Abstract
Enterotoxigenic Escherichia coli (ETEC) strains with K88 fimbriae are often associated with the outbreaks of diarrhea in newborn and weaned piglets worldwide. In the present study, we observed that 108 CFU/ml of K88+ ETEC strain JG280 caused more death of pig intestinal IPEC-J2 cells than did 109 CFU/ml, suggesting that ETEC-induced cell death was cell density dependent and that quorum sensing (QS) may play a role in pathogenesis. Subsequent investigations demonstrated a positive correlation between autoinducer 2 (AI-2) activity of JG280 and death of IPEC-J2 cells during the infection for up to 3 h. However, there was a negative correlation between AI-2 activity and expression of the JG280 enterotoxin genes estA and estB when IPEC-J2 cells were exposed to the pathogen at 108 CFU/ml. We therefore cloned the luxS gene (responsible for AI-2 production) from JG280 and overexpressed it in E. coli DH5α, because deletion of the luxS gene was retarded by the lack of suitable antibiotic selection markers and the resistance of this pathogen to a wide range of antibiotics. The addition of culture fluid from E. coli DH5α with the overexpressed luxS reduced cell death of IPEC-J2 cells by 108 CFU/ml JG280. The addition also reduced the estA expression by JG280. Nonpathogenic K88+ strain JFF4, which lacks the enterotoxin genes, caused no death of IPEC-J2 cells, although it produced AI-2 activity comparable to that produced by JG280. These results suggest the involvement of AI-2-mediated quorum sensing in K88+ ETEC pathogenesis, possibly through a negative regulation of STa production.
Enterotoxigenic Escherichia coli (ETEC) is an important pathogen causing severe watery diarrhea in farm animals, especially postweaning diarrhea in pigs (12). In humans, ETEC is the most common cause of traveler's diarrhea and can be fatal for children under 5 years of age (6, 9, 16). ETEC strains are characterized by the production of adhesins that mediate bacterial adherence to the intestine and enterotoxins that elicit diarrhea. The most common and severe infections in pigs are caused by ETEC strains carrying F4 (K88) and F18 fimbrial adhesins (21). After binding of the fimbriae to enterocytes, ETEC strains proliferate rapidly to attain massive numbers and produce one or more types of enterotoxin, including heat-stable enterotoxins (STs) and heat-labile enterotoxin (LT) (22), which stimulate fluid and electrolyte secretion by intestinal cells, thus leading to diarrhea. Other virulence factors, including SepA (sepA), which belongs to a family of proteins from enteropathogenic (32) and enterohemorrhagic (7) E. coli, and Paa (paa), porcine attaching-effacing-associated protein (5), may also contribute to ETEC pathogenesis.
Previous in vitro studies with pig intestinal epithelial (IPEC-J2) cells showed that infection with a wild-type (wt) ETEC strain (expressing LT and STb toxins) (3) significantly increased cell death and phosphatidylserine (PS) exposure on the outer leaflet of IPEC-J2 cells (14). In addition, infection with both the ΔeltAB (an isogenic mutant deficient in LT expression) and ΔeltAB/pLT (ΔeltAB mutant complemented by plasmid-based eltAB expression) ETEC strains also modestly increased death of IPEC-J2 cells (14). Another ETEC strain, B41M, that expresses STa toxin was also reported to induce IPEC-J2 cell damage (14). Previous in vivo studies also showed that enterotoxins LT and STb secreted by ETEC contributed significantly to the severity of diarrhea in gnotobiotic piglets, and deletion of estB and eltAB could prevent the development of electrolyte imbalances and dehydration of piglets (11). However, it is still unclear how much each of the enterotoxins contributes to ETEC pathogenesis.
Diarrheagenic E. coli strains regulate their virulence gene expression in response to a variety of environmental factors and use quorum sensing (QS) to respond to their cell population to coordinate virulence gene expression (10). QS is a bacterial cell-to-cell communication mechanism, involving the production and detection of autoinducers (AIs). QS may be involved in the regulation of virulence factors, including the type III secretion system in enterohemorrhagic E. coli (EHEC) and enteropathogenic E. coli (EPEC) (29). A breakthrough discovery of a new signaling molecule, AI-3, whose synthesis is not dependent on LuxS (35), indicates that this molecule may be involved in EHEC cross talk with the epinephrine-norepinephrine host signaling system (30). QseBC (a two-component system) was reported to be activated by QS through the AI-3 system (31) and is responsible for the regulation of flagella and motility (31).
To our knowledge, the role of QS in ETEC pathogenesis has not been reported. In the present study, we observed that K88+ ETEC strain-induced damage of cultured pig intestinal epithelial cells was bacterial cell density dependent, which suggests the involvement of QS. Further studies were carried out to determine possible roles of AI-2 of K88+ ETEC in causing the cell damage and regulating the expression of enterotoxin genes.
MATERIALS AND METHODS
Bacterial strains, growth, and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. ETEC O149:K88(F4) strain JG280 is a porcine isolate, possessing the virulence genes elt, estA, estB, and astA and antibiotic resistance to ampicillin, apramycin, ceftiofur, gentamicin, neomycin, spectinomycin, tetracycline, and trimethoprim-sulfonamide (22). K88+ strain JFF4 is also a porcine isolate with F4/K88 fimbriae but lacking elt, estA, and estB virulence genes (E. Nadeau and J. Fairbrother, U.S. patent application 2007/0218035 A1). The K88+ strains were grown without shaking in 3 ml of tryptic soy broth (TSB) in 12-ml capped sterile plastic tubes (Fisher Scientific, Nepean, ON, Canada). After 12 h of incubation at 37°C, bacterial cells were pelleted through centrifugation (9,000 × g for 10 min) and washed with phosphate-buffered saline (Invitrogen, Carlsbad, CA) followed by resuspension in either Dulbecco's modified Eagle's medium (DMEM)-F-12 or DMEM without antibiotics prior to mixing with IPEC-J2 cells for assays.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| JG280 | ETEC O149: K88(F4) strain, positive for K88, elt, estA, and estB | 22 |
| JFF4 | E. coli strain, positive for K88 but negative for elt, estA, and estB | Nadeau and Fairbrother, U.S. patent application 2007/0218035 A1 |
| DH5α-pBAD | E. coli DH5α harboring plasmid pBAD, used as a cloning host | This study |
| JZ-luxS | E. coli DH5α-pBAD with a luxS gene cloned from JG280 | This study |
| Vibrio harveyi MM32 | AI-2 biosensor strain | E. Allen-Vercoe (personal communication) |
| Vibrio harveyi BB152 | luxM::Tn5 (AI-1− AI-2+) | 1 |
| Plasmids | ||
| pBAD-TOPO | Cloning expression vector PBAD; Kanr | Invitrogen |
| pBAD-luxS | pBAD-TOPO harboring a luxS gene from JG280 | This study |
AI-2 biosensor strain Vibrio harveyi MM32 and its positive control, Vibrio harveyi BB152, which produces AI-2 but not AI-1, were used for AI-2 assays. V. harveyi strains were grown at 30°C on marine agar (MA; Difco, MI) for routine maintenance.
