Abstract
Natural transformation by competence is a major mechanism of horizontal gene transfer in bacteria. Competence is defined as the genetically programmed physiological state that enables bacteria to actively take up DNA from the environment. The conditions that signal competence development are multiple and elusive, complicating the understanding of its evolutionary significance. We used expression of the competence gene comEA as a reporter of competence development and screened several hundred molecules for their ability to induce competence in the freshwater living pathogen Legionella pneumophila. We found that comEA expression is induced by chronic exposure to genotoxic molecules such as mitomycin C and antibiotics of the fluoroquinolone family. These results indicated that, in L. pneumophila, competence may be a response to genotoxic stress. Sunlight-emitted UV light represents a major source of genotoxic stress in the environment and we found that exposure to UV radiation effectively induces competence development. For the first time, we show that genetic exchanges by natural transformation occur within an UV-stressed population. Genotoxic stress induces the RecA-dependent SOS response in many bacteria. However, genetic and phenotypic evidence suggest that L. pneumophila lacks a prototypic SOS response and competence development in response to genotoxic stress is RecA independent. Our results strengthen the hypothesis that competence may have evolved as a DNA damage response in SOS-deficient bacteria. This parasexual response to DNA damage may have enabled L. pneumophila to acquire and propagate foreign genes, contributing to the emergence of this human pathogen.
Bacterial competence is a genetically programmed physiological state that confers the ability to take up DNA from the environment and allows subsequent genetic and phenotypic transformation. The competent state involves expression of a dedicated, well-conserved DNA uptake machinery required to actively import exogenous DNA (9). Bacterial competence allows for uptake and recombination of DNA released by bacteria and can promote genetic exchange among individuals within a population. Most bacteria possess competence genes but the conditions or signals triggering competence are multiple and often species specific (25). Expression of competence is influenced by the growth phase, cell density, metabolic activity, and nutritional stress, and in the well-studied Gram-positive organisms Bacillus subtilis and Streptococcus pneumoniae it is believed to be a general response to stress (10). In S. pneumoniae, competence is induced by antibiotic stress, including DNA-damaging antibiotics (36). Competence development in Gram-negative bacteria is less understood and is induced by different conditions, for example, by starvation for Haemophilus influenzae (21) or by growth on a specific carbon source (chitin) for Vibrio cholerae (31). For a specific organism it is therefore difficult to predict and identify the conditions that lead to competence development. Legionella pneumophila is a Gram-negative pathogen that targets the alveolar macrophage in its human host or the unicellular protozoa in its natural environment. Sequencing of several Legionella pneumophila genomes revealed high genetic diversity within the species and the presence of multiple virulence-associated eukaryotic-like genes thought to have been acquired by horizontal gene transfer (4, 5, 8, 10a). These genes are often found within hypervariable regions (34, 46), and the ability of L. pneumophila to import foreign DNA may offer a plausible mechanism for the acquisition and spread of DNA of eukaryotic origin (16). L. pneumophila has been shown to be naturally transformable at low frequency under microaerophilic conditions (41). However, the frequency of transformation can be increased after genetic alterations in the comR, proQ, and rnr genes, suggesting that L. pneumophila is capable of high transformability under conditions that have yet to be found (6, 40). Transcriptional profiling of constitutively competent L. pneumophila mutants (comR, proQ, and rnr) revealed comEA as the most differentially expressed gene in the competence state (6). ComEA is a small DNA-binding periplasmic protein important for DNA uptake (7, 35) and is indeed required for the natural transformation of L. pneumophila (6).
We show here that a comEA-gfp transcriptional fusion can be used as an indicator of competence development. This transcriptional reporter allowed us to systematically test hundreds of natural and synthetic molecules for their capacity to induce competence. We found that the human pathogen L. pneumophila expresses competence genes when exposed to DNA-damaging agents and conditions that can lead to genome instability. We show that exposure to UV radiation strongly stimulated the bacterium's ability to take up and integrate exogenous DNA. In addition, the expression of competence following genotoxic stress effectively promotes genetic exchange and recombination within a stressed population. We discuss the benefit of importing and integrating exogenous DNA when chromosome integrity is compromised.
MATERIALS AND METHODS
Strains, media, and plasmids.
