Abstract
The interplay between pathogens and their hosts has been studied for decades using targeted approaches, such as the analysis of mutants and host immunological responses. Although much has been learned from such studies, they focus on individual pathways and fail to reveal the global effects of infection on the host. To alleviate this issue, high-throughput methods, such as transcriptomics and proteomics, have been used to study host-pathogen interactions. Recently, metabolomics was established as a new method to study changes in the biochemical composition of host tissues. We report a metabolomic study of Salmonella enterica serovar Typhimurium infection. Our results revealed that dozens of host metabolic pathways are affected by Salmonella in a murine infection model. In particular, multiple host hormone pathways are disrupted. Our results identify unappreciated effects of infection on host metabolism and shed light on mechanisms used by Salmonella to cause disease and by the host to counter infection.
Salmonella enterica serovar Typhimurium has been used as a model organism to study host-pathogen interactions for decades (12, 18, 37). Although much is known regarding the effects of Salmonella on the host, most studies have focused on the analysis of individual host metabolic pathways. A few global studies on the effect of Salmonella infection on the host have been performed, using both transcriptomic and proteomic techniques (29, 40). However, the effect of Salmonella on the biochemical composition of host tissues remains unknown. Recently, techniques that detect and quantify multiple small metabolites in complex biological samples have been developed, giving rise to the field of metabolomics (19, 25, 39). Metabolomic studies have relied mainly on the use of chromatography coupled to mass spectrometry (MS) or nuclear magnetic resonance spectroscopy to identify and quantify metabolites in biological samples. Because these techniques require extensive sample preparation and, in some cases, have limited sensitivity, high-throughput studies have been impractical. More powerful techniques, such as direct-infusion ultrahigh-resolution Fourier transform ion cyclotron resonance (DI-FT-ICR) MS, have been developed with the potential to identify and quantify hundreds of metabolites with higher mass accuracy and without the need for extensive sample preparation, thus allowing comprehensive metabolic fingerprinting (11). We used DI-FT-ICR MS to investigate the impact of Salmonella infection on host metabolism using a murine typhoid infection model. To our knowledge, this is the first comprehensive metabolomic analysis of the effect of bacterial infection on host metabolism. Our study revealed a profound impact of Salmonella on host metabolism, with dozens of pathways being affected. Interestingly, some of the most impacted pathways are involved in host hormone signaling. Hormones are important mammalian signaling molecules and are fundamental for host metabolic and immune homeostasis (5, 9). The disruption of such pathways by Salmonella reveals the impact of bacterial infection on host metabolic function and may shed light on the molecular mechanisms used by pathogens to cause disease, as well as those used by host defenses.
MATERIALS AND METHODS
Chemical reagents.
Haloperidol, reserpine, acetonitrile, water, formic acid, ammonium hydroxide, streptomycin, dimethyl sulfoxide, and pregnenolone were purchased from Sigma-Aldrich (St. Louis, MO). ES tuning mix was purchased from Agilent Technologies (Santa Clara, CA).
Salmonella infection.
Salmonella enterica serovar Typhimurium SL1344 (42) was grown overnight in Luria-Bertani (LB) broth containing 100 μg/ml of streptomycin at 37°C with shaking. Age- and gender-matched C57BL/6 mice (The Jackson Laboratory, Bar Harbor, ME) between 8 and 20 weeks old were infected with approximately 7.5 × 107 bacterial cells from an overnight culture by oral gavage. To determine the impact of Salmonella infection on the metabolite composition of the intestinal tract, fresh feces from four uninfected mice were collected and stored at −80°C. These mice were then infected, and 5 days later, fresh feces were collected and stored at −80°C. Additionally, mice were sacrificed and colon sections were collected and homogenized in 1 ml of sterile Dulbecco's phosphate-buffered saline (PBS; HyClone, Logan, UT) using a tungsten bead and a mixer mill MM 301 (Retsch, Haan, Germany). Serial dilutions of the homogenates were plated on LB agar plates containing 100 μg/ml of streptomycin and incubated at 37°C overnight for bacterial enumeration. As expected, Salmonella colonized the gastrointestinal tract of all mice to high levels (Table 1). To investigate the impact of Salmonella infection on the metabolite composition of livers, four groups of three to four mice each were either infected with Salmonella as described above or kept uninfected (total of 11 mice per treatment group). Five days after infection, both uninfected and infected mice were sacrificed, and liver sections were collected and stored at −80°C. Additionally, liver sections were collected and processed for bacterial enumeration as described above. All mice showed high levels of Salmonella colonization of livers (Table 1). We chose to analyze samples from mice infected for 5 days, because at this time point the infection is at its maximum, allowing us to identify as many changes as possible in host metabolism caused by infection. All animal experiments were approved by the Animal Care Committee of the University of British Columbia and performed in accordance with institutional guidelines.
TABLE 1.
Bacterial loads on colon and liver sections from mice used for FT-ICR MS experiments
| Sample | Bacterial load/g of tissue |
|---|---|
| Colon | |
| 1 | 1.0 × 107 |
| 2 | 2.9 × 107 |
| 3 | 8.7 × 107 |
| 4 | 1.2 × 108 |
| Liver | |
| 1 | 1.5 × 108 |
| 2 | 1.3 × 106 |
| 3 | 3.8 × 105 |
| 4 | 8.2 × 106 |
| 5 | 1.2 × 106 |
| 6 | 3.0 × 105 |
| 7 | 5.5 × 105 |
| 8 | 2.3 × 106 |
| 9 | 1.7 × 106 |
| 10 | 5.1 × 106 |
| 11 | 7.7 × 105 |
Metabolite extraction.
To extract metabolites from livers and feces, acetonitrile was added to samples (approximately 1 μl of acetonitrile per 1 μg of tissue), which were then homogenized as described above. The samples were then cleared by centrifugation, and the supernatant was collected into a new tube and dried at room temperature using a centrifuge equipped with a vacuum pump. All extracts were kept at −80°C until further use.
DI-FT-ICR MS experiments.
For metabolic profiling, the dried extracts from mouse feces and livers were suspended in 50% acetonitrile (in water), vortexed, and cleared by centrifugation. Supernatants were collected and used as described herein. Because fecal samples from the same mouse before and after infection can be compared directly (because the mice do not need to be sacrificed), fecal extracts were used individually in the subsequent steps, i.e., without the mixing of samples from different mice. Liver extracts, however, cannot be compared within the same animal because mice need to be sacrificed for organ collection. Therefore, to ameliorate the inherent issue of intersubject variability, liver extracts were further prepared by mixing equal volumes of extracts from three to four mice, generating three liver extract mixtures per treatment (uninfected and infected), which were used in the subsequent steps. This also avoids that any potential differences in metabolite levels are masked by differences in bacterial loads in the organs of different animals. All extracts were diluted 1:20 with 50% acetonitrile containing either 0.2% formic acid (for positive-ion mode) or 0.5% ammonium hydroxide (for negative-ion mode) and spiked with predefined amounts of haloperidol, reserpine, and the ES tuning mix as the internal standards for mass calibration. The solutions were then infused through a syringe pump (KDS Scientific, Holliston, MA) at a flow rate of 2.5 μl per minute into a 12-T Apex-Qe hybrid quadrupole-FT-ICR mass spectrometer (Bruker Daltonics, Billerica, MA) equipped with an Apollo II electrospray ionization source, a quadrupole mass filter, and a hexapole collision cell. Data were recorded in positive- and negative-ion modes with broadband mode detection and an FT acquisition size of 1,024 kilobytes per second, within a range of m/z 150 to 1,000. Under these settings, a mass resolution of ca. 100,000 (full width at half maximum [FWHM]) at m/z 400 and a mass accuracy within 2 ppm or less for all detected components, following internal mass calibration, were observed. Other experimental parameters used were a capillary electrospray voltage of 3,600 to 3,750 V, spray shield voltage of 3,300 to 3,450 V, source ion accumulation time of 0.1 s, and collision cell ion accumulation time of 0.2 s. To increase detection sensitivity, survey scan mass spectra in positive- and negative-ion modes were acquired from the accumulation of 200 scans per spectrum, and duplicate acquisitions per sample were carried out to ensure data reproducibility. As an indication of the analytical reproducibility of the methods employed, we report that more than 90% of the peaks detected showed less than 10% variation in intensity between the analytical replicates. Additionally, the majority of the mass values analyzed in uninfected fecal (73.7%) and liver (71.9%) samples showed a variation in intensity between samples of 25% or less (standard errors of the means).