Pig epithelial cell culture.
The neonatal jejunal epithelial cell line IPEC-J2 (27) cells were grown in DMEM-Ham's F-12 (1:1) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), penicillin (100 IU/ml), streptomycin (100 μg/ml), and 0.25 μg of amphotericin B per milliliter and maintained in an atmosphere of 5% CO2 at 37°C for cultures and assays. Cell cultures were routinely tested with the VenorGeM-mycoplasma detection kit (Minerva Biolabs, Berlin, Germany) and confirmed to be free of mycoplasma contamination for all the assays.
Cytotoxicity assay.
The release of lactate dehydrogenase (LDH) from IPEC-J2 cells was determined as described previously (18) with some modifications. Briefly, IPEC-J2 cells were seeded in six-well plates at approximately 2.5 × 105 cells per well in a 1.0-ml volume and grown overnight (12 to 16 h) to reach approximately 50% confluence. The cells were rinsed twice with prewarmed DMEM (with 1% FBS) without antibiotics and were then exposed to the K88+ E. coli for selected lengths of time by incubation at 37°C with 1 × 108 CFU or 1 × 109 CFU of the bacteria in 1 ml of DMEM (with 1% FBS). The use of 1% FBS was to reduce the background of the assay. Each sample had triplicates in the assay. The supernatants of the assay mixtures were collected after incubation by two consecutive centrifugations, firstly at 300 × g for 5 min to remove tissue cell debris and then at 21,000 × g for 5 min to remove bacterial cells. A 100-μl aliquot of each supernatant was dispensed in triplicate wells of a 96-well plate, and the activity of LDH was determined according to the manufacturer's protocol (Roche, Indianapolis, IN). The plates were incubated at 22°C, and the absorbance at 490 and 655 nm was measured after 5 to 10 min of incubation using a plate reader. IPEC-J2 cell lysates and DMEM were used as positive and negative controls, respectively, for each experiment. Percent cytotoxicity values were determined as follows: percent cytotoxicity = (Aexp − ADMEM)/(AIPEC-J2 cell lysates − ADMEM) × 100, where Aexp is the absorbance of test samples, ADMEM is the absorbance of DMEM, and AIPEC-J2 cell lysates is the absorbance of IPEC-J2 cell lysates. The DMEM was selected for the LDH assay due to the absence of pyruvate from the medium, which can inhibit the LDH reaction.
Cell viability assay.
Cell death of IPEC-J2 induced by different concentrations of JG280 cells (106 to 109 CFU) was initially assessed by microscopy. Trypan blue staining was then used for quantitative analysis of cell viability as described previously (37). Briefly, IPEC-J2 cells grown in six-well plates as described above were mixed with different numbers of K88+ E. coli cells (108 or 109 CFU) in 1 ml of DMEM (with 10% FBS) without antibiotics. Each sample had triplicates in the assays and was incubated at 37°C for up to 3 h. At different time intervals (0.5, 1.0, 1.5, 2.0, 2.5, and 3.0 h), IPEC-J2 cells either floating in the supernatant or detached by trypsin treatment from the plates were collected and pooled for trypan blue staining. To assess cell viability, aliquots of the collected IPEC-J2 cells were mixed with trypan blue at a final concentration of 0.2% for 5 min at 22°C and loaded onto a hemocytometer. The total numbers of cells and blue cells (dead cells) were counted. Cell viability was presented as the percentage of blue cells versus total cells.
Cloning and expression of the luxS gene.
The PCR primers (Table 2) for cloning the luxS gene were designed based on the E. coli MG1655 genome sequence. The high-fidelity PCR was conducted with genomic DNA of strain JG280 as the template. The PCR program consisted of a 4-min initiation at 94°C for denaturing the template DNA and 32 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s for amplification, followed by a final extension at 72°C for 10 min. The PCR amplicons were 936 bp in size (with the primer pair of luxS-K88-F4′ and luxS-K88-R3) and contained the promoter region of the luxS gene. The PCR amplicons were purified using the QIAquick PCR purification kit (Qiagen, Mississauga, Ontario, Canada) and cloned into the pBAD/Thio-TOPO vector according to the manufacturer's instructions (pBAD202 directional TOPO expressions kit; Invitrogen, Carlsbad, CA) after the addition of the pBAD sequence (5′-CACC) for directional cloning of blunt-end DNA fragments. The colonies with recombinant pBAD/ThioFusion plasmids were selected by plating the transformants on kanamycin (50 μg/ml)-Luria broth agar plates. Positive clones containing an luxS-pBAD/Thio plasmid with the luxS gene in the correct orientation were determined by PCR using a pair of primers, luxS-K88-R3-pBAD and TrxFus (5′-GATTTAATCTGTATCAGG-3′). The sequence of the TrxFus primer is part of the vector and provided in the Invitrogen kit. The sequence of the luxS gene (516 bp) was confirmed with 100% homology with the luxS gene of strain JG280. The expression of the luxS gene was verified by examining AI-2 activity with a V. harveyi-based autoinducer bioassay (AB). One positive clone, named JZ-luxS, was subsequently used in the experiments to examine possible roles of AI-2 in the cytotoxicity caused by ETEC strain JG280 and its expression of enterotoxin genes. The clone has GenBank accession number HQ538844.
TABLE 2.
