Abstract
Phagocytosis resistance is an important virulence factor in Klebsiella pneumoniae. Dictyostelium has been used to study the interaction between phagocytes and bacteria because of its similarity to mammalian macrophages. In this study, we used a Dictyostelium model to investigate genes for resistance to phagocytosis in NTUH-K2044, a strain of K. pneumoniae causing pyogenic liver abscess that is highly resistant to phagocytosis. A total of 2,500 transposon mutants were screened by plaque assay, and 29 of them permitted phagocytosis by Dictyostelium. In the 29 mutants, six loci were identified; three were capsular synthesis genes. Of the other three, one was related to carnitine metabolism, one encoded a subunit of protease (clpX), and one encoded a lipopolysaccharide O-antigen transporter (wzm). Deletion and complementation of these genes showed that only ΔclpX and Δwzm mutants became susceptible to Dictyostelium phagocytosis, and their complementation restored the phagocytosis resistance phenotype. These two mutants were also susceptible to phagocytosis by human neutrophils and revealed attenuated virulence in a mouse model, implying that they play important roles in the pathogenesis of K. pneumoniae. Furthermore, we demonstrated that clpP, which exists in an operon with clpX, was also involved in resistance to phagocytosis. The transcriptional profile of ΔclpX was examined by microarray analysis and revealed a 3-fold lower level of expression of capsular synthesis genes. Therefore, we have identified genes involved in resistance to phagocytosis in K. pneumoniae using Dictyostelium, and this model is useful to explore genes associated with resistance to phagocytosis in heavily encapsulated bacteria.
Phagocytes are a first line of defense against pathogenic organisms and play a key role in the host's innate immune response to bacterial infection. At the same time, bacteria that have developed resistance to phagocytosis or intracellular killing should be more virulent and more likely to succeed in establishing infection. Recently, a powerful model system involving Dictyostelium discoideum was introduced for studying the interactions between phagocytes and bacteria. The amoebal form of D. discoideum shares many unique traits with human phagocytes, especially the ability to ingest and kill bacteria. In addition, bacteria are thought to use similar mechanisms to resist both Dictyostelium in the environment and human phagocytes; therefore, Dictyostelium provides a useful system to study both cellular response and bacterial virulence. Many aspects of phagocytosis have been studied in Dictyostelium, such as the dynamics of the actin cytoskeleton, cellular adhesion, phagosome maturation, and intracellular killing (13, 33, 35). Dictyostelium has also been used to analyze virulence in different bacterial species, including extracellular and intracellular bacteria, such as Pseudomonas (11, 42), Yersinia (50), Vibrio (43, 44), Legionella (22, 25, 28), and Mycobacterium (20, 41). In addition, host genes required for the killing of a laboratory strain of Klebsiella pneumoniae by phagocytes were also identified with the Dictyostelium model (4).
Traditionally, K. pneumoniae is an opportunistic hospital-acquired pathogen which usually causes urinary tract infections, pneumonia, and septicemia (1, 40). Over the past 2 decades, community-acquired K. pneumoniae associated with pyogenic liver abscess has been found globally (9, 14, 26, 36, 37, 45, 51). This emerging disease is often complicated by septic meningitis and endophthalmitis. The capsule, allantoin metabolic genes, and the ferric iron uptake system were reported to be virulence factors that contribute to pathogenesis (6, 24, 32). The capsule was thought to play a crucial role in resistance to phagocytosis. Moreover, in a previous study we have identified genes determining resistance to phagocytosis and serum by screening of mucoviscosity and found that capsular mutants with the low-mucoviscosity phenotype became extremely sensitive to serum, susceptible to phagocytosis, and avirulent in mice (14). In the current study, we used the Dictyostelium model to screen for phagocytosis resistance genes in NTUH-K2044, a strain of K. pneumoniae isolated from a patient with pyogenic liver abscess (14).
MATERIALS AND METHODS
Transposon mutant library.
A mutant library of strain NTUH-K2044 constructed by use of a randomly inserted mini-Tn5 transposon was used in this study (14). The insertion sites of 30 randomly selected mutants were determined by semirandom PCR (8). The insertion sites of the mutants were mapped to 28 unique locations (26 mutants had unique insertion sites; the remaining four mutants had two insertion sites, with each two having the same insertion site). In the unique insertions, 23 (82%) were within the coding regions, while 5 (18%) were outsides the coding regions.
Plaque assay to screen for mutants susceptible to Dictyostelium phagocytosis.
D. discoideum AX-2 cells were grown at 23°C in HL5 medium (10 g proteose peptone, 10 g glucose, 5 g yeast extract, 0.965 g Na2HPO4·7H2O, 0.485 g KH2PO4, and 0.03 g dihydrostreptomycin in a final volume of 1 liter, pH 6.5) (10). A transposon mutant library of K. pneumoniae strain NTUH-K2044 (14) was used for screening. To test Dictyostelium growth on Klebsiella mutant strains, log-phase cultured bacteria were plated on SM agar (2.31 g KH2PO4, 1 g K2HPO4, 5 g glucose, 5 g Bacto peptone, and 5 g yeast extract in a final volume of 1 liter) (2). Five thousand Dictyostelium cells were added to the bacterial lawn, and the formation of phagocytic plaques was observed after 5 days at 23°C.
Identification of transposon mutants by semirandom PCR.
Transposon insertional sites were determined by semirandom PCR (8) and DNA sequencing. Briefly, genomic DNA was extracted and subjected to PCR amplification with a transposon-specific primer (primer P1) and an anchored random primer (primer P2). The PCR conditions have been previously described (8). The products of the first PCR were used as the template for a second PCR with a nested transposon-specific primer (primer P3) and a primer (primer P4) with a sequence identical to the conserved portion of the P2 primer (Fig. 1 B). Finally, the DNA sequence of the nested PCR product was resolved; and the transposon insertional site was defined accordingly. Furthermore, the results were confirmed by PCR with transposon-specific primers and primers for the disrupted genes.
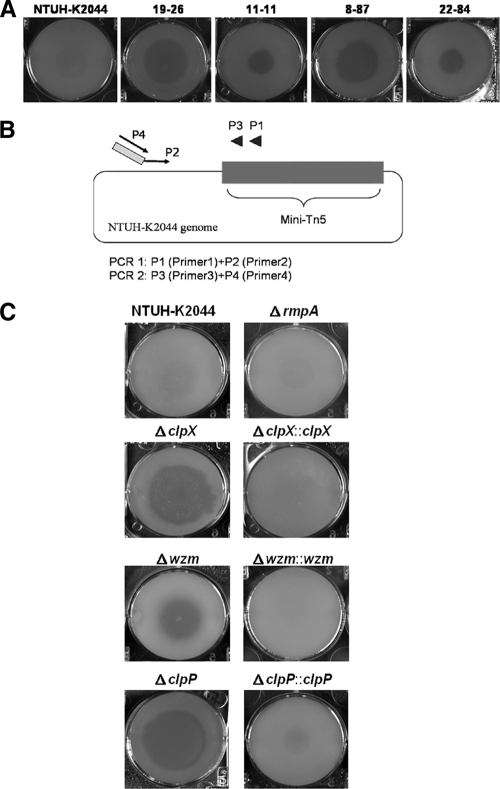
FIG. 1.
Identification of genes for resistance to phagocytosis in K. pneumoniae NTUH-K2044 using Dictyostelium. (A) Screening of K. pneumoniae NTUH-K2044 mutant library by plaque assay. By plaque assay of the wild type, 4 of the 29 transposon mutants were shown to be permissive for Dictyostelium growth. These mutants, 19-26, 11-11, 8-87, and 22-84, were further identified as wzc (capsular synthesis gene), wzm, clpX, and KP2125 insertional mutants, respectively. (B) Primers used for semirandom PCR. Primers P1 and P2 are used for the first PCR, and primers P3 and P4 are used for nested PCR. (C) Confirmation of the deletion mutants and complementation strains by plaque assay. ΔclpX, Δwzm, and ΔclpP indicate deletion mutants of the clpX, wzm, and clpP genes, respectively; ΔclpX::clpX, Δwzm::wzm, and ΔclpP::clpP, chromosome-complemented strains of the ΔclpX, Δwzm, and ΔclpP mutants, respectively.
Construction of unmarked deletion mutants of K. pneumoniae.