DI-FT-ICR MS data processing.
Raw mass spectrometry data were processed using a custom-developed software package, as described elsewhere (11). First, raw mass spectra acquired from each sample group were batch processed using the instrument vendor's data analysis software, DataAnalysis, but with a home-written Visual Basic for Applications (VBA) script to do automatic internal mass calibration with the reference masses of the spiked calibration standards and a known contaminant, N-butylbenzensulfonamide. Monoisotopic peaks corresponding to the isotopic pattern distributions were then automatically determined, and those with a signal/noise ratio above 3 were picked. Their m/z values were converted to neutral masses by subtracting 1.007276 for positive-ion mode or adding 1.007276 for negative-ion mode. Next, the resulting mass lists from all the mass spectra within each set of uninfected/infected groups detected in positive- or negative-ion modes were further processed with another customized software program developed with LabVIEW (National Instruments, Austin, TX). The first step of this software is to remove the adduct ions (M + Na)+ and (M + K)+ in positive-ion mode and (M + Cl)− in negative-ion mode from the mass lists based on the expected mass differences for these ions within 2 ppm to yield a list of unique biochemical component masses together with their peak intensities. The peak intensities of all the monoisotopic neutral masses are subsequently normalized to the intrasample total ion intensity. Masses observed in at least three of four samples (feces) or two of three samples (livers) of one of the conditions (infected or uninfected) were aligned and combined into unique metabolite features from the masses that matched within 2 ppm across all the data. Finally, a two-dimensional data matrix (mass versus relative intensity) was generated for each sample group and saved in a format amenable for further data analysis. Heat maps were then created using the freely available softwares Cluster and Java TreeView (http://rana.lbl.gov/EisenSoftware.htm). To identify differences in metabolite compositions between samples from uninfected and infected mice, we first filtered our list of masses for metabolites that were present on one set of samples (uninfected or infected) but not the other. Additionally, we averaged the mass intensities of metabolites in each group and calculated the ratios between averaged intensities of metabolites from uninfected and infected mice. To assign possible metabolite identities to the masses present in only one of the sample groups or showing at least a 2-fold change in intensities between the sample groups, the monoisotopic neutral masses of interest were queried against MassTrix (http://masstrix.org), a software designed to incorporate mass queries into metabolic pathways using information from the Kyoto Encyclopedia of Genes and Genomes (KEGG), the Human Metabolome Database (HMDB), and LipidMaps (34). Masses were searched against the Mus musculus database within a mass error of 3 ppm.
Enzyme-linked immunosorbent assays.
Fresh feces were collected from uninfected mice and mice that had been infected for 4 days. Feces were collected in PBS and extracted as described above. Samples were cleared by centrifugation, and commercially available enzyme-linked immunosorbent assay (ELISA) kits were used to determine fecal concentrations of prostaglandin E2 (PGE2), cortisol (Cayman Chemical Company, Ann Arbor, MI), 15-deoxy-Δ12,14-PGJ2, thromboxane B2 (TXB2), and aldosterone (Assay Designs, Ann Arbor, MI).
Tissue culture.
RAW 264.7 macrophage-like cells (26) and AML12 hepatocyte-like cells (43) (henceforth referred to as macrophages and hepatocytes, respectively) were obtained from the American Type Culture Collection (Manassas, VA). RAW 264.7 is a tumorigenic macrophage-like cell line obtained from murine tumors induced with Abelson leukemia virus (26). AML12 is a nontumorigenic, differentiated, hepatocyte-like cell line derived from livers of transgenic mice overexpressing transforming growth factor α (43). AML12 cells have typical hepatocyte features, including peroxisomes and bile canalicular-like structures. Macrophages were grown in Dulbecco's modified Eagle medium (DMEM; HyClone, Waltham, MA) supplemented with 10% fetal bovine serum (FBS; HyClone), 1% nonessential amino acids (Gibco, Carlsbad, CA), and 1% GlutaMAX (Gibco). Hepatocytes were grown in DMEM-nutrient mixture F-12 (Gibco) supplemented with 10% FBS (HyClone). For cocultures, cells were cultured as described above and mixed at a 1:1 ratio immediately before seeding. Cells were seeded 20 h before experiments in 24-well plates at a density of 105 cells per well. Pregnenolone was dissolved in dimethyl sulfoxide, and the final concentration used was 10 μM. Controls without pregnenolone contained the same amounts of dimethyl sulfoxide (DMSO), which was always used at 0.1%. For infection assays, cells in mid-logarithmic growth were spun down and resuspended in PBS and diluted in tissue culture medium. Cells were infected at a multiplicity of infection of 100 for 10 min at 37°C and 5% CO2. Subsequently, cells were washed twice with PBS and incubated at 37°C and 5% CO2 in growth medium containing 50 μg/ml gentamicin (Sigma-Aldrich) for 1 h and 50 min. After a total of 2 h of infection, either cells were used in subsequent experiments or the medium was replaced to decrease the gentamicin concentration to 5 μg/ml for later time points. At the appropriate times, cells were lysed in 250 μl of 1% Triton X-100 (BDH, Yorkshire, United Kingdom), 0.1% sodium dodecyl sulfate (Sigma-Aldrich). Serial dilutions were plated on LB plates containing 100 μg/ml of streptomycin (Sigma-Aldrich) for bacterial enumeration.
Real-time PCR.
Liver sections were collected in RNAlater (Qiagen, Hilden, Germany) and kept at −80°C until used. Tissue was homogenized as described above. Tissue RNA was isolated using the RNeasy minikit from Qiagen, with the on-column DNA digestion, followed by DNA digestion using DNase I (Invitrogen, Carlsbad, CA) and further purification. RNA from cultured cells was isolated in the same way, except that the additional DNase I treatment was not performed. cDNA was synthesized using the QuantiTect reverse transcription kit (Qiagen). Tissue cDNA was purified using the MinElute PCR purification kit (Qiagen) and quantified, and 5 ng was used in real-time (RT) PCRs. Because of the low levels of transcripts for some hormone enzymes in cultured cells, cDNA from cells was not purified but used directly on RT PCRs. For RT PCR, we used the QuantiTect SYBR green PCR kit (Qiagen) and the Applied Biosystems (Foster City) 7500 system. Reaction mixtures contained forward and reverse primers at 0.4 μM each. All results were normalized using the mRNA levels of the acidic ribosomal phosphoprotein PO as baseline. Averages of the data obtained with untreated samples were normalized to 1, and the data from each sample (untreated or treated) were normalized accordingly. Primer sequences are available upon request.
Statistical analysis.
Data were analyzed by unpaired t tests with 95% confidence intervals using GraphPad Prism version 4.0 (GraphPad Software Inc., San Diego, CA). Outliers were detected using the Grubbs' test and removed from data sets when indicated.
DI-FT-ICR MS data accession number.
The mass spectrometry data have been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE21720.
RESULTS
Salmonella infection has a profound effect on fecal and hepatic metabolite profiles.
