-PCR analysis of atl transcription, which was in turn supported by the findings of gel shift experiments, demonstrating that recombinant AtlR protein bound specifically to the atl promoter. Thus, our data reveal that AtlR acts as a repressor of Atl-mediated biofilm formation in BH1CC and may suggest that temperature regulates Atl expression in part via AltR.
Unprocessed, wall-anchored Atl proprotein contains Ami and GL domains, which are proteolytically cleaved to Ami and GL extracellular lytic enzymes (25, 49, 62). To probe the involvement of the proprotein and lytic enzymes in Atl-dependent biofilm production, we employed the serine protease inhibitor PMSF, which increases the release of extracellular autolytic enzymes (19), and PAS, which inhibits autolytic activity (69, 70). Interestingly, both PMSF and PAS blocked Atl-dependent biofilm production in isolate BH1CC, indicating that pro-Atl and the Ami and GL lytic enzymes are required for Atl-dependent biofilm production. In addition, mutation of the H265 residue to alanine in the active site of the Ami enzyme significantly interfered with Atl-mediated lytic activity, primary attachment, and biofilm production. Consistent with these data, BH1CC biofilm development was inhibited by DNase I, implicating eDNA in this biofilm phenotype. Furthermore, pretreatment of BH1CC cells with DNase I significantly reduced primary attachment rates, whereas DNase I did not disperse mature BH1CC biofilms, implicating Atl-mediated eDNA release in the early stages of Atl/FnBP-dependent biofilm development. In S. epidermidis, AtlE-promoted lysis leads to eDNA release, which then acts as a component of the biofilm matrix (51). Rice et al. demonstrated that in S. aureus the CidA/LrgAB system regulates cell lysis, eDNA release, and biofilm development possibly by regulating access of murein hydrolases to cell wall substrates (52). Our results support a relationship between Atl-dependent lytic activity, eDNA release, primary attachment, and biofilm production in S. aureus.
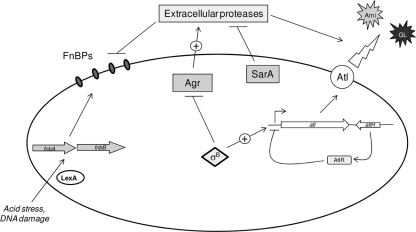
In summary we propose a model for S. aureus biofilm production in which the activities of AtlR, Agr, SarA, and σB work in concert to regulate Atl expression and activity and ultimately facilitate initial attachment and eDNA release during the early stages of ica-independent, FnBP-mediated biofilm development (Fig. 10). Unprocessed wall-anchored Atl appears to promote primary attachment to surfaces, following which proteolytic cleavage of Atl leads to some cell lysis, eDNA release, and cell accumulation. Following these early biofilm events, the FnBPs are required for biofilm maturation (Fig. 10) (47, 48).
FIG. 10.
Proposed model of Atl/FnBP-mediated biofilm formation in S. aureus. The major autolysin, Atl, which is proteolytically cleaved to release Ami and GL lytic enzymes, promotes primary attachment and early biofilm development. Either FnBPA or FnBPB is subsequently required for biofilm maturation. Atl/FnBP-mediated biofilm formation is triggered by acid stress or DNA damage and is influenced by the global regulators Agr, SarA, and σB. Agr and SarA regulate expression of extracellular proteases, which negatively affect FnBP and Atl activity. σB negatively regulates the agr locus and can positively influence transcription of the atl gene. Mutation of lexA, which represses fnbB transcription, increases biofilm formation.
Acknowledgments
This study was funded by grants from the Health Research Board (Ireland) and the Hospital Infection Society (United Kingdom) to J. P. O'Gara. E. M. Waters is supported by an Irish Research Council for Science, Engineering, and Technology postgraduate scholarship.
We are grateful to M. Sugai for strain JT1392 (Δatl::Cmr), S. J. Foster for strains PC400 (sigB::Tcr) and PC6911 (agr::Tcr), M. Smeltzer for UAMS-240 (sarA::Tcr), F. Vandenesch for WA400 (ΔRNAIII::Cmr), T. Maira Litran and G. B. Pier for rabbit anti-PIA/PNAG serum, and T. J. Foster for phage 80α. We thank H. McCarthy for experimental assistance and E. O'Neill, L. Holland, and B. Conlon for advice throughout the study.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 28 December 2010.
