Abstract
Orientia tsutsugamushi, the causative agent of scrub typhus, is an obligate intracellular pathogen whose mechanism of cellular adhesion and invasion is poorly characterized. Bioinformatic analyses of two O. tsutsugamushi genomes revealed the presence of a group of genes that encode autotransporter proteins. In this study, we identified 10 autotransporter gene products and categorized them into five groups of orthologs (ScaA to ScaE) based on their sequence similarities. Sequence homology was highest between members of ScaC group, suggesting the functional conservation of bacterium-host interactions. ScaC was actively expressed on the surface of O. tsutsugamushi and induced antibody responses in scrub typhus patients. Experiments using microbeads conjugated to recombinant ScaC or a surrogate Escherichia coli expression system showed that ScaC was sufficient to mediate attachment to, but not invasion of, nonphagocytic mammalian cells. In addition, preincubation of host cells with recombinant ScaC significantly inhibited their interaction with O. tsutsugamushi. Finally, fibronectin was identified as a potential receptor for ScaC by using yeast two-hybrid screening, and this was confirmed using a glutathione S-transferase (GST) pulldown assay. Taken together, these results demonstrate that ScaC is involved in the interaction of O. tsutsugamushi with mammalian host cells and suggest that ScaC may play a critical role in bacterial pathogenesis.
Orientia tsutsugamushi is an obligate intracellular organism and the causative agent of scrub typhus (31), a disease characterized by fever, rash, eschar, pneumonitis, meningitis, and disseminated intravascular coagulation. If left untreated, scrub typhus can lead to multiorgan failure, with mortality rates ranging from 1% to 40% depending on the strain of O. tsutsugamushi encountered (36). Scrub typhus is geographically confined to southeastern Asia and is found in many countries in this region, including South Korea, Japan, China, and India (31). An estimated 1 billion people in this area are at risk from scrub typhus, with an estimated 1 million new cases occurring annually (36). The rapid increase in scrub typhus cases (17), coupled with new outbreaks within some areas of disease endemicity (38) in which the disease has not been seen previously, is becoming a public health issue. Even though scrub typhus is effectively treated with antibiotics such as doxycycline and chloramphenicol, reinfections are common because of the wide variety of antigenically distinct serotypes (15). In addition, decreasing efficacy of antibiotic treatments has been reported in several cases (23, 35). In spite of an increasing number of patients and recurrent outbreaks of scrub typhus in areas of disease endemicity (17, 21, 23), an effective vaccine has not yet been developed (4).
Bacterial invasion of host cells is mediated primarily by interactions between bacterial surface components and complementary host receptors. As an obligate intracellular organism, O. tsutsugamushi must be internalized into host cells in order to survive and replicate. The bacterium infects several types of nonphagocytic cells, such as endothelial cells and fibroblasts, as well as macrophages and polymorphonuclear leukocytes (PMN) (9, 24, 29, 31). After entry into the host cells, the intracellular pathogens escape from vacuoles and move to the perinuclear region, where they replicate (16). However, the molecular basis of intracellular invasion by O. tsutsugamushi is poorly characterized. Previously, we reported that O. tsutsugamushi could bind to host fibronectin and utilize it for internalization via interactions with the outer membrane protein TSA56 (7, 18). Fibronectin is known to facilitate bacterial entry into host cells, potentially via its interaction with integrins. O. tsutsugamushi exploits integrin-mediated signaling and rearrangement of the actin cytoskeleton, which mediate “induced phagocytosis” in nonphagocytic host cells (7).
Bacterial entry into host cells can be divided into two distinct stages: adherence and invasion. Recently, it was reported that Rickettsia spp. utilize multiple outer membrane proteins to adhere to and invade nonphagocytic host cells. The Rickettsia conorii autotransporter protein Sca1 mediates bacterial adherence to, but not invasion of, a panel of epithelial and endothelial cells (30), whereas the Sca2 autotransporter protein can mediate both adherence to and invasion of nonphagocytic host cells (2). Also, two rickettsial surface proteins, rickettsial outer membrane protein A (rOmpA) and rickettsial outer membrane protein B (rOmpB), participate in adhesion to and invasion of mammalian cells in vitro (3, 20, 34). rOmpB mediates bacterial adhesion to mammalian cells by binding to its mammalian receptor, Ku70, and subsequently activating host cell signaling pathways that may ultimately induce actin polymerization at the site of bacterial contact (3, 22). Therefore, rickettsial entry into host cells occurs sequentially via the initial interaction between bacterial adhesins and host receptors, the activation of downstream host signaling, and finally, active invasion (defined as induced phagocytosis).
Interestingly, all of the identified outer membrane proteins involved in rickettsial entry belong to a family of autotransporter proteins that contain an N-terminal signal sequence and a highly conserved C-terminal β-barrel, or autotransporter, domain (1, 14). The signal sequence targets the protein to the bacterial periplasm, where the autotransporter domain inserts itself into the outer membrane to form a conduit through which the central passenger domain is transported and exposed to the extracellular surface. A recent bioinformatic analysis of the rickettsial genome showed that at least 15 autotransporter genes, denoted surface cell antigen (sca) genes, are present in the bacterial genome and that some of them appear to have evolved under positive selection and are conserved within the genomes of most rickettsial species (1). This suggests that the positively selected passenger domains may be involved in host cell interactions and may have conserved functions.
Even though accumulating evidences have supported the hypothesis that rickettsial Sca proteins may play a critical role in the bacterium-host interactions, there has been no study on the role of Sca proteins in the pathogenesis of O. tsutsugamushi thus far. Here, we identified multiple sca genes in the genomes of two O. tsutsugamushi strains, Boryong (8) and Ikeda (26), and characterized them through bioinformatic approaches. Moreover, we selected one sca gene, scaC, which is highly conserved between the two strains, and performed functional analysis to investigate its role in the bacterial adherence and/or invasion of nonphagocytic host cells.
MATERIALS AND METHODS
Sequence analysis and alignment.
O. tsutsugamushi sca genes were searched in the two fully sequenced genomes of O. tsutsugamushi strains Boryong (GenBank accession number AM494475.1) and Ikeda (GenBank accession number AP008981.1). We used the BlastP program (E value < 1 × 10−10) with Sca proteins previously identified in Boryong strain (8) and their autotransporter domain to search against the whole proteomes of two O. tsutsugamushi strains. Ten sca gene sequences were finally extracted and aligned for phylogenetic analysis using ClustalW embedded within MegAlign software (DNAstar Inc., Madison, WI). The degrees of identity and divergence among the Sca sequences were determined by using calculated alignment. The divergence between two genes is calculated by comparing sequence pairs (i, j) in relation to the constructed phylogeny as follows: divergence (i, j) = 100 × [distance (i, j)]/total distance. Distance equals the sum of the calculated branch length between two sequences, i and j, and the total distance is the sum of all branch lengths in the phylogenic tree. The identification and annotation of protein domains were performed using the SMART (Simple Modular Architecture Research Tool) program (http://smart.embl-heidelberg.de/) (19).
Reverse transcription-PCR.
To detect O. tsutsugamushi scaC gene expression, total RNA was isolated from O. tsutsugamushi-infected L929 cells by using an RNeasy minikit (Qiagen, Hilden, Germany). The RNA samples were then digested with RNase-free DNase (Qiagen) at room temperature to remove any contaminating DNA. Reverse transcriptase PCR (RT-PCR) was performed using a reverse transcription system (Promega, Madison, WI) and an Accupower Pfu PCR kit (Bioneer, Daejon, South Korea). The reactions were performed according to the manufacturers' instructions by using 1 μg total RNA and 10 μM concentrations of the relevant primers (forward primer 5′-AAGTGTAAATATCCTCAGGCAGAGG and reverse primer 5′-AGCGCTTGCTCTAGCTTTACTTC).
Cloning, expression, and purification of ScaC.