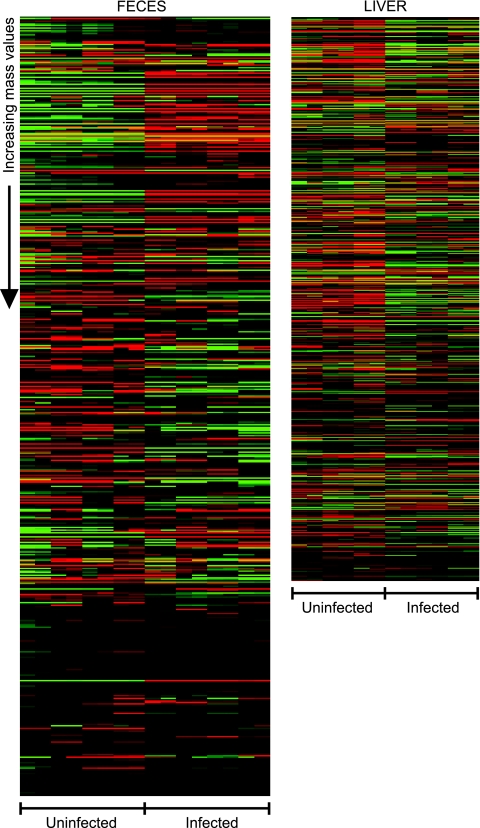
Because Salmonella colonizes the gastrointestinal tract of mice to high levels (Table 1) (22), we expected that infection would cause significant changes in the chemistry of the intestinal environment. In order to identify such changes, we prepared acetonitrile extracts from fresh feces from mice before and 5 days after infection with Salmonella. DI-FT-ICR MS analysis of fecal extracts yielded a total of 1,575 different masses from both uninfected and infected mice (Table 2). To investigate which of these masses were present in different levels in samples from uninfected and infected mice, we first selected those that were present only in uninfected or infected samples. Additionally, we calculated the average intensities of all masses and compared values from uninfected and infected samples. Masses that showed changes of 2-fold or more were combined with those that were observed only in infected or uninfected mice for further analyses. As a result, we found that Salmonella infection altered the levels of 924 masses (Table 2 and Fig. 1). This represents over 58% of all masses detected, showing that Salmonella infection has a great impact on the biochemical composition of feces.
TABLE 2.
Overview of DI-FT-ICR MS results and the impact of Salmonella infection on the biochemical composition of liver and feces
| Section | No. of masses detected |
Masses changed |
||||||
|---|---|---|---|---|---|---|---|---|
| With negative ionization | With positive ionization | Overlapa | Total | Uninfected > infectedb | Infected > uninfectedc | Total no. changed | % total | |
| Feces | 883 | 809 | 117 | 1,575 | 398 | 526 | 924 | 58.7 |
| Liver | 565 | 666 | 82 | 1,149 | 395 | 341 | 736 | 64.1 |
Masses were considered to overlap when they differed by 0.001 Da or less.
Number of metabolites showing higher levels in uninfected samples.
Number of metabolites showing higher levels in infected samples.
FIG. 1.
Heat maps showing the effect of Salmonella infection (5 days) on the levels of metabolites from mouse feces and livers. Data were median centered using Cluster (7), and heat maps were constructed using Java TreeView (http://rana.lbl.gov/EisenSoftware.htm). Masses are presented from lowest (top) to highest (bottom). Green represents masses with signal intensities higher than the median, whereas red indicates signal intensities lower than the median. Black indicates missed values or values with no difference from the median signal intensity.
Besides colonizing the intestinal tract, Salmonella causes a systemic infection in mice and can colonize multiple organs, most notably the liver and spleen (Table 1) (22). To further our understanding of the effects of Salmonella infection on host metabolism, we examined the changes in the chemical composition of liver extracts upon infection. We collected liver sections from uninfected and infected mice after 5 days of infection and analyzed acetonitrile extracts by DI-FT-ICR MS. Such analysis yielded a total of 1,149 masses (Table 2). To identify masses present at different levels in uninfected and infected samples, we examined our data set as described above. In total, 736 masses from livers were affected (Table 2 and Fig. 1). This represents over 64% of all masses detected, showing that Salmonella infection also causes a major change in the biochemical composition of liver tissue.
Multiple host metabolic pathways are affected by infection.
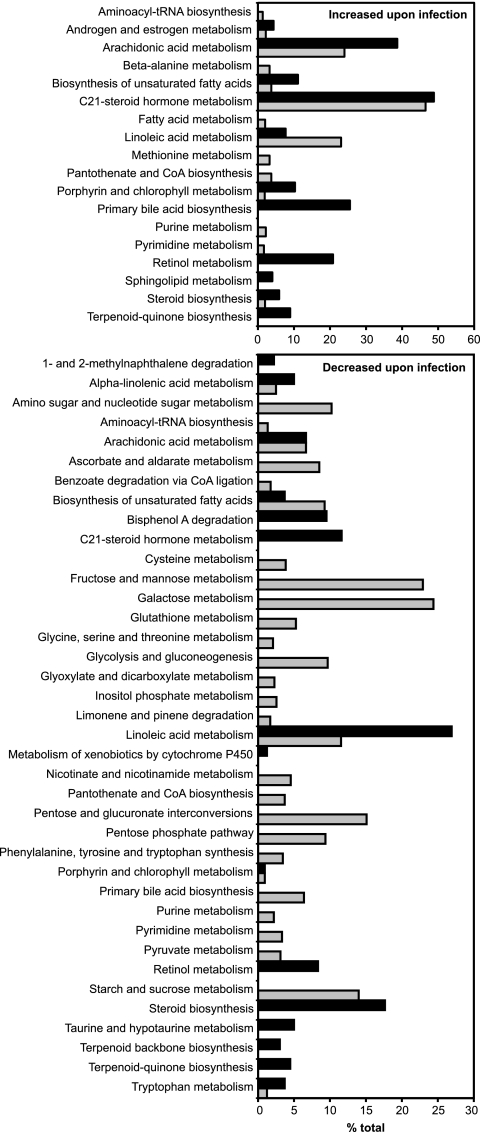
In order to identify the individual pathways in which the masses of interest are involved, we queried the MassTrix database (http://masstrix.org) (34). Figure 2 shows the categories of metabolites affected by Salmonella infection. The metabolic pathways identified involve steroids, eicosanoids, bile acids, carbohydrates, and porphyrin, among others. Some of the most deeply impacted pathways involve the metabolism of steroid and eicosanoid hormones. The effect of Salmonella infection on these pathways was pursued in more detail.
FIG. 2.
Metabolic pathways affected by Salmonella infection (5 days). Masses of interest were searched against the KEGG database using the MassTrix software (http://masstrix.org) (34). Bars indicate the percentage of metabolites from each KEGG pathway that was affected by infection. Black bars indicate metabolites from feces, and gray bars indicate metabolites from livers.
Salmonella infection disturbs eicosanoid metabolism.
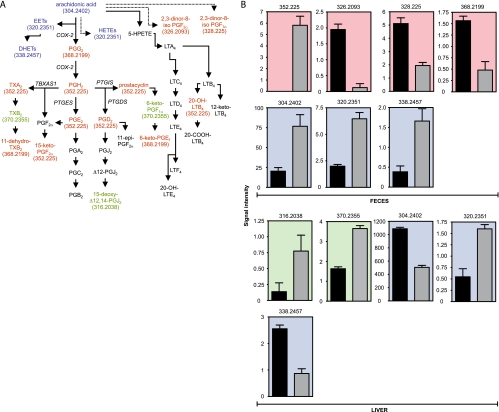
One of the metabolic pathways extensively affected by Salmonella was that of arachidonic acid, the main precursor of eicosanoid hormones (Fig. 2 and 3) (9). Infection caused an increase in the levels of 29 potential metabolites in feces and 18 in the liver, for a total of 32 potential metabolites affected in this pathway (due to the overlap between the two data sets). All of these metabolites were matched within mass errors ranging from −0.4 to +0.65 ppm, based on the comparison of their calculated monoisotopic masses with the FT MS-measured accurate masses (data not sown). Additionally, infection caused a decrease in the levels of five potential metabolites in feces and five in the liver, for a total of 10 potential metabolites (Fig. 3A and B). Because multiple metabolites in any given pathway may have identical masses, we manually screened the potential metabolites assigned by MassTrix to determine the number of different masses that were affected by infection. This analysis revealed that six different masses were increased upon infection, whereas five masses were decreased (Fig. 3B).