REFERENCES
- 1.Allignet, J., S. Aubert, K. G. Dyke, and N. El Solh. 2001. Staphylococcus caprae strains carry determinants known to be involved in pathogenicity: a gene encoding an autolysin-binding fibronectin and the ica operon involved in biofilm formation. Infect. Immun. 69:712-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., and O. Schneewind. 1998. Targeting of muralytic enzymes to the cell division site of Gram-positive bacteria: repeat domains direct autolysin to the equatorial surface ring of Staphylococcus aureus. EMBO J. 17:4639-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beenken, K. E., et al. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benito, Y., G. Lina, T. Greenland, J. Etienne, and F. Vandenesch. 1998. trans-Complementation of a Staphylococcus aureus agr mutant by Staphylococcus lugdunensis agr RNAIII. J. Bacteriol. 180:5780-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisognano, C., et al. 2004. A recA-LexA-dependent pathway mediates ciprofloxacin-induced fibronectin binding in Staphylococcus aureus. J. Biol. Chem. 279:9064-9071. [DOI] [PubMed] [Google Scholar]
- 7.Biswas, R., et al. 2006. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 259:260-268. [DOI] [PubMed] [Google Scholar]
- 8.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boles, B. R., and A. R. Horswill. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruckner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 11.Cerca, N., J. L. Brooks, and K. K. Jefferson. 2008. Regulation of the intercellular adhesin locus regulator (icaR) by SarA, sigmaB, and IcaR in Staphylococcus aureus. J. Bacteriol. 190:6530-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor sigmaB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung, A. L., and S. J. Projan. 1994. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 176:4168-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chien, Y., A. C. Manna, and A. L. Cheung. 1998. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol. Microbiol. 30:991-1001. [DOI] [PubMed] [Google Scholar]
- 15.Christensen, G. D., et al. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corrigan, R. M., D. Rigby, P. Handley, and T. J. Foster. 2007. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153:2435-2446. [DOI] [PubMed] [Google Scholar]
- 17.Fitzpatrick, F., H. Humphreys, and J. P. O'Gara. 2005. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J. Clin. Microbiol. 43:1973-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 19.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gill, S. R., et al. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene, C., et al. 1995. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17:1143-1152. [DOI] [PubMed] [Google Scholar]
- 22.Gross, M., S. E. Cramton, F. Gotz, and A. Peschel. 2001. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 69:3423-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reference deleted.
- 24.Heilmann, C., J. Hartleib, M. S. Hussain, and G. Peters. 2005. The multifunctional Staphylococcus aureus autolysin aaa mediates adherence to immobilized fibrinogen and fibronectin. Infect. Immun. 73:4793-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 26.Hell, W., H. G. Meyer, and S. G. Gatermann. 1998. Cloning of aas, a gene encoding a Staphylococcus saprophyticus surface protein with adhesive and autolytic properties. Mol. Microbiol. 29:871-881. [DOI] [PubMed] [Google Scholar]
- 27.Hennig, S., S. Nyunt Wai, and W. Ziebuhr. 2007. Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int. J. Med. Microbiol. 297:117-122. [DOI] [PubMed] [Google Scholar]
- 28.Horsburgh, M. J., et al. 2002. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janzon, L., and S. Arvidson. 1990. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 9:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reference deleted.
- 31.Khan, S. A., and R. P. Novick. 1983. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid 10:251-259. [DOI] [PubMed] [Google Scholar]
- 32.Koprivnjak, T., et al. 2006. Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus. J. Bacteriol. 188:3622-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreiswirth, B. N., et al. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 34.Lasa, I., and J. R. Penades. 2006. Bap: a family of surface proteins involved in biofilm formation. Res. Microbiol. 157:99-107. [DOI] [PubMed] [Google Scholar]
- 35.Lauderdale, K. J., B. R. Boles, A. L. Cheung, and A. R. Horswill. 2009. Interconnections between sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 77:1623-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101-105. [DOI] [PubMed] [Google Scholar]
- 37.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186:722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maki, H., T. Yamaguchi, and K. Murakami. 1994. Cloning and characterization of a gene affecting the methicillin resistance level and the autolysis rate in Staphylococcus aureus. J. Bacteriol. 176:4993-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mani, N., P. Tobin, and R. K. Jayaswal. 1993. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J. Bacteriol. 175:1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marti, M., et al. 2010. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 12:55-64. [DOI] [PubMed] [Google Scholar]
- 41.Massey, R. C., et al. 2001. Fibronectin-binding protein A of Staphylococcus aureus has multiple, substituting, binding regions that mediate adherence to fibronectin and invasion of endothelial cells. Cell. Microbiol. 3:839-851. [DOI] [PubMed] [Google Scholar]
- 42.Mazmanian, S. K., G. Liu, E. R. Jensen, E. Lenoy, and O. Schneewind. 2000. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. U. S. A. 97:5510-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merino, N., et al. 2009. Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 191:832-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novick, R. P., et al. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Gara, J. P. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270:179-188. [DOI] [PubMed] [Google Scholar]
- 46.O'Neill, E., H. Humphreys, and J. P. O'Gara. 2009. Carriage of both the fnbA and fnbB genes and growth at 37oC promote FnBP-mediated biofilm development in meticillin-resistant Staphylococcus aureus clinical isolates. J. Med. Microbiol. 58:399-402. [DOI] [PubMed] [Google Scholar]
- 47.O'Neill, E., et al. 2008. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 190:3835-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Neill, E., et al. 2007. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 45:1379-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oshida, T., et al. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. U. S. A. 92:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reference deleted.