The full-length scaC gene was PCR amplified from the genomic DNA of the O. tsutsugamushi Boryong strain by using the primer pair 5′-GGCGGATCCAAAAGTATAACTCCAGAAAAGTA (forward) and 5′-GGTCGACTTTAATTTAGCACGATTTAT (reverse) (restriction sites underlined). It was then cloned into a pET28a vector via the BamHI and SalI sites to yield pScaC. To generate anti-ScaC antibodies, a gene fragment corresponding to the ScaC passenger domain (amino acids 33 to 232; ScaC33-232) was amplified from the genomic DNA by using the primer pair 5′- GGCGGATCCAAAAGTATAACTCCAGAAAAGTA (forward) and 5′-CGGTCGACCTAAAAATTAGTT CCTATATG (reverse). The amplified fragment was then directionally cloned into pET28a via the BamHI and SalI sites to yield pScaC33-232. For the expression and purification of ScaC proteins, Escherichia coli BL21(DE3) (Novagen) was transformed with pScaC33-232 and grown in LB broth containing ampicillin (100 μg/ml). Protein expression was induced by adding 0.4 mM isopropyl β-d-thiogalactoside (IPTG; Duchefa, Zwijndrecht, Netherlands) for 4 h at 37°C. Bacteria were harvested by centrifugation at 1,000 × g for 10 min, resuspended in binding buffer (300 mM NaCl, 50 mM sodium phosphate buffer, 10 mM imidazole) containing 1 mg/ml of lysozyme, incubated at 4°C for 30 min, and disrupted by sonication on ice for 5 min. The sonicated lysates were centrifuged at 1,600 × g for 20 min at 4°C and the supernatants applied to a Ni-nitrilotriacetic acid (NTA) His-binding resin (Novagen) preequilibrated with binding buffer. His-tagged proteins bound to the Ni-NTA resin were eluted with elution buffer (300 mM NaCl, 50 mM sodium phosphate buffer, 250 mM imidazole) and serially dialyzed against elution buffer to remove any free imidazole.
pGST-ScaC33-232 was constructed by restriction enzyme-mediated insertion into pGEX4T-1 (GE Healthcare, Piscataway, NJ). The ScaC fragment was amplified from O. tsutsugamushi genomic DNA by using the primer pair GGCGGATCCAAAAGTATAACTCCAGAAAAGTG (forward) and TCTGAATTCCTAACTGATATAGTTTAA (reverse) (restriction sites underlined). It was then cloned into pGEX4T-1 via the BamHI and EcoRI sites. Recombinant glutathione S-transferase (GST)-ScaC33-232 and GST proteins were expressed in E. coli BL21(DE3) harboring either pGST-ScaC33-232 or pGEX4T-1. Following induction with IPTG, the proteins were purified using glutathione columns according to the manufacturer's instructions (GE Healthcare). Finally, the identity and purity of proteins were assessed by Western blotting and Coomassie blue staining, respectively.
Cell culture.
HeLa cells (ATCC CCL-2; American Type Culture Collection), L929 cells (NCTC929; ATCC), Vero cells (ATCC CCL-81), and ECV304, an endothelial cell-like cell line (33), were maintained in Dulbecco's modified Eagle's medium (DMEM; Welgene, Daegu, South Korea) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Welgene), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco BRL, Gaithersburg, MD) at 37°C in 5% CO2.
Preparation of O. tsutsugamushi.
The Boryong strain of O. tsutsugamushi was purified by using a modified Percoll gradient purification method (7). O. tsutsugamushi was propagated in L929 cells. At 3 to 4 days postinfection, infectivity was determined using an indirect immunofluorescence assay (see below). When an infection rate of >90% was achieved, the cells were harvested by centrifugation at 6,000 × g for 20 min. The cell pellet was resuspended with 6.5 ml of Tris-sucrose (TS) buffer (33 mM Tris-Cl [pH 7.4], 0.25 M sucrose) and the cells homogenized using 100 strokes of a Polytron homogenizer (Wheaton Inc., Millville, NJ) followed by centrifugation at 200 × g for 5 min. The supernatant was then mixed with 40% Percoll (Pharmacia Fine Chemicals, Uppsala, Sweden) in TS buffer and centrifuged at 25,000 × g for 60 min. The bacterial band was collected and centrifuged at 77,000 × g for 30 min. The bacterial pellet was washed 3 times in TS buffer, resuspended in DMEM, and stored in liquid nitrogen until use. The infectivity titer of the inoculum was determined as previously described (10), with minor modifications. Infected-cell counting units (ICU) were calculated as follows: [(total number of cells used for infection) × (percentage of infected cells) × (dilution of the O. tsutsugamushi suspension)]/100 (10). For infection assays, 2.5 × 106 ICU of O. tsutsugamushi was used to infect cells cultured in 24-well plates.
Antibodies and reagents.
Both preimmune rabbit serum and anti-ScaC polyclonal rabbit serum (produced by immunization with purified ScaC33-232 protein; Koma Biotech, Seoul, South Korea) were used for the experiments. Human sera were prepared from scrub typhus patients, control patients with an acute febrile illness not diagnosed as scrub typhus, or healthy volunteers, following institutional review board approval and the receipt of informed consent from all subjects. Horseradish peroxidase (HRP)-conjugated anti-mouse, anti-rabbit, or anti-human IgG secondary antibodies (Santa Cruz Biotech Inc., Santa Cruz, CA) were used for immunoblotting. The Alexa Fluor 488- or Alexa Fluor 594-conjugated anti-human, -mouse, and -rabbit antibodies used in the immunofluorescence assays were purchased from Molecular Probes (Invitrogen). For the bead-binding assay, Fluoresbrite microparticles (1 μm; Polyscience Inc., Warrington, PA) containing rhodamine were conjugated to GST or GST-ScaC33-232 proteins by using a PolyLink protein coupling kit (Polyscience Inc.) in accordance with the manufacturer's instructions.
Immunofluorescence microscopy.
Immunofluorescence microscopy was used to visualize O. tsutsugamushi (18). Infected monolayers of L929 cells in 24-well tissue culture plates were collected by trypsin treatment 1 h postinfection. Extracellular or surface-bound bacteria were removed by washing three times with phosphate-buffered saline (PBS). The infected cells were fixed in PBS containing 4% paraformaldehyde for 15 min at room temperature and permeabilized in 0.2% Triton X-100 for 15 min. Cells infected with O. tsutsugamushi were incubated with human serum or anti-ScaC immune serum for 1 h, followed by incubation with Alexa Fluor 488-conjugated goat anti-human IgG and Alexa Fluor 594-conjugated mouse anti-rabbit IgG (Molecular Probes). In some experiments, recombinant E. coli was fixed with 4% paraformaldehyde and stained with preimmune rabbit serum, anti-ScaC serum, or anti-E. coli serum, followed by incubation with Alexa Fluor 488-conjugated mouse anti-rabbit IgG (Molecular Probes). Cells were examined under an Olympus FV1000 laser scanning confocal microscope (Olympus, Tokyo, Japan). Images of cell sections were captured every 200 nm, and all images were analyzed and processed using Olympus Fluoview software (Olympus).
ELISA.
Immunoassay plates (96-well plates; Nunc, Rochester, NY) were coated with 100 μl of purified His-tagged ScaC33-232 at a concentration of 5 μg/ml in 0.05 M bicarbonate buffer (pH 9.5) overnight and blocked with 3% bovine serum albumin (BSA) at room temperature for 1 h. Human sera, from 10 scrub typhus patients (indirect immunofluorescence assay [IFA] titer ≥ 1:1,280) or from 10 control patients with an acute febrile illness not diagnosed as scrub typhus, were serially diluted (2-fold dilutions from 1:100 to 1:51,200) in PBS containing 3% BSA and 0.1% Tween 20, added to the immunoassay wells, and incubated at 37°C for 1 h. After washing six times with PBS containing 0.05% Tween 20, an anti-human IgG-HRP conjugate (1:10,000) in PBS containing 3% BSA and 0.1% Tween 20 was added and incubated for 1 h. TMB microwell peroxidase substrate solution (KPL, Gaithersburg, MD) was then added to develop color for 7 min, and the reaction was stopped by the addition of 1 M H3PO4 solution. Absorbance was measured at 450 nm by using an enzyme-linked immunosorbent assay (ELISA) plate reader (Beckman). Titers were expressed as the inverse of the highest dilution in which a net optical density (absorbance with antigen − absorbance without antigen) was greater than the mean net absorbance plus 2 standard deviations of 20 negative-control sera collected from healthy volunteers.