FIG. 3.
The effect of Salmonella infection on eicosanoid hormone metabolism. (A) Schematic of the eicosanoid hormone metabolic pathway. Colors indicate the organs in which a given hormone was affected by infection (5 days). Red indicates metabolites affected in feces, green indicates those affected in liver, and blue indicates those affected in both samples. Black indicates metabolites not detected or unchanged. Masses (Da) for metabolites affected are shown in parentheses. Solid arrows indicate direct steps, and dashed arrows indicate multiple steps that are not shown. Enzymes involved in some of the reactions displayed are shown in italics. (B) Levels of masses affected by infection (5 days) in the eicosanoid pathway. Masses are shown at the top of each graph. The y axis indicates mass signal intensity. Signal/noise ratios for metabolites shown varied between 14.2 and 14,641.2. Color scheme is the same as that described for panel A. Black bars represent uninfected samples, and gray bars indicate infected samples. Averages with standard errors of means are shown. Four samples were used for feces and three for livers, with duplicate measurements (n = 8 for feces and n = 6 for livers). Ions presumptively neutralized to a mass of 304.2402 Da using our method were detected in feces in both negative- and positive-ionization modes, and all results were used (n = 16). All P values were ≤0.0001, except for the metabolites in feces with masses of 338.2457 (P = 0.0023) and 304.2402 (P = 0.0008) Da and the metabolites in livers with masses of 316.2038 (P = 0.0492) and 320.2351 (P = 0004) Da. PG, prostaglandin; LT, leukotriene; TX, thromboxane; EET, epoxyeicosatrienoic acid; HETE, hydroxyeicosatetraenoic acid; HPETE, hydroperoxyeicosatetraenoic acid; DHET, dihydroxyeicosatrienoic acid.
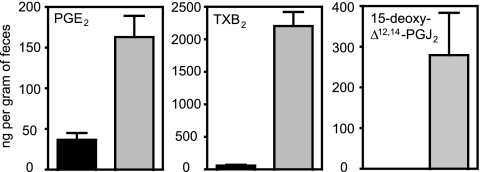
Eicosanoids are divided into several classes, including prostaglandins (PG), leukotrienes (LT), thromboxanes (TX), prostacyclins, and others (9). In order to obtain further evidence that eicosanoids are affected by Salmonella infection, we measured fecal concentrations of PGE2, 15-deoxy-Δ12,14-PGJ2, and TXB2 using an ELISA. Confirming our DI-FT-ICR MS results, these three hormones were significantly increased upon infection (Fig. 4).
FIG. 4.
Fecal levels of eicosanoids in uninfected (black bars) and infected (4 days; gray bars) samples, determined by ELISAs. Averages with standard errors of the means are shown. The numbers of uninfected mice used were 4 (15-deoxy-Δ12,14-PGJ2) and 5 (TBX2 and PGE2). The number of infected mice used was 4 in all cases. All differences were statistically significant (P < 0.05). Outliers were detected using the Grubbs' test and removed.
Transcript levels of eicosanoid synthesis enzymes are affected by infection.
Although the entire arachidonic acid pathway was affected by infection, the PG-TX-prostacyclin branch was particularly disrupted (Fig. 3A). The first reaction in this branch is the conversion of arachidonic acid into PGG2 (9). PGG2 is then converted into PGH2, which acts as a substrate for several enzymes that produce PGs, TXs, and prostacyclins. The enzymes responsible for the synthesis of PGG2 and PGH2 are prostaglandin-endoperoxide synthases (COX). Two main forms of COX exist. COX-1 is constitutively expressed, whereas COX-2 is an inducible enzyme (9). To confirm that Salmonella infection affects the production of eicosanoids, we analyzed the transcript levels of COX-2 in the livers of uninfected and infected mice. Salmonella infection caused an induction of over 90-fold in COX-2 transcript levels (Fig. 5). Next, we studied the mRNA levels of enzymes involved in the conversion of PGH2 to prostacyclin, PGE2, PGD2, and TXA2 (9). The transcript levels of prostaglandin E synthase (PTGES) and thromboxane A synthase (TBXAS1), the enzymes responsible for the production of PGE2 and TXA2, were increased over 21- and 5-fold upon infection, respectively (Fig. 5). Transcripts for prostaglandin-H2 D-isomerase (PTGDS), the enzyme involved in PGD2 synthesis, were repressed over 36-fold upon infection (Fig. 5). PTGIS, the prostacyclin synthase, was unaffected (Fig. 5).
FIG. 5.
Relative transcript levels of enzymes involved in eicosanoid synthesis in uninfected (black bars) and infected (5 days; gray bars) livers. Averages of the results obtained from uninfected tissues were normalized to 1, and the levels for individual mice, uninfected and infected, were adjusted accordingly. Averaged results are shown. Bars indicate the standard errors of the means. Ten mice were used in all cases except for COX-2 determinations from uninfected mice (n = 7). All P values were <0.002, except for PTGDS (P = 0.0191) and PTGIS (P = 0.8963).
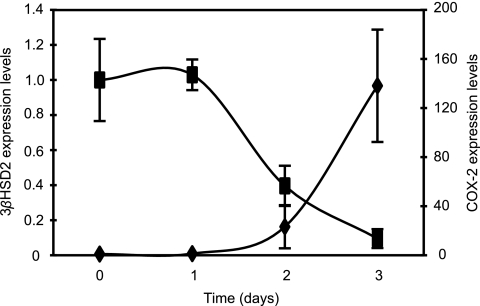
To investigate whether the effect of Salmonella infection on eicosanoid metabolism is a consequence of an overt bacterial presence at day 5 after infection, we tracked the levels of COX-2 transcripts in the liver during the initial stages of infection. The induction of COX-2 by Salmonella starts 2 days after infection, when an average 23.2-fold increase in the levels of COX-2 transcript is observed (although not statistically significant). At 3 days postinfection, full induction of COX-2 expression is achieved, accounting for a 138.1-fold increase in its transcript levels (Fig. 6). At days 2 and 3 postinfection, the numbers of bacteria within livers are approximately 5,000- and 100-fold lower than those found on day 5 (22). This suggests that the effect of Salmonella on host hormone gene expression is probably not simply an indirect consequence of an overt bacterial presence but rather is due to specific host responses to infection.
FIG. 6.
Transcript levels of COX-2 (diamonds) and 3βHSD2 (squares) in mouse livers throughout the course of Salmonella infection. Averages of the results obtained from uninfected tissues were normalized to 1, and the levels for individual mice, uninfected and infected, were adjusted accordingly. Averaged results are shown. Bars indicate the standard errors of the means. Numbers of mice used for COX-2 determinations were 7 (day 0), 3 (day 1), 3 (day 2), and 4 (day 3). Numbers of mice used for 3βHSD2 determinations were 10 (day 0), 4 (day 1), 4 (day 2), and 4 (day 3). Compared to results from uninfected samples (day 0), data from days 1 and 2 were not statistically significant (P > 0.05). Data from day 3 were statistically significant for both 3βHSD2 (P = 0.0351) and COX-2 (P = 0.0025).
