Abstract
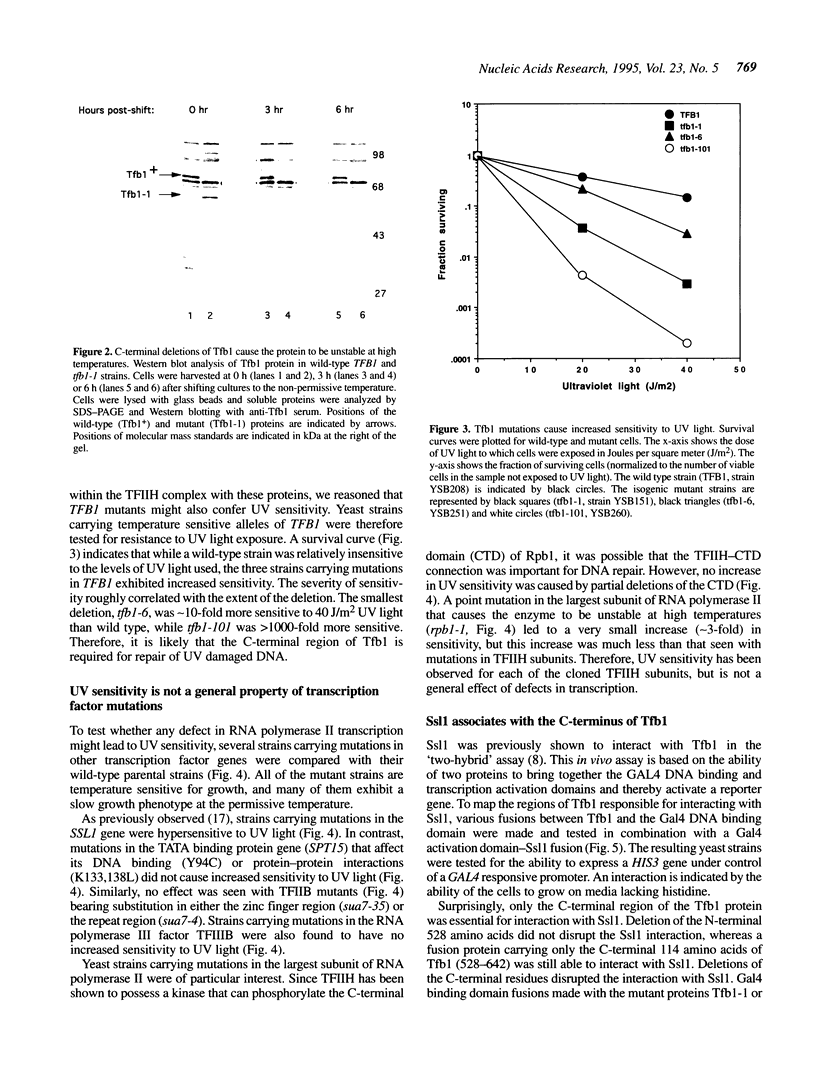
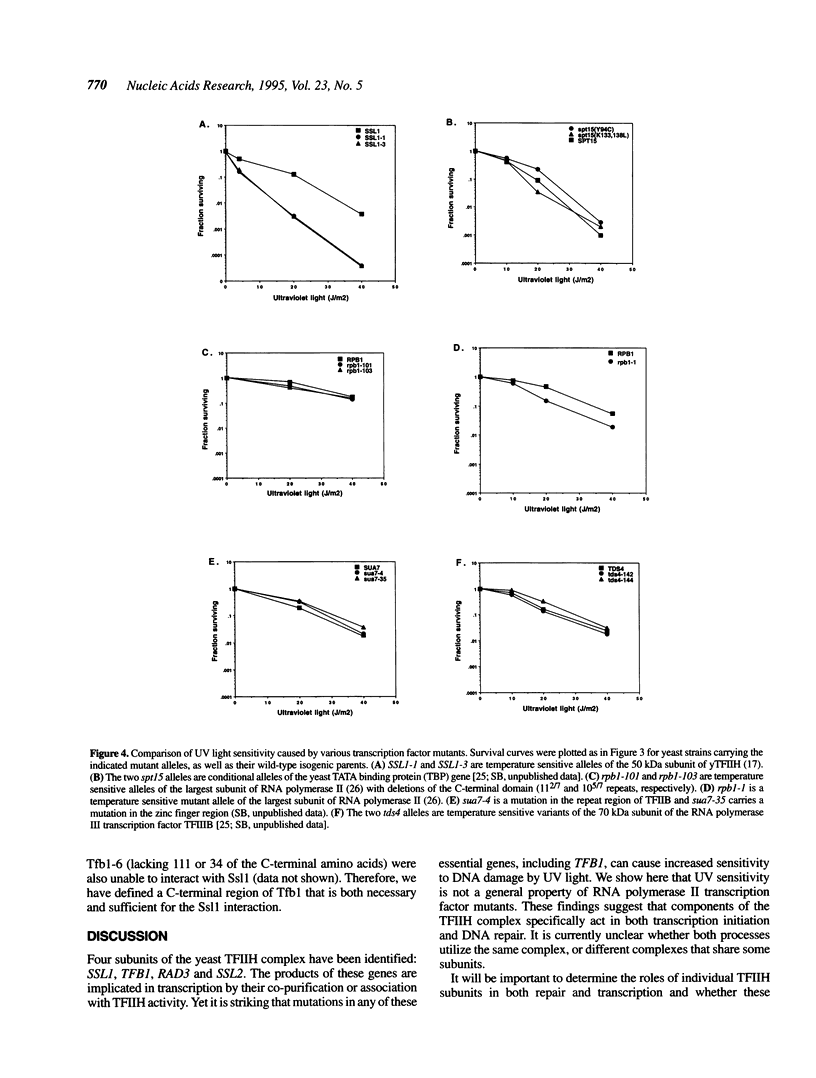
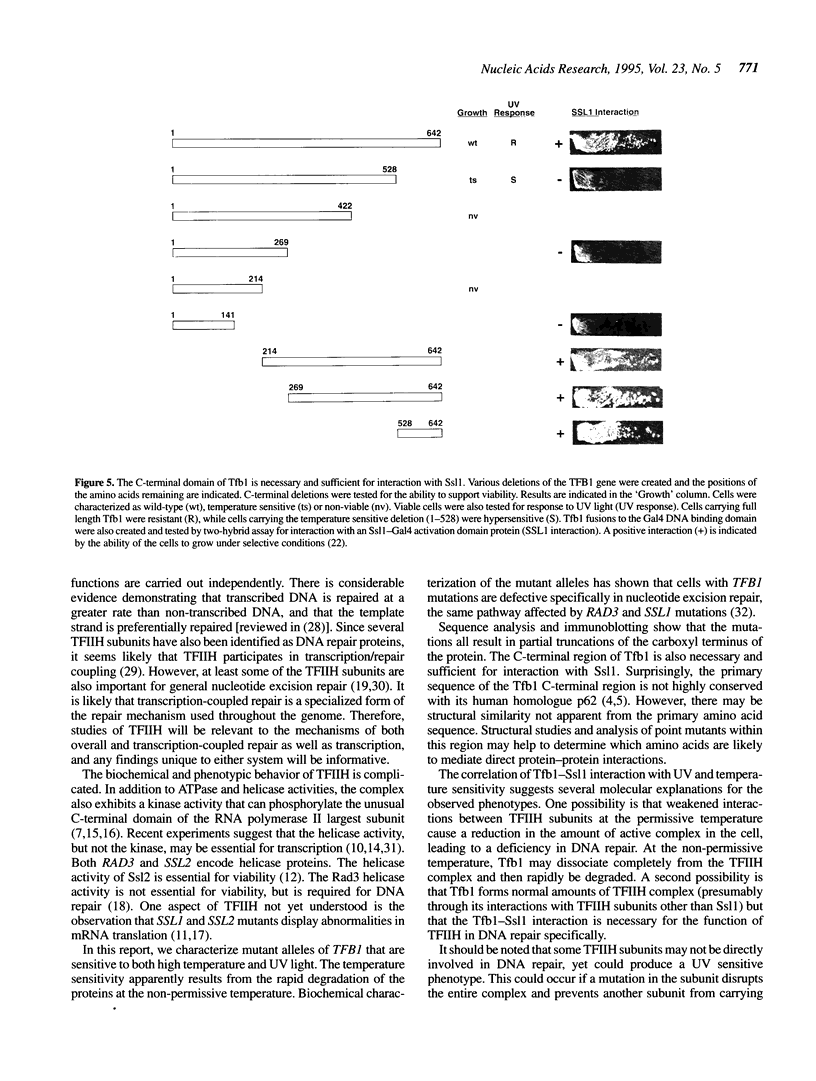
Yeast transcription factor TFIIH (also known as factor b) is a component of the RNA polymerase II initiation complex. Several TFIIH subunits (RAD3, SSL2 and SSL1) have also been implicated in DNA repair. Ssl1 interacts directly with another TFIIH subunit, Tfb1, which has not previously been shown to have a role in DNA repair. We isolated mutations in TFB1 that lead to a temperature sensitive phenotype. These mutations result in C-terminal truncations of the Tfb1 protein and disrupt its interaction with Ssl1. The C-terminal 114 amino acids of Tfb1 are necessary and sufficient for this interaction. Interestingly, cells carrying these truncations in Tfb1 cause sensitivity to ultraviolet (UV) light induced DNA damage, as previously observed for mutations in RAD3, SSL1 and SSL2. Many other mutations in RNA polymerase II basal factors were tested and found not to cause an increase in UV sensitivity, indicating that this phenotype is not due to a general defect in transcription.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Buratowski S. DNA repair and transcription: the helicase connection. Science. 1993 Apr 2;260(5104):37–38. doi: 10.1126/science.8465198. [DOI] [PubMed] [Google Scholar]
- Buratowski S. The basics of basal transcription by RNA polymerase II. Cell. 1994 Apr 8;77(1):1–3. doi: 10.1016/0092-8674(94)90226-7. [DOI] [PubMed] [Google Scholar]
- Buratowski S., Zhou H. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992 Oct 16;71(2):221–230. doi: 10.1016/0092-8674(92)90351-c. [DOI] [PubMed] [Google Scholar]
- Buratowski S., Zhou H. Transcription factor IID mutants defective for interaction with transcription factor IIA. Science. 1992 Feb 28;255(5048):1130–1132. doi: 10.1126/science.1546314. [DOI] [PubMed] [Google Scholar]