Bead-binding assay.
HeLa cells (1.2 × 105 cells in a 24-well plate) were incubated with Fluoresbrite microparticles (530 μg/well) conjugated to GST or GST-ScaC for 1 h, washed extensively with PBS, and fixed with 4% paraformaldehyde for 15 min. Cells were subsequently incubated with ToPro-3 (Molecular Probes) for nuclear staining and observed under a confocal microscope or analyzed using a FACScan (Becton Dickinson).
Membrane fractionation of E. coli.
The outer membrane of recombinant E. coli was fractionated as previously described (2). Briefly, 10 ml of induced E. coli cultures was pelleted and resuspended in 1 ml of lysis buffer (PBS plus protease inhibitor cocktail [Sigma Chemicals]). Cells were lysed by sonication for 3 s and incubated further for 10 s on ice. Unbroken cells were removed by centrifugation for 10 min at 4°C at 1,000 × g. The supernatant was transferred to a new tube, and inner membrane protein was extracted with Sarkosyl (final concentration, 0.5%) at room temperature for 5 min. Outer membrane fractions were pelleted by centrifugation at maximum speed for 30 min at 4°C, resuspended in 2× sample buffer, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and subsequently analyzed by immunoblotting with anti-ScaC rabbit serum (1:5,000) and goat anti-rabbit IgG-HRP conjugate (1:10,000). Immunoreactive bands were revealed with chemiluminescence substrates (Pierce) and exposure of membranes to an LAS-4000 reader (GE Healthcare).
Cellular adhesion and invasion assays.
Bacterial adhesion and invasion assays were performed as previously described (7, 18, 30). Briefly, E. coli strains harboring pET28a or pScaC were induced with IPTG and added to confluent monolayers of ECV304, HeLa, and Vero cells in serum-free media. Portions of the bacterium-containing media were plated to determine the number of CFU added to each host cell monolayer. Contact between bacteria and the mammalian cells was synchronized by centrifugation at 200 × g, and the preparations were incubated at 37°C for either 20 min or 60 min for the adherence and invasion assays, respectively. For the invasion assays, infected cells were washed extensively with PBS and incubated for 2 h with complete medium supplemented with 100 μg/ml of gentamicin to kill any extracellular bacteria. For all E. coli assays, infected cells were washed extensively with PBS and the bacteria liberated by incubation with 0.1% Triton X-100 in sterile H2O. The lysate was then plated on LB agar to enumerate the cell-associated bacteria. The results were expressed as the percentages of bacteria recovered relative to the number of bacteria in the initial inoculum.
To assess O. tsutsugamushi infection, HeLa cells were cultured on 12-mm-diameter glass coverslips in 24-well plates and inoculated with O. tsutsugamushi. For the inhibition assays, cells were preincubated with 100 μg/ml of GST or GST-ScaC33-232 at 37°C for 1 h. Thirty minutes after bacterial inoculation, infected cells were washed three times with PBS, fixed with 4% paraformaldehyde, and permeabilized in a 0.2% Triton X-100 solution for use in immunofluorescence assays incorporating an anti-TSA56 monoclonal antibody (MAb), followed by Alexa Fluor 488-conjugated rabbit anti-mouse IgG to stain host cell-associated bacteria. One hundred cells were randomly selected by using an Olympus FV1000 laser scanning confocal microscope and analyzed using the Fluoview software.
Yeast two-hybrid screening.
Yeast two-hybrid screening with ScaC33-232 was performed using the human aorta cDNA activation domain (AD) library. Saccharomyces cerevisiae (yeast) strain PBN240 (Panbionet Inc., Pohang, South Korea) was cotransformed with two hybrid plasmids, a bait plasmid, pBCT-ScaC33-232 (which encodes a GAL4 DNA binding domain [BD]-fused to ScaC cDNA), and a pACT plasmid that encodes human aorta cDNA fused to GAL4 AD. During the screening process, three different reporter genes (ura3, ade2, and lacZ), each under the control of a different GAL4 binding site, were used to minimize any false positives. After sequential screening using the selective media, we obtained 40 genuine positive clones. To reconfirm the specific interaction between BD-ScaC33-232 and the AD-prey proteins, the AD-prey DNAs were amplified by PCR using total DNA purified from the transformants. The amplified PCR fragment, together with a linearized prey vector, was reintroduced into yeast strain PBN204 expressing BD-ScaC33-232. The specific interactions were confirmed in all the transformants by checking the expression levels of URA3, ADE2, and LacZ. pBCT-polypyrimidine tract-binding protein (PTB) and pACT2-PTB served as positive controls for the protein-protein interactions. pBCT (Panbionet Inc.) and pACT2 (Clontech Laboratories Inc.) were used as the negative controls.
GST pulldown assay.
The GST or GST-ScaC33-232 fusion protein was purified from E. coli BL21 after induction with IPTG. The recombinant proteins were bound to glutathione beads and incubated with fibronectin (20 μg/ml) in binding buffer (20 mM HEPES [pH 7.4], 100 mM NaCl, 1% NP-40, protease inhibitors) at 4°C overnight. The glutathione beads were then washed four times with binding buffer, and the fibronectin associated with the beads was analyzed by SDS-PAGE and subjected to immunoblotting using an antifibronectin antibody (Takara Bio Inc., Shiga, Japan).
Statistical analysis.
Statistical analysis was performed using Student's t test and SigmaPlot (Jandel, San Rafael, CA). Data were expressed as means ± standard deviations, and P values of <0.01 were considered to be statistically significant.
RESULTS
ScaC is highly conserved among O. tsutsugamushi strains.
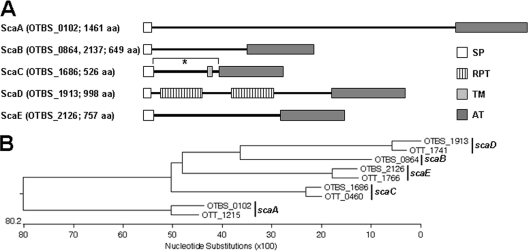
Analysis of the genomic sequences of O. tsutsugamushi revealed the presence of six genes encoding a conserved autotransporter domain in the Boryong strain and four genes in Ikeda strain (Fig. 1) (8, 26). One of the Sca proteins (ScaB; OTBS_0864 and OTBS_2137 [GenBank accession no. CAM79930.1 and CAM81232.1, respectively]) was duplicated in the Boryong strain but absent from the Ikeda strain. Eight Sca proteins were classified into four groups of orthologs (ScaA, ScaC, ScaD, and ScaE) on the basis of the nucleotide sequence similarity of the genes (Fig. 1B). The genes surrounding three orthologous sca genes (scaC, scaD, and scaE) were found in colinear order within the genomes of the two strains (data not shown), further suggesting orthologous relationships. The scaA gene is surrounded by mobile genetic elements or repetitive sequences in the two bacterial genomes. All the Sca proteins were likely to be functional because they possessed both the C-terminal autotransporter domain and the N-terminal signaling peptide required for export across the inner membrane (Fig. 1A). The passenger domain of the ScaD protein contained two conserved, repeated motifs containing internal repeat sequences, whereas the other proteins lacked any known protein motif (Fig. 1A). The ScaC protein was predicted to have a transmembrane domain close to the autotransporter domain.
FIG. 1.
Analysis of O. tsutsugamushi autotransporter proteins. (A) Schematic representation of the five Sca proteins identified in the genome of the O. tsutsugamushi Boryong strain. The domain structures were predicted using SMART (Simple Modular Architecture Research Tool; http://smart.embl-heidelberg.de/). *, cloned region of ScaC33-232 used in this study; SP, signal peptide; RPT, internal repeat sequence; TM, transmembrane; AT, autotransporter domain; aa, amino acids. (B) Phylogenetic tree of the sca genes identified in the genome of Boryong (OTBS) and Ikeda (OTT) strains. The five groups of ortholog genes are indicated on the right-hand side.