Salmonella infection disturbs steroid metabolism.
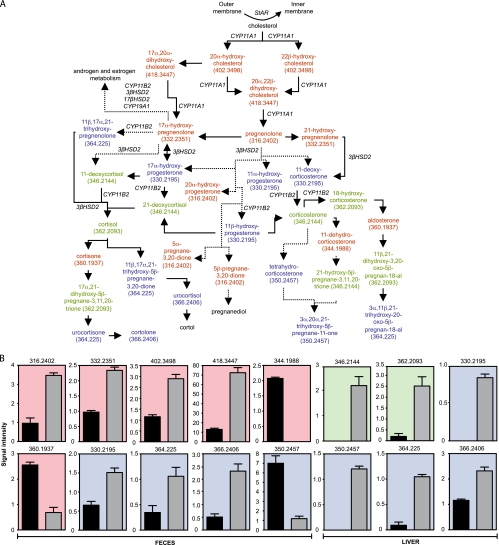
Of the many metabolic pathways affected by Salmonella infection, the metabolism of steroid hormones was the most notably impacted (Fig. 2 and 7). The levels of 21 potential metabolites predicted to be part of the C21 steroid hormone metabolic pathway were increased in feces during infection. In the liver, 20 potential metabolites involved in this pathway were increased during infection. In total, 31 potential metabolites were increased (due to the overlap between the two data sets). Additionally, the levels of five potential metabolites in feces were decreased by infection, whereas no metabolites were decreased in the liver (Fig. 7A and B). Further analysis revealed that metabolites of 10 different masses were increased upon infection, whereas metabolites of three masses were decreased (Fig. 7B).
FIG. 7.
The effect of Salmonella infection on steroid hormone metabolism. (A) Schematic of the C21 steroid hormone metabolic pathway. Colors indicate the organs in which a given hormone was affected by infection (5 days). Color coding is the same as that described for Fig. 2. The outer and inner membranes mentioned are mitochondrial. Masses (Da) for metabolites affected are shown in parentheses. Solid arrows indicate direct steps, and dashed arrows indicate multiple steps that are not shown. Enzymes involved in some of the reactions displayed are shown in italics. (B) Levels of masses affected by infection (5 days) in the steroid pathway. Masses are shown at the top of each graph. The y axis indicates mass signal intensity. Signal/noise ratios for the metabolites shown varied between 7.4 and 125.9. Color coding is the same as that described for Fig. 2. Black bars represent uninfected samples, and gray bars indicate infected samples. Averages with standard errors of the means are shown. Four samples were used for feces and three for livers, with duplicate measurements (n = 8 for feces and n = 6 for livers). Ions presumptively neutralized to masses of 362.2093 and 364.225 Da using our method were detected in livers in both negative- and positive-ionization modes, and all results were used (n = 12). All P values were ≤0.0001, except for the metabolite with a mass of 364.225 Da in feces (P = 0.0065).
To provide further evidence that steroid hormones are affected by Salmonella infection, we also measured fecal levels of aldosterone and cortisol using an ELISA. Both hormones were significantly increased in infected mice, confirming our DI-FT-ICR MS results (Fig. 8).
FIG. 8.
Fecal levels of steroids in uninfected (black bars) and infected (4 days; gray bars) samples, determined by ELISAs. Averages with standard errors of the means are shown. Results from five uninfected mice are shown. For infected samples, the number of mice was 4 for aldosterone and 3 for cortisol. All differences were statistically significant (P < 0.05). Outliers were detected using the Grubbs' test and removed.
Transcript levels of steroid synthesis enzymes are affected by infection.
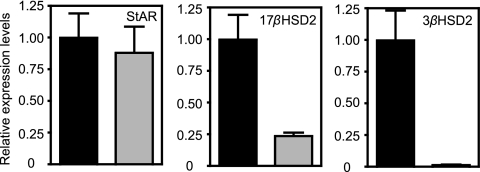
To determine whether the effect of Salmonella on steroid hormone levels was due to differential transcription of enzymes in this pathway, we analyzed the transcript levels of several of these enzymes. Steroid synthesis begins with the conversion of cholesterol to pregnenolone (10). Cholesterol is found in the mitochondrial outer membrane and is transferred to the inner membrane by the steroidogenic acute regulatory protein (StAR), where it is made available for steroid synthesis (Fig. 7A) (10). We analyzed StAR transcript levels in livers from uninfected and infected mice. No significant difference was detected (Fig. 9). We then studied the mRNA levels of several enzymes downstream of StAR. Although some of these enzymes were undetectable by our assays (CYP11A1, CYP11B2, and CYP19A1), we found that transcript levels of 3-β-hydroxy-δ-5-steroid dehydrogenase (3βHSD2) were decreased over 66-fold upon infection (Fig. 9). Additionally, 17-β-hydroxysteroid dehydrogenase 2 (17βHSD2) transcript levels were decreased over 4-fold in infected samples (Fig. 9). Both 3βHSD2 and 17βHSD2 are key enzymes in the synthesis of C18, C19, and C21 steroids (Fig. 7A) (10).
FIG. 9.
Relative transcript levels of enzymes involved in steroid synthesis in uninfected (black bars) and infected (5 days; gray bars) livers. Averages of the results obtained from uninfected tissues were normalized to 1, and the levels for individual mice, uninfected and infected, were adjusted accordingly. Averaged results are shown. Bars indicate the standard errors of the means. Ten mice were used in all cases except for StAR determinations from uninfected mice (n = 7) and 3βHSD2 determinations from infected mice (n = 8). P values were <0.002, except for StAR (P = 0.6972).
To investigate whether the effect of Salmonella infection on steroid metabolism is due to overt bacterial infection, we tracked the levels of 3βHSD2 transcripts in livers throughout the initial stages of infection. 3βHSD2 expression is repressed 2.5-fold at day 2 after infection (although not statistically significant). Additionally, a 10.5-fold repression of 3βHSD2 is achieved 3 days after infection (Fig. 6). As previously mentioned, at this stage of infection Salmonella loads within livers are significantly lower than on day 5 (22), suggesting that the effect of Salmonella on host hormone gene expression is not due to an overt bacterial presence in mouse livers.
Eicosanoid and steroid pathways are disrupted by Salmonella infection in vitro.