PCR primers
| Primer name | Amplicon (bp) | Sequence (5′ to 3′) | Function | Source or reference |
|---|---|---|---|---|
| luxS-K88-F4′ | 940 | TGCGCACTAAGTACAACTAAG | For luxS cloning | This study |
| luxS-K88-R3 | TCTATGATTGATACTGGTATTG | For luxS cloning | This study | |
| luxS-K88-R3-pBAD | CACCTCTATGATTGATACTGGTATTGa | |||
| estA-F | 158 | CAACTGAATCACTTGACTCTT | For qPCR analysis | 22 |
| estA-R | TTAATAACATCCAGCACAGG | For qPCR analysis | 22 | |
| estB-F | 113 | TGCCTATGCATCTACACAAT | For qPCR analysis | 22 |
| estB-R | CTCCAGCAGTACCATCTCTA | For qPCR analysis | 22 | |
| elt-F | 322 | TCTCTATGTGCATACGGAGC | For qPCR analysis | 25 |
| elt-R | CCATACTGATTGCCGCAAT | For qPCR analysis | 25 | |
| gapA-F | 299 | TCCGTGCTGCTCAGAAACG | Housekeeping gene | 8 |
| gapA-R | CACTTTCTTCGCACCAGCG | Housekeeping gene | 8 | |
| faeG-F | 215 | ACTGGTGATT TCAATGGTTCG | For qPCR analysis | This study |
| faeG-R | GTTACTGGCGTAGCAAATGC | For qPCR analysis | This study |
To enable directional cloning, the forward PCR primer pair must contain the sequence CAAC (underlined) at the 5′ end. The four nucleotides CACC base pair with the overhang sequence GTGG in the pBAD202/D-TOPOR vector.
Preparation of bacterial cell-free culture supernatants.
To prepare bacterial cell-free culture supernatants from IPEC-J2 cultures, either alone or mixed with K88+ strains, individual cultures were subjected to centrifugation (21,000 × g, 5 min) followed by filtration through Millipore membrane filters with a pore size of 0.2 μm (Bioscience Division, Cambridge, Canada). To prepare the extracellular culture fluid (ECF) from the cultures of V. harveyi and E. coli DH5α that harbored the luxS gene (clone JZ-luxS) or pBAD vector only, 16-h-grown cultures of the bacteria, with E. coli grown in DMEM-F-12 (with 10% FBS), were subjected to centrifugation and membrane filtration with the Millipore filters as described above.
AI-2 bioassay.
AB medium (19) was used in the bioassays with V. harveyi for measuring AI-2 activity. V. harveyi MM32 was grown in AB medium at 30°C with agitation for 16 h followed by a dilution with fresh AB medium (1:5,000). The diluted V. harveyi culture (180 μl) was mixed with the cell-free ECF (20 μl each) in a black 96-well microtiter plate with a clear bottom (Corning Costar, Fisher Scientific, Canada) to test AI-2 activity. All assays were conducted with each sample in triplicate. Positive-control wells contained 20 μl of the ECF from V. harveyi strain BB152, and the blank-control wells contained 20 μl of sterile DMEM-F-12 (with 10% FBS). The assay mixtures were incubated at 30°C for up to 8 h. The luminescence and turbidity (optical density at 620 nm [OD620]) of the cultures were measured at 30-min intervals using a Victor Wallac 1420 plate reader (Wallac, Turku, Finland). AI-2-induced bioluminescence was given in counts per second (CPS) per unit of optical density at 620 nm (OD620) (CPS/OD620). AI-2 activity is expressed as a relative activity, which was calculated as the ratio of CPS/OD620 of the test samples to that of the control (negative) samples.
RNA extraction.
Bacterial total RNA was extracted from JG280 cells that had been treated in RNAlater solution (Ambion, TX) for 16 h after incubation with IPEC-J2 cells. The Ambion RiboPure bacteria kit (Ambion, TX) was used for RNA extraction, and the procedure described in the manufacturer's manual was followed. For each RNA sample, genomic DNA contamination was examined by a PCR assay with primers specific to a housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (gapA) (8), after DNase I (Ambion, TX) digestion. RNA integrity was verified by visualization in an agarose gel (38). The concentration of total RNA was determined with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Reverse transcription and real-time qPCR analysis.
Bacterial gene expression was determined by reverse transcription and quantitative PCR (qPCR) analysis as described previously (28) with some modifications. Briefly, 1.7 μg of total RNA was used for first-strand cDNA synthesis using the SuperScript first-strand synthesis system (Invitrogen, Carlsbad, CA) according to the manufacturer's instruction. Housekeeping gene gapA was used to normalize input amounts of RNA and the levels of estA, estB, elt, and faeG expression. qPCR was subsequently performed using a Stratagene MX4000 thermal cycler and brilliant SYBR green qPCR master mix (Stratagene, La Jolla, CA). Previously published PCR primers specific to each of the estA (STa), estB (STb), elt (LT), faeG (F4), and gapA genes (Table 2) were experimentally validated and used for qPCR assays. One microliter of each cDNA sample was included in a 13-μl PCR mixture that contained 1× master mix, 150 nM each primer, and 30 nM ROX (6-carboxy-X-rhodamine). The PCR programs included a 10-min initiation at 95°C, 40 cycles of 95°C for 30 s, 55 to 63°C (depending on the primers used) for 1 min, and 72°C for 30 s to 2 min (depending on the primers used) for amplification. Fluorescence was measured after each step of annealing during the cycles.
qPCR data were analyzed as described previously (13) by using the 2−ΔΔCT method to determine the relative abundance (fold changes) of target genes. The cycle threshold, CT, is the point at which fluorescence above the background is statistically significant. CT values were determined with the MX4000 software based on a threshold line that was manually defined above the noninformative fluorescent data. ΔCT represents the difference between the CT value of the primers to a target gene and the CT value of the primers to the housekeeping gene. ΔΔCT represents the difference between the ΔCT value of each time point after incubation and the ΔCT value of the zero time point. The values derived from 2−ΔΔCT represent fold changes in the abundance of the samples relative to that of the reference samples. The reference samples (zero time point) have a 2−ΔΔCT value of 1.
Examination of the relationship between virulence gene expression and AI-2 activity.
To examine the expression of K88+ ETEC virulence genes in relation to AI-2 activity, JG280 total RNA was extracted after incubation with IPEC-J2 cells (approximately 50% confluence) in six-well plates at 37°C with different treatments. In the experiments to test JG280, 1 × 109 CFU or 1 × 108 CFU JG280 cells were incubated with IPEC-J2 cells in 1 ml of the DMEM-F-12 medium supplemented with 10% FBS but no antibiotics after two washes with the same medium (prewarmed). Samples were collected at different time intervals during the incubation, and bacterial cells were treated immediately with the RNAlater solution (Ambion, Austin, TX) after the harvest through centrifugation, firstly at 300 × g for 5 min to remove cell debris and then at 6,000 × g for 5 min to pellet bacterial cells. In the experiments to test clone JZ-luxS, 1 × 108 CFU JG280 cells were incubated with IPEC-J2 cells for 2 h, either alone or simultaneously with 100 μl ECF (containing approximately 35,000 relative AI-2 activity values) from a 16-h-grown culture of clone JZ-luxS or from an E. coli DH5α culture harboring pBAD vector only, in 1 ml of the DMEM-F-12 medium with 10% FBS but no antibiotics. Incubation of the ECF from a clone JZ-luxS culture with IPEC-J2 cells only in parallel served as a control in the assays. The wash procedure of IPEC-J2 cells before the assays, the sampling and harvesting of cells, and the treatment of bacterial cells with the RNAlater were as described above. All the assays were conducted with each sample in triplicate.