Since autotransporter domains are highly conserved between Rickettsia Sca proteins (1), we further examined the degrees of sequence similarity between the passenger domains of the Orientia Sca proteins. The passenger domains, including the signal peptide sequences, of the Orientia Sca proteins showed 9 to 23% amino acid sequence identity between paralogs (Table 1). Similar levels of identity between Rickettsia Sca proteins were observed (1). However, the amino acid sequences of Orientia Sca proteins are also highly conserved between orthologs (73 to 84% identical at the amino acid level), implying functional conservation. Of the Orientia Sca paralogs, the ScaC protein showed the highest sequence identity (84%) between the Boryong and Ikeda strains. The high degrees of sequence identity and conservation seen in the ScaC proteins from these different strains suggest that ScaC may play a critical role during O. tsutsugamushi infection.
TABLE 1.
Degrees of identity and divergence among Sca sequencesa
| Protein | % identity or divergenceb |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| BR |
IK |
||||||||
| ScaA (1,185 aa) | ScaB (374 aa) | ScaC (280 aa) | ScaD (718 aa) | ScaE (494 aa) | ScaA (1,232 aa) | ScaC (274 aa) | ScaD (552 aa) | ScaE (492 aa) | |
| BR | |||||||||
| ScaA | — | 12.3 | 16.1 | 12.7 | 11.7 | 75.3 | 19.7 | 16.8 | 11.4 |
| ScaB | 386.0 | — | 10.7 | 17.4 | 19.8 | 11.2 | 9.9 | 15.5 | 23.0 |
| ScaC | 159.9 | 323.0 | — | 15.4 | 9.3 | 18.9 | 83.6 | 18.6 | 10.7 |
| ScaD | 284.0 | 311.0 | 323.0 | — | 12.8 | 13.5 | 17.2 | 79.5 | 13.4 |
| ScaE | 422.0 | 198.0 | 313.0 | 357.0 | — | 11.5 | 13.9 | 11.9 | 73.2 |
| IK | |||||||||
| ScaA | 27.1 | 426.0 | 174.6 | 282.0 | 466.0 | — | 16.4 | 15.8 | 10.4 |
| ScaC | 155.9 | 313.0 | 18.6 | 332.0 | 298.0 | 165.2 | — | 14.6 | 15.3 |
| ScaD | 296.0 | 325.0 | 292.0 | 23.2 | 362.0 | 305.0 | 285.0 | — | 11.6 |
| ScaE | 430.0 | 186.5 | 292.0 | 317.0 | 26.9 | 482.0 | 285.0 | 335.0 | — |
The amino acid sequences of the signal peptide and passenger domain from each of the Sca proteins were used for analysis, and the number of amino acids of (aa) each protein is presented.
Values to the left of the dashes are divergences (see Materials and Methods), and those to the right of the dashes are percent identities. BR, Boryong strain; IK, Ikeda strain. Boldface and underlined values are the degrees of identity or divergence between orthologs.
ScaC is expressed by O. tsutsugamushi.
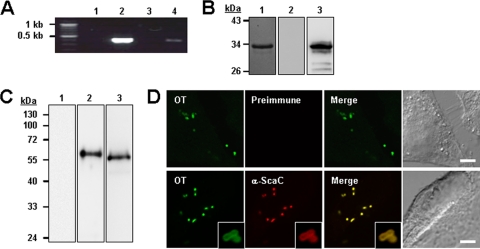
To examine whether the scaC gene was expressed during bacterial infection, we isolated total RNA from infected host cells, removed any contaminating bacterial or host cell genomic DNA, and examined scaC expression by RT-PCR. As shown in Fig. 2 A, agarose gel electrophoresis of the PCR products confirmed the amplification of the scaC gene from infected host cells. The PCR products were cloned and verified by sequencing to confirm that the correct gene had been amplified. No PCR products were detected in the control reactions incorporating RNA from uninfected cells or in those lacking reverse transcriptase.
FIG. 2.
Expression of ScaC by O. tsutsugamushi. (A) RT-PCR of scaC mRNA from L929 cells infected with O. tsutsugamushi. scaC DNA was amplified from total RNA (without reverse transcription) isolated from infected cells (lane 1), from genomic DNA (lane 2), from cDNA from uninfected cells (lane 3), and from cDNA from infected cells (lane 4). (B) Specificity of anti-ScaC antiserum. Purified His-tagged ScaC33-232 protein was resolved by SDS-PAGE, stained with Coomassie blue (lane 1), and immunoblotted with rabbit preimmune serum (lane 2) or anti-ScaC serum (lane 3). (C) Immunoblot analysis of total O. tsutsugamushi proteins by using rabbit preimmune serum (lane 1) or anti-ScaC serum (lane 2). Anti-ScaC serum detected a protein with a molecular mass of approximately 60 kDa. Preimmune serum did not react with the bacterial lysate. Immunoblotting using anti-TAS56 was performed as a control (lane 3). (D) Immunofluorescence confocal microscopy using preimmune serum or anti-ScaC serum (α-ScaC) showed ScaC in the O. tsutsugamushi-infected L929 cells. The left-hand panels show bacteria (O. tsutsugamushi [OT]) stained with pooled serum from scrub typhus patients. Magnified images are shown in the lower panels (inset boxes). Scale bars, 5 μm.
In order to examine ScaC protein expression, we generated a polyclonal anti-ScaC serum by immunizing a rabbit with the purified ScaC passenger domain (amino acids 33 to 232). The specificity of this antiserum was tested using the recombinant ScaC33-232 protein after separation by SDS-PAGE and subsequent immunoblotting. As shown in Fig. 2B, the anti-ScaC serum reacted with the recombinant ScaC protein (lane 3), but preimmune serum showed no specific reactivity with the recombinant protein. The full-length ScaC protein was predicted to have a mass of ∼60 kDa. To identify endogenous ScaC protein in O. tsutsugamushi, the anti-ScaC serum was reacted with the O. tsutsugamushi lysates. Immunoblot analysis shows that an ∼60-kDa protein was recognized by the anti-ScaC serum but not by preimmune serum (Fig. 2C). The TSA56 protein, a major outer membrane protein of O. tsutsugamushi, was used as a positive control. These results indicate that the ScaC protein is expressed by O. tsutsugamushi isolated from infected host cells. To further confirm the specificity of the anti-ScaC antiserum for O. tsutsugamushi, intracellular bacteria were stained using the pooled sera of scrub typhus patients together with anti-ScaC serum or preimmune rabbit serum. As shown in Fig. 2D, anti-ScaC serum readily detected the bacteria within the host cells, whereas the preimmune serum did not. In addition, we found that the ScaC proteins were located on the periphery of bacterial cells (Fig. 2D, lower panels, inset boxes). Taken together, these results confirm that scaC gene is actively translated in O. tsutsugamushi within eukaryotic host cells and that the protein might be expressed on the outer membrane of the bacteria.
We next tested whether scrub typhus patients produced antibodies to ScaC, thereby confirming expression of the scaC gene during in vivo infection. Pooled sera from 10 scrub typhus patients (IFA titer ≥ 1,280) or control patients with an acute febrile illness not diagnosed as scrub typhus were used in an immunoblot analysis using the recombinant ScaC passenger domain. As shown in Fig. 3 A, the sera from scrub typhus patients (lane 3) readily reacted with the ScaC protein, whereas the pooled sera from control patients (lane 2) did not. Moreover, eight sera out of 10 scrub typhus patients' sera were reactive to ScaC antigen (IgG titer ≥ 1:100) when assessed by anti-ScaC IgG ELISA (Fig. 3B). In contrast, only one serum from a control patient showed reactivity to the ScaC protein (IgG titer = 1:100), and the other nine sera were nonreactive in the test. These results indicate that the ScaC protein is expressed during in vivo infection by O. tsutsugamushi and induces an antibody response.