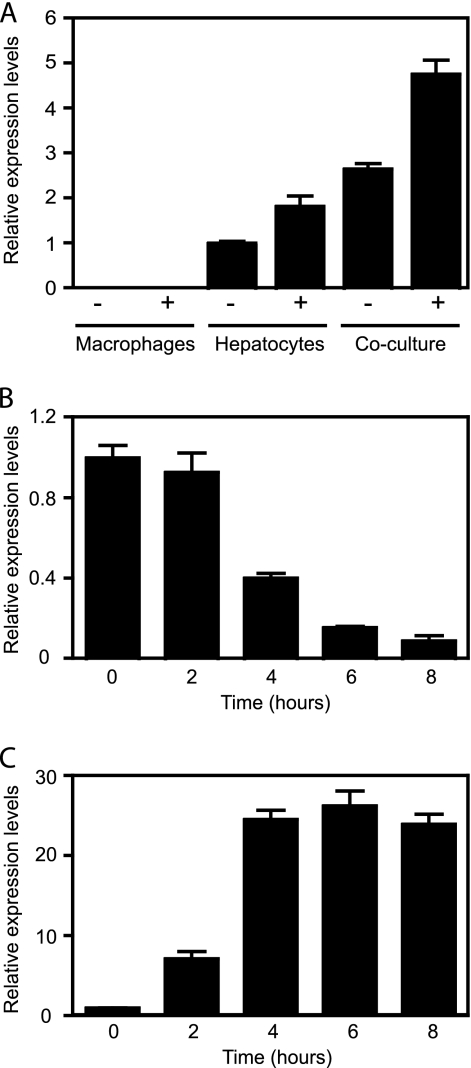
Salmonella infection is accompanied by an influx of inflammatory cells (22). To investigate whether the changes observed in the steroid and eicosanoid pathways were due to the altered cellular composition due to inflammation, we analyzed the effects of Salmonella infection on cultured cells. For this purpose, we assessed transcript levels of 3βHSD2 and COX-2 as indicators of pathway disruption. While studying the transcript levels of 3βHSD2 in cultured cells, we found that such transcripts are undetectable in macrophage-like cells and that transcript levels of this gene are 1,000-fold lower in total RNA isolated from hepatocyte-like cells than in RNA isolated from liver sections (in reference to transcript levels of the acidic ribosomal phosphoprotein PO; data not shown). We attempted to restore 3βHSD2 expression through a number of treatments. First, we reasoned that the substrate for this enzyme would be a good candidate to induce its expression. We compared 3βHSD2 transcript levels in both macrophages and hepatocytes with or without pregnenolone, the major precursor of all steroid hormones. Indeed, the addition of pregnenolone caused a modest (2-fold) but significant increase in 3βHSD2 levels in hepatocytes (Fig. 10 A). Pregnenolone addition had no effect on 3βHSD2 expression by macrophages. We also hypothesized that interactions between different cell types in the liver could be a determining factor in 3βHSD2 expression. Therefore, we used cocultures of macrophages and hepatocytes to find that 3βHSD2 expression in cocultures was approximately 3-fold higher than in hepatocytes alone. Moreover, the addition of pregnenolone to cocultures caused a further increase in expression, resulting in levels 5-fold higher than those found in hepatocytes (Fig. 10A). Next, we used this coculture system to study the impact of Salmonella on both COX-2 and 3βHSD2 expression. Salmonella infection caused a 10-fold repression of 3βHSD2 transcript levels, reminiscent of the effect during mouse infections (Fig. 10B). Additionally, infection caused a significant induction of COX-2 expression (Fig. 10C). These results suggest that at least some of the effects of Salmonella on hormone metabolism are not linked to the migration of inflammatory cells to sites of infection but, rather, due to specific and localized changes in host metabolism.
FIG. 10.
Salmonella infection disrupts steroid and eicosanoid metabolism in vitro. (A) Pregnenolone and cocultures of macrophages (RAW 264.7) and hepatocytes (AML12) increase 3βHSD2 expression. Cells were grown with (+) or without (−) 10 μM pregnenolone. The average of the relative mRNA levels in cultured hepatocytes with no additions was normalized to 1, and individual results for all treatments were adjusted accordingly. Averages with standard errors are shown. In all groups, the number of mice used was 6. Pregnenolone addition caused a significant increase in 3βHSD2 expression in hepatocytes (P = 0.0048) and cocultures (P < 0.0001) but not in macrophages (P = 0.6648). Coculturing caused a statistically significant increase in 3βHSD2 expression compared with that of macrophages or hepatocytes, with or without the addition of pregnenolone (all P values of <0.0001). Cells were incubated under each condition for 20 h. (B) Salmonella infection represses 3βHSD2 expression during coculture of macrophages and hepatocytes in the presence of pregnenolone. Averages of the relative mRNA levels in uninfected wells (time zero) were normalized to 1, and the results for individual wells, uninfected and infected, were adjusted accordingly. Averages and standard errors of the means are shown. Five wells were used for the first time point (time zero), and three wells were used for all other time points. Comparisons of the 4-h, 6-h, and 8-h time points with time zero yielded P values of <0.0005. The difference between the 0-h and 2-h time points was not statistically significant (P = 0.5178). (C) Salmonella infection induces COX-2 expression during coculture of macrophages and hepatocytes in the presence of pregnenolone. Averages of the relative mRNA levels in uninfected wells (time zero) were normalized to 1, and the results for individual wells, uninfected and infected, were adjusted accordingly. Averaged results are shown. Bars indicate the standard errors of the means. Five wells were used for the first time point (time zero), and three wells were used for all other time points. Comparisons between the 0-h time point and all other time points yielded P values of <0.0001.
Other pathways significantly affected by infection.
Besides steroid and eicosanoid hormone metabolism, many other metabolic functions were deeply affected by Salmonella infection. One of the most significantly affected pathways was that of primary bile acid synthesis. Out of the 47 metabolites annotated by KEGG in the primary bile acid synthesis pathway, 12 potential metabolites showed increased levels in the feces of infected animals (Fig. 2). Bile acids are synthesized in the liver, stored in the gallbladder, and excreted in the lumen of the small intestine. That the levels of primary bile acids in feces were increased by infection suggests that these compounds are being either synthesized or excreted at higher rates upon infection. The mechanisms leading to increased bile acid levels in feces in response to Salmonella infection are currently being investigated.
Another metabolic function that was drastically affected by Salmonella infection is sugar metabolism. At least 8 KEGG categories of sugar metabolism showed significant changes upon infection. All of the categories were affected only in livers and showed decreases in metabolite levels in infected samples (Fig. 2). Although the reason for such is unclear, we hypothesize that decreased sugar levels in the livers of mice infected for 5 days may reflect the high energy expenditure that is required to fight the infectious process. Additionally, it is possible that changes in sugar metabolism were a consequence of the effect of Salmonella infection on hormone signaling, since some of the hormones affected are known regulators of carbohydrate metabolism (38).
DISCUSSION
In the recent past, microbes have been studied using several “omics” techniques, such as genomics, transcriptomics, and proteomics. More recently, metabolomics has been established as a new omics methodology that is based on the analysis of small metabolites to study changes in biological properties (46). Undeniably, the use of omics methodologies has provided us with important information about many biological systems. However, although each of these approaches has its strengths, each and every one of them also has particular drawbacks. For this report, we performed a metabolomic study of the effect of Salmonella infection on host metabolism. Our results indicated that Salmonella infection causes a significant disturbance in the hormonal balance of the host system. Many hormones were affected by infection, and some of these effects could be attributed to the transcriptional regulation of enzymes involved in hormone synthesis. However, in one case the results from our transcriptional data were the opposite of what one would expect based on the metabolomic analyses. Although the levels of many steroid hormones were increased by Salmonella infection, many of the steroid synthesis enzymes studied were unaffected. Even more striking, 3βHSD2, a major enzyme involved in the synthesis of all steroids, was strongly repressed by infection. These results highlight that, when used alone, transcriptional data can be deceiving, and it is important to study the end products of pathways in order to fully understand variations in metabolic activity. Our results also indicate that it is imperative that multiple methodologies be applied if one is to gain a complete understanding of the impact of infection on host metabolism. If we had approached our research simply by using a transcriptome analysis, we would have found that steroid synthesis is mostly unaffected (or even repressed) during Salmonella infection, while we now know, based on our metabolomics results, that steroid synthesis is actually significantly induced upon infection. In that sense, DI-FT-ICR MS represents a powerful technique to be used either as confirmation of other analyses or for the identification of phenotypes that could be missed by other techniques. As with any other methodology, DI-FT-ICR MS does have its weaknesses. For instance, although we filtered our data to eliminate some ion adducts from our final mass lists, many adducts exist, and it is likely that some of the masses identified in our study represent such adducts. Additionally, DI-FT-ICR MS relies solely on accurate mass determination for presumptive metabolite identification. Because several metabolites can have identical masses, it is possible that some of the metabolites that were identified in our analysis as affected by infection were not truly affected but represent false positives generated during our database searches from other compounds with identical masses that were truly affected. This is exemplified by a few discrepancies between our MS and ELISA data on hormone levels. Limitations notwithstanding, the use of DI-FT-ICR MS provided us with hypotheses that could be tested through other methodologies. DI-FT-ICR MS results cannot be used as definite measurements of metabolite abundance but rather as platforms for the design of additional experiments that address specific questions about metabolic pathway disturbances.