Plasmid pKO3-Km was used to generate deletion mutants (29). The flanking regions of target genes wzm, clpX, and clpP were amplified with specific primers (Table 1) and cloned into the pKO3-Km plasmid. The resulting plasmid was used to generate deletion mutants in the NTUH-K2044 strain as previously described (24).
TABLE 1.
Primers used in this study
| Primer name | Sequence | Position | Purpose (reference) |
|---|---|---|---|
| P1 | GCGCGCTGCGCAGGGCTTTATTGATTCCATTTTTAC | EZ-Tn5 (KAN-2) transposon | Semirandom PCR (8) |
| P2 | GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT | Randomly anchored on genome | Semirandom PCR (8) |
| P3 | GTACCGAGCTCGAATTCGGC | EZ-Tn5 (KAN-2) transposon | Semirandom PCR (8) |
| P4 | GGCCACGCGTCGACTAGTAC | Partially complementary to P2 | Semirandom PCR (8) |
| 22416F | CTTTGCCATACCTTCACGGT | Upstream of wzm gene | wzm mutant construct |
| wzt-R | TTAAGCCATTGCTTGCTTG | Downstream of wzm gene | wzm mutant construct |
| wzm-IR | AAATTATTCTGCAAATATCCACTGAATGAG | Upstream of wzm gene | wzm mutant construct |
| wzt-F | ATGCACCCAGTTATTAAC | wzt | wzm mutant construct |
| wzm-R | TTACAAGATCTCTGCAAATC | wzm | wzm complementation |
| clpX-750F | CGGTGAACGTTCTTTTG | Upstream of clpX gene | clpX mutant construct |
| clpX + 750R | TCAACCTGCAGCAGATC | lon | clpX mutant construct and junction PCR |
| clpX-F | TTAACCATTTCACTCAAGC | Downstream of clpX gene | clpX mutant construct |
| clpX-R | GAGTCCAAACCTCTTTTTAAG | Upstream of clpX gene | clpX mutant and clpP complementation |
| clpP-P | TTTCGCCGCTATCGTTTAAG | Upstream of clpP gene | clpX and clpP complementation |
| clpP5′R | TTCCGTCTCCTGGAGTAATTTT | Upstream of clpP gene | clpX complementation |
| clpP3′F | TGCCCAAGGCGCAAGGG | Downstream of clpP gene | clpX complementation |
| clpX-deR | TTATTCACCAGATGCCTG | clpX | clpX complementation |
| clpP-750F | AAATATGGAACGCGAGC | tig | clpP mutant construct and junction PCR |
| clpP + 750R | ATATAAACGATGCCGCG | clpX | clpP mutant construct |
| clpP-R-TUD | TCGTTCGCCACTGTATGAC | clpP | Junction PCR |
| clpX-R-TUD | CCGCCAACACCTTCTTC | clpX | Junction PCR |
| clpP-F-TUD | TTGACCCATCGTAATTG | clpP | Junction PCR |
| clpX-F-TUD | TCAGCAGGCATCTGGTG | clpX | Junction PCR |
| galF-145F | GCCGGGATCAAAGAGATTGTT | galF | Real-time PCR |
| galF-217R | AGGAGGTGTCGAAGTGGTTTTC | galF | Real-time PCR |
Construction of chromosomal complementation strains of K. pneumoniae.
For chromosomal complementation, the intact wzm, clpX, and clpP genes with their promoters were amplified by PCR (Table 1) and cloned into the intergenic region of pgpA and yajO in a PKO3-Km-pgpA-yajO recombinant vector. The resulting plasmid was used to generate complemented strains as previously described (23).
Silver stain for lipopolysaccharide O antigen.
Extracellular polysaccharides were isolated as previously reported (7). Samples were separated by SDS-PAGE. Gels were then placed in fixation solution (40% ethanol, 5% acetic acid) overnight at 4°C, and the fixer was replaced with oxidation solution (40% ethanol, 5% acetic acid, 0.7% periodic acid) for 5 min with gentle agitation. After oxidation, gels were washed three times in double-distilled H2O (ddH2O) for 15 min. The gels were then stained with freshly prepared staining reagent (9.3 ml 0.1 N NaOH, 0.66 ml NH4OH, and 1.66 ml 20% AgNO3 with ddH2O added to a final volume of 50 ml) for 10 min and washed three times in ddH2O for 15 min. For color development, developing solution (5 mg citric acid in 100 ml ddH2O mixed with 50 μl 37% formaldehyde) was added for 10 to 20 min, and bands slowly appeared. Finally, the reaction was terminated by washing in stop solution (0.8% acetic acid).
Determination of transcriptional unit by RT-PCR.
Total RNA was extracted from strain NTUH-K2044 as previously described (6, 24). Four micrograms of total RNA was reverse transcribed with 0.1 mmol of random hexamer primer (Roche, Indianapolis, IN) and 5 μl of reverse transcriptase II (Invitrogen, Carlsbad, CA) in a reaction mixture of 20 μl. Reaction mixtures without reverse transcriptase were used as negative controls. PCR was performed with 1 μl of each reverse transcription (RT) reaction mixture as template and subjected to the following program: 96°C for 3 min, followed by 30 cycles of 96°C for 30 s, 54°C for 15 s, and 72°C for 1 min.
Phagocytosis assay in Dictyostelium.
Plasmid pCRII-TOPO with a gene encoding green fluorescence protein (GFP) was electroporated into wild-type and mutant strains. An inoculum containing 108 CFU of bacteria carrying this plasmid was incubated with 106 Dictyostelium cells at 23°C for 1 h. Cells were washed three times with phosphate-buffered saline (PBS), fixed with 2% paraformaldehyde for 15 min, treated with 0.1% Triton X-100 for 1 min, and stained for 15 min with rhodamine-phalloidin, which was diluted 1:100 in PBS. After preparation, the cells were observed by confocal microscopy under ×630 image magnification, and the numbers of intracellular bacteria in five fields were counted (≈50 cells in each field). The sum of the intracellular bacteria was divided by the total number of cells in these fields and multiplied by 100, to calculate the number of intracellular bacteria in 100 cells.
Phagocytosis assay in human neutrophils.
Human neutrophils were freshly isolated from peripheral blood donated by healthy volunteers (14). An inoculum containing 108 CFU of bacteria was opsonized with 25% normal human serum for 15 min on ice and incubated with 106 human neutrophils at 37°C for 15 min for observation of the cell-bacterium association or 45 min for phagocytosis. Cells were also stained with rhodamine-phalloidin for observation. The numbers of intracellular bacteria and cells were counted as described above.
Confocal microscopy.
Images were capture by a Leica SP5 confocal microscope with a 488-nm argon laser and a 543-nm laser, which emitted the excitation wavelengths of GFP and rhodamine, respectively.
A serial confocal Z stack taken with a step of 0.5 μm was obtained by scanning a representative number of cells from top to bottom, and the images were combined for observation of cell-associated bacteria; a representative confocal section through the middle of the cell was shown for observation of intracellular bacteria.
Analysis of transcriptome of ΔclpX strain by microarray.
Total RNA was isolated from log-phase cultured bacteria. A total of 40 μg RNA was reverse transcribed using random hexamers (Roche) and labeled for microarray hybridization (6, 24). Densitometry was performed and analyzed using Image J software, and the 23S rRNA signal was used as an internal standard for normalization.
Quantitative real-time reverse-transcription quantitative PCR (RT-qPCR) for quantification of relative RNA expression.
RNA was reverse transcribed with random hexamers (Roche) and reverse transcriptase II (Invitrogen), according to the manufacturer's instruction. Amplification of the cDNAs was monitored using a SYBR green dye mixture in an ABI Prism 7900 thermocycler (Applied Biosystems, Foster City, CA). The relative quantities of RNA were analyzed by normalizing the calculated threshold cycle (ΔCT) of the detected gene to the CT of 23S rRNA from the same sample and calculation of the fold change with the ΔΔCT method (3).
Neutrophil killing assay.
Human neutrophils were isolated as described above. An inoculum containing 103 CFU of bacteria was opsonized with 25% normal human serum for 15 min on ice and incubated with or without 105 human neutrophils in 1× PBS at 37°C for 45 min. Percent survival of wild-type and mutant strains was calculated on the basis of the viable counts relative to those for no-neutrophil controls.
Serum resistance assay.
The serum resistance assay of K. pneumoniae strains was performed as previously described (14). Briefly, 2.5 × 104 CFU bacteria was mixed with serum from healthy human volunteers at a 1:3 volume ratio. The mixture was incubated at 37°C for 3 h, and bacterial numbers were determined.