FIG. 3.
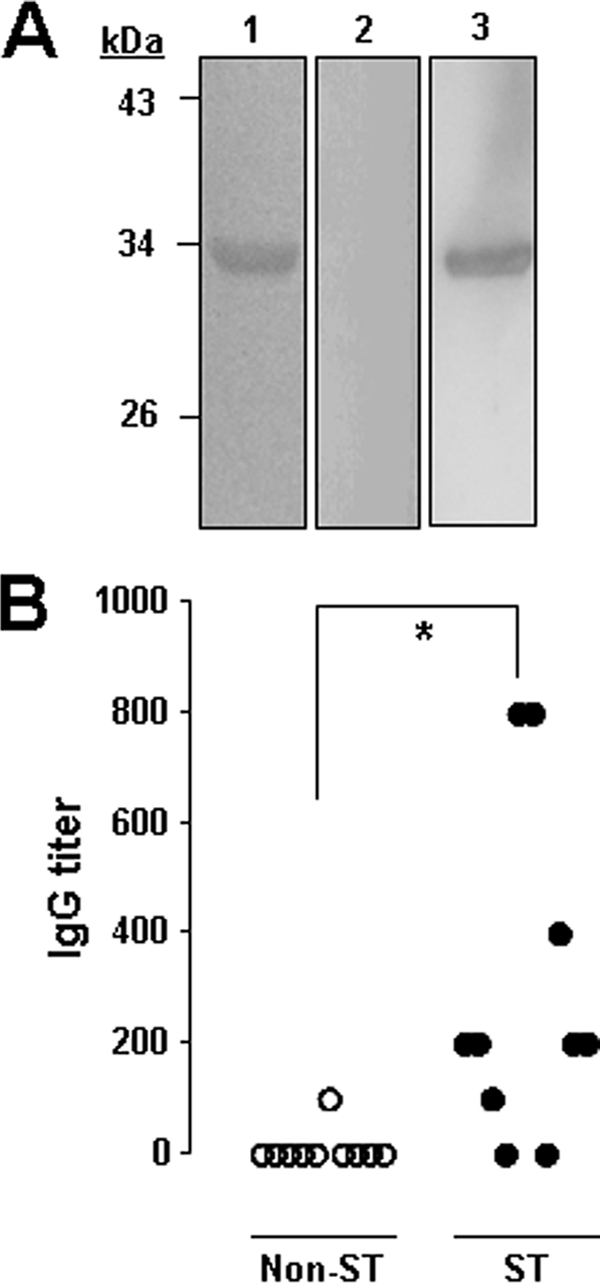
Induction of anti-ScaC antibodies in scrub typhus patients. (A) Purified ScaC33-232 protein was separated by SDS-PAGE and stained with Coomassie blue (lane 1) and immunoblotted with pooled sera from 10 control patients with a non-scrub typhus, acute febrile illness (lane 2) or from 10 scrub typhus patients (lane 3). (B) Anti-ScaC antibody responses in 10 scrub typhus (ST) patients (black circles) and in 10 control patients (white circles) were measured by ELISA and are presented as anti-ScaC IgG titers. *, P = 0.007.
ScaC protein mediates adherence to, but not invasion of, nonphagocytic host cells.
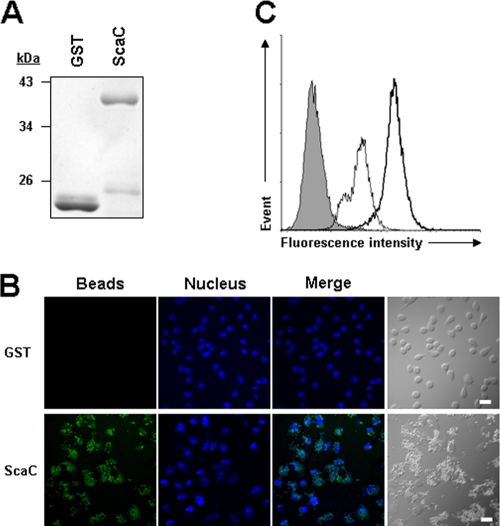
Recently, several studies reported that rickettsial Sca proteins could mediate bacterial adherence to and/or invasion into mammalian host cells (2, 3, 30). Therefore, we examined whether the O. tsutsugamushi ScaC protein could function as a virulence factor for bacterial adhesion and/or invasion. First, we performed a bead-binding assay using fluorescent microbeads (1 μm in diameter) covalently conjugated to either purified GST or GST-ScaC (Fig. 4 A). Incubation of HeLa cells with GST-ScaC-conjugated beads resulted in marked binding to the host cells, even after extensive washing. The control beads linked to GST alone interacted only weakly with the HeLa cells (Fig. 4B). The interaction of the fluorescent beads with the host cells was quantified using flow cytometry (Fig. 4C). After fixation, the mean fluorescence intensity (MFI) of the HeLa cells incubated with beads conjugated to GST-ScaC dramatically increased (MFI = 211.5) compared to that of cells incubated with beads conjugated to GST (MFI = 42.2) or that of untreated cells.
FIG. 4.
Adhesion of ScaC-coated microbeads to HeLa cells. (A) Purity of recombinant GST or GST-ScaC (ScaC) protein for the conjugation with fluorescent microbeads was assessed by SDS-PAGE followed by Coomassie blue staining. (B) Cells were incubated with fluorescent microbeads coated with GST or GST-ScaC (ScaC) for 1 h, washed extensively, and fixed. Cell-bound microbeads (green) were visualized by fluorescence microscopy after staining of cell nuclei (blue). Scale bars, 10 μm. (C) Relative binding of the microbeads coated with GST (thin line) or GST-ScaC (thick line) to HeLa cells was quantified directly using fluorescence-activated cell sorter (FACS) analysis. The gray histogram represents unbound cells (cells not incubated with microbeads).
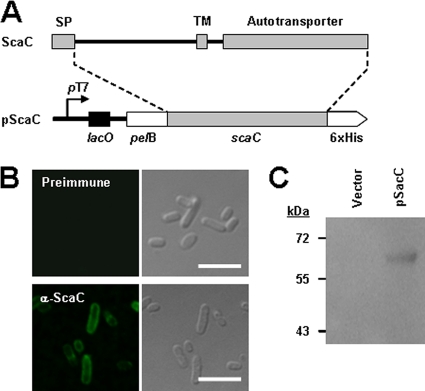
To further verify the role of the scaC gene in bacterial adherence to host cells, we utilized a heterologous E. coli expression system. The entire O. tsutsugamushi scaC open reading frame was cloned into the IPTG-inducible expression vector, pET-28a, to yield the plasmid pScaC (Fig. 5 A). ScaC was expressed in the E. coli strain BL21(DE3) and analyzed using anti-ScaC serum and confocal microscopy after fixation with 4% paraformaldehyde. As shown in Fig. 5B, ScaC was readily detectable on the surface of recombinant E. coli cells by anti-ScaC serum (lower panels) but not by preimmune serum (upper panels), as was observed for O. tsutsugamushi. To further confirm the expression of ScaC on the bacterial surface, we biochemically prepared the outer membrane fraction from E. coli transformed with pET-28a vector or pScaC plasmid after induction for protein expression. Immunoblot analysis using anti-ScaC antibody resulted in the presence of a ∼60-kDa product in the isolated outer membrane fraction from E. coli harboring pScaC but not that harboring pET-28a (Fig. 5C).
FIG. 5.
Expression of O. tsutsugamushi ScaC on the surface of E. coli. (A) Schematic diagram of the scaC-containing pET28a plasmid (pScaC). This vector encodes a recombinant protein fusion containing an N-terminal E. coli PelB signal sequence, O. tsutsugamushi ScaC, and a C-terminal 6×His tag. (B) Immunofluorescence microscopy using an anti-ScaC antibody revealed the presence of ScaC on the surface of the recombinant E. coli (lower panels). Preimmune serum did not detect the recombinant protein (upper panels). Scale bars, 5 μm. (C) Immunoblot analysis of outer membrane fractions of induced E. coli harboring the empty pET28a (vector) or pScaC was performed using an anti-ScaC serum.