Statistical analysis.
All analyses were performed with an SPSS software program (version 17.0; SPSS), and the significance was evaluated by one-way analysis of variance (ANOVA). P values of ≤0.05 were considered to differ significantly.
RESULTS
IPEC-J2 cell damage caused by ETEC strain JG280.
At the initial stage of the current study, we observed that strain JG280 at 1 × 106, 1 × 107, and 1 × 108 CFU/ml caused significantly more cell death of IPEC-J2 than at 1 × 109 CFU/ml, which was revealed by light microscopy after incubation for up to 3.5 h (data not shown). The effect of two bacterial concentrations (1 × 108 and 1 × 109 CFU/ml) on the cell death of IPEC-J2 was investigated. As shown in Fig. 1 A and B, JG280 at 1 × 108 CFU/ml caused a significantly higher level of death of IPEC-J2 than at 1 × 109 CFU/ml as determined by trypan blue staining and LDH release assay for cytotoxicity.
FIG. 1.
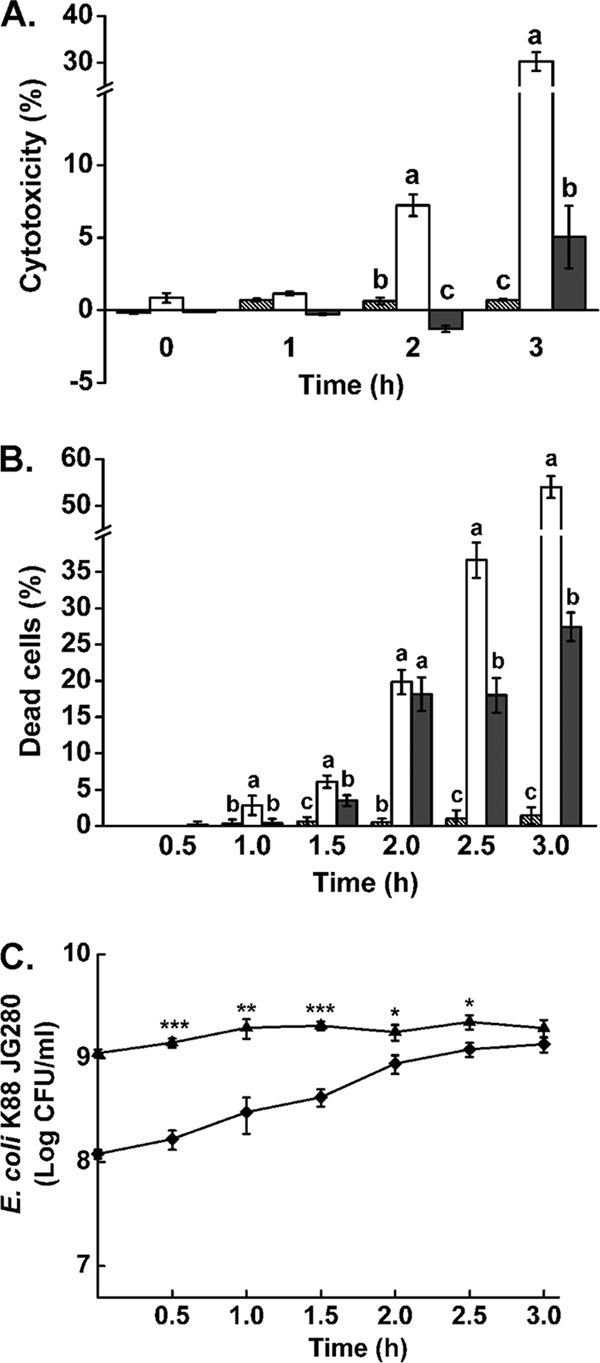
Cell viability and cytotoxicity of IPEC-J2 cells caused by ETEC strain JG280 and growth of the pathogen during the assays. (A) Cytotoxicity of IPEC-J2 cells determined by LDH release. (B) Accumulated death of IPEC-J2 cells determined by trypan blue staining. Hatched bars represent IPEC-J2 cells alone. White bars represent IPEC-J2 with 1 × 108 JG280. Gray bars represent IPEC-J2 cells with 1 × 109 JG280. (C) Growth of JG280 during a 3-h incubation with IPEC-J2 cells. Data are presented as means ± standard deviations (SD). Means marked with different letters (a, b, c) differ significantly at P values of <0.05. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
With cytotoxicity assays, JG280 at 1 × 108 CFU/ml induced 7.2% ± 0.7% and 30.3% ± 2.0% release of LDH from IPEC-J2 cells at 2 h and 3 h, respectively, which were significantly higher than −1.3% ± 0.2% and 5.0% ± 2.2% by 1 × 109 CFU/ml, respectively (Fig. 1A). A similar observation was also made with trypan blue staining of IPEC-J2 cells (Fig. 1B). However, the nonenterotoxigenic K88+ JFF4 strain generated little cell death or cytotoxicity to IPEC-J2 cells in the assays, parallel to those with JG280 (data now shown). The growth of JG280 during the assays in DMEM-F-12 was investigated. While the pathogen in the assay mixtures with an initial inoculum at 1 × 109 CFU/ml showed no significant cell proliferation during incubation, there was a steady growth of JG280 during the first 2 h of incubation in the assay mixtures inoculated initially with 1 × 108 CFU/ml (Fig. 1C). By 2 h, these mixtures had a level of the bacterial population close to 1 × 109 CFU/ml, similar to the level of the assay mixtures with the initial inoculation at 1 × 109 CFU/ml. After 3 h of incubation, there was no significant difference in the population sizes of the assay mixtures regardless of the initial levels of the inoculation.
Production of AI-2 activity by ETEC strain JG280.
To determine whether the bacterial density-dependent cell death of IPEC-J2 involved quorum sensing, we examined AI-2 activity produced by JG280 in the presence or absence of IPEC-J2 cells. In the assay mixtures containing JG280 at 1 × 108 CFU/ml and IPEC-J2 cells, AI-2 activity was not detectable after 1 h of incubation. The activity increased gradually after 1.5 h of incubation and reached the highest level at 3 h of incubation (Fig. 2 A). However, this observation was not obtained with the assay mixtures inoculated initially with 1 × 109 CFU/ml of JG280 (Fig. 2E). The presence and absence of IPEC-J2 cells in the assays made no significant difference in the production of AI-2 activity (data not shown). The production of AI-2 activity by the nonenterotoxigenic strain JFF4 was also examined in parallel. The strain produced AI-2 activity with a level comparable to that produced by JG280, indicating the presence of a functional luxS gene (data not shown).