Hypermucoviscosity test.
The string test was performed as previously described (14). Briefly, the tested strains were inoculated on blood agar plates and incubated at 37°C overnight. A standard bacteriologic loop was used to stretch a mucoviscous string from the colony. A positive string test was defined by the formation of viscous strings >5 mm in length when a loop was used to stretch the colony. To further differentiate the subtle difference in the levels of capsule production in different strains, low-speed centrifugation was performed, since bacteria with thick and mucoid capsules were more difficult to pellet. Briefly, equal numbers of bacteria in 1.1 ml LB medium were centrifuged at 2,000 × g for 5 min. After centrifugation, 1 ml of the supernatant was harvested and subjected to measurement of the optical density at 600 nm (OD600).
Immunoblot.
Capsule was extracted from wild-type and mutant strains with equal numbers of bacteria by the water-phenol extraction method (7). The capsule extracted from same amount of bacteria was serially diluted, subjected to immunoblotting (38), and detected with serotype K1-specific antiserum (Statens Serum Institute, Copenhagen, Denmark).
Animal inoculation.
Five-week-old female BALB/cByl mice were intraperitoneally inoculated with 103 CFU of bacteria prepared from mid-log phase. Eight mice for each group were monitored for 30 days. The rates of survival of the different groups of mice were compared and analyzed with the Kaplan-Meier method. To determine the bacterial load in vivo, an inoculation dose of 103 CFU of wild-type or mutant bacteria was administered to each of four mice by intraperitoneal injection. Mice were killed at 24 h or 48 h after inoculation. Liver, spleen, and blood were harvested, and tissues were further homogenized for quantitation of bacterial numbers. The number of bacteria detected in tissues was standardized per 0.1 g of wet organ weight.
Microarray data accession number.
Microarray data for the transcriptome of ΔclpX have been deposited in the Center for Information Biology Gene Expression Database (http://cibex.nig.ac.jp) under accession no. CBX130.
RESULTS
Screening virulence genes essential for resistance to phagocytosis in K. pneumoniae using Dictyostelium.
We had constructed a transposon mutant library of a K. pneumoniae strain (NTUH-K2044) causing pyogenic liver abscess (14). In the current study, we used a Dictyostelium system to investigate phagocytosis resistance genes in NTUH-K2044. A total of 2,500 transposon mutants were screened by plaque assay. Wild-type NTUH-K2044, which is resistant to Dictyostelium phagocytosis, does not permit Dictyostelium to form plaques on the bacterial lawn. In contrast, 29 transposon mutants permitted phagocytosis and showed clear plaques (Fig. 1A), implying that the genes disrupted by the transposon might be involved in resistance to phagocytosis. The locations of the inserted transposon, determined by semirandom PCR and DNA sequencing, are shown in Table 2. Twenty-one of these mutants had transposons inserted in the genes for capsule synthesis (mutants 3-1 to 3-16 were disrupted in the wza gene; mutants 3-18 and 3-23 were disrupted in magA; and mutants 3-22, 16-5, and 19-26 were disrupted in wzc). Six mutants were disrupted in a gene encoding the O-antigen synthesis transporter (wzm) (mutants 10-21, 11-11, 11-84, 12-28, 13-67, and 19-77). One mutant (mutant 22-84) was disrupted in a gene related to carnitine metabolism (KP2125), and for the remaining mutant (mutant 8-87), which created plaques comparable to those of the capsular mutants, the transposon was inserted in the gene encoding a protease subunit (clpX). Because the capsule has been well-known to play a role in resistance to phagocytosis, in the remainder of this study we focused on the three mutants that were not involved in capsular synthesis.
TABLE 2.
Transposon mutants permissive for Dictyostelium growth
| Transposon mutant(s) | Disrupted gene name | Gene function | Reference or source |
|---|---|---|---|
| 3-1 to 3-16 | wza | Putative capsule polysaccharide export protein | 14 |
| 3-18, 3-23 | magA | Capsular polysaccharide synthesis | 14 |
| 3-22, 16-5, 19-26 | wzc | Tyrosine-protein kinase, capsular polysaccharide synthesis | 14 |
| 10-21, 11-11, 11-84, 12-28, 13-67, 19-77 | wzm | Lipopolysaccharide O-antigen transporter | This study |
| 8-87 | clpX | Protease subunit | This study |
| 22-84 | KP2125 | Carnitine metabolism | This study |
Two deletion mutants and complemented strains of noncapsular genes were generated and confirmed by plaque assay.
The resistance to phagocytosis conferred by the three noncapsular synthesis genes was confirmed by construction of deletion mutants and complemented strains. The wzm and clpX deletion mutants showed defects in resistance to phagocytosis like the transposon mutants, and complementation of these strains restored the resistance to phagocytosis (Fig. 1C). In contrast, the KP2125 deletion mutant was not susceptible to Dictyostelium phagocytosis, implying that there might be other mutations present in the transposon mutant (data not shown).
Observation of phagocytosis by confocal microscopy.
We used confocal microscopy to observe phagocytosis of wild-type and mutant bacteria by Dictyostelium and to calculate the number of intracellular bacteria. The numbers of intracellular Δwzm and ΔclpX mutants were significantly higher than the number of the wild-type strain after coculture of bacteria and Dictyostelium (Fig. 2 A and B). These results indicated that the two mutants were more easily phagocytosed by Dictyostelium than the wild type was.
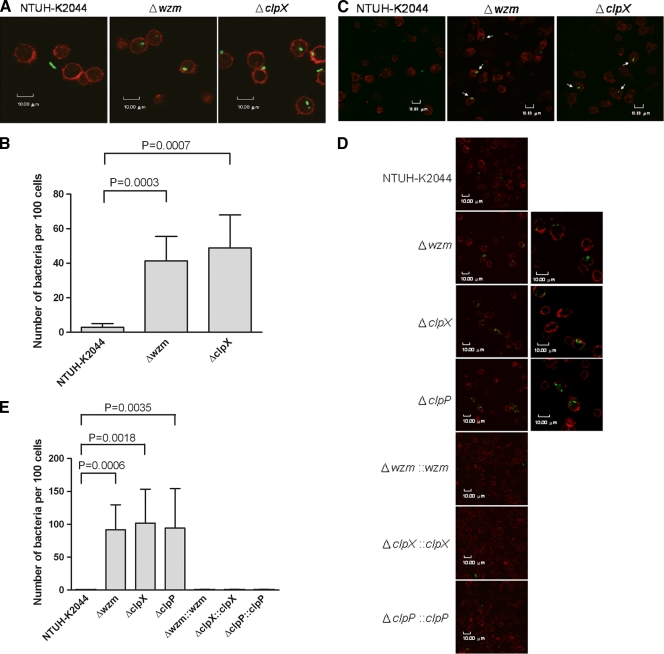
FIG. 2.
Observation of phagocytosis using confocal microscopy. (A) Phagocytosis by Dictyostelium. The wild-type strain and clpX and wzm deletion mutants carrying the GFP plasmid were incubated with Dictyostelium and observed under a confocal microscope. (B) The numbers of bacteria of wild type and Δwzm and ΔclpX mutants phagocytosed by Dictyostelium were counted under a confocal microscope. The P values for the Δwzm and ΔclpX mutants were 0.0003 and 0.0007, respectively, by t-test analysis. (C) Observation of the association of bacteria and human neutrophils. Bacteria carrying the GFP plasmid were incubated with human neutrophils for 15 min and observed under a confocal microscope. Image magnifications, ×630 with a zoom-in factor of 2×. Serial confocal Z stacks taken with a step of 0.5 μm were obtained by scanning a representative number of cells from top to bottom, and the images were combined. Arrows denote cell-associated bacteria. (D) Determination of phagocytosis resistance of NTUH-K2044 and isogenic mutants by human neutrophils. Bacteria carrying the GFP plasmid were incubated with human neutrophils for 45 min and observed under a confocal microscope. A representative confocal section through the middle of the cell was shown for observation of intracellular bacteria. Image magnifications, ×630 with a zoom-in factor of 2× (left panels) and with a zoom-in factor of 4× (right panels). (E) Bacteria phagocytosed by human neutrophils were counted under a confocal microscope. For wild type and Δwzm mutant, P = 0.0006; for wild type and ΔclpX mutant, P = 0.0018; for wild type and ΔclpP mutant, P = 0.0035; for Δwzm mutant and Δwzm::wzm strain, P = 0.0006; for ΔclpX mutant and ΔclpX::clpX strain, P = 0.0018; for ΔclpP mutant and ΔclpP::clpP strain, P = 0.0036 (all comparisons were by t test).