The L. pneumophila strains used in the present study are derived from Philadelphia-1, Paris, or Lens isolates (Table 1). L. pneumophila strains were grown in liquid ACES [N-(2-acetamido)-2-aminoethanesulfonic acid]-buffered yeast extract (AYE) medium or ACES-buffered charcoal yeast extract (CYE) plates. For JR32-derived strains, chloramphenicol, kanamycin, and gentamicin were used at 5, 50, and 10 μg/ml, respectively. Kanamycin and gentamicin were used at 15 and 5 μg/ml, respectively, for Paris-derived strains. Deletion mutant of the JR32 and Paris strains were respectively generated by allelic exchange (43) and natural transformation (1). The gfp+ reporter plasmid pXDC42 was constructed by cloning an omega terminator followed by a promoterless gfp+ gene in a pMMB207C derivative. Plasmid pX3 is an improved version of pXDC42 with two additional omega terminators upstream of the gfp+ gene. Reporter constructs for the comEA, pilE, rplU, and rpmB genes were constructed by cloning the corresponding predicted open reading frame and putative promoter sequence (200-nucleotide upstream sequence) in front of the promoterless gfp+ gene in pXDC42 (comEA) or pX3 (pilE, rplU, and rpmB).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description and/or genotypea | Source or reference |
|---|---|---|
| Strains | ||
| Philadelphia-1 | Isolate from the 1976 Philadelphia outbreak | |
| JR32 | Philadelphia-1; Smr, r− m+ | |
| Paris | Outbreak isolate CIP107629 | CNR Lyon |
| Lens | Outbreak isolate CIP108286 | CNR Lyon |
| Paris ΔcomEC | ΔcomEC::aptII; Knr | This study |
| Paris ΔcomEC | ΔcomEC::aac1; Gmr | This study |
| Paris ΔrecA | ΔrecA::aptII; Knr | This study |
| Paris Δlpp3026 | Δlpp3026::aptII; Knr | This study |
| Paris Δlpp2764 | Δlpp2764::aac1; Gmr | This study |
| Plasmids | ||
| pXDC42 | RSF1010 derivative with promoterless gfp+ (Cmr; ΔmobA) | |
| pXDC91 | pXDC42 with comEA driving expression of gfp+ | |
| pX3 | pXDC42 with two Ω terminators upstream of gfp+ (Cmr; ΔmobA) | |
| pX3-pilE | pX3 with pilE driving expression of gfp+ | |
| pX3-rplU | pX3 with rplU driving expression of gfp+ | |
| pX3-rpmB | pX3 with rpmB driving expression of gfp+ |
Gmr, gentamicin resistance; Cmr, chloramphenicol resistance; Smr, streptomycin resistance; Knr, kanamycin resistance.
Screening of small molecules and detection of comEA-gfp expression.
Freshly grown L. pneumophila strains carrying the comEA-gfp reporter plasmid pXDC91 were resuspended from CYE plates in distilled water and overlaid on fresh CYE plates. Small molecules in dimethyl sulfoxide (ICCB Known Bioactives Library; Biomol) were spotted (1 μl) onto the plates. The plates were incubated at 37°C for 24 h, followed by exposure to long-range UV light (365 nm) to examine the expression of comEA-gfp. Additional molecules on paper disks were tested similarly. Induction of comEA-gfp by small molecules in liquid media was tested in 96-well plates containing serial dilutions of the tested molecules. Reporter strains from CYE plates were diluted to an initial optical density (OD) of 0.1 in AYE (100 μl/well), and the 96-well plate was placed inside a temperature-controlled plate reader (M200; Tecan) with intermittent shaking (100 rpm for 60 s every 10 min). Growth and expression of the green fluorescent protein (GFP) gene fusions were monitored by measuring the absorbance at 600 nm (A600) and fluorescence (F520; excitation, 485 nm; emission, 520 nm), respectively, every 10 min. Fold change in expression corresponds to the ratio of the maximal relative fluorescence (F520/A600) recorded under treated and untreated conditions. For exposure to UV radiation, reporter strains (JR32/pXDC91 or Paris/pXDC91) were resuspended at an OD of 2 in distilled water, after which, 250-μl aliquots of the suspension were placed in a sterile petri dish (without a lid) and exposed to various doses of 254-nm UV light (Stratalinker; Stratagene). An equal amount of 2× AYE broth was added to the irradiated cells, and the suspension was distributed in a 96-well plate (100 μl/well) at an OD of 1 or 0.1. The expression of comEA-gfp over time was monitored as described above and is reported as the ratio of fluorescence per cell density (F520/A600).
Natural transformation experiments.
Freshly grown L. pneumophila strains (24 h) from CYE plates were resuspended in distilled water at an OD of 1, and 250-μl aliquots were exposed to various doses of 254-nm UV light (Stratalinker; Stratagene). An equal amount of 2× AYE broth was added to the irradiated cells, and the suspension was placed in 24-well plates (1 ml/well), followed by incubation with shaking (150 rpm) at 37°C. After 6 h of incubation, 1 μg of a 5-kb PCR product of the lpp3026 gene interrupted with a kanamycin resistance marker and including 2 kb of each of the upstream and downstream flanking regions was added to the well. The culture was incubated at 37°C for another 18 h (24 h total). Serial dilutions of the cultures were then plated on CYE plates with or without selective agent. The transformation frequency represents the ratio of total CFU determined from plating on selective medium by the total CFU determined from plating on nonselective media. When indicated, DNase I was used at 100 μg/ml. The growth medium for the L. pneumophila culture was prepared by different laboratories with various sources of iron (iron sulfate, ferric nitrate, and iron pyrophosphate) and with the optional addition of alpha-ketoglutaric acid. Increases in transformation frequency caused by UV radiation were not significantly affected by either the iron source or the presence of alpha-ketoglutaric acid (see Fig. S3 in the supplemental material). The transformation frequency is affected by the freshness of the liquid AYE medium; for this reason, transformation experiments were performed with freshly prepared 2× AYE medium.