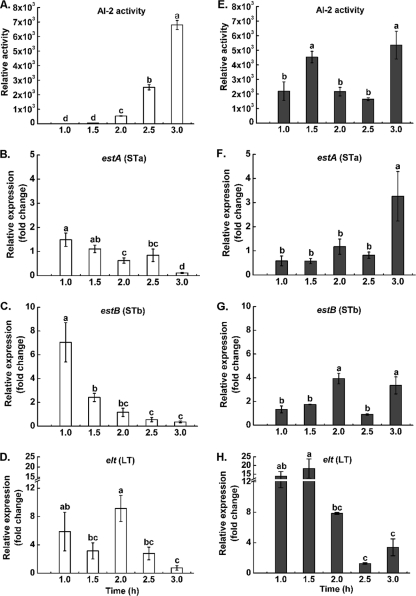
FIG. 2.
AI-2 activity and expression of enterotoxin genes from ETEC strain JG280. (A, B, C, and D) Levels of AI-2 activity and expression of estA (STa), estB (STb), and elt (LT), respectively, produced by JG280 at 1 × 108 CFU/ml (white bars) during incubation with IPEC-J2 cells. (E, F, G, and H) Corresponding data for JG280 at 1 × 109 CFU/ml (gray bars) in parallel to panels A, B, C, and D. Relative expression was calculated as the ratio of gene transcript level of each time point to that of the zero time point and is expressed as the fold change. Data are presented as means ± SD. Means marked with different letters (a, b, c, d) are significantly different at P values of <0.05.
Relationship between AI-2 activity and expression of enterotoxin genes of JG280.
Possible correlations between AI-2 activity and expression of major virulence genes of JG280 during the incubation with IPEC-J2 cells were investigated. As shown in Fig. 2B and C, the levels of estA (encoding STa) or estB (encoding STb) transcripts of JG280 in the assay mixtures inoculated initially with 1 × 108 CFU/ml decreased during the 3-h incubation and were negatively correlated with the increase in AI-2 production (Fig. 2A). Such a clear trend was not observed for elt (encoding LT) during the incubation (Fig. 2D). In contrast, the assay mixtures with an initial inoculum at 109 CFU/ml showed no decreased patterns of estA and estB expression (Fig. 2F and G), although the expression of elt appeared to be reduced during the incubation (Fig. 2H).
Production of AI-2 activity by clone JZ-luxS.
The luxS gene expression by clone JZ-luxS that contains a luxS gene in the pBAD-TOPO plasmid was investigated by measuring production of AI-2 activity. The clone produced a significant amount of AI-2 activity in its ECF recovered from 12-h cultures, which was comparable to the level produced by JG280 at 1 × 108 CFU/ml after the 3-h incubation. The production of AI-2 activity was not affected by the types of media used and presence or absence of arabinose in the media (data not shown). The parent strain of the clone (E. coli DH5α-pBAD) was also examined, and no AI-2 activity was detected. Therefore, the ECF from 12-h cultures of clone JZ-luxS was used for subsequent assays for exogenous luxS gene expression.
Effect of AI-2 activity from clone JZ-luxS on the cytotoxicity induced by JG280.
Knockout of the luxS gene from strain JG280 was attempted but failed, due to the lack of selective antibiotic markers. Therefore, an approach with the addition of the exogenously expressed luxS gene product (AI-2) was taken to investigate the effects of AI-2 on the cytotoxicity to IPEC-J2 cells caused by JG280 at 1 × 108 CFU/ml. As demonstrated in Fig. 3 A, little release of LDH activity from IPEC-J2 cells was detected for all assay mixtures with the various treatments after 1 h of incubation. All the assay mixtures, except for the treatment of IPEC-J2 cells with the ECF of clone JZ-luxS only, showed a high level of cytotoxicity (>20% release of LDH), but there was no significant difference among them after 3 h of incubation. However, after 2 h of incubation, the addition of the ECF (containing approximately 35,000 AI-2 relative activity values) from the culture of clone JZ-luxS significantly reduced LDH release from IPEC-J2 cells (from 20.1% ± 1.0% to 6.3% ± 1.8%, P = 0.00016). The original level of AI-2 in the assay mixtures prior to the addition of the ECF was approximately 540 AI-2 relative activity values. The addition of the ECF from the culture of E. coli DH5α-pBAD (parent strain of clone JZ-luxS) had no effect on the degree of JG280-induced cytotoxicity. Incubation of the ECF from the culture of clone JZ-luxS caused little release of LDH from IPEC-J2 cells in the absence of JG280, regardless of the length of incubation.
FIG. 3.