We also analyzed the susceptibilities of these mutants to phagocytosis by healthy human neutrophils by confocal microscopy. Compared to the level of association observed with wild-type strain NTUH-K2044, which is rarely associated with neutrophils, more association was observed between mutants and neutrophils with an incubation time of 15 min (Fig. 2C). Moreover, when the incubation time was extended to 45 min, the numbers of Δwzm and ΔclpX mutants were significantly increased in the human neutrophils. However, wild-type strain NTUH-K2044 and complemented strains were rarely present inside the human neutrophils (Fig. 2D and E).
Neutrophil-mediated killing of Δwzm and ΔclpX strains.
Neutrophil-mediated killing of the Δwzm and ΔclpX mutants was examined. Compared to the number of wild-type bacteria, the numbers of Δwzm and ΔclpX mutants were significantly decreased after incubation with human neutrophils, and the complemented strains restored the resistance to neutrophil killing (Fig. 3).
FIG. 3.
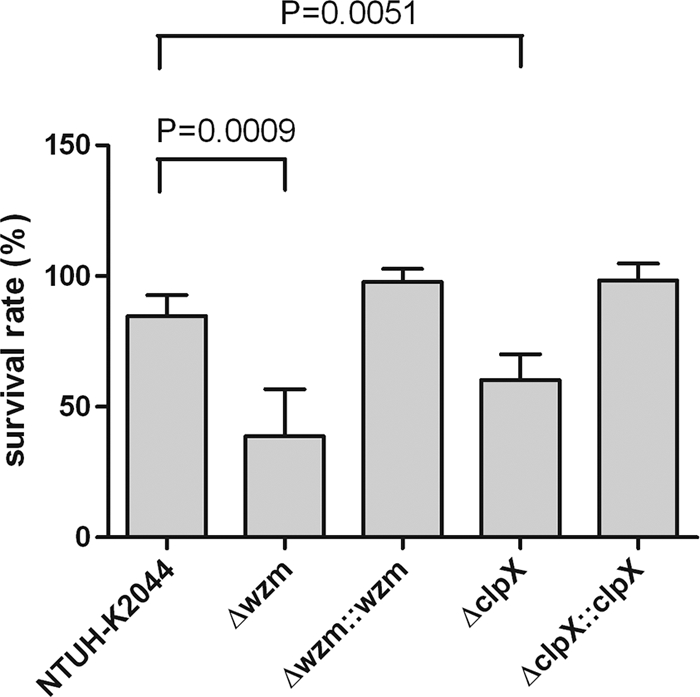
Neutrophil-mediated killing of Δwzm and ΔclpX. Bacterial susceptibilities to killing by human neutrophils of wild-type, Δwzm, ΔclpX, and complemented strains after 45 min of incubation are presented. Survival rate indicates percent survival of wild-type or mutant strains calculated on the basis of viable counts relative to those for the no-neutrophil controls. For wild type and Δwzm mutant, P = 0.0009; for wild type and ΔclpX mutant, P = 0.0051; for Δwzm mutant and Δwzm::wzm strain, P = 0.0055; fir ΔclpX mutant and ΔclpX::clpX strain, P = 0.0049 (all comparisons were by t test).
Silver stain of wzm deletion mutant.
The wzm gene is located in the wb gene cluster, which is responsible for O-antigen lipopolysaccharide synthesis and which has been reported to be an O-antigen transporter (12). Therefore, we further analyzed whether the Δwzm mutant in Klebsiella had a defect in O-antigen synthesis. The O antigen, revealed as a ladder form on silver staining, was absent in the Δwzm mutant and restored in the complemented strain (Fig. 4).
FIG. 4.
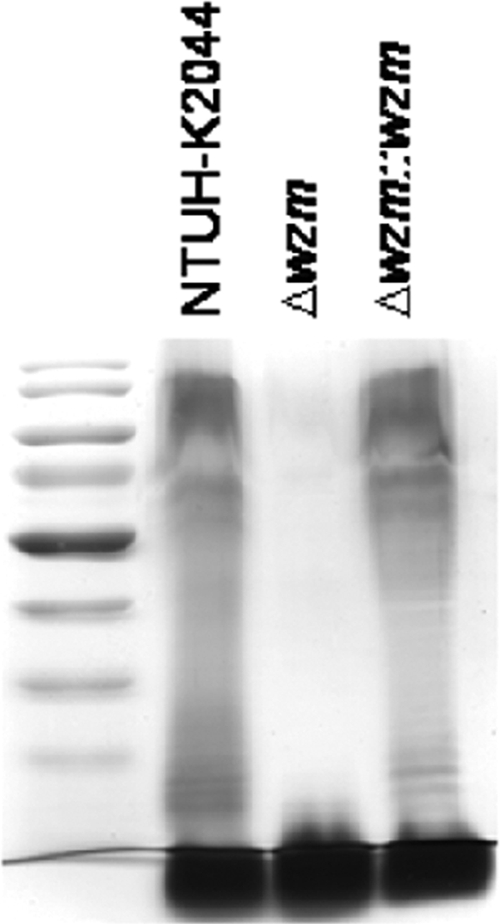
Effect of mutation in wzm gene on lipopolysaccharide O-antigen expression. Extracellular polysaccharides of the wild-type, Δwzm, and Δwzm::wzm strains were purified, and lipopolysaccharide O antigen was detected by silver stain.
Genetic organization of Klebsiella clpX and transcriptional unit determination.
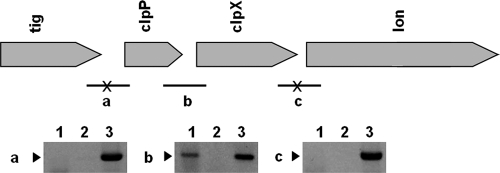
It has been documented that ClpX is a component of the ClpPX protease system and ClpX is responsible for substrate specificity (47). Moreover, in Salmonella enterica serovar Typhimurium, the clpX gene is located in an operon with clpP, and both of them are required for growth of S. enterica serovar Typhimurium in macrophages and virulence in mice (52). The genomic alignment surrounding clpX in Klebsiella was similar to the genetic organization of S. enterica serovar Typhimurium (Fig. 5). When RT-PCR was used to define the clpX transcriptional unit, mRNA was detected between clpX and clpP but not between tig and clpP, clpX, and lon (Fig. 5). The results indicated that clpP and clpX constitute a transcriptional unit.
FIG. 5.
Genetic organization of clpX and transcriptional unit in K. pneumoniae NTUH-K2044. (Top) Genetic organization. Transcriptional units of clpX in NTUH-K2044 were defined by RT-PCR. (a to c) The corresponding PCR products for primers located at each open reading frame (ORF) junction. (a) primers clpP-R-TUD and clpP-750F; (b) primers clpX-R-TUD and clpP-F-TUD; (c) primers clpX-F-TUD and clpX + 750R. Bar, RT-PCR-positive junction; bars with an X, RT-PCR-negative junctions. (Bottom) RT-PCR results. Lane 1, RT-PCR products of each junction; lane 2, RT-PCR without reverse transcriptase, as a negative control; lane 3, PCR with genomic DNA as a template, as a positive control. Arrowheads denote the expected sizes of the RT-PCR products.
Role of clpP in K. pneumoniae for phagocytosis resistance.
To further explore whether clpP is important for resistance to phagocytosis, a clpP deletion mutant and the complemented strain were generated and subjected to a plaque assay with Dictyostelium. This mutant was susceptible to Dictyostelium phagocytosis (Fig. 1C), and it became more susceptible to phagocytosis by human neutrophils (Fig. 2D and E). These phenotypes were restored in the complemented strain (Fig. 1C; Fig. 2D and E).
Microarray analysis of ΔclpX mutant.