- Conaway R. C., Conaway J. W. General initiation factors for RNA polymerase II. Annu Rev Biochem. 1993;62:161–190. doi: 10.1146/annurev.bi.62.070193.001113. [DOI] [PubMed] [Google Scholar]
- Durfee T., Becherer K., Chen P. L., Yeh S. H., Yang Y., Kilburn A. E., Lee W. H., Elledge S. J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993 Apr;7(4):555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Feaver W. J., Gileadi O., Li Y., Kornberg R. D. CTD kinase associated with yeast RNA polymerase II initiation factor b. Cell. 1991 Dec 20;67(6):1223–1230. doi: 10.1016/0092-8674(91)90298-d. [DOI] [PubMed] [Google Scholar]
- Feaver W. J., Svejstrup J. Q., Bardwell L., Bardwell A. J., Buratowski S., Gulyas K. D., Donahue T. F., Friedberg E. C., Kornberg R. D. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell. 1993 Dec 31;75(7):1379–1387. doi: 10.1016/0092-8674(93)90624-y. [DOI] [PubMed] [Google Scholar]
- Fischer L., Gerard M., Chalut C., Lutz Y., Humbert S., Kanno M., Chambon P., Egly J. M. Cloning of the 62-kilodalton component of basic transcription factor BTF2. Science. 1992 Sep 4;257(5075):1392–1395. doi: 10.1126/science.1529339. [DOI] [PubMed] [Google Scholar]
- Flores O., Lu H., Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Identification and characterization of factor IIH. J Biol Chem. 1992 Feb 5;267(4):2786–2793. [PubMed] [Google Scholar]
- Gerard M., Fischer L., Moncollin V., Chipoulet J. M., Chambon P., Egly J. M. Purification and interaction properties of the human RNA polymerase B(II) general transcription factor BTF2. J Biol Chem. 1991 Nov 5;266(31):20940–20945. [PubMed] [Google Scholar]
- Gileadi O., Feaver W. J., Kornberg R. D. Cloning of a subunit of yeast RNA polymerase II transcription factor b and CTD kinase. Science. 1992 Sep 4;257(5075):1389–1392. doi: 10.1126/science.1445600. [DOI] [PubMed] [Google Scholar]