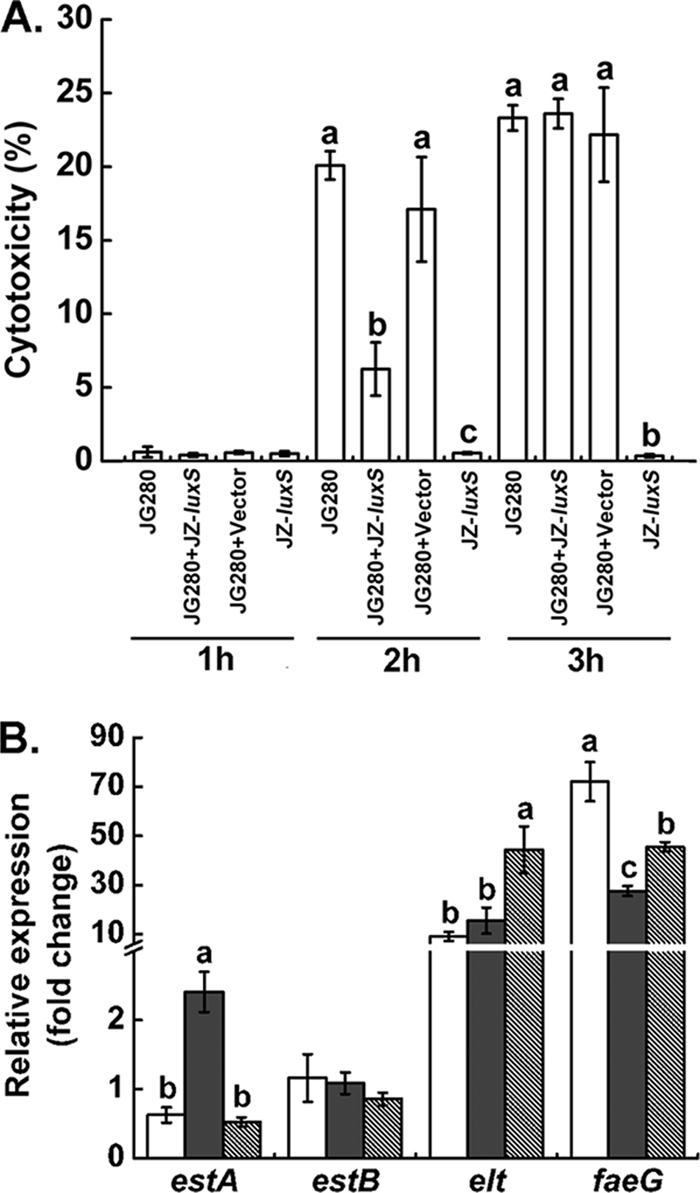
Effect of AI-2 from clone JZ-luxS on the cytotoxicity to IPEC-J2 and virulence gene expression of ETEC strain JG280. The bacterial density of JG280 was 1 × 108 CFU/ml for the assays. (A) Cytotoxicity to IPEC-J2 cells. JG280, treatment of IPEC-J2 with JG280 only; JG280+JZ-luxS, treatment of IPEC-J2 with JG280 and AI-2-containing ECF of clone JZ-luxS; JG280+Vector, treatment of IPEC-J2 with JG280 and the ECF of DH5α without AI-2; JZ-luxS, treatment of IPEC-J2 with AI-2-containing ECF of clone JZ-luxS only. Different hours indicate different lengths of incubation. (B) Expression of virulence genes by JG280 after 2 h of incubation with IPEC-J2. White bars represent the treatment of IPEC-J2 with JG280 only. Gray bars represent the treatment of IPEC-J2 with JG280 and the ECF of DH5α without AI-2. Hatched bars represent the treatments of IPEC-J2 with JG280 and AI-2-containing ECF of clone JZ-luxS. Data are presented as means ± SD. Means marked with different letters (a, b, c) are significantly different at P values of <0.05.
Effect of AI-2 activity from clone JZ-luxS on virulence gene expression in JG280.
Since the addition of AI-2 activity from clone JZ-luxS reduced JG280-induced cytotoxicity significantly after 2 h of incubation, the expression of enterotoxin genes of JG280 after 2 h of incubation was specifically examined. As shown in Fig. 3B, the expression of the estA gene was reduced significantly by the AI-2 activity of clone JZ-luxS compared with that of its parent strain (E. coli DH5α-pBAD), which produces no AI-2 activity (P = 0.0000011). The addition of the ECF from the DH5α-pBAD culture produced significantly more estA transcripts than the assay mixtures containing JG280 and IPEC-J2 cells only. Conversely, the expression of elt and of faeG that plays an important role in colonization of the pathogen to intestinal epithelia (12) was increased by AI-2-containing ECF from the JZ-luxS culture compared to that of the ECF of DH5α-pBAD culture without AI-2 (P = 0.001 and P = 0.003, respectively). No difference in estB expression of JG280 was observed with all tested samples. Putative virulence factors, paa (porcine AE associated) (17) and sepA (encoding a secreted serine protease of the autotransporter family) (12), were also investigated, and their gene expression was not affected by the addition of AI-2-containing ECF (data not shown).
DISCUSSION
In the present study, K88+ ETEC strain JG280 at 106 to 109 CFU/ml induced IPEC-J2 cell death. Interestingly, the pathogen caused significantly more IPEC-J2 cell death with the initial inoculums of 106 to 108 CFU/ml than at 109 CFU/ml. ETEC induces fluid and electrolyte secretion in the intestine due to the production of enterotoxins ST and LT (20). LT and STa stimulate adenylate cyclase and guanylate cyclase (GC-C) activity, respectively, resulting in increased intracellular cyclic AMP (cAMP) and cyclic GMP (cGMP) and leading to hypersecretion (12). In addition, ETEC was reported to cause cytotoxicity to the J774 macrophage (15). The activation of GC-C and accumulation of cGMP induced by ST have been shown to inhibit cell proliferation and/or apoptosis, and this effect was dependent on the cell surface expression of GC-C (23, 34). It has also been demonstrated that STa inhibits cell proliferation on T84 and COLO-205 colon carcinoma cells (24, 26). Recently, Johnson et al. reported that ETEC-induced cell damage was associated with an early stage of apoptosis of IPEC-J2 cells (14). A late stage of apoptosis (DNA fragmentation) was not observed in their study, possibly due to the bacterial cell numbers used (∼105 CFU) (14). Based on our observation of the cell death of IPEC-J2 induced by ETEC, apoptosis might also be cell density dependent. Activation of apoptosis enhanced bacterial adherence; inducing apoptosis could therefore be an important virulence mechanism of ETEC (14).
In line with the role of ST in activation of GC-C and antiproliferation (23, 34), and the observation by Johnson et al. that the effect of ETEC on the early stage of apoptosis was independent of LT (14), the effect of ETEC on cell death/apoptosis might be due mainly to the action of ST. It has been demonstrated that STb damaged CHO, IEC-18, and HT-29 cells, resulting in a trypan blue dye intake level of up to 51% (2). Consistent with the observation, we found that the higher frequency of cell death by 108 CFU/ml ETEC than by 109 CFU/ml was concomitant with higher relative expression levels of STa and STb during the first 1.5 h of infection (Fig. 2B, C, F, and G). After 2 h postinfection, the trend was not pronounced, because both doses of the pathogen reached similar peak levels (Fig. 1C). On the other hand, relative expression of LT with 109 CFU/ml was higher than that with 108 CFU/ml during the first 1.5 h of infection (Fig. 2D and H). Furthermore, the nonpathogenic K88+ E. coli strain JFF4 (toxin negative) caused no cytotoxicity to IPEC-J2 cells, implying the effect of toxins on cell damage/death. Future investigation is required to confirm whether the cell death observed with ETEC was due to apoptosis or necrosis. It also appears from our data that the effect of ETEC strain JG280 on the cell damage/death of IPEC-J2 cells was due to its accumulation during infection, which could lead IPEC-J2 cells to a nonreversible cell damage/death when it reaches a certain amount. The irreversibility of cell damage/death could be due to continuous enterotoxin secretion and the lack of a mature immune system of mammalian cells. A similar observation was reported previously by Johnson et al. (14), in which the percentage of calcein-negative and annexin V-positive IPEC-J2 cells increased gradually from 1 to 4 h after the exposure of the cells to ETEC. This suggests a low metabolic activity and slow loss of cell viability of IPEC-J2. Although the “accumulated effect” could explain our observation over the early stage of ST toxin gene expression by ETEC strain JG280 in relation to the cytotoxicity of IPEC-J2 to a certain degree, knockout mutants of ST toxins from ETEC strain JG280 would make a clear conclusion.