To identify the genes affected by ClpX, we compared the transcriptome of the ΔclpX mutant with that of its parent strain, K. pneumoniae NTUH-K2044, by microarray analysis. Fold changes in transcription (increased and decreased) were ranked in duplicate experiments. Genes that were reproducibly represented in the top 5%, according to the level of decreased transcription, are shown in Table 3. No genes for which an increase in the level of transcription exceeded 0.3-fold were found. Among the genes with decreased transcription, the expression of the capsular synthesis gene was reduced in the ΔclpX mutant. We further validated this result by real-time RT-qPCR. Consistently, compared to the wild-type strain, the expression of a capsular synthesis gene, galF, was decreased approximately 3-fold in the ΔclpX mutant. The RNA level was 70% restored when the mutant was complemented with the wild-type clpX gene (for wild type and ΔclpX mutant, P < 0.0001; for ΔclpX and ΔclpX::clpX, P = 0.0235; paired t test for both comparisons) (Fig. 6).
TABLE 3.
Gene expression decreases in ΔclpX of K. pneumoniae NTUH-K2044
| Putative function (gene name) | Avg fold decrease of RNAa |
|---|---|
| Putative inner membrane protein | 114.0 |
| Threonine dehydratase catabolic operon genes (tdcDE) | 40.5 |
| Putative transcriptional regulator | 29.8 |
| Putative transcription activator | 26.5 |
| Possible lipoprotein | 24.6 |
| Threonine dehydratase catabolic operon genes (tdcCDE) | 22.8 |
| Putative oxidoreductase (aegA) | 22.3 |
| DNA repair protein (recO) | 21.6 |
| Alcohol dehydrogenase | 21.4 |
| Threonine dehydratase catabolic operon genes (tdcABC) | 19.8 |
| Cadaverine/lysine decarboxylase operon genes (cadBC) | 18.5 |
| Superoxide dismutase (sodB) | 17.3 |
| Siroheme synthetase (cysF) | 17.2 |
| Permeases of the major facilitator superfamily | 16.2 |
| Outer membrane protein S1 | 16.0 |
| Putative outer membrane protein | 15.2 |
| Putative ligase (yjfG) | 14.8 |
| Glyoxalase family protein | 14.7 |
| Transcriptional activator protein (LysR) | 14.6 |
| Ecotin precursor (eco) | 14.3 |
| Synthesis of vitamin B12 adenosyl cobalamide precursor | |
| (cbiKJHG) | 14.3 |
| Tagatose-1-phosphate kinase (tagK) | 14.0 |
| Protein kinase | 13.6 |
| Putative sarcosine oxidase (solA) | 13.5 |
| Putative diguanylate cyclase/phosphodiesterase domain 1 | 13.3 |
| Possible lipoprotein | 13.2 |
| Acid shock protein | 13.1 |
| CPS synthesis (wzi) | 13.1 |
| Lysine-accepting tRNA synthetase (lysS) | 12.8 |
| Sensor protein (rcsC) | 12.5 |
| Putative hydrolase (yjcS) | 11.8 |
| Transport of long-chain fatty acids (fadL) | 11.5 |
| Carbon starvation protein A (cstA) | 11.4 |
| Iron ABC transporter ATP-binding protein | 11.3 |
| Putative sulfatase (yejM) | 11.2 |
| Heat shock protein A (hslT) | 11.1 |
| O-antigen synthesis (wzm) | 11.0 |
| Paral putative membrane protein (yhiI) | 10.8 |
| Cold shock protein (cspA) | 10.6 |
| Outer membrane protein TolC precursor | 10.6 |
| Putative ferrous iron transport protein | 10.5 |
| Two-component system (rstAB) | 9.9 |
| Putative cytoplasmic protein | 9.8 |
| Putative protease (yegQ) | 9.8 |
| Adenylosuccinate synthetase (purA) | 9.7 |
| Putative lipase | 9.3 |
| Putative membrane protein | 9.0 |
| Putative membrane protein | 9.0 |
| Putative membrane protein | 8.9 |
| Putative UDP-glucose dehydrogenase (ugd) | 8.5 |
| Hypothetical protein | 8.3 |
The data represent averages of the fold decrease in the level of RNA expression from two independent experiments.
FIG. 6.
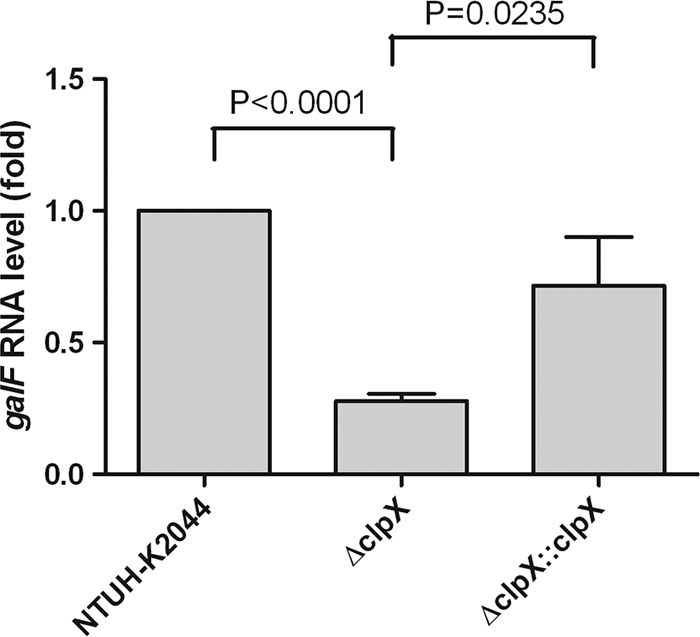
Determination of RNA level of capsular synthesis gene by RT-qPCR. The transcriptional level of galF, a gene responsible for capsular synthesis, in the wild type, the ΔclpX mutant, and complemented strain ΔclpX::clpX was analyzed using real-time RT-qPCR. For wild type and ΔclpX, P < 0.0001; for ΔclpX mutant and ΔclpX::clpX strain, P = 0.0235 (both comparisons were by paired t test).
Serum sensitivity of Δwzm and ΔclpX mutants.
After it was incubated with serum for 3 h, the numbers of CFU of the Δwzm mutant decreased to about 1% of the initial inoculum, suggesting that the mutant became serum sensitive. However, the ΔclpX mutant was as resistant to serum as the wild type.
Capsule production of Δwzm and ΔclpX mutants.
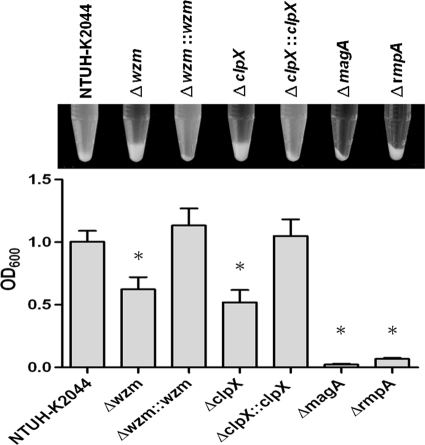
Although the Δwzm and ΔclpX mutants were positive by the string test, which indicates their mucoid phenotype, the reduced production of capsule in these two mutants was evident by low-speed centrifugation. Compared to the wild-type strain, a capsular mutant (the ΔmagA mutant) which lost its mucoviscosity, was easily pelleted and resulted in a tight pellet after centrifugation. The Δwzm and ΔclpX mutants did not present tight pellets and were also more easily pelleted than the wild type, indicating that the two mutants were partially impaired in capsular production (Fig. 7). However, compared to the two mutants, a rmpA (regulator of capsule synthesis) deletion mutant whose mutation which resulted in an apparently lower level of capsule production (P < 0.01) appeared to be resistant to phagocytosis (Fig. 7 and Fig. 1C). Moreover, immunoblotting was performed to determine the reaction of serotype K1-specific antiserum and different dilutions of capsule extracts. As the results indicated, the capsular (ΔmagA) mutant lost its antigenicity; in contrast, the Δwzm, ΔclpX, and ΔrmpA mutants with mucoid phenotypes all preserved their antigenicity (Fig. 8); however, the signal intensity suggested that the ΔrmpA mutant produced a smaller capsule amount than the amounts produced by the Δwzm and ΔclpX mutants.
FIG. 7.
Capsule production of Δwzm and ΔclpX mutants. Capsule production of the wild-type, Δwzm mutant, ΔclpX mutant, and complemented strains was assessed by low-speed centrifugation. The ΔmagA mutant, a capsular mutant, was used as a control. Results for the ΔrmpA mutant, with mutation of a regulator of capsule synthesis, are also presented. OD600 measurements of the supernatant were shown below. ζ, P < 0.01 compared to wild-type strain. For wild type and Δwzm mutant, P = 0.0073; for wild type and ΔclpX mutant, P = 0.0031; for Δwzm mutant and Δwzm::wzm strain, P = 0.0061; for ΔclpX mutant and ΔclpX::clpX strain, P = 0.0051; for Δwzm mutant and ΔrmpA mutant, P = 0.0006; for ΔclpX mutant and ΔrmpA mutant, P = 0.0014 (all comparisons were by t test).
