Abstract
Inflammasomes are cytosolic multiprotein complexes that assemble in response to infectious or noxious stimuli and activate the CASPASE-1 protease. The inflammasome containing the nucleotide binding domain-leucine-rich repeat (NBD-LRR) protein NLRC4 (interleukin-converting enzyme protease-activating factor [IPAF]) responds to the cytosolic presence of bacterial proteins such as flagellin or the inner rod component of bacterial type III secretion systems (e.g., Salmonella PrgJ). In some instances, such as infection with Legionella pneumophila, the activation of the NLRC4 inflammasome requires the presence of a second NBD-LRR protein, NAIP5. NAIP5 also is required for NLRC4 activation by the minimal C-terminal flagellin peptide, which is sufficient to activate NLRC4. However, NLRC4 activation is not always dependent upon NAIP5. In this report, we define the molecular requirements for NAIP5 in the activation of the NLRC4 inflammasome. We demonstrate that the N terminus of flagellin can relieve the requirement for NAIP5 during the activation of the NLRC4 inflammasome. We also demonstrate that NLRC4 responds to the Salmonella protein PrgJ independently of NAIP5. Our results indicate that NAIP5 regulates the apparent specificity of the NLRC4 inflammasome for distinct bacterial ligands.
The innate immune system initiates defense against infectious agents by employing germ line-encoded receptors to detect microbial molecules (also called pathogen-associated molecular patterns, or PAMPs) (10). Examples of PAMPs include lipopolysaccharide, cell wall components, bacterial nucleic acids, and flagellin. The mammalian Toll-like receptors (TLRs) are transmembrane receptors that are capable of detecting microbial products at the cell surface and within intracellular compartments (26). Signaling downstream of TLR stimulation results in the activation of NF-κB and the induction of proinflammatory cytokines, chemokines, and other antimicrobial defenses. In addition to TLRs, there are several types of receptors that lack transmembrane domains and function to sense PAMPs within the cytosol (26). For example, the cytosolic presence of RNA is detected by the MDA5/RIG-I family of RNA helicases. It is believed that cytosolic immunosurveillance permits host cells to make specialized or unique responses to intracellular pathogens and thus to distinguish these pathogens from extracellular microbes that do not access the host cell cytosol (27).
The nucleotide binding domain (NBD)-leucine-rich repeat (LRR)-containing proteins constitute an important family of cytosolic immunosensors. Certain NBD-LRR proteins, including NAIP5 and NLRC4, are involved in orchestrating the assembly and activation of multiprotein complexes called inflammasomes (24). The primary function of inflammasomes is to activate the cysteine protease CASPASE-1 (CASP-1), which is produced initially as an inactive proprotein and requires recruitment to inflammasomes to become activated (16). Once activated, CASP-1 is required for the proteolytic processing and secretion of the proinflammatory cytokines interleukin-1β (IL-1β) and IL-18. In addition, activated CASP-1 can induce a rapid, inflammatory cell death termed pyroptosis (2).
Several distinct NBD-LRR proteins are expressed by host cells and appear to dictate inflammasome assembly in response to specific stimuli. For example, the NBD-LRR protein NLRP3 (NALP3; cryopyrin) orchestrates the assembly of an inflammasome that responds to a wide variety of stimuli, including crystalline substances such as uric acid, asbestos, and alum (24). In the mouse, the related NBD-LRR protein NLRP1B (NALP1B) activates a distinct inflammasome in response to anthrax lethal toxin (3). NBD-LRR proteins are not the only sensor-scaffold proteins responsible for CASP-1 activation. For example, the PYHIN family member AIM2 binds cytosolic DNA and is responsible for CASP-1 activation in response to infection with bacterial pathogens (7, 11, 12, 21, 23, 28).
The inflammasome containing the NBD-LRR protein NLRC4 (interleukin-converting enzyme protease-activating factor [IPAF]) is one of the most well-characterized inflammasomes, and it has been shown to activate CASP-1 specifically in response to a conserved domain within the bacterial protein flagellin (1, 9, 14, 17), as well as to the conserved inner rod component of the type III secretion system, called PrgJ in Salmonella (18). Interestingly, in some instances, the activation of the NLRC4 inflammasome requires the presence of a second NBD-LRR protein, NAIP5 (formerly BIRC1E) (14, 19, 22, 30). NAIP5 is believed to heterooligomerize with NLRC4 (5, 30), but the precise biochemical function of NAIP5 in NLRC4 activation remains enigmatic. It was previously shown that NAIP5 is required for CASP-1 activation in response to some, but not all, NLRC4-dependent stimuli (14). For example, NAIP5 was required for the NLRC4-dependent activation of CASP-1 in response to Legionella pneumophila, whereas Naip5 deficiency had only partial effects on NLRC4-dependent responses to Salmonella and Pseudomonas (14). NAIP5/NLRC4/CASP-1 activation by L. pneumophila is dependent largely on amino acids within the C terminus of flagellin (14), and accordingly the retrovirus-mediated expression of a minimal C-terminal peptide from flagellin activates CASP-1-mediated pyroptotic cell death in a manner completely dependent on NLRC4 and NAIP5 (14). Unexpectedly, however, the retrovirus-mediated expression of full-length flagellin activated NLRC4- and CASP-1-dependent pyroptosis independently of NAIP5 (14).