Our observations over the differences in cell damage/death of IPEC-J2 and the ST production by ETEC strain JG280 at different bacterial densities suggested a possible negative correlation. We therefore speculated that QS might be involved in the regulation of STa and STb production and thus the cell death. Consistent with our speculation, ETEC strain JG280 at 108 CFU/ml produced less AI-2 than that at 109 CFU/ml during the early 2-h infection (Fig. 2A and E), implying that AI-2 might be a negative regulator of STa and STb. To test this proposal, deletion of the luxS gene (responsible for AI-2 production) was attempted, but due to multiantibiotic resistance by this bacterium and lack of a suitable antibiotic resistance selection marker, mutation of the luxS gene was not successful. Therefore, we cloned and overexpressed the luxS gene from strain JG280 in E. coli DH5α. The ECF of DH5α with overexpressed luxS (AI-2) (clone JZ-luxS) significantly reduced IPEC-J2 cell death at 2 h postinfection (Fig. 3A), confirming the negative effect of AI-2 on ETEC-induced cell death. However, this effect was not observed at 3 h postinfection, probably due to the degradation of the exogenous AI-2 molecule (33). The effect of overexpressed AI-2 on virulence gene expression of ETEC at 2 h postinfection was then examined. As demonstrated in Fig. 3B, expression of estA was significantly reduced by the addition of the ECF of clone JZ-luxS with overexpressed luxS (AI-2) compared to that with the ECF of DH5α without AI-2, indicating that AI-2 repressed estA transcription. The increased expression of estA by the ECF without AI-2, compared to that by the control without the addition of the DH5α ECF, might result from the effect of unknown factors from DH5α which activated estA expression. It has been shown that a luxS mutation decreased LEE promoter transcription (36), which is in contrast to the LEE promoter activation in the E. coli K-12 background (29). This has led the authors to conclude that the difference between these two backgrounds might be the additional uncharacterized regulators specific to EHEC (36). Thus, we suggested that there might be other regulators specific to ETEC that influence estA expression. Whether this speculation can be applied to our observation described above remains to be determined. The expression of estB was not affected by AI-2-containing ECF. Interestingly, AI-2-containing ECF increased the level of elt transcripts. It is unclear whether AI-2 directly affects expression of STa and LT or indirectly via other regulators. Since LT was not responsible for the IPEC-J2 cell death by ETEC (14), consistently, our data suggested that STa might be the primary determinant of cell death. These results indicate that AI-2 may have differential effects on the regulation of ST and LT. Similar to the pattern by cAMP-cyclic AMP receptor protein (CRP), which positively regulates estA but represses elt expression (4), AI-2 negatively regulates estA while it activates elt expression, which counteracts the effect of cAMP-CRP, suggesting that STa and LT may be spatially and temporally regulated and coordinated during the course of infection. In Vibrio cholerae, it has been shown that QS negatively modulates virulence gene expression in a reciprocal manner in vivo (39). We reasoned that AI-2 may block the virulence gene expression of 109 CFU/ml ETEC before 1.5 h postinoculation, resulting in the lower toxin production. However, virulence genes of 108 CFU/ml ETEC were steadily expressed before 1.5 h due to the lack of AI-2, thus the high production of enterotoxins, which further induced the IPEC-J2 cell damage/death. Since the profile of AI-2 regulation and ETEC virulence gene expression have not been well studied, this variation in the regulatory circuit could be unique to ETEC JG280. To reach a clear conclusion on the role of AI-2 in regulating the production of enterotoxins and ETEC pathogenesis, a luxS mutant is required for pursuing the studies.
In conclusion, the present study showed that ETEC strain JG280 caused IPEC-J2 cell death that was bacterial cell density dependent. When IPEC-J2 cells were infected with ETEC strain JG280 at 108 CFU/ml, more cell death/damage was observed than at 109 CFU/ml, and this was correlated with AI-2 production. Further analysis showed that the addition of overexpressed AI-2 reduced the cell death/damage of JPEC-J2. Concurrently, estA expression was also reduced while elt expression was increased, suggesting that AI-2 regulates virulence of ETEC via controlling estA and elt expression, and STa may play a major role in ETEC-induced cell death/damage.
Acknowledgments
This research was supported by Agriculture and Agri-Food Canada. J. Zhu was a visiting graduate student financially supported by the China Scholarship Council through the MOE-AAFC Ph.D. Research Program.
IPEC-J2 was a gift from A. T. Blikslager, North Carolina State University. ETEC strain JG280 was provided by C. Gyles, University of Guelph, and E. coli strain JFF4 was a gift from J. Fairbrother, University of Montreal. AI-2 biosensor strain Vibrio harveyi MM32 and its corresponding positive control, Vibrio harveyi BB152, were gifts from E. Allen-Vercoe and M. W. Griffiths at the University of Guelph, respectively. We are grateful to E. Allen-Vercoe for her kindly permission to use the Victor Wallac 1420 for the AI-2 assay.
Editor: S. M. Payne
Footnotes
Published ahead of print on 7 February 2011.