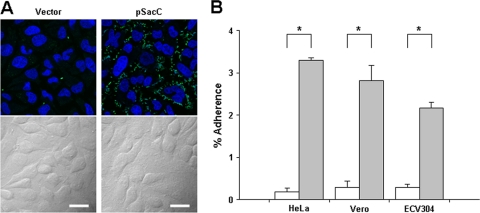
We next examined the ability of ScaC-expressing E. coli to adhere to monolayers of nonphagocytic host cells. Epithelial (HeLa and Vero) and endothelial (ECV304) cells were incubated with recombinant E. coli harboring a control vector or pScaC. The cells were then washed extensively to remove nonadherent bacteria, fixed, and analyzed under a confocal microscope after staining with an anti-E. coli antibody and the nuclear stain ToPro-3. Immunofluorescence analysis revealed that ScaC expression resulted in an increase in the number of adherent E. coli bacteria (Fig. 6 A). This ScaC-mediated adhesion was verified by removing the adherent bacteria from live host cells and counting them using a CFU-based quantification assay. The assay confirmed that ScaC expression significantly increased bacterial adherence to both epithelial and endothelial cells (Fig. 6B). The degree of adherence was comparable to that seen for Rickettsia autotransporter proteins (2, 3, 30). Therefore, the expression of O. tsutsugamushi ScaC on the outer surface of E. coli enhances adherence to nonphagocytic host cells.
FIG. 6.
Adherence of ScaC-expressing E. coli to host cells. (A) E. coli transformed with the pET28a vector or with pScaC was induced with IPTG and incubated with HeLa cells. After being washed to remove adherent bacteria, the cells were fixed, permeabilized, and stained with an anti-E. coli antibody (green) and ToPro-3 for nuclear staining (blue). Scale bars, 10 μm. (B) CFU-based quantification of adherent E. coli transformed with the vector (white bars) or pScaC (gray bars) was performed for different host cells (HeLa, Vero, and ECV304 cell lines). The results are presented as the percentages of adherent bacteria relative to the total bacterial input. Data are representative of three independent assays for each of the host cells. *, P < 0.01.
Several reports showed that the expression of rickettsial autotransporter proteins in E. coli is sufficient to mediate adherence to, and invasion into, nonphagocytic host cells (2, 3, 34). Therefore, we investigated whether the expression of the O. tsutsugamushi ScaC protein was able to mediate bacterial invasion of host cells. E. coli harboring a control vector or pScaC was added to cultured monolayers of HeLa, Vero, or ECV304 cells. Bacterial internalization was quantified using a gentamicin protection assay. No colonies were obtained (data not shown), indicating that ScaC is not able to mediate invasion of nonphagocytic host cells.
ScaC polypeptide inhibits the interaction between O. tsutsugamushi and host cells.
Next, we determined whether ScaC could inhibit O. tsutsugamushi-host cell interactions. HeLa cells were preincubated with 100 μg/ml of soluble GST or GST-ScaC protein for 1 h and then infected with O. tsutsugamushi for 30 min. After extensive washing, bacterial interactions with the host cells were examined by using confocal microscopy and the O. tsutsugamushi/host cell ratio was determined. As shown in Fig. 7, preexposure of HeLa cells to GST-ScaC significantly inhibited O. tsutsugamushi-host cell interactions. The number of bacteria per cell was reduced by approximately 80% compared with the control group (GST alone). This shows that excess ScaC can competitively inhibit the binding of the “native” bacterial ScaC protein to its cognate mammalian ligand and that ScaC-mediated binding to host cells is required for efficient invasion.
FIG. 7.
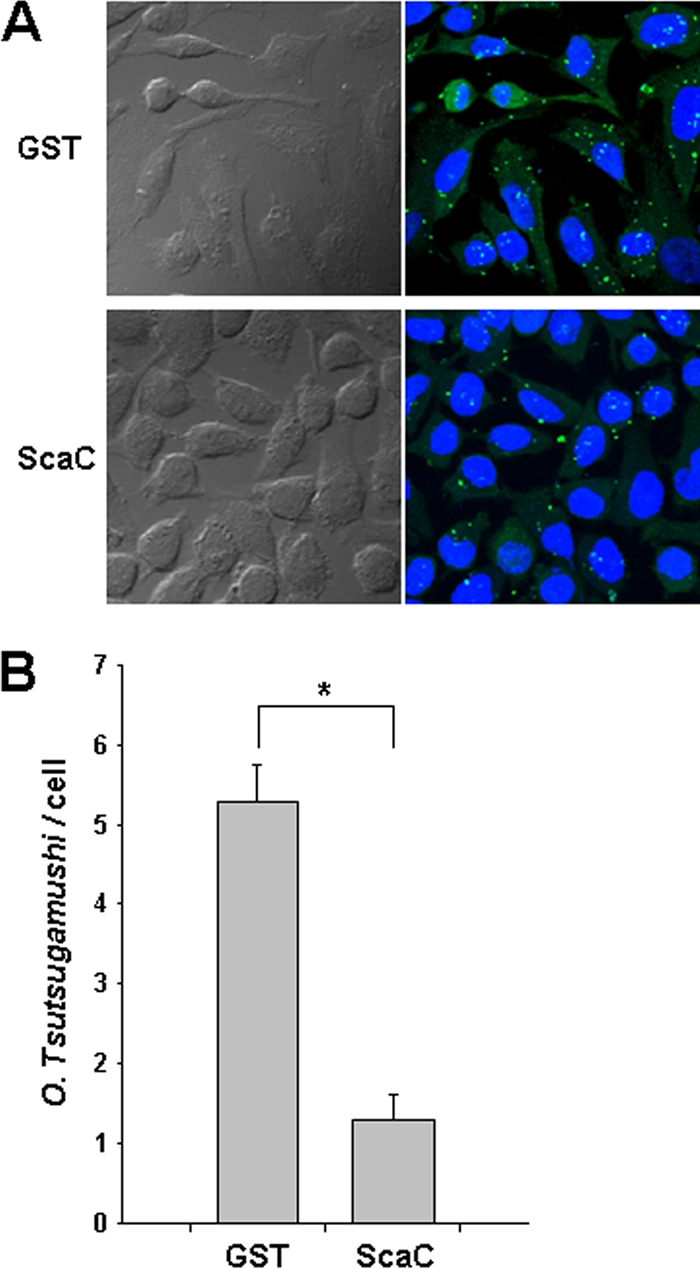
Inhibition of O. tsutsugamushi interactions with host cells by the ScaC polypeptide. (A) Preincubation of HeLa cells with GST-ScaC (lower panels) significantly inhibited O. tsutsugamushi interactions with host cells compared with preincubation with GST (upper panels). After incubation of the polypeptides with the host cells, O. tsutsugamushi was added, and incubation continued for a further 30 min. Cells were visualized by immunofluorescence microscopy. Green, O. tsutsugamushi; blue, nuclei. (B) The numbers of bacteria associated with each of 100 randomly selected host cells. The bars indicate the means ± standard deviations of triplicate experiments. *, P < 0.01.
Fibronectin is a potential host receptor for bacterial ScaC.
To identify the ScaC receptor on host cells, we performed yeast two-hybrid screening analysis using a GAL4 transcription activation domain-fused human aorta cDNA library. Using the GAL4 DNA binding domain-fused to the ScaC passenger domain (amino acids 33 to 232) as the bait, we identified 40 independent, genuine positive clones from the 6.6 × 106 transformants; the genuine positive clones were positive for lacZ, ura3, and ade2 expression (data not shown). Sequence analysis of the cDNAs obtained from the 40 positive colonies identified 25 known cDNA fragments (see Table S1 in the supplemental material). Interestingly, one of the clones (clone 27) encoded fibronectin, which is known to interact with O. tsutsugamushi via the outer membrane protein TSA56 (7, 18). The specific interaction between ScaC and fibronectin was confirmed by using GST pulldown assays (Fig. 8). These results suggest that ScaC may function as another bacterial ligand that mediates interaction with host cells by binding to fibronectin.