The gastrointestinal tract is a highly complex environment, and both the host and its resident microbiota contribute to the shaping of its chemical composition. Salmonella infection has been shown to significantly impact the microbial ecology of the gastrointestinal tract (28, 33). Therefore, we expect that some of the changes in host metabolism caused by infection are a consequence of the impact of infection on the intestinal microbiota. Supporting an important role of the intestinal microbiota in shaping the chemical composition of the intestinal environment, we have recently reported that antibiotic treatment elicits major changes in the mammalian intestinal metabolome (1). Teasing out the metabolic changes that are direct consequences of Salmonella infection from those that are caused by a disturbance of the microbial ecology of the gut will require significant effort.
Hormones have crucial functions in maintaining host metabolic homeostasis. Eicosanoids have many important biological functions, such as the control of vasoconstriction, platelet aggregation, immune responses, and others (3, 9). The effect of infection on eicosanoid metabolism has been studied in considerable detail. For instance, Staphylococcus aureus has been shown to induce the production of PGE2 in primary osteoblasts (31). Chlamydia trachomatis infection also induces COX-2 expression and PGE2 production in cultured epithelial cells (8). The infection of cultured macrophages with Salmonella has also been shown to induce the expression of COX-2 mRNA and protein, as well as the production of PGE2 and PGI2 (29, 36). Although some information regarding the impact of Salmonella infection on eicosanoid metabolism is available, our results show that this effect is much more profound than previously appreciated. In our studies, Salmonella disrupted virtually every branch of the eicosanoid metabolic pathway. Interestingly, specific enzymes were targeted in unique ways, with some being induced and some repressed. Although bacterial endotoxins are capable of stimulating prostaglandin production (45), a Salmonella mutant with a reduced capacity to affect PG metabolism has been identified (36), suggesting that this effect is specific and not a generalized response to infection.
The consequences of the effects of Salmonella infection on PG metabolism are not completely known. PGs can have both pro- and anti-inflammatory effects (6, 20, 24, 27, 44). One likely reason for the effect of Salmonella on these hormone pathways is to modulate inflammatory responses. Whether this is a host effort to control infection or a bacterial effort to benefit from host activities remains to be determined. The fact that one particular type three-secreted effector in the Salmonella pathogenicity island 2 is involved in this process (36) suggests that Salmonella actively induces this phenomenon, although the molecular details of this process are still unknown. Also, the addition of a COX-2 inhibitor to macrophages infected with Salmonella causes a reduction in bacterial loads (36), suggesting that Salmonella can benefit from the activation of PG production.
Steroid hormones are involved in a variety of physiological processes, including sex differentiation, sugar metabolism, cholesterol metabolism, immune regulation, and others (35, 38). Many steroids have anti-inflammatory properties, and steroid production during inflammation is part of an important negative feedback circuit that prevents overt host immune responses (35). Nevertheless, our current knowledge of the relationship between bacterial infections and steroid hormone metabolism is lacking. For instance, lipopolysaccharide (LPS) can induce the production of steroid hormones (41), and Shigella flexneri infection of human volunteers causes significant changes in fecal steroid profiles (16). However, human volunteers infected with Vibrio cholerae or pathogenic Escherichia coli showed no changes in steroid levels (14, 15). Likewise, the effects of steroid hormones on the progression and outcome of bacterial infections are varied. Treatment with the glucocorticoid analogue dexamethasone increases Salmonella colonization during infection of mice (30), although it caused reduced mortality in humans with typhoid fever (13). Conversely, Cooles has shown a significant association between the use of steroids during the treatment of typhoid fever and increased relapse rates (4). Our studies indicate that Salmonella disturbs steroid metabolism profoundly. Findings from our metabolomics and ELISAs suggest that fecal and hepatic steroid levels increase upon Salmonella infection. However, we found that the expression of two major enzymes involved in steroid synthesis is repressed by infection in mouse livers. This suggests that the increased steroid levels observed in livers and feces are caused by increased steroid production at other sites. The repression of 3βHSD2 may represent a strategy employed by the host or Salmonella to halt local hormone synthesis to compensate for the increased systemic levels of steroids.
To our knowledge, this is the first comprehensive study of the effect of Salmonella infection on host hormones. The consequences of these host-pathogen interactions are mostly unknown. They could be seen as host mechanisms to control immune responses against the bacterial infection as well as bacterial mechanisms to modulate inflammation. Intriguingly, studies have shown that Salmonella and other intestinal pathogens can benefit from host inflammatory responses (21, 33). Additionally, it is possible that changes in the levels of these hormones can have a direct impact on bacterial cells. A few mammalian hormones have been shown to affect Salmonella gene expression through bacterial adrenergic receptors (2, 17, 23, 32). Whether Salmonella can also sense eicosanoid and steroid hormones remains to be determined. Nevertheless, our results have raised interesting questions about the role of hormone pathways in host-pathogen interactions during Salmonella infection. We speculate that the balance between the effects of infection on these two pathways is important in determining the severity of disease. On one hand, the host needs to activate inflammatory responses to combat infection. However, it also needs to protect itself from the destructive effects of an uncontrolled inflammatory response. At this point, it is unclear which of these hormonal responses is a consequence of active mechanisms used by Salmonella to cause disease or by the host to defend itself. This work provides a platform for more detailed studies of the molecular interactions between Salmonella and these pathways, as well as many others.
Acknowledgments
We thank the anonymous scientists who reviewed the manuscript for their constructive criticism and members of the Finlay lab for critical reading of the manuscript.
This work was funded by the Canadian Institutes of Health Research, Crohn's and Colitis Foundation of Canada, Genome Canada, and Genome British Columbia. L.C.M.A. is supported by postdoctoral fellowships from the Department of Foreign Affairs and International Trade Canada and the Canadian Institutes of Health Research. E.T.A. is supported by the University Graduate Fellowship and the Armauer Hansen Memorial Fellowship. A.M. is supported by postdoctoral fellowships from the Michael Smith Foundation for Health Research, Genome British Columbia, and the Natural Sciences and Engineering Research Council of Canada. R.B.R.F. is funded by a postdoctoral fellowship from the Canadian Institutes of Health Research. M.M.C.B. is supported by a graduate scholarship from the National Sciences and Engineering Research Council of Canada. B.B.F. is an HHMI International Research Scholar and the University of British Columbia Peter Wall Distinguished Professor.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 14 February 2011.