FIG. 8.
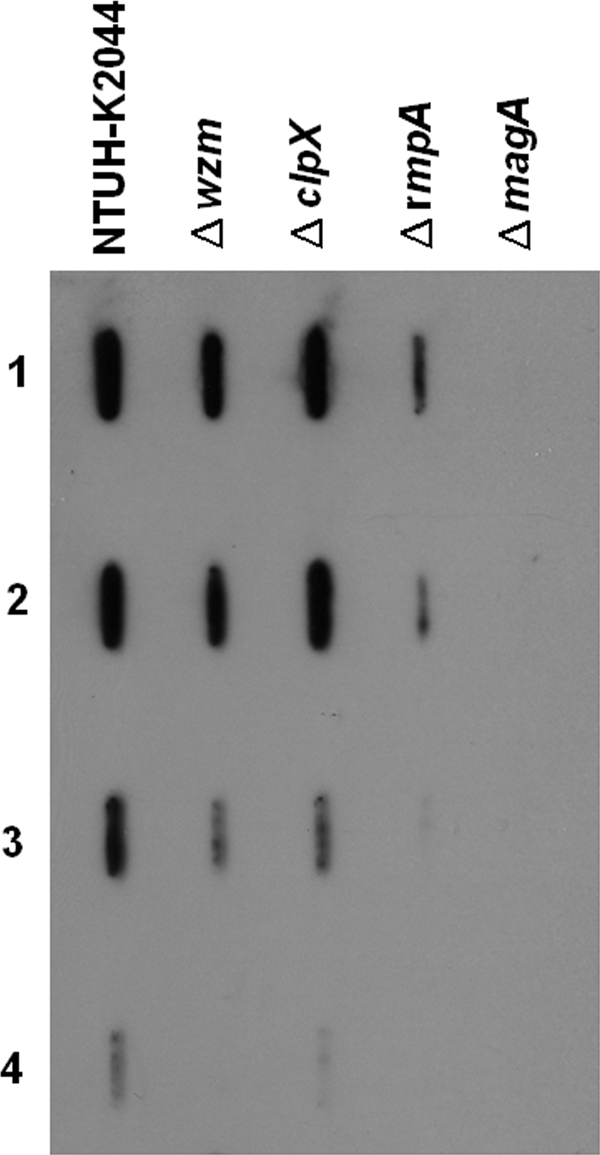
Immunoblot. Capsule extracts from wild-type and mutant strains with equal numbers of bacteria were diluted 1/5, 1/10, 1/100, and 1/1,000 (rows 1 to 4, respectively) in water and detected with serotype K1-specific antiserum.
Animal inoculation.
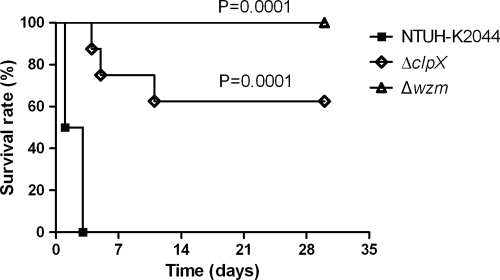
Mice infected by an intraperitoneal inoculation with 103 CFU of the wild-type strain died within 3 days. In contrast, five out of eight mice (∼60%) infected with the ΔclpX mutant and all of the mice infected with the Δwzm mutant survived and appeared to be healthy after 30 days. Statistical analysis of survival rates revealed that both the ΔclpX and Δwzm mutants were attenuated in virulence (Fig. 9). To further investigate the bacterial clearance in vivo, we examined the bacterial loads in liver, spleen, and blood of mice inoculated with 103 CFU of the wild-type, Δwzm, or ΔclpX strain 24 h and 48 h after inoculation. The results indicated that Δwzm mutants were rarely recovered from tissue at either 24 or 48 h, suggesting that Δwzm mutants were rapidly cleared in vivo, and the ΔclpX group had significantly reduced colony counts in the liver and spleen compared with the wild-type group (Fig. 10 A and B). Whereas bacteria were detected in the blood of only a few mice (at 24 h, one from the wild-type group with ∼5 × 102 CFU per 100 μl and one from the ΔclpX group with ∼2 × 102 CFU per 100 μl; at 48 h, one from the wild-type group with ∼2 × 102 CFU per 100 μl). Therefore, the difference between the counts of mutants and the wild type in blood was not obvious at the two time points.
FIG. 9.
In vivo virulence of ΔclpX and Δwzm mutants. Eight BALB/cByl mice in each group were infected with 103 CFU bacteria and monitored for 30 days. x axis, number of days after inoculation; y axis, survival rate of mice. Statistical analysis of the wild type and mutants was performed using the Kaplan-Meier method. For ΔclpX, P = 0.0001; for Δwzm, P = 0.0001 (log-rank test for both comparisons).
FIG. 10.
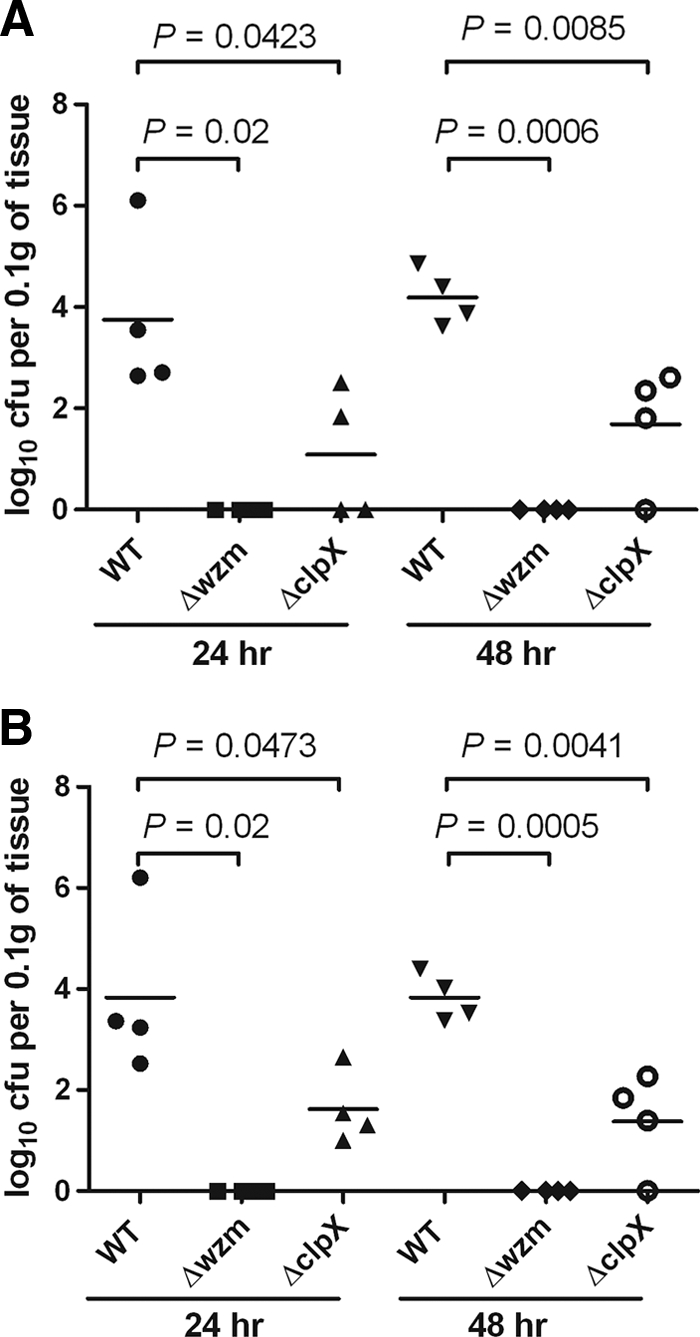
Bacterial clearance in vivo. An inoculation dose of 103 CFU of the wild-type (WT), Δwzm, or ΔclpX strain was administered to each of four mice by intraperitoneal injection. Bacterial levels in tissue were determined at 24 and 48 h. The number of log10 CFU was standardized per 0.1 g of wet organ weight. (A) Bacterial load in liver (at 24 h, for wild type and Δwzm mutant, P = 0.02; for wild type and ΔclpX mutant, P = 0.0423; at 48 h, for wild type and Δwzm mutant, P = 0.0006; for wild type and ΔclpX mutant, P = 0.0085; comparisons were by t test). (B) Bacterial load in spleen (at 24 h, for wild type and Δwzm mutant, P = 0.02; for wild type and ΔclpX mutant, P = 0.0473; at 48 h, for wild type and Δwzm mutant, P = 0.0005; for wild type and ΔclpX mutant, P = 0.0041; comparisons were by t test).