FIG. 8.
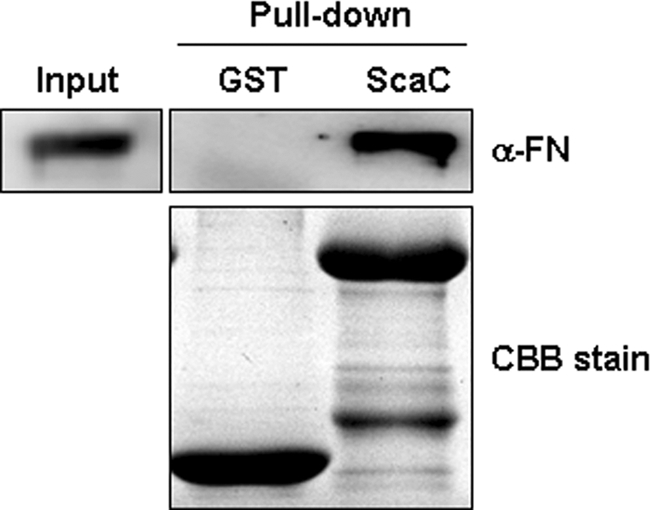
Interaction of ScaC with fibronectin. Direct interaction of ScaC with fibronectin was assessed by a GST pulldown assay incorporating purified GST fusion proteins and fibronectin. After GST pulldown using glutathione Sepharose beads, the interacting proteins were analyzed by immunoblotting using an antifibronectin (α-FN) antibody. Ten percent of the input fibronectin was used for pulldown experiments (Input lane). CBB stain, Coomassie brilliant blue staining of GST or GST-ScaC protein.
DISCUSSION
Autotransporter proteins are found in a wide range of Gram-negative bacteria and constitute a family of outer membrane/secreted proteins that are often associated with virulent functions (14, 37). Structurally, autotransporter proteins are characterized by the presence of three distinct domains: (i) an N-terminal signal sequence that mediates export of the proteins across the cytoplasmic membrane, (ii) a surface-localized passenger domain, and (iii) a C-terminal translocation domain that facilitates the secretion of the passenger domain through the outer membrane (37). While the translocation domain is highly homologous between autotransporter proteins, the secreted passenger domain shows considerable sequence variation. As a result, autotransporter protein passenger domains may confer many different virulence-related phenotypes, including adhesion, invasion, biofilm formation, and cytotoxicity (37). The apparent role of autotransporter proteins in virulence and host cell interactions suggests that they may represent rational targets for the design of novel vaccines directed against human pathogens (37). For example, pertactin, a major virulence factor of Bordetella pertussis, is an autotransporter protein that mediates bacterial binding to the lung epithelium in mammalian hosts (12) and has been successfully used to provide the acellular components of a pertussis vaccine (5, 28). The passenger domain of the Haemophilus influenzae autotransporter protein, Hap, which mediates attachment and entry into epithelial cells (32) as well as attachment to extracellular matrix proteins (13), elicits significant antibody responses and protects preimmunized mice from nasopharyngeal colonization (11). Genome sequencing has identified numerous genes predicted to encode autotransporter proteins in various human pathogens (14, 37). A search of the autotransporter superfamily proteins in the NCBI database identified more than 10,000 proteins. The majority of these putative autotransporter proteins are uncharacterized with respect to their bacterial virulence. Therefore, the characterization of potential virulent autotransporter proteins encoded by bacterial pathogens may provide a valuable basis for their utilization as vaccine targets.
In the present study, we analyzed the autotransporter proteins encoded by the genomes of two O. tsutsugamushi strains and categorized them into five different groups of orthologs (ScaA to ScaE) based on their sequence similarities. To date, 17 autotransporter proteins (Sca0 to Sca16) have been identified from nine fully sequenced Rickettsia genomes (1). Even though the autotransporter domain is generally well conserved between rickettsial paralogs, with an average of 31% sequence identity, the passenger domains are less well conserved between the different subgroups, with the average level of amino acid identity being 9 to 18%. In this study, we also observed a similar level of sequence variation in the O. tsutsugamushi autotransporter proteins; however, the sequences of each gene were highly conserved between orthologs found in the two different strains (Table 1), suggesting a functional conservation. Recently, it has been reported that the core gene set of the family Rickettsiaceae is highly conserved between the two strains, although amplified repetitive sequences have induced extensive genome shuffling and duplications and deletions of many genes (6, 25). Considering the average nucleotide identity of the 541 core genes in the two strains is 97.5% (25), that of four sca gene orthologs is slightly lower (90.8%). This could be explained by the positive diversifying selection observed in the passenger domains of rickettsial Sca proteins (1), which might be coevolved through evasion-recognition cycles within host, i.e., the surface-exposed Sca proteins may have been involved in host adhesion processes and they also represent potential targets for host defense. Indeed, the nucleotide sequence identity of a gene encoding a major outer membrane protein of O. tsutsugamushi, TSA56, in the two strains is even lower (84.4%) than those of sca genes. Interestingly, the O. tsutsugamushi Boryong strain genome encodes two copies of the ScaB protein, which is absent from the Ikeda strain. Considering that different O. tsutsugamushi strains show different levels of virulence in mouse models of infection (27), the presence or absence of potential virulence factors within the genome of each strain may explain the clinical and epidemiological strain-specific differences suggested by a recent comparative genomic study (25). Thus, further investigation of the role of ScaB in bacterial virulence would be interesting.
Of the O. tsutsugamushi Sca genes, scaC was selected for further functional analysis, since it showed the highest level of conservation between the two different strains (Table 1). The ScaC autotransporter protein is actively expressed at both the transcriptional and translational levels within infected host cells and induces specific antibody responses in scrub typhus patients. Using immunofluorescence confocal microscopy, we showed that ScaC might be localized on the O. tsutsugamushi cell surface, and we were able to confirm this by using a surrogate E. coli system (Fig. 5).
During the initial stages of O. tsutsugamushi invasion into host cells, the intracellular pathogen may utilize multiple receptor-ligand interactions in a manner similar to those observed for R. conorii (2, 30). The bacterium then uses host intracellular signaling pathways to manipulate the host cytoskeletal environment at the sites of infection on nonphagocytic host cells (7). In the present study, phenotypic analysis of ScaC by using a recombinant protein and a surrogate expression system showed that it can mediate bacterial adherence but not cell invasion. We did not observe any rearrangement of the host actin cytoskeleton at the contact sites, suggesting that ScaC may simply enhance bacterial affinity for the host cells during initial contact. Competitive inhibition of bacterial binding to host cells was achieved using recombinant ScaC protein (Fig. 7). This further supports the idea that this protein is likely to participate in the bacterium-host cell interactions during infection. Recently, it was reported that an R. conorii autotransporter protein, Sca1, mediates a similar phenotypic function during the bacterial infection process (30).
Finally, we identified fibronectin as the host receptor for the ScaC ligand by using yeast two-hybrid screening and confirmed the specificity of the interaction by using GST pulldown analysis (Fig. 8). Interestingly, we previously showed that O. tsutsugamushi directly binds to fibronectin, which significantly enhances bacterial invasion into the host cells when added to the infection media (18). The major O. tsutsugamushi outer membrane protein, TSA56, is the bacterial ligand responsible for this interaction with fibronectin. Antigenic domain III and the adjacent C-terminal region of TSA56 both interact with fibronectin and eventually induce bacterial invasion via the activation of integrin signaling (7, 18). The identification of ScaC as another ligand for fibronectin suggests that autotransporter proteins may play an important role during O. tsutsugamushi infection, complementing adhesion and/or invasion mediated by the fibronectin-integrin pathway. It is possible that ScaC may work in concert with TSA56 via sequential or concomitant interactions with fibronectin to facilitate bacterial invasion. Identifying the region within the fibronectin molecule that is responsible for binding to each of the bacterial ligands may provide valuable clues regarding their contribution to the initial stages of infection and remains the focus of further study.