- Gulyas K. D., Donahue T. F. SSL2, a suppressor of a stem-loop mutation in the HIS4 leader encodes the yeast homolog of human ERCC-3. Cell. 1992 Jun 12;69(6):1031–1042. doi: 10.1016/0092-8674(92)90621-i. [DOI] [PubMed] [Google Scholar]
- Hanawalt P., Mellon I. Stranded in an active gene. Curr Biol. 1993 Jan;3(1):67–69. doi: 10.1016/0960-9822(93)90156-i. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J. H. Nucleotide excision repair I: from E. coli to yeast. Trends Genet. 1993 May;9(5):173–177. doi: 10.1016/0168-9525(93)90164-d. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J. H. Nucleotide excision repair. II: From yeast to mammals. Trends Genet. 1993 Jun;9(6):211–217. doi: 10.1016/0168-9525(93)90121-w. [DOI] [PubMed] [Google Scholar]
- Humbert S., van Vuuren H., Lutz Y., Hoeijmakers J. H., Egly J. M., Moncollin V. p44 and p34 subunits of the BTF2/TFIIH transcription factor have homologies with SSL1, a yeast protein involved in DNA repair. EMBO J. 1994 May 15;13(10):2393–2398. doi: 10.1002/j.1460-2075.1994.tb06523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Zawel L., Fisher L., Egly J. M., Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992 Aug 20;358(6388):641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- Nonet M. L., Young R. A. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989 Dec;123(4):715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Guzder S. N., Koken M. H., Jaspers-Dekker I., Weeda G., Hoeijmakers J. H., Prakash S., Prakash L. RAD25 (SSL2), the yeast homolog of the human xeroderma pigmentosum group B DNA repair gene, is essential for viability. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11416–11420. doi: 10.1073/pnas.89.23.11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I., Bell B., Broad P., Hollis M. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992 Sep 1;118(1):137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- Schaeffer L., Moncollin V., Roy R., Staub A., Mezzina M., Sarasin A., Weeda G., Hoeijmakers J. H., Egly J. M. The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription factor. EMBO J. 1994 May 15;13(10):2388–2392. doi: 10.1002/j.1460-2075.1994.tb06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer L., Roy R., Humbert S., Moncollin V., Vermeulen W., Hoeijmakers J. H., Chambon P., Egly J. M. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993 Apr 2;260(5104):58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- Serizawa H., Conaway J. W., Conaway R. C. Phosphorylation of C-terminal domain of RNA polymerase II is not required in basal transcription. Nature. 1993 May 27;363(6427):371–374. doi: 10.1038/363371a0. [DOI] [PubMed] [Google Scholar]
- Serizawa H., Conaway R. C., Conaway J. W. A carboxyl-terminal-domain kinase associated with RNA polymerase II transcription factor delta from rat liver. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7476–7480. doi: 10.1073/pnas.89.16.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa H., Conaway R. C., Conaway J. W. Multifunctional RNA polymerase II initiation factor delta from rat liver. Relationship between carboxyl-terminal domain kinase, ATPase, and DNA helicase activities. J Biol Chem. 1993 Aug 15;268(23):17300–17308. [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Higgins D., Prakash L., Prakash S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 1988 Oct;7(10):3263–3269. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H., Miller S. P., Pabich E. K., Donahue T. F. SSL1, a suppressor of a HIS4 5'-UTR stem-loop mutation, is essential for translation initiation and affects UV resistance in yeast. Genes Dev. 1992 Dec;6(12B):2463–2477. doi: 10.1101/gad.6.12b.2463. [DOI] [PubMed] [Google Scholar]