The molecular basis for the differential requirements for NAIP5 in NLRC4 activation is not well understood. Legionella and Salmonella both activate NLRC4 primarily via the translocation of flagellin to the cytosol of host cells, but the flagellin molecules translocated exhibit amino acid differences in the critical C-terminal domain. Moreover, while Salmonella translocates flagellin into host cells via its SPI-I type III secretion system (T3SS) (25), Legionella lacks a T3SS and instead utilizes the evolutionarily unrelated Dot/Icm type IV secretion system (T4SS) to translocate flagellin into host cells. T3SSs, but not T4SSs, contain homologs of the bacterial protein PrgJ, another known activator of NLRC4 (18). Thus, differences in flagellin itself, and/or the flagellin-translocating apparatus, may underlie the differential requirements for NAIP5 in NLRC4 activation.
In this paper, we dissect the molecular features that dictate the requirement for NAIP5 in the activation of the NLRC4 inflammasome. We demonstrate that L. pneumophila engineered to express Salmonella flagellin activates NLRC4 in a strictly NAIP5-dependent manner, thus ruling out polymorphisms within flagellin as an explanation for the differential requirement for NAIP5 in the activation of NLRC4 by the two bacterial species. We further demonstrate that the N terminus of flagellin, while not sufficient itself to activate NLRC4, nevertheless is able to transform the C terminus of flagellin from an NAIP5-dependent activator of the NLRC4 inflammasome to an NAIP5-independent activator. Lastly, we show that the PrgJ protein can activate NLRC4 without a requirement for NAIP5. Taken together, our results suggest a model in which NAIP5 functions to dictate the specificity of NLRC4 for distinct stimuli.
MATERIALS AND METHODS
Mice.
Wild-type (WT) C57BL/6J (B6) mice were from Jackson Laboratories. Nlrc4−/− mice on a pure B6 background (15) were obtained from S. Mariathasan and V. Dixit (Genentech). Naip5−/− mice on a pure B6 background mice were described previously (14).
Bacterial strains.
The Salmonella enterica serovar Typhimurium strain LT2 and the flagellin mutant S. Typhimurium LT2FliC/FljB− were gifts from A. Van Der Velden and M. Starnbach. The pLIV2-L.p.FlaA and pLIV2-S.t.PrgJ constructs were constructed as isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible variants of previously described strains. A construct fusing the amino-terminal 300 bp of the actA gene with the flaA gene from L.p.FlaA was previously described and used to facilitate the ectopic secretion of Legionella pneumophila FlaA (J.-D. Sauer et al., submitted for publication). The actA-L.p.FlaA fusion was amplified from the pPL2-L.p.FlaA plasmid with the primers AAAAGCGGCCGGTGGGATTAAATAGATTTATGCGTGC and AAAAGTCGACCAGAAATCGAAGTGCAGTTG using Pfu Ultra II (Agilent Technologies, Santa Clara, CA), where underlining indicates the restriction sites. The resulting PCR product and the IPTG-inducible variant of pPL2, pLIV2 (8), were double digested with EagI and SalI (New England Biosystems, Waltham, MA) and ligated using the NEB quick ligation kit. The resulting vector, pLIV2-L.p.FlaA, subsequently was integrated at the tRNAArg locus on the L. monocytogenes 10403s chromosome as previously described (13). pLIV2-S.t.PrgJ was constructed similarly to the previously described actA-prgJ fusion construct (Sauer et al., submitted). The fusion was amplified from the pPL2-S.t.PrgJ plasmid using the primers AAAAGCGGCCGCAGGAGGGAGTATAAGTGGGATTAAATAG and TTTTGTCGACTCATGAGCGTAATAGCGTTTC, digested, and ligated into pLIV2, and then it was integrated into the 10403s chromosome as previously described. Legionella strain LP02 is a streptomycin-resistant thymidine auxotroph derivative of Legionella pneumophila strain LP01. LP02ΔflaA contains an unmarked deletion of flaA and was described previously (22). LP02ΔflaA::fliC expresses Salmonella fliC from the chromosome under the control of the endogenous Legionella flaA promoter. To generate this strain, the flaA promoter from LP02 was amplified by PCR using primers pflaAFwd (ATAGTCGACTTAATGCCTCTTTCTCTCCTGTCG) and pflaARev (AATGACTTGTGCCATAATTTTAGTCTCCTCAGACCTGAATCC), while fliC was amplified from S. Typhimurium using primers fliCfwd (GAGGAGACTAAAATTATGGCACAAGTCATTAATACAAACAGC) and fliCrev (ATAGGATCCTTAACGCAGTAAAGAGAGGACGTTTTG). These amplicons were mixed and spliced in a second round of PCR using pflaAFwd and fliCrev and cloned into the allelic exchange vector pSR47S. This suicide construct was introduced onto the LP02ΔflaA chromosome by a single crossover and selection with kanamycin (50 μg/ml).
Retroviral constructs and transductions.