REFERENCES
- 1.Bassler, B., E. Greenberg, and A. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beausoleil, H., V. Labrie, and J. Dubreuil. 2002. Trypan blue uptake by Chinese hamster ovary cultured epithelial cells: a cellular model to study Escherichia coli STb enterotoxin. Toxicon 40:185-191. [DOI] [PubMed] [Google Scholar]
- 3.Berberov, E., et al. 2004. Relative importance of heat-labile enterotoxin in the causation of severe diarrheal disease in the gnotobiotic piglet model by a strain of enterotoxigenic Escherichia coli that produces multiple enterotoxins. Infect. Immun. 72:3914-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodero, M. D., and G. P. Munson. 2009. Cyclic AMP receptor protein-dependent repression of heat-labile enterotoxin. Infect. Immun. 77:791-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boerlin, P., et al. 2005. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl. Environ. Microbiol. 71:6753-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouckenooghe, A. R., Z. D. Jiang, F. J. Cabada, C. D. Ericsson, and H. L. DuPont. 2002. Enterotoxigenic Escherichia coli as cause of diarrhea among Mexican adults and US travelers in Mexico. J. Travel Med. 9:137-140. [DOI] [PubMed] [Google Scholar]
- 7.Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157: H7 cleaves human coagulation factor V. Mol. Microbiol. 24:767-778. [DOI] [PubMed] [Google Scholar]
- 8.Carey, C., M. Kostrzynska, S. Ojha, and S. Thompson. 2008. The effect of probiotics and organic acids on Shiga-toxin 2 gene expression in enterohemorrhagic Escherichia coli O157:H7. J. Microbiol. Methods 73:125-132. [DOI] [PubMed] [Google Scholar]
- 9.Dalton, C. B., E. D. Mintz, J. G. Wells, C. A. Bopp, and R. V. Tauxe. 1999. Outbreaks of enterotoxigenic Escherichia coli infection in American adults: a clinical and epidemiologic profile. Epidemiol. Infect. 123:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Kievit, T., and B. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erume, J., et al. 2008. Comparison of the contributions of heat-labile enterotoxin and heat-stable enterotoxin b to the virulence of enterotoxigenic Escherichia coli in F4ac receptor-positive young pigs. Infect. Immun. 76:3141-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fairbrother, J., E. Nadeau, and C. Gyles. 2005. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev. 6:17-39. [DOI] [PubMed] [Google Scholar]
- 13.Feng, Y., et al. 2010. Identification of changes in the composition of ileal bacterial microbiota of broiler chickens infected with Clostridium perfringens. Vet. Microbiol. 140:116-121. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, A. M., R. S. Kaushik, N. J. Rotella, and P. R. Hardwidge. 2009. Enterotoxigenic Escherichia coli modulates host intestinal cell membrane asymmetry and metabolic activity. Infect. Immun. 77:341-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai, X. H., J. G. Xu, S. Melgar, and B. E. Uhlin. 1999. An apoptotic response by J774 macrophage cells is common upon infection with diarrheagenic Escherichia coli. FEMS Microbiol. Lett. 172:29-34. [DOI] [PubMed] [Google Scholar]
- 16.Lasaro, M., et al. 2006. Production and release of heat-labile toxin by wild-type human-derived enterotoxigenic Escherichia coli. FEMS Immunol. Med. Microbiol. 48:123-131. [DOI] [PubMed] [Google Scholar]
- 17.Leclerc, S., et al. 2007. paa, originally identified in attaching and effacing Escherichia coli, is also associated with enterotoxigenic E. coli. Res. Microbiol. 158:97-104. [DOI] [PubMed] [Google Scholar]
- 18.Maldonado, Y., J. C. Fiser, C. H. Nakatsu, and A. K. Bhunia. 2005. Cytotoxicity potential and genotypic characterization of Escherichia coli isolates from environmental and food sources. Appl. Environ. Microbiol. 71:1890-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medellin-Peña, M. J., H. Wang, R. Johnson, S. Anand, and M. W. Griffiths. 2007. Probiotics affect virulence-related gene expression in Escherichia coli O157:H7. Appl. Environ. Microbiol. 73:4259-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagy, B., and P. Z. Fekete. 2005. Enterotoxigenic Escherichia coli in veterinary medicine. Int. J. Med. Microbiol. 295:443-454. [DOI] [PubMed] [Google Scholar]
- 21.Nagy, B., R. A. Wilson, and T. S. Whittam. 1999. Genetic diversity among Escherichia coli isolates carrying f18 genes from pigs with porcine postweaning diarrhea and edema disease. J. Clin. Microbiol. 37:1642-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noamani, B. N., J. M. Fairbrother, and C. L. Gyles. 2003. Virulence genes of O149 enterotoxigenic Escherichia coli from outbreaks of postweaning diarrhea in pigs. Vet. Microbiol. 97:87-101. [DOI] [PubMed] [Google Scholar]
- 23.Pitari, G. M., M. D. Di Guglielmo, J. Park, S. Schulz, and S. A. Waldman. 2001. Guanylyl cyclase C agonists regulate progression through the cell cycle of human colon carcinoma cells. Proc. Natl. Acad. Sci. U. S. A. 98:7846-7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pitari, G. M., et al. 2003. Bacterial enterotoxins are associated with resistance to colon cancer. Proc. Natl. Acad. Sci. U. S. A. 100:2695-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reischl, U., M. Youssef, H. Wolf, E. Hyytia-Trees, and N. Strockbine. 2004. Real-time fluorescence PCR assays for detection and characterization of heat-labile I and heat-stable I enterotoxin genes from enterotoxigenic Escherichia coli. J. Clin. Microbiol. 42:4092-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha, S., P. Chowdhury, A. Pal, and M. Chakrabarti. 2008. Downregulation of human colon carcinoma cell (COLO-205) proliferation through PKG-MAP kinase mediated signaling cascade by E. coli heat stable enterotoxin (STa), a potent anti-angiogenic and anti-metastatic molecule. J. Appl. Toxicol. 28:475-483. [DOI] [PubMed] [Google Scholar]
- 27.Schierack, P., et al. 2006. Characterization of a porcine intestinal epithelial cell line for in vitro studies of microbial pathogenesis in swine. Histochem. Cell Biol. 125:293-305. [DOI] [PubMed] [Google Scholar]
- 28.Si, W., et al. 2007. Quantification of cell proliferation and alpha-toxin gene expression of Clostridium perfringens in the development of necrotic enteritis in broiler chickens. Appl. Environ. Microbiol. 73:7110-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sperandio, V., J. Mellies, W. Nguyen, S. Shin, and J. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sperandio, V., A. Torres, B. Jarvis, J. Nataro, and J. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. U. S. A. 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sperandio, V., A. Torres, and J. Kaper. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809-821. [DOI] [PubMed] [Google Scholar]
- 32.Stein, M., B. Kenny, M. Stein, and B. Finlay. 1996. Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J. Bacteriol. 178:6546-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waldman, S. A., et al. 1998. Heterogeneity of guanylyl cyclase C expressed by human colorectal cancer cell lines in vitro. Cancer Epidemiol. Biomarkers Prev. 7:505-514. [PubMed] [Google Scholar]
- 35.Walters, M., M. Sircili, and V. Sperandio. 2006. AI-3 synthesis is not dependent on luxS in Escherichia coli. J. Bacteriol. 188:5668-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walters, M., and V. Sperandio. 2006. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect. Immun. 74:5445-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weng, L., et al. 1999. PTEN suppresses breast cancer cell growth by phosphatase activity-dependent G1 arrest followed by cell death. Cancer Res. 59:5808-5814. [PubMed] [Google Scholar]
- 38.Yin, X., et al. 2009. Adherence of Escherichia coli O157:H7 mutants in vitro and in ligated pig intestines. Appl. Environ. Microbiol. 75:4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu, J., et al. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]