- 51.Qin, Z., et al. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153:2083-2092. [DOI] [PubMed] [Google Scholar]
- 52.Rice, K. C., et al. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 104:8113-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rice, K. C., J. B. Nelson, T. G. Patton, S. J. Yang, and K. W. Bayles. 2005. Acetic acid induces expression of the Staphylococcus aureus cidABC and lrgAB murein hydrolase regulator operons. J. Bacteriol. 187:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson, D. A., and M. C. Enright. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 186:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson, D. A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roche, F. M., et al. 2004. The N-terminal A domain of fibronectin-binding proteins A and B promotes adhesion of Staphylococcus aureus to elastin. J. Biol. Chem. 279:38433-38440. [DOI] [PubMed] [Google Scholar]
- 57.Rohde, H., et al. 2007. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 28:1711-1720. [DOI] [PubMed] [Google Scholar]
- 58.Sadovskaya, I., E. Vinogradov, S. Flahaut, G. Kogan, and S. Jabbouri. 2005. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain, Staphylococcus epidermidis RP62A. Infect. Immun. 73:3007-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schlag, M., et al. 2010. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol. Microbiol. 75:864-873. [DOI] [PubMed] [Google Scholar]
- 60.Schroeder, K., et al. 2009. Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PLoS One 4:e7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shanks, R. M., et al. 2008. Genetic evidence for an alternative citrate-dependent biofilm formation pathway in Staphylococcus aureus that is dependent on fibronectin binding proteins and the GraRS two-component regulatory system. Infect. Immun. 76:2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sugai, M., et al. 1989. Purification of a 51 kDa endo-beta-N-acetylglucosaminidase from Staphylococcus aureus. FEMS Microbiol. Lett. 52:267-272. [DOI] [PubMed] [Google Scholar]
- 63.Takahashi, J., et al. 2002. Molecular characterization of an atl null mutant of Staphylococcus aureus. Microbiol. Immunol. 46:601-612. [DOI] [PubMed] [Google Scholar]
- 64.Valle, J., et al. 2003. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 65.Vergara-Irigaray, M., et al. 2008. Wall teichoic acids are dispensable for anchoring the PNAG exopolysaccharide to the Staphylococcus aureus cell surface. Microbiology 154:865-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vergara-Irigaray, M., et al. 2009. Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect. Immun. 77:3978-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 68.Wann, E. R., S. Gurusiddappa, and M. Hook. 2000. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem. 275:13863-13871. [DOI] [PubMed] [Google Scholar]
- 69.Wecke, J., M. Lahav, I. Ginsburg, E. Kwa, and P. Giesbrecht. 1986. Inhibition of wall autolysis of staphylococci by sodium polyanethole sulfonate “liquoid.” Arch. Microbiol. 144:110-115. [DOI] [PubMed] [Google Scholar]
- 70.Yabu, K., and S. Kaneda. 1995. Salt-induced cell lysis of Staphylococcus aureus. Curr. Microbiol. 30:299-303. [DOI] [PubMed] [Google Scholar]
- 71.Yamada, S., et al. 1996. An autolysin ring associated with cell separation of Staphylococcus aureus. J. Bacteriol. 178:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ziebuhr, W., et al. 1997. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zoll, S., et al. 2010. Structural basis of cell wall cleavage by a staphylococcal autolysin. PLoS Pathog. 6:e1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]