Retroviral constructs were generated and transductions were performed as previously described (14). Briefly, retroviral particles were generated by the transient transfection of Phoenix-Eco packaging cells with MSCV2.2-based retroviral vectors. Bone marrow-derived cells (1 × 106) were cultured for 48 h in a 6-well plate in medium containing macrophage colony-stimulating factor (M-CSF) and then were transduced with 1 ml of retrovirus-containing packaging cell supernatant. Macrophages were analyzed 3 to 4 days posttransduction and analyzed for GFP expression on a Beckman Coulter FC-500 flow cytometer. More than 10,000 cells were analyzed per sample. When we simultaneously transduced cells with multiple constructs (see Fig. 2), equal volumes of each retroviral supernatant were used. These macrophages were analyzed on a BD Influx cell sorter for green fluorescent protein (GFP) and mCherry expression. The retroviral constructs used in this paper were generated as follows: GFP-C65 was constructed by amplifying the C-terminal 65 amino acids of flagellin using the primers pC65fwd (GACGAGCTGTACAAGTTTGAATCAACGATAG) and pFlaA65rev (TTGCGGCCGCCTATCGACCTAACAAAGATAATACAGATTGCG), while GFP was amplified using the primers pGFPfwd (ATAAGATCTCCACCATGGTGAGCAAGGGCGAGGA) and pGFP65rev (CTATCGTTGATTCAAACTTGTACAGCTCGTC). These amplicons were mixed and spliced in a second round of PCR using the primers pGFPfwd and pFlaArev and were cloned into the retroviral expression vector pMSCV2.2 using the enzymes BglII and NotI. Similarly, the construct N65-GFP-C65 was generated by amplifying GFP-C65 with the primers pN65GFPfwd (GGATGAACCAAGCCGTTATGGTGAGCAAGGGC) and pFlaA65rev, while the N-terminal 65 amino acids of flagellin were amplified with the primers pN65fwd (ATAAGATCTCCACCATGGCTCAAGTAATCAACACTAATGTGG) and pN65GFPrev (GCCCTTGCTCACCATAACGGCTTGGTTCATCC). These amplicons were spliced in a second round of PCR using the primers pN65fwd and pFlaA65rev. N65-GFP was generated by amplifying the N-terminal 65 amino acids of flagellin with the primers pN65fwd and pN65GFPrev while amplifying GFP with the primers pN65GFPfwd and pGFPrev (TTGCGGCCGCTTACTTGTACAGCTCGTC). These amplicons were mixed and spliced in a second round of PCR using the primers pN65fwd and pGFPrev. The constructs N65-GFP-C65 L12L32::II and N65-GFP-C65 were generated using site-directed mutagenesis on the parental N65-GFP-C65 construct, pL12fwd (CACTAATGTGGCGTCGatcACAGCCCAACGTAATTTGGG), pL12rev (CCCAAATTACGTTGGGCTGTgatCGACGCCACATTAGTG), pL32fwd (CATCGATCCAGCGTatcTCATCGGGATTAAGG), and pL32rev (CCTTAATCCCGATGAgatACGCTGGATCGATG) for L12L32::II and pL12Afwd (CACTAATGCAGCGTCGGCAACAGCCCAACG), pL12Arev (CGTTGGGCTGTT GCCGACGCTGCATTAGTG), pI5V9Aafwd (GGCTCAAGTAGCAAACACTAATGCAGCGTCGCTCAC), and pI5V9AArev(GTGAGCGACGCTGCATTAGTGTTT GCTACTTGAGCC) for I5V9L12:AAA. mCherry-C65 was generated by amplifying mCherry using pmCherryfwd (ATAAGATCTCCACCATGGTGAGCAAGGGCGAGG) and pmCherry65rev (GGCTATCGTTGATTCAAACTTGTACAGCTCGTCC), while the C-terminal 65 amino acids of flagellin were amplified using the primers pmCherry65fwd (GGACGAGCTGTACAAGTTTGAATCAACGATGCC) and pFlaA65rev. These amplicons were mixed and spliced during a second round of PCR with the primers pmCherryfwd and pFlaA65rev and cloned into pMSCV2.2 with BglII and NotI.
Cytotoxicity assays.
Cytotoxicity was measured as described previously (6) by determining the activity of lactate dehydrogenase (LDH) released by cells. Bone marrow-derived macrophages (5 × 104 to 1 × 105) were seeded onto tissue culture-treated 96-well plates and infected with bacteria at the indicated multiplicity of infection. Plates were centrifuged at 400 × g to ensure the equivalent infectivity of WT and flagellin mutant strains. At 30 min to 1 h postinfection, medium was removed and replaced with medium containing 10 to 100 μg/ml gentamicin to kill extracellular bacteria. Six hours after infection, plates were centrifuged at 400 × g, and supernatant was collected and assayed for LDH release. Legionella strains used for infection were grown overnight to an optical density of >3.8. Salmonella cultures were grown overnight with shaking at 37°C and were diluted 3 h prior to infection at 1:40 in 1 ml of LB and regrown with shaking until reaching late-exponential phase. Listeria strains were grown as standing cultures at 37°C in brain heart infusion (BHI) medium. In the experiments with Listeria, infected cells were treated with IPTG to induce the expression of flaA or prgJ. The percent specific lysis of infected cells was calculated as the following: (amount of LDH released) − (background release by uninfected cells)/(amount of LDH released by detergent lysed cells minus background) × 100.
Growth curves.
Bacterial growth was determined as previously described (4). Macrophages were plated at a density of 5 × 105 cells per ml in 24-well plates, allowed to adhere overnight, and infected with stationary-phase Legionella (OD, >3.8) at a multiplicity of infection (MOI) of 0.01. CFU were determined by lysing washed macrophage monolayers with sterile distilled water and plating lysates on buffered charcoal yeast extract plates.
Protein transfection.
Salmonella FliC, PrgJ, and SsaI proteins were purified as described previously (18). Macrophages were plated at 5 × 104 per well in a 96-well plate, allowed to adhere overnight, and primed with tripalmitoyl cysteinyl seryl tetralysine lipopeptide (Pam3CSK4; 0.5 μg/ml) for 3.5 h prior to protein transfection to induce the expression of pro-interleukin-1β (pro-IL-1β). Protein (0.2 μg/well) was transfected using the Profect P1 (Targeting Systems) transfection reagent according to the manufacturer's instructions. Supernatants were collected for analysis 3 h posttransfection and analyzed by enzyme-linked immunosorbent assay (ELISA) for IL-1β (R&D Systems).