DISCUSSION
In this study, we used a model system incorporating wild-type Dictyostelium to investigate bacterial genes important for resistance to phagocytosis in a pathogenic clinical isolate of K. pneumoniae which is heavily encapsulated and highly resistant to phagocytosis. Moreover, we validated the results by testing the phagocytosis of the mutants by human neutrophils, and we examined the mutants' virulence in vivo. In our screening, capsular mutants that became permissive for phagocytosis were found, as predicted; but we also found two additional genes involved in resistance to phagocytosis: one was the lipopolysaccharide O-antigen transporter, and the other was a protease subunit. Our mutant library covered approximately 2,325 (2,500 × 93%) diverse mutants and displayed 7% redundancy. Of the 28 unique insertions from randomly selected mutants, 23 (82%) were within the coding regions, while 5 (18%) were outside the coding regions. Accordingly, among these 2,500 transposon mutants there were 1,906 (2325 × 82%) mutants which were disrupted in different genes. However, the real coverage was difficult to estimate, since insertions close to an operon can cause phenotypic changes due to polar effects. On the other hand, essential genes, although they were less likely to be specific virulence genes, could not be identified by transposon mutagenesis. Therefore, there could be a few genes missed by screening of this library.
A Dictyostelium model has been used previously to study virulence in extracellular bacteria, such as Yersinia (50), Pseudomonas (2), and Vibrio (44), and in intracellular bacteria, such as Legionella (28, 48) and Mycobacterium (20, 41). The results of these investigations demonstrated that the Dictyostelium model is useful for investigating both phagocytosis and bacterial virulence. This model was also used with Klebsiella in a previous study which focused on discovering host genes required for intracellular killing of a nonpathogenic laboratory strain of K. pneumoniae (4). Two Dictyostelium mutants, phg1 and kil1 mutants, were found to be incapable of growing on K. pneumoniae due to a defect in efficient intracellular killing. They also tried to identify bacterial genes for resistance to intracellular killing in a Dictyostelium phg1 mutant. A total of 800 bacterial mutants were screened, and 6 of them were permissive for Dictyostelium phg1 mutant growth. Genes for intracellular killing resistance were clustered into three categories: capsule or lipopolysaccharide biosynthesis-related genes, genes involved in amino acid biosynthesis, and DNA adenine methylase genes. The virulence of the mutants was assessed in a mouse pneumonia model, and for four of them, the result was a higher survival rate for mice compared with that achieved with the parent strain. In the current study, we used a wild-type strain of Dictyostelium which is more similar to natural phagocytes than the mutants are to investigate bacterial genes important for resistance to phagocytosis in a pathogenic clinical isolate of K. pneumoniae that is highly resistant to phagocytosis.
Our observation that the lipopolysaccharide O-antigen transporter of Klebsiella is important for resistance to phagocytosis and intracellular killing is consistent with the results of the previous study in which a Dictyostelium model was used to identify host genes required for intracellular killing of a nonpathogenic laboratory strain of Klebsiella described above (4). The lipopolysaccharide O antigen of Klebsiella has also been implicated in resistance to killing by murine neutrophils in a previous report (31). In our study, we have proved that a similar phenomenon was operating in human neutrophils. Although the mechanism is still unknown, we conclude that the lipopolysaccharide biosynthetic genes in Klebsiella and intact lipopolysaccharide O antigen appear to be among the factors associated with resistance to phagocytosis and bacterial virulence in vivo. Besides, we also found that the Δwzm mutant became sensitive to serum killing, which is consistent with the observation that O antigen is important for serum resistance (34). Therefore, the rapid clearance of the Δwzm mutant from mice may have resulted from its susceptibility to serum.
Our findings with the ΔclpP and ΔclpX mutants suggest that the protease complex ClpPX is critical for resistance to phagocytosis in K. pneumoniae. ClpX was reported to be a component of the ClpPX protease system that accounts for substrate specificity (47). Previous reports also indicated that clpP was important for degradation of misfolded proteins and tolerance to stresses, such as heat shock, acid, and oxidative stress, in different bacteria (17, 19, 21, 46). In addition, clpP is required for the expression of the virulence factor listeriolysin O and plays a crucial role in virulence and intracellular survival of Listeria monocytogenes (18). Although the ClpPX protease has been studied in several bacteria, there are few reports of the study of ClpPX protease in K. pneumoniae. Furthermore, despite the reports that clpP is related to the intracellular survival of bacteria in macrophages (18, 27, 30, 52), there are no previous reports examining the relationship between resistance to phagocytosis and the ClpPX complex.
It has been reported that inactivation of clpP and clpX may alter the levels of transcription of some genes (15, 39, 46). We also identified several genes related to the cell surface, stress response, transport, and metabolism, the transcription of which was altered in ΔclpX mutants. Interestingly, the expression of capsular synthesis genes and capsule production was reduced in the ΔclpX mutant, indicating that ClpX could regulate capsule synthesis in K. pneumoniae. Although the exact mechanism still needs further studies, according to the results of microarray analysis, a two-component system, RcsBC, involved in cps gene expression, was also downregulated to a similar level as the cps genes in the ΔclpX mutant (Table 3). Therefore, ClpPX-mediated proteolysis was probably important for the normal existence of some mediator(s) which was involved in RNA expression of rcsBC, cps genes, and finally, capsule production. However, although disruption of wzm and clpX partially affected capsule production, a defect in the amount of capsule could not be the critical factor resulting in attenuation of phagocytosis resistance and virulence because a rmpA mutant, which produced a smaller amount of capsule than the amounts produced by the Δwzm and ΔclpX mutants, was not attenuated in phagocytosis resistance and in vivo virulence in mice (unpublished data).
According to the results of the microarray analysis, membrane proteins, proteases, and transcriptional factors were downregulated in the ΔclpX mutant. It has been documented that bacteria have different strategies to prevent phagocytosis. For example, the Yersinia type III secretion system injects effector protein to inhibit the host actin required for engulfment (49). Proteases and membrane or secreted proteins have also been found to interfere with deposition of opsonins or to directly inactivate antibody and complement in several bacterial species (5, 16). Although more experiments are still needed to reveal the mechanism of the resistance to phagocytosis contributed by clpX, our results provide some insight. In one hypothesis, the K. pneumoniae ClpPX protease system may be responsible for regulation of downstream genes, at least the capsular gene, resulting in resistance to phagocytosis by human neutrophils, and this could play some role in K. pneumoniae virulence.
In conclusion, we have identified capsular genes and two noncapsular virulence genes in K. pneumoniae that were related to resistance to phagocytosis in a Dictyostelium model, suggesting that the model is useful for evaluation of heavily encapsulated bacteria. Documentation of the role of clpPX in resistance to phagocytosis may help provide a further understanding of the virulence mechanism of K. pneumoniae.
Acknowledgments
We thank Mei-Yu Chen (Institute of Biochemistry and Molecular Biology, National Yang-Ming University, Taipei, Taiwan) for kindly providing a Dictyostelium strain.
This study was supported by grants from the National Science Council, National Taiwan University, and the Liver Disease Prevention and Treatment Research Foundation in Taiwan.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 20 December 2010.
REFERENCES
- 1.Abbot, S. L. 2003. Klebsiella, Enterobacter, Citrobacter, Serratia, Plesiomonas, and other Enterobacteriaceae, p. 684-700. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, DC.
- 2.Alibaud, L., et al. 2008. Pseudomonas aeruginosa virulence genes identified in a Dictyostelium host model. Cell. Microbiol. 10:729-740. [DOI] [PubMed] [Google Scholar]
- 3.Applied Biosystems. 2001. User bulletin. Relative quantitation of gene expression. Applied Biosystems, Foster City, CA.