Mutagenesis experiments.
The frequency of mutation to rifampin resistance was determined under conditions found to induce competence. The L. pneumophila Paris and E. coli W3110 strains were exposed to various doses of UV light and inoculated into AYE medium and LB broth, respectively. After 24 h at 37°C, serial dilutions were plated on nonselective media to determine the total CFU and on plates containing rifampin (25 μg/ml for L. pneumophila and 100 μg/ml for E. coli). Mutation frequencies represent the ratio of CFU on selective versus nonselective media. Mutagenesis during chronic exposure to nalidixic acid was measured similarly. L. pneumophila strain Paris was inoculated at a starting OD of 0.2 and grown for 24 h in 1 ml of AYE broth containing various doses of nalidixic acid. Serial dilutions were then plated to determine the frequency of rifampin resistance.
Detection of comEA transcription by Northern blotting.
Northern blot detection of comEA expression was determined after UV exposure or during chronic exposure to norfloxacin under the same conditions used to determine mutation frequencies. After 24 h of growth in 1 ml of AYE, UV-irradiated cells or norfloxacin-exposed cells were collected by centrifugation, and the total RNA was extracted with 1 ml of TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Northern blot analyses were performed with denaturing Tris-borate-EDTA (TBE)-urea 6% acrylamide gel as previously described (6). Briefly, 2 μg of total RNA in denaturing buffer was loaded per lane and run in TBE buffer. The RNA was transferred to a nylon membrane (Immobilon-NY+; Millipore Corp., Billerica, MA) by electrophoretic transfer in 0.5× TBE buffer. The RNA was cross-linked to the membrane by UV irradiation. Membranes were hybridized at 42°C with a 45-nucleotide 5′-biotinylated oligonucleotide probe (5 nM) in ULTRAhyb ultrasensitive hybridization buffer (Ambion, Austin, TX) and then washed according to the manufacturer's instructions (Ambion). Membranes were developed using horseradish peroxidase-conjugated streptavidin and enhanced luminol substrate (chemiluminescent nucleic acid detection module; Pierce, Rockford, IL) and Biomax films (Kodak).
RESULTS
Identification of small molecules inducing expression of the competence gene comEA.
A transcriptional fusion of comEA to the green fluorescent protein gene gfp+ on a RSF1010 plasmid accurately reports the expression of comEA in previously described constitutively competent comR and proQ mutants (40) and can be detected on solid media (see Fig. S1 in the supplemental material). We sought to gain information about the conditions of competence development by testing the effects of small organic molecules with known targets on the expression of comEA-gfp fusion. When spotted onto solid media, small molecules conveniently form a gradient of concentration, allowing rapid screening of a wide range of concentration of specific molecules on a small surface. A total of 506 small molecules were spotted onto solid media previously overlaid with L. pneumophila carrying the comEA-gfp reporter construct and scored for their ability to inhibit growth and to induce comEA-gfp expression. The tested molecules include a collection of 480 bioactives molecules, as well as additional antibiotics, heavy metals, and other toxic molecules. Of all 506 tested molecules, 64 (12%) were found to be toxic to L. pneumophila and produced a zone of growth inhibition in a manner similar to an antibiogram (see Table S1 in the supplemental material). These toxic molecules are inducers of a wide range of stress, such as oxidative stress, inhibition of DNA replication, transcription, protein synthesis, cell wall synthesis, and proton and metal homeostasis (see Table S1 in the supplemental material). Of the 64 toxic molecules, only 6 (mitomycin C, norfloxacin, ofloxacin, nalidixic acid, bicyclomycin, and hydroxyurea) induced strong expression of the comEA-gfp (Fig. 1 A and see Table S1 in the supplemental material). All other tested antibiotics, including beta-lactams (ampicillin), aminoglycosides (kanamycin, streptomycin), and macrolides (erythromycin), were inactive, although they generated an inhibition zone (Fig. 1A and see Table S1 in the supplemental material). Also, no nontoxic molecules could induce comEA-gfp expression. Induction of comEA-gfp occurs at the edge of the inhibition zone and can be observed in the independent L. pneumophila isolates Philadelphia-1, Paris, and Lens (not shown), revealing a conserved response to exposure to these antibiotics (Fig. 1A and data not shown).
FIG. 1.