Statistics.
Statistical significance was assessed by the t test or one-way analysis of variance (ANOVA) with Bonferroni posttests. Graph Pad Prism 5 software was used for statistical analyses.
RESULTS
The N terminus of flagellin can relieve the requirement for NAIP5 in flagellin sensing.
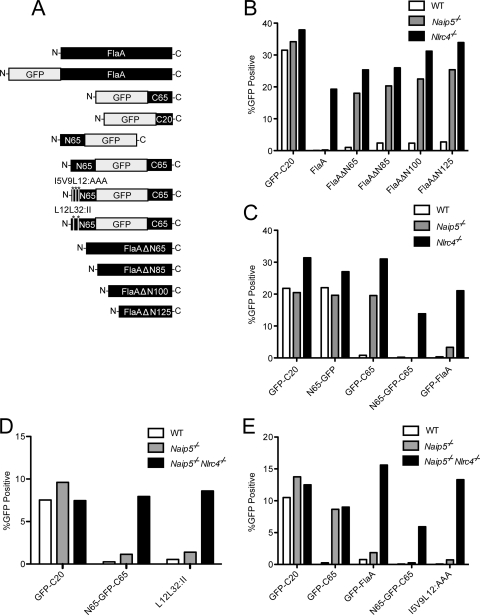
To better understand the molecular basis for NAIP5/NLRC4 activation, we first sought to determine what regions of flagellin enable full-length flagellin to be toxic to Naip5−/− macrophages in the retroviral lethality assay (14). In this assay, flagellin is expressed downstream of a mammalian promoter in macrophages using a retrovirus-based construct. The transduction of macrophages with a construct expressing a stimulatory flagellin molecule results in the rapid pyroptotic cell death of the macrophages and thereby prevents the expression of a coexpressed GFP reporter (14). Conversely, the transduction of a construct encoding a nonstimulatory flagellin mutant results in GFP+ cells (14). Previously, the retroviral lethality assay was used to establish that NLRC4 and NAIP5 both are required for the response to the C-terminal domain of flagellin, whereas pyroptotic cell death in response to full-length flagellin required NLCR4 but not NAIP5 (14). Thus, to determine what regions in full-length flagellin relieve the requirement for NAIP5, we began by making a series of retroviral constructs expressing N-terminally deleted flagellin molecules (Fig. 1A). These constructs were transduced into primary macrophages. As shown previously (14), retroviral expression constructs encoding full-length flagellin (FlaA) did not stably transduce C57BL/6 (WT) or isogenic Naip5−/− macrophages, as evidenced by a lack of GFP+ cells 3 days after transduction (Fig. 1B). However, we found that a retroviral construct that expresses a flagellin molecule lacking the N-terminal 65 amino acids from flagellin (FlaANΔ65) could be transduced into Naip5−/− macrophages (Fig. 1B). Consistently with our previous observation that the C terminus of flagellin is sufficient to activate NLRC4, FlaANΔ65 (which retains the normal flagellin C terminus) could not be transduced into WT macrophages. Further, N-terminal deletions of FlaA (ΔN85, ΔN100, and ΔN125) resembled ΔN65 and could be transduced into Naip5−/− macrophages (Fig. 1B). These results suggest that the N terminus of flagellin is important for the ability of full-length flagellin to activate NLRC4 independently of NAIP5.
FIG. 1.
Role for the N terminus of flagellin in NLRC4 activation. (A) Diagram of all retroviral constructs used in transduction experiments. Note that all constructs expressed GFP, either as a direct fusion to L. pneumophila flagellin (FlaA; as shown) or downstream of an internal ribosome entry site (IRES) (not shown). C65, C-terminal 65 amino acids of FlaA; C20, C-terminal 20 amino acids of FlaA; N65, N-terminal 65 amino acids of FlaA. FlaAΔN constructs lack the indicated number of amino acids from the N terminus. (B and C) Wild-type (C57BL/6) or isogenic Naip5−/− or Nlrc4−/− macrophages were transduced with the indicated constructs, and the percentage of GFP+ cells was enumerated by flow cytometry 3 to 4 days after transduction. (D and E) Wild-type, Naip5-deficient, or Naip5−/− Nlrc4−/− doubly deficient macrophages were transduced with the indicated constructs, and the percentage of GFP+ cells was enumerated by flow cytometry 3 to 4 days after transduction. More than 10,000 cells were analyzed for each experiment. For each panel, data shown are from a single representative experiment of at least three that produced similar results.
Molecular basis by which the N terminus affects the requirement for Naip5.
We next sought to address whether the N terminus of flagellin is sufficient to convert the C terminus of flagellin into an NAIP5-independent activator of NLRC4. Consistently with our previous studies (14), we found that the C-terminal 65 amino acids of flagellin, fused to GFP, is an NAIP5-dependent activator of NLRC4 (Fig. 1C). In contrast, full-length flagellin (whether or not it is fused to GFP) is an NAIP5-independent activator of NLRC4 (Fig. 1B and C). To determine the role of the N terminus in NAIP5/NLRC4 activation, we created an expression construct that consisted of GFP flanked by the N-terminal and C-terminal 65 amino acids of FlaA (Fig. 1A). The resulting N65-GFP-C65 construct behaved just like full-length flagellin; namely, it was cytotoxic to macrophages in a manner requiring NLRC4 but independent of the presence or absence of NAIP5 (Fig. 1C). The N terminus of flagellin fused to GFP was itself noncytotoxic and could be transduced into WT (Nlrc4+) macrophages (Fig. 1C). Thus, the N terminus of flagellin is not sufficient to activate NLRC4. However, the N terminus of flagellin appears to enhance or alter NLRC4-dependent responses to the C terminus of flagellin in such a way that NAIP5 is no longer required.