- 4.Benghezal, M., et al. 2006. Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell. Microbiol. 8:139-148. [DOI] [PubMed] [Google Scholar]
- 5.Blom, A. M., T. Hallstrom, and K. Riesbeck. 2009. Complement evasion strategies of pathogens—acquisition of inhibitors and beyond. Mol. Immunol. 46:2808-2817. [DOI] [PubMed] [Google Scholar]
- 6.Chou, H. C., et al. 2004. Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect. Immun. 72:3783-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuang, Y. P., C. T. Fang, S. Y. Lai, S. C. Chang, and J. T. Wang. 2006. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 193:645-654. [DOI] [PubMed] [Google Scholar]
- 8.Chun, K. T., H. J. Edenberg, M. R. Kelley, and M. G. Goebl. 1997. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13:233-240. [DOI] [PubMed] [Google Scholar]
- 9.Chung, D. R., et al. 2007. Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. J. Infect. 54:578-583. [DOI] [PubMed] [Google Scholar]
- 10.Cornillon, S., R. A. Olie, and P. Golstein. 1998. An insertional mutagenesis approach to Dictyostelium cell death. Cell Death Differ. 5:416-425. [DOI] [PubMed] [Google Scholar]
- 11.Cosson, P., et al. 2002. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 184:3027-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuthbertson, L., M. S. Kimber, and C. Whitfield. 2007. Substrate binding by a bacterial ABC transporter involved in polysaccharide export. Proc. Natl. Acad. Sci. U. S. A. 104:19529-19534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cvrckova, F., F. Rivero, and B. Bavlnka. 2004. Evolutionarily conserved modules in actin nucleation: lessons from Dictyostelium discoideum and plants. Review article. Protoplasma 224:15-31. [DOI] [PubMed] [Google Scholar]
- 14.Fang, C. T., Y. P. Chuang, C. T. Shun, S. C. Chang, and J. T. Wang. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flynn, J. M., S. B. Neher, Y. I. Kim, R. T. Sauer, and T. A. Baker. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11:671-683. [DOI] [PubMed] [Google Scholar]
- 16.Foster, T. J. 2005. Immune evasion by staphylococci. Nat. Rev. Microbiol. 3:948-958. [DOI] [PubMed] [Google Scholar]
- 17.Frees, D., and H. Ingmer. 1999. ClpP participates in the degradation of misfolded protein in Lactococcus lactis. Mol. Microbiol. 31:79-87. [DOI] [PubMed] [Google Scholar]
- 18.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286-1294. [DOI] [PubMed] [Google Scholar]
- 19.Gerth, U., E. Kruger, I. Derre, T. Msadek, and M. Hecker. 1998. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28:787-802. [DOI] [PubMed] [Google Scholar]
- 20.Hagedorn, M., K. H. Rohde, D. G. Russell, and T. Soldati. 2009. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science 323:1729-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hensel, M., et al. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 22.Hilbi, H., S. S. Weber, C. Ragaz, Y. Nyfeler, and S. Urwyler. 2007. Environmental predators as models for bacterial pathogenesis. Environ. Microbiol. 9:563-575. [DOI] [PubMed] [Google Scholar]
- 23.Hsieh, P. F., H. H. Lin, T. L. Lin, and J. T. Wang. 2010. CadC regulates cad and tdc operons in response to gastrointestinal stresses and enhances intestinal colonization of Klebsiella pneumoniae. J. Infect. Dis. 202:52-64. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh, P. F., T. L. Lin, C. Z. Lee, S. F. Tsai, and J. T. Wang. 2008. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 197:1717-1727. [DOI] [PubMed] [Google Scholar]
- 25.Jules, M., and C. Buchrieser. 2007. Legionella pneumophila adaptation to intracellular life and the host response: clues from genomics and transcriptomics. FEBS Lett. 581:2829-2838. [DOI] [PubMed] [Google Scholar]
- 26.Ko, W. C., et al. 2002. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg. Infect. Dis. 8:160-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon, H. Y., et al. 2004. The ClpP protease of Streptococcus pneumoniae modulates virulence gene expression and protects against fatal pneumococcal challenge. Infect. Immun. 72:5646-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Z., et al. 2009. The amoebal MAP kinase response to Legionella pneumophila is regulated by DupA. Cell Host Microbe 6:253-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loughlin, M. F., V. Arandhara, C. Okolie, T. G. Aldsworth, and P. J. Jenks. 2009. Helicobacter pylori mutants defective in the clpP ATP-dependant protease and the chaperone clpA display reduced macrophage and murine survival. Microb. Pathog. 46:53-57. [DOI] [PubMed] [Google Scholar]
- 31.Lugo, J. Z., et al. 2007. Lipopolysaccharide O-antigen promotes persistent murine bacteremia. Shock 27:186-191. [DOI] [PubMed] [Google Scholar]
- 32.Ma, L. C., C. T. Fang, C. Z. Lee, C. T. Shun, and J. T. Wang. 2005. Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection. J. Infect. Dis. 192:117-128. [DOI] [PubMed] [Google Scholar]
- 33.Maniak, M., R. Rauchenberger, R. Albrecht, J. Murphy, and G. Gerisch. 1995. Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein tag. Cell 83:915-924. [DOI] [PubMed] [Google Scholar]
- 34.Merino, S., et al. 2000. Cloning and sequencing of the Klebsiella pneumoniae O5 wb gene cluster and its role in pathogenesis. Infect. Immun. 68:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noegel, A. A., and M. Schleicher. 2000. The actin cytoskeleton of Dictyostelium: a story told by mutants. J. Cell Sci. 113(Pt 5):759-766. [DOI] [PubMed] [Google Scholar]
- 36.Ohmori, S., et al. 2002. Septic endophthalmitis and meningitis associated with Klebsiella pneumoniae liver abscess. Hepatol. Res. 22:307-312. [DOI] [PubMed] [Google Scholar]
- 37.Okano, H., et al. 2002. Clinicopathological analysis of liver abscess in Japan. Int. J. Mol. Med. 10:627-630. [PubMed] [Google Scholar]
- 38.Pan, Y. J., et al. 2008. Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J. Clin. Microbiol. 46:2231-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pederson, K. J., S. Carlson, and D. E. Pierson. 1997. The ClpP protein, a subunit of the Clp protease, modulates ail gene expression in Yersinia enterocolitica. Mol. Microbiol. 26:99-107. [DOI] [PubMed] [Google Scholar]
- 40.Podschun, R., and U. Ullmann. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11:589-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pozos, T. C., and L. Ramakrishnan. 2004. New models for the study of Mycobacterium-host interactions. Curr. Opin. Immunol. 16:499-505. [DOI] [PubMed] [Google Scholar]
- 42.Pukatzki, S., R. H. Kessin, and J. J. Mekalanos. 2002. The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc. Natl. Acad. Sci. U. S. A. 99:3159-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pukatzki, S., A. T. Ma, A. T. Revel, D. Sturtevant, and J. J. Mekalanos. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U. S. A. 104:15508-15513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pukatzki, S., et al. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A. 103:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahimian, J., T. Wilson, V. Oram, and R. S. Holzman. 2004. Pyogenic liver abscess: recent trends in etiology and mortality. Clin. Infect. Dis. 39:1654-1659. [DOI] [PubMed] [Google Scholar]
- 46.Robertson, G. T., W. L. Ng, J. Foley, R. Gilmour, and M. E. Winkler. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 184:3508-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin, Y., et al. 2009. Single-molecule denaturation and degradation of proteins by the AAA+ ClpXP protease. Proc. Natl. Acad. Sci. U. S. A. 106:19340-19345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solomon, J. M., A. Rupper, J. A. Cardelli, and R. R. Isberg. 2000. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 68:2939-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urban, C. F., S. Lourido, and A. Zychlinsky. 2006. How do microbes evade neutrophil killing? Cell. Microbiol. 8:1687-1696. [DOI] [PubMed] [Google Scholar]
- 50.Vlahou, G., et al. 2009. Yersinia outer protein YopE affects the actin cytoskeleton in Dictyostelium discoideum through targeting of multiple Rho family GTPases. BMC Microbiol. 9:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, J. H., et al. 1998. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 26:1434-1438. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto, T., et al. 2001. Disruption of the genes for ClpXP protease in Salmonella enterica serovar Typhimurium results in persistent infection in mice, and development of persistence requires endogenous gamma interferon and tumor necrosis factor alpha. Infect. Immun. 69:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]