In summary, we showed that a highly conserved autotransporter protein, ScaC, is actively expressed by O. tsutsugamushi and induces antibody responses during in vivo infection. Expression of ScaC on the surface of E. coli enhances bacterial adherence to various nonphagocytic host cells, potentially via the interaction with fibronectin. Moreover, the association of O. tsutsugamushi with host cells was significantly reduced by the addition of purified recombinant ScaC. Taken together, our results suggest that the ScaC autotransporter protein may be a potential target for vaccine development against scrub typhus.
Supplementary Material
Acknowledgments
This study was supported by grant A090053 from the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 31 January 2011.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Blanc, G., et al. 2005. Molecular evolution of rickettsia surface antigens: evidence of positive selection. Mol. Biol. Evol. 22:2073-2083. [DOI] [PubMed] [Google Scholar]
- 2.Cardwell, M. M., and J. J. Martinez. 2009. The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infect. Immun. 77:5272-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, Y. G., M. M. Cardwell, T. M. Hermanas, T. Uchiyama, and J. J. Martinez. 2009. Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell. Microbiol. 11:629-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chattopadhyay, S., and A. L. Richards. 2007. Scrub typhus vaccines: past history and recent developments. Hum. Vaccines 3:73-80. [DOI] [PubMed] [Google Scholar]
- 5.Cherry, J. D., J. Gornbein, U. Heininger, and K. Stehr. 1998. A search for serologic correlates of immunity to Bordetella pertussis cough illnesses. Vaccine 16:1901-1906. [DOI] [PubMed] [Google Scholar]
- 6.Cho, B. A., et al. 2010. Global gene expression profile of Orientia tsutsugamushi. Proteomics 10:1699-1715. [DOI] [PubMed] [Google Scholar]
- 7.Cho, B. A., N. H. Cho, S. Y. Seong, M. S. Choi, and I. S. Kim. 2010. Intracellular invasion by Orientia tsutsugamushi is mediated by integrin signaling and actin cytoskeleton rearrangements. Infect. Immun. 78:1915-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, N. H., et al. 2007. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc. Natl. Acad. Sci. U. S. A. 104:7981-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, N. H., S. Y. Seong, M. S. Choi, and I. S. Kim. 2001. Expression of chemokine genes in human dermal microvascular endothelial cell lines infected with Orientia tsutsugamushi. Infect. Immun. 69:1265-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, N. H., et al. 2000. Expression of chemokine genes in murine macrophages infected with Orientia tsutsugamushi. Infect. Immun. 68:594-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cutter, D., et al. 2002. Immunization with Haemophilus influenzae Hap adhesin protects against nasopharyngeal colonization in experimental mice. J. Infect. Dis. 186:1115-1121. [DOI] [PubMed] [Google Scholar]
- 12.Everest, P., et al. 1996. Role of the Bordetella pertussis P. 69/pertactin protein and the P. 69/pertactin RGD motif in the adherence to and invasion of mammalian cells. Microbiology 142(Pt. 11):3261-3268. [DOI] [PubMed] [Google Scholar]
- 13.Fink, D. L., B. A. Green, and J. W. St. Geme III. 2002. The Haemophilus influenzae Hap autotransporter binds to fibronectin, laminin, and collagen IV. Infect. Immun. 70:4902-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly, D. J., P. A. Fuerst, W. M. Ching, and A. L. Richards. 2009. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin. Infect. Dis. 48(Suppl. 3):S203-S230. [DOI] [PubMed] [Google Scholar]
- 16.Kim, S. W., et al. 2001. Microtubule- and dynein-mediated movement of Orientia tsutsugamushi to the microtubule organizing center. Infect. Immun. 69:494-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kweon, S. S., et al. 2009. Rapid increase of scrub typhus, South Korea, 2001-2006. Emerg. Infect. Dis. 15:1127-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, J. H., et al. 2008. Fibronectin facilitates the invasion of Orientia tsutsugamushi into host cells through interaction with a 56-kDa type-specific antigen. J. Infect. Dis. 198:250-257. [DOI] [PubMed] [Google Scholar]
- 19.Letunic, I., T. Doerks, and P. Bork. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37:D229-D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, H., and D. H. Walker. 1998. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb. Pathog. 24:289-298. [DOI] [PubMed] [Google Scholar]
- 21.Liu, Y. X., et al. 2009. Clinical characteristics of the autumn-winter type scrub typhus cases in south of Shandong province, northern China. BMC Infect. Dis. 9:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez, J. J., S. Seveau, E. Veiga, S. Matsuyama, and P. Cossart. 2005. Ku70, a component of DNA-dependent protein kinase, is a mammalian receptor for Rickettsia conorii. Cell 123:1013-1023. [DOI] [PubMed] [Google Scholar]
- 23.Mathai, E., et al. 2003. Outbreak of scrub typhus in southern India during the cooler months. Ann. N. Y. Acad. Sci. 990:359-364. [DOI] [PubMed] [Google Scholar]
- 24.Moron, C. G., V. L. Popov, H. M. Feng, D. Wear, and D. H. Walker. 2001. Identification of the target cells of Orientia tsutsugamushi in human cases of scrub typhus. Mod. Pathol. 14:752-759. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama, K., et al. 2010. Genome comparison and phylogenetic analysis of Orientia tsutsugamushi strains. DNA Res. 17:281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakayama, K., et al. 2008. The whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 15:185-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohashi, N., et al. 1996. Demonstration of antigenic and genotypic variation in Orientia tsutsugamushi which were isolated in Japan, and their classification into type and subtype. Microbiol. Immunol. 40:627-638. [DOI] [PubMed] [Google Scholar]
- 28.Poolman, J. T., and H. O. Hallander. 2007. Acellular pertussis vaccines and the role of pertactin and fimbriae. Expert Rev. Vaccines 6:47-56. [DOI] [PubMed] [Google Scholar]
- 29.Rikihisa, Y., and S. Ito. 1982. Entry of Rickettsia tsutsugamushi into polymorphonuclear leukocytes. Infect. Immun. 38:343-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riley, S. P., et al. 2010. The Rickettsia conorii autotransporter protein Sca1 promotes adherence to nonphagocytic mammalian cells. Infect. Immun. 78:1895-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seong, S. Y., M. S. Choi, and I. S. Kim. 2001. Orientia tsutsugamushi infection: overview and immune responses. Microbes Infect. 3:11-21. [DOI] [PubMed] [Google Scholar]
- 32.St. Geme, J. W., III, M. L. de la Morena, and S. Falkow. 1994. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol. Microbiol. 14:217-233. [DOI] [PubMed] [Google Scholar]
- 33.Suda, K., B. Rothen-Rutishauser, M. Gunthert, and H. Wunderli-Allenspach. 2001. Phenotypic characterization of human umbilical vein endothelial (ECV304) and urinary carcinoma (T24) cells: endothelial versus epithelial features. In Vitro Cell. Dev. Biol. 37:505-514. [DOI] [PubMed] [Google Scholar]
- 34.Uchiyama, T., H. Kawano, and Y. Kusuhara. 2006. The major outer membrane protein rOmpB of spotted fever group rickettsiae functions in the rickettsial adherence to and invasion of Vero cells. Microbes Infect. 8:801-809. [DOI] [PubMed] [Google Scholar]
- 35.Watt, G., et al. 1996. Scrub typhus infections poorly responsive to antibiotics in northern Thailand. Lancet 348:86-89. [DOI] [PubMed] [Google Scholar]
- 36.Watt, G., and P. Parola. 2003. Scrub typhus and tropical rickettsioses. Curr. Opin. Infect. Dis. 16:429-436. [DOI] [PubMed] [Google Scholar]
- 37.Wells, T. J., J. J. Tree, G. C. Ulett, and M. A. Schembri. 2007. Autotransporter proteins: novel targets at the bacterial cell surface. FEMS Microbiol. Lett. 274:163-172. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, S., et al. 2010. Scrub typhus in previously unrecognized areas of endemicity in China. J. Clin. Microbiol. 48:1241-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.