Although the C-terminal region of flagellin is thought to be unstructured when flagellin is in its monomeric form, the structure of flagellin within the flagellin filament (29) suggests that the N and C termini of flagellin interact weakly via a coiled-coil interaction. Moreover, the crystal structure of GFP reveals that its N and C termini are near each other (20), and it therefore is possible that the appended N and C termini of flagellin interact with each other in the N65-GFP-C65 construct. Thus, we hypothesized that a weak coiled-coil interaction with the N terminus of flagellin resulted in a stabilized or structurally altered C-terminal region that no longer required NAIP5 for the stimulation of NLRC4. To test this idea, we mutated the N terminus of flagellin within the N65-GFP-C65 construct to disrupt the putative interaction between the N and C termini. Two leucines (L12 and L32) in the N terminus of flagellin were mutated to more bulky isoleucines, which would not be easily accommodated within the core of a coiled coil and thus should result in a disruption of putative coiled-coil interactions. In addition, a series of mutations predicted to be even more disruptive to the putative coiled-coil interaction were made, namely, isoleucine 5, valine 9, and leucine 12 all were mutated to alanines (I5V9L12:AAA). Neither series of mutations was sufficient to transform the parental N65-GFP-C65 stimulus from an NAIP5-independent stimulus (N65-GFP-C65) into an NAIP5-dependent one (Fig. 1D and E). Thus, we were unable to find evidence that a coiled-coil interaction between the N and C termini of flagellin is important for the ability of full-length flagellin to activate NLRC4 independently of NAIP5. These results do not rule out the possibility that an interaction between the N and C termini of flagellin (that we did not disrupt with the mutations we made) is important in determining the requirement for NAIP5 in NLRC4 activation. However, a lack of a role for a coiled-coil interaction would not be surprising, given that the interaction between the N and C termini of flagellin likely is very weak (29) and probably not significant when flagellin is in its monomeric form, which is the form that appears to be recognized by NLRC4 (17).
The N and C termini of flagellin can act in trans to activate NLRC4.
Because coiled-coil interactions did not appear to be required for the NAIP5-independent activation of NLRC4 by flagellin, we questioned whether the N and C termini of flagellin needed to be expressed on the same molecule or whether they could cooperate in trans to activate NLRC4 without NAIP5. To address this question, a construct encoding the N-terminal 65 amino acids of flagellin fused to GFP (N65-GFP), and a separate construct encoding the C-terminal 65 amino acids of flagellin fused to mCherry (mCherry-C65), were expressed simultaneously in wild-type, Naip5-deficient, and Nlrc4-deficient macrophages via retroviral transduction. As expected, mCherry-C65 was cytotoxic to wild-type macrophages but not Naip5−/− or Nlrc4−/− macrophages (Fig. 2A to C). While a robust population of brightly double-positive macrophages was detected in transduced Nlrc4−/− macrophages, Naip5−/− macrophages harbored only a proportionally smaller, less bright population of double-positive macrophages (Fig. 2B and C). These results indicate that, at the relatively high expression levels achieved by retroviral transduction, the N terminus of flagellin can act in trans to convert the C terminus of flagellin into an NAIP5-independent stimulus. We were unable to detect a physical interaction between mCherry-C65 and N65-GFP by immunoprecipitation (data not shown). Taken together, our data raise the possibility that the N and C termini cooperate to activate NLRC4 without physically interacting with each other, although a role for a transient or weak physical interaction cannot be rule out.
FIG. 2.
N terminus of flagellin can relieve requirement for NAIP5 in flagellin sensing even when expressed on a distinct molecule from the C terminus. A retroviral construct expressing the N-terminal 65 amino acids of flagellin fused to GFP, and a separate retroviral construct expressing the C-terminal 65 amino acids of flagellin fused to mCherry, were transduced simultaneously into wild-type (A), Naip5−/− (B), or Nlrc4−/− (C) macrophages, and the percentage of cells expressing GFP and/or mCherry was enumerated by flow cytometry 4 days posttransduction. The percentage of double-positive cells among all transduced cells is indicated in parentheses. The results are representative of two representative experiments.
NAIP5-independent detection of Salmonella.
We previously reported that while the activation of the inflammasome by Legionella was strictly flagellin and NAIP5 dependent, the activation of the inflammasome by Salmonella was partially NAIP5 independent. It is difficult to compare infections between two different bacterial species when there may be differences in infectivity, translocation levels of flagellin, or flagellin molecules themselves. Thus, to begin to determine if there is in fact a qualitative difference between Salmonella and Legionella with respect to the activation of NLRC4, we assayed macrophage cell death at a wide range of MOIs for both bacterial species (Fig. 3). We centrifuged bacteria onto the macrophages to minimize differences in infectivity between motile and nonmotile strains and carefully monitored the infectivity of the various strains by determining gentamicin-resistant cell-associated CFU 1 h after infection. We found that the infection of macrophages with Legionella causes macrophage cell death in a manner dependent upon both the host expression of NAIP5 and NLRC4 and the bacterial expression of flagellin across a broad range of MOIs, from 0.3 to 10 (Fig. 3A). In contrast, we found that Salmonella causes a partially flagellin-independent cell death at higher bacterial loads (e.g., MOI, >2) (Fig. 3A), which is consistent with previous results (18). As previously described (14), we also found that at these higher MOIs, Salmonella also induces a partially NAIP5-independent (but NLCR4-dependent) cell death. Some of the NAIP5-independent death appeared to be flagellin dependent, but some appeared to be flagellin independent. The flagellin-dependent activation of NLRC4 could not be explained by a reduced infectivity of the ΔfliC Salmonella mutant, since the CFU per cell were quantified and not found to be significantly different. Thus, the activation of NLRC4 by Salmonella seems to be only partially NAIP5 and flagellin dependent. This contrasts with the activation of NLRC4 by Legionella, which was more uniformly NAIP5 and flagellin dependent.