REFERENCES
- 1.Antunes, L. C. M., et al. 7 February 2011. The effect of antibiotic treatment on the intestinal metabolome. Antimicrob. Agents Chemother. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed]
- 2.Bearson, B. L., and S. M. Bearson. 2008. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb. Pathog. 44:271-278. [DOI] [PubMed] [Google Scholar]
- 3.Calder, P. C. 2009. Polyunsaturated fatty acids and inflammatory processes: new twists in an old tale. Biochimie 91:791-795. [DOI] [PubMed] [Google Scholar]
- 4.Cooles, P. 1986. Adjuvant steroids and relapse of typhoid fever. J. Trop. Med. Hyg. 89:229-231. [PubMed] [Google Scholar]
- 5.Daynes, R. A., B. A. Araneo, J. Hennebold, E. Enioutina, and H. H. Mu. 1995. Steroids as regulators of the mammalian immune response. J. Investig. Dermatol. 105:14S-19S. [DOI] [PubMed] [Google Scholar]
- 6.Dugo, L., M. Collin, S. Cuzzocrea, and C. Thiemermann. 2004. 15d-prostaglandin J2 reduces multiple organ failure caused by wall-fragment of Gram-positive and Gram-negative bacteria. Eur. J. Pharmacol. 498:295-301. [DOI] [PubMed] [Google Scholar]
- 7.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuda, E. Y., S. P. Lad, D. P. Mikolon, M. Iacobelli-Martinez, and E. Li. 2005. Activation of lipid metabolism contributes to interleukin-8 production during Chlamydia trachomatis infection of cervical epithelial cells. Infect. Immun. 73:4017-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funk, C. D. 2001. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 294:1871-1875. [DOI] [PubMed] [Google Scholar]
- 10.Ghayee, H. K., and R. J. Auchus. 2007. Basic concepts and recent developments in human steroid hormone biosynthesis. Rev. Endocr. Metab. Disord. 8:289-300. [DOI] [PubMed] [Google Scholar]
- 11.Han, J., et al. 2008. Towards high-throughput metabolomics using ultrahigh-field Fourier transform ion cyclotron resonance mass spectrometry. Metabolomics 4:128-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haraga, A., M. B. Ohlson, and S. I. Miller. 2008. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6:53-66. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman, S. L., et al. 1984. Reduction of mortality in chloramphenicol-treated severe typhoid fever by high-dose dexamethasone. N. Engl. J. Med. 310:82-88. [DOI] [PubMed] [Google Scholar]
- 14.Huang, C. T., M. M. Levine, G. S. Daoud, D. R. Nalin, and B. L. Nichols. 1982. Fecal steroids in diarrhea: IV. cholera. Lipids 17:612-616. [DOI] [PubMed] [Google Scholar]
- 15.Huang, C. T., M. M. Levine, G. S. Daoud, D. R. Nalin, and B. L. Nichols. 1980. Fecal steroids in diarrhea. III. Experimentally-induced travelers' diarrhea. Am. J. Clin. Nutr. 33:40-44. [DOI] [PubMed] [Google Scholar]
- 16.Huang, C. T., W. E. Woodward, R. B. Hornick, J. T. Rodriguez, and B. L. Nichols. 1976. Fecal steroids in diarrhea. I. Acute shigellosis. Am. J. Clin. Nutr. 29:949-955. [DOI] [PubMed] [Google Scholar]
- 17.Hughes, D. T., M. B. Clarke, K. Yamamoto, D. A. Rasko, and V. Sperandio. 2009. The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC). PLoS Pathog. 5:e1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibarra, J. A., and O. Steele-Mortimer. 2009. Salmonella—the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell. Microbiol. 11:1579-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansson, J., et al. 2009. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS One 4:e6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawahito, Y., et al. 2000. 15-Deoxy-delta(12,14)-PGJ(2) induces synoviocyte apoptosis and suppresses adjuvant-induced arthritis in rats. J. Clin. Invest. 106:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupp, C., et al. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:119-129. [DOI] [PubMed] [Google Scholar]
- 22.Menendez, A., et al. 2009. Salmonella infection of gallbladder epithelial cells drives local inflammation and injury in a model of acute typhoid fever. J. Infect. Dis. 200:1703-1713. [DOI] [PubMed] [Google Scholar]
- 23.Moreira, C. G., D. Weinshenker, and V. Sperandio. 2010. QseC mediates Salmonella enterica serovar Typhimurium virulence in vitro and in vivo. Infect. Immun. 78:914-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morishita, M., M. Miyagi, and Y. Iwamoto. 1999. Effects of sex hormones on production of interleukin-1 by human peripheral monocytes. J. Periodontol. 70:757-760. [DOI] [PubMed] [Google Scholar]
- 25.Olszewski, K. L., et al. 2009. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe 5:191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raschke, W. C., S. Baird, P. Ralph, and I. Nakoinz. 1978. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell 15:261-267. [DOI] [PubMed] [Google Scholar]
- 27.Rocklin, R. E., K. Bendtzen, and D. Greineder. 1980. Mediators of immunity: lymphokines and monokines. Adv. Immunol. 29:55-136. [DOI] [PubMed] [Google Scholar]
- 28.Sekirov, I., et al. 2008. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76:4726-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi, L., et al. 2009. Proteomic investigation of the time course responses of RAW 264.7 macrophages to infection with Salmonella enterica. Infect. Immun. 77:3227-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smyth, T., et al. 2008. Dexamethasone modulates Salmonella enterica serovar Typhimurium infection in vivo independently of the glucocorticoid-inducible protein annexin-A1. FEMS Immunol. Med. Microbiol. 54:339-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Somayaji, S. N., S. Ritchie, M. Sahraei, I. Marriott, and M. C. Hudson. 2008. Staphylococcus aureus induces expression of receptor activator of NF-κB ligand and prostaglandin E2 in infected murine osteoblasts. Infect. Immun. 76:5120-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spencer, H., et al. 2010. Genome-wide transposon mutagenesis identifies a role for host neuroendocrine stress hormones in regulating the expression of virulence genes in Salmonella. J. Bacteriol. 192:714-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stecher, B., et al. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5:2177-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suhre, K., and P. Schmitt-Kopplin. 2008. MassTRIX: mass translator into pathways. Nucleic Acids Res. 36:W481-W484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tait, A. S., C. L. Butts, and E. M. Sternberg. 2008. The role of glucocorticoids and progestins in inflammatory, autoimmune, and infectious disease. J. Leukoc. Biol. 84:924-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchiya, K., and T. Nikai. 2004. Salmonella enterica serovar Typhimurium infection induces cyclooxygenase 2 expression in macrophages: involvement of Salmonella pathogenicity island 2. Infect. Immun. 72:6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valdez, Y., R. B. Ferreira, and B. B. Finlay. 2009. Molecular mechanisms of Salmonella virulence and host resistance. Curr. Top. Microbiol. Immunol. 337:93-127. [DOI] [PubMed] [Google Scholar]
- 38.Vinson, G. P. 2009. The adrenal cortex and life. Mol. Cell. Endocrinol. 300:2-6. [DOI] [PubMed] [Google Scholar]
- 39.Waldram, A., et al. 2009. Top-down systems biology modeling of host metabotype-microbiome associations in obese rodents. J. Proteome Res. 8:2361-2375. [DOI] [PubMed] [Google Scholar]
- 40.Wang, Y., et al. 2008. Analysis of porcine transcriptional response to Salmonella enterica serovar Choleraesuis suggests novel targets of NFkappaB are activated in the mesenteric lymph node. BMC Genomics 9:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolff, J., and G. H. Cook. 1975. Endotoxic lipopolysaccharides stimulate steroidogenesis and adenylate cyclase in adrenal tumor cells. Biochim. Biophys. Acta 413:291-297. [DOI] [PubMed] [Google Scholar]
- 42.Wray, C., and W. J. Sojka. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25:139-143. [PubMed] [Google Scholar]
- 43.Wu, J. C., G. Merlino, and N. Fausto. 1994. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc. Natl. Acad. Sci. U. S. A. 91:674-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue, L., et al. 2005. Prostaglandin D2 causes preferential induction of proinflammatory Th2 cytokine production through an action on chemoattractant receptor-like molecule expressed on Th2 cells. J. Immunol. 175:6531-6536. [DOI] [PubMed] [Google Scholar]
- 45.Yaron, M., I. Yaron, O. Smetana, E. Eylan, and U. Zor. 1980. Stimulation of prostaglandin E production by bacterial endotoxins in cultured human synovial fibroblasts. Arthritis Rheum. 23:921-925. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, W., F. Li, and L. Nie. 2010. Integrating multiple ‘omics’ analysis for microbial biology: application and methodologies. Microbiology 156:287-301. [DOI] [PubMed] [Google Scholar]