FIG. 3.
Differential requirement for NAIP5 in sensing Legionella and Salmonella is dictated by the bacterium and not the flagellin itself. (A) Cell death was assessed by the measurement of lactate dehydrogenase (LDH) release from wild-type (B6), isogenic Naip5−/−, or isogenic Naip5−/− Nlrc4−/− doubly deficient macrophages infected with either wild-type (WT) Legionella (LP02), isogenic flagellin-deficient Legionella (ΔflaA), wild-type Salmonella (LT2), or isogenic flagellin-deficient Salmonella (designated LT2 fliC/fljB−). Gentamicin (100 μg/ml) was added 30 min after infection to kill extracellular bacteria. (B) Cell death was assessed by LDH release from B6, Naip5−/−, or Nlrc4−/− macrophages infected with WT Legionella (LP02), Legionella ΔflaA, or Legionella expressing Salmonella fliC from the Legionella chromosome in place of its native flaA gene (ΔflaA::fliC). Gentamicin (100 μg/ml) was added 30 min after infection to kill extracellular bacteria. (C) IL-1β release assayed from macrophages infected with the indicated strains, as shown in panel B. Macrophages were pretreated with tripalmitoyl cysteinyl seryl tetralysine lipopeptide (Pam3CSK4; 0.5 μg/ml) for 3.5 h to induce the expression of pro-IL-1β prior to infection. (D) Growth of ΔflaA or ΔflaA::fliC Legionella in wild-type B6, isogenic Naip5−/−, or isogenic Nlrc4−/− macrophages was assayed by determining the CFU at the time points indicated. *, P < 0.00. Two-way ANOVA and Bonferroni's test were used to determine the statistical significance (P) of differences in IL-1β and LDH release (B and C) against infection with ΔflaA Legionella. For each panel, data shown are from a single representative experiment of at least two that produced similar results.
To further assess the underlying differences in NLRC4 activation between Salmonella and Legionella, we asked whether the NAIP5-independent death initiated by Salmonella is due to an intrinsic difference between Legionella and Salmonella flagellin. Thus, we generated a strain of Legionella that expresses Salmonella flagellin (fliC) in place of its own flagellin. This strain (LP02 ΔflaA::fliC) was capable of activating the NLRC4 inflammasome almost as efficiently as wild-type Legionella (22), and interestingly, it did so in a fully NAIP5-dependent manner (Fig. 3B). In addition, complementing flagellin-deficient Legionella with Salmonella fliC restored the Naip5-dependent IL-1β secretion in response to infection with Legionella (Fig. 3C) and the growth restriction of Legionella within macrophages (Fig. 3D). Thus, when expressed from Legionella, Salmonella flagellin activates NLRC4 in an entirely NAIP5-dependent manner, similarly to Legionella's own flagellin. For reasons that remain unclear, we were unable to generate a strain of Salmonella that was capable of translocating Legionella flagellin into host cells. Nevertheless, our data strongly suggest that there are not intrinsic differences between the flagellins that underlie the differing abilities of Legionella and Salmonella to activate the NLRC4 inflammasome independently of NAIP5. These results are consistent with previous experiments that demonstrated that the C termini of Legionella and Salmonella flagellin behaved identically in the retroviral lethality assay (14).
PrgJ activates Nlrc4 in an Naip5-independent manner.
Recently, it was reported that Salmonella PrgJ activates NLRC4 (18). PrgJ is an essential inner rod component of the SPI-1 type III secretion system. Legionella utilizes a type IV secretion system to translocate flagellin into host cells and lacks both a type III secretion system and a PrgJ homolog. Thus, one hypothesis to explain the ability of Salmonella to activate NLRC4 independently of NAIP5 is that Salmonella PrgJ can activate NLRC4 independently of NAIP5. We therefore utilized the retroviral lethality assay to test whether PrgJ activated NLRC4 independently of NAIP5 (Fig. 4A). We found that in contrast to the control retrovirus expressing the Naip5-dependent GFP-C65 stimulus, the PrgJ-expressing retrovirus was not efficiently transduced into Naip5−/− macrophages. This result is consistent with PrgJ being an NAIP5-independent activator of NLRC4; however, the interpretation of the result is complicated by the fact that full-length flagellin itself behaves as an NAIP5-independent activator of NLRC4 in the retroviral lethality assay (Fig. 1B) (14). Thus, the lack of NAIP5 dependence for PrgJ in the retroviral lethality assay may be due in part to a peculiarity of the assay (for example, the very high expression levels obtained from the retroviral promoter). Therefore, to test whether PrgJ could stimulate NLRC4 independently of NAIP5, it was necessary to utilize an assay in which full-length flagellin behaved as an NAIP5-dependent stimulus. Two such assays were employed. In the first, full-length recombinant FliC and PrgJ proteins were transfected into the macrophage cytosol using protein transfection as described previously (17, 18). The activation of NLRC4 was monitored by measuring the NLRC4/CASP-1-dependent release of IL-1β into the supernatant (Fig. 4B). Although IL-1β release in response to transfected FliC protein required NAIP5, IL-1β release in response to PrgJ was nearly entirely NAIP5 independent (Fig. 4B).
FIG. 4.
Sensing of Salmonella PrgJ is independent of NAIP5. (A) Flow cytometry of wild-type (WT; C57BL/6), isogenic Naip5−/−, or Nlrc4−/− macrophages transduced with retroviruses expressing N65-GFP, GFP-C65, or Salmonella PrgJ. The PrgJ construct expressed GFP downstream of an internal ribosome entry site. (B) IL-1β release by wild-type, Naip5−/−, or Nlrc4−/− macrophages transfected with purified Salmonella flagellin (FliC), PrgJ, or SsaI proteins. (C) Cell death assessed by LDH released from wild-type, Naip5−/−, or Nlrc4−/− macrophages infected with wild-type Listeria, Listeria expressing PrgJ, or Listeria expressing Legionella FlaA. The expression of PrgJ or FlaA was induced by the addition of the indicated concentrations of IPTG. *, P < 0.001. For B and C, two-way ANOVA and Bonferroni's test were used to determine the statistical significance (P) of differences in IL-1β and LDH release. Significance tests were done versus WT macrophages or versus both Naip5−/− and WT macrophages when transfecting with PrgJ (B) or infecting with Listeria expressing PrgJ (C). For each panel, data shown are from a single representative experiment of at least two that produced similar results.
In the second assay to assess whether PrgJ is an NAIP5-independent activator of NLRC4, we utilized flagellin- and PrgJ-expressing strains of the Gram-positive bacterium Listeria to activate NLRC4. Listeria strains engineered to express Legionella flagellin (FlaA) strongly activate NLRC4-dependent cell death (Sauer et al., submitted). Although the cell death induced by Listeria flaA was NAIP5 dependent, cell death induced by Listeria prgJ was entirely NAIP5 independent (Fig. 4C). Thus, we conclude that PrgJ can activate NLRC4 independently of NAIP5.
DISCUSSION
Our results demonstrate that NAIP5 is required for the activation of the NLRC4 inflammasome only in response to certain stimuli. These stimuli include infection with Legionella (expressing Legionella or Salmonella flagellin) or transduction with a retrovirus expressing the C terminus of flagellin fused to GFP. Other stimuli, such as constructs expressing both the N and C termini of flagellin, or PrgJ, appear to activate NLRC4 without a requirement for NAIP5. Lastly, infection with Salmonella activates the NLRC4 inflammasome in a partially NAIP5-dependent, partially NAIP5-independent manner.
The NLRC4 inflammasome has yet to be reconstituted or purified, thus the biochemical role of NAIP5 in the activation of NLRC4 remains poorly understood. Nevertheless, our results indicate that NAIP5 affects the ability of the NLRC4 inflammasome to respond to specific stimuli. There are several possible models that account for the ability of NAIP5 to regulate the apparent specificity of NLRC4. One model (the potentiation model) is that NAIP5 functions as a potentiator of NLRC4 activation. In this model, the NLRC4 stimuli that exhibit a requirement for NAIP5 are intrinsically weak activators of NLRC4 and thus require NAIP5 to activate NLRC4. For example, the C terminus of flagellin might be poorly structured in the absence of the N terminus of flagellin and therefore may be a relatively poor activator of NLRC4. The presence of NAIP5 could stabilize the C terminus of flagellin and thus improve its ability to stimulate NLRC4. NAIP5 also might potentiate NLRC4 responses by acting downstream of flagellin recognition and act to amplify the NLRC4-dependent activation of CASP-1 in response to a weak stimulus. We found that the N terminus of flagellin can relieve the requirement for NAIP5 in NLRC4 activation. In the potentiation model, the N terminus might function to stabilize the C terminus, thereby improving its signaling capacity and thus relieving the requirement for NAIP5. A direct biochemical interaction between flagellin and NLRC4 or NAIP5 has yet to be demonstrated; thus, these models are difficult to distinguish at present. It also is not clear whether Legionella is in fact a weaker activator of NLRC4 than is Salmonella, as would be implied under the potentiation model. In fact, at similar MOIs, Legionella and Salmonella appear to induce comparable levels of NLRC4-dependent cell death (Fig. 3). It also is not clear whether PrgJ, an NAIP5-independent activator of NLRC4, is a substantially stronger activator of NLRC4 than is flagellin. In fact, the transfection of wild-type macrophages with equivalent amounts of each protein activates NLRC4 similarly, but PrgJ appears to be a much better activator of NLRC4 specifically in Naip5−/− macrophages. Thus, it is not clear whether our results can be readily explained by a model in which NAIP5 functions primarily to potentiate NLRC4 signaling.
Although it is widely assumed that NLRC4 is the proximal sensor of the ligands that lead to its activation, this has never been formally demonstrated. In fact, it is possible that NAIP5, or another host protein, is the true direct cytosolic sensor (or receptor) of flagellin. Thus, another model to explain our data is that NAIP5 functions not as a general potentiator of NLRC4 activation/signaling but as a specific sensor of certain bacterial ligands. We call the latter model the specificity model. For example, NAIP5 may specifically recognize the C terminus of flagellin, whereas another host protein may specifically recognize full-length flagellin or PrgJ. This model could explain the differential NAIP5 dependence for NLRC4 activation by Legionella versus Salmonella, since Salmonella expresses PrgJ, an NAIP5 independent stimulus, whereas Legionella does not. The specificity model is difficult to address until direct evidence is obtained that NAIP5, NLRC4, or another host protein is the true receptor for flagellin and/or PrgJ.
It was interesting that full-length flagellin delivered by protein transfection or Listeria activated NLRC4 in an NAIP5-dependent manner (Fig. 4B and C), whereas full-length flagellin expressed from a retroviral construct activated NLRC4 in an NAIP5-independent manner (Fig. 1B) (14). Although there are many possible explanations for this result, one plausible idea is that the very high levels of flagellin expression attained via retroviral transduction are responsible for relieving the requirement for NAIP5. Thus, both the expression levels of the stimulus and its molecular characteristics may influence the requirement for NAIP5.
Regardless of the underlying molecular mechanism, our data illustrate the ability of NLRC4 to respond to distinct bacterial ligands. In future studies, it will be important to determine the specific molecular role that NAIP5 plays in NLRC4 activation.
Acknowledgments
This work was supported by Investigator Awards to R.E.V. from the Burroughs Wellcome Fund and the Cancer Research Institute as well as by NIH grants (AI075039 and AI080749). J.P. was supported by Stiftelsen Olle Engkvist Byggmästare through the Swedish Research Council. J.-D.S. was supported by the American Cancer Society (PF-07-066-01-LIB).
We thank Tom Alber for help with the analysis of the putative coiled coil in the D0 region of flagellin.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 31 January 2011.
REFERENCES
- 1.Amer, A., et al. 2006. Regulation of Legionella phagosome maturation and infection through flagellin and host IPAF. J. Biol. Chem. 281:35217-35223. [DOI] [PubMed] [Google Scholar]
- 2.Bergsbaken, T., S. L. Fink, and B. T. Cookson. 2009. Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 7:99-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyden, E. D., and W. F. Dietrich. 2006. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 38:240-244. [DOI] [PubMed] [Google Scholar]
- 4.Coers, J., R. E. Vance, M. F. Fontana, and W. F. Dietrich. 2007. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell Microbiol. 9:2344-2357. [DOI] [PubMed] [Google Scholar]
- 5.Damiano, J. S., V. Oliveira, K. Welsh, and J. C. Reed. 2004. Heterotypic interactions among NACHT domains: implications for regulation of innate immune responses. Biochem. J. 381:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decker, T., and M. L. Lohmann-Matthes. 1988. A quick and simple method for the quantitation of lactate dehydrogenase release in measurements of cellular cytotoxicity and tumor necrosis factor (TNF) activity. J. Immunol. Methods 115:61-69. [DOI] [PubMed] [Google Scholar]
- 7.Fernandes-Alnemri, T., et al. 2010. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 11:385-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischetti, V. A., and American Society for Microbiology. 2006. Gram-positive pathogens, 2nd ed. ASM Press, Washington, DC.
- 9.Franchi, L., et al. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat. Immunol. 7:576-582. [DOI] [PubMed] [Google Scholar]
- 10.Janeway, C. A., Jr. 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54(Pt 1):1-13. [DOI] [PubMed] [Google Scholar]
- 11.Jones, J. W., et al. 2010. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl. Acad. Sci. U. S. A. 107:9771-9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, S., et al. 2010. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur. J. Immunol. 40:1545-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauer, P., M. Y. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lightfield, K. L., et al. 2008. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat. Immunol. 9:1171-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mariathasan, S., et al. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430:213-218. [DOI] [PubMed] [Google Scholar]
- 16.Martinon, F., K. Burns, and J. Tschopp. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10:417-426. [DOI] [PubMed] [Google Scholar]
- 17.Miao, E. A., et al. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7:569-575. [DOI] [PubMed] [Google Scholar]
- 18.Miao, E. A., et al. 2010. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. U. S. A. 107:3076-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molofsky, A. B., et al. 2006. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J. Exp. Med. 203:1093-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ormö, M., et al. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science 273:1392-1395. [DOI] [PubMed] [Google Scholar]
- 21.Rathinam, V. A., et al. 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11:395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren, T., D. S. Zamboni, C. R. Roy, W. F. Dietrich, and R. E. Vance. 2006. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauer, J. D., et al. 2010. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe 7:412-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroder, K., R. Zhou, and J. Tschopp. 2010. The NLRP3 inflammasome: a sensor for metabolic danger? Science 327:296-300. [DOI] [PubMed] [Google Scholar]
- 25.Sun, Y. H., H. G. Rolan, and R. M. Tsolis. 2007. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype typhimurium. J. Biol. Chem. 282:33897-33901. [DOI] [PubMed] [Google Scholar]
- 26.Takeuchi, O., and S. Akira. 2010. Pattern recognition receptors and inflammation. Cell 140:805-820. [DOI] [PubMed] [Google Scholar]
- 27.Vance, R. E., R. R. Isberg, and D. A. Portnoy. 2009. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe 6:10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warren, S. E., et al. 2010. Cutting edge: cytosolic bacterial DNA activates the inflammasome via Aim2. J. Immunol. 185:818-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yonekura, K., S. Maki-Yonekura, and K. Namba. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424:643-650. [DOI] [PubMed] [Google Scholar]
- 30.Zamboni, D. S., et al. 2006. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 7:318-325. [DOI] [PubMed] [Google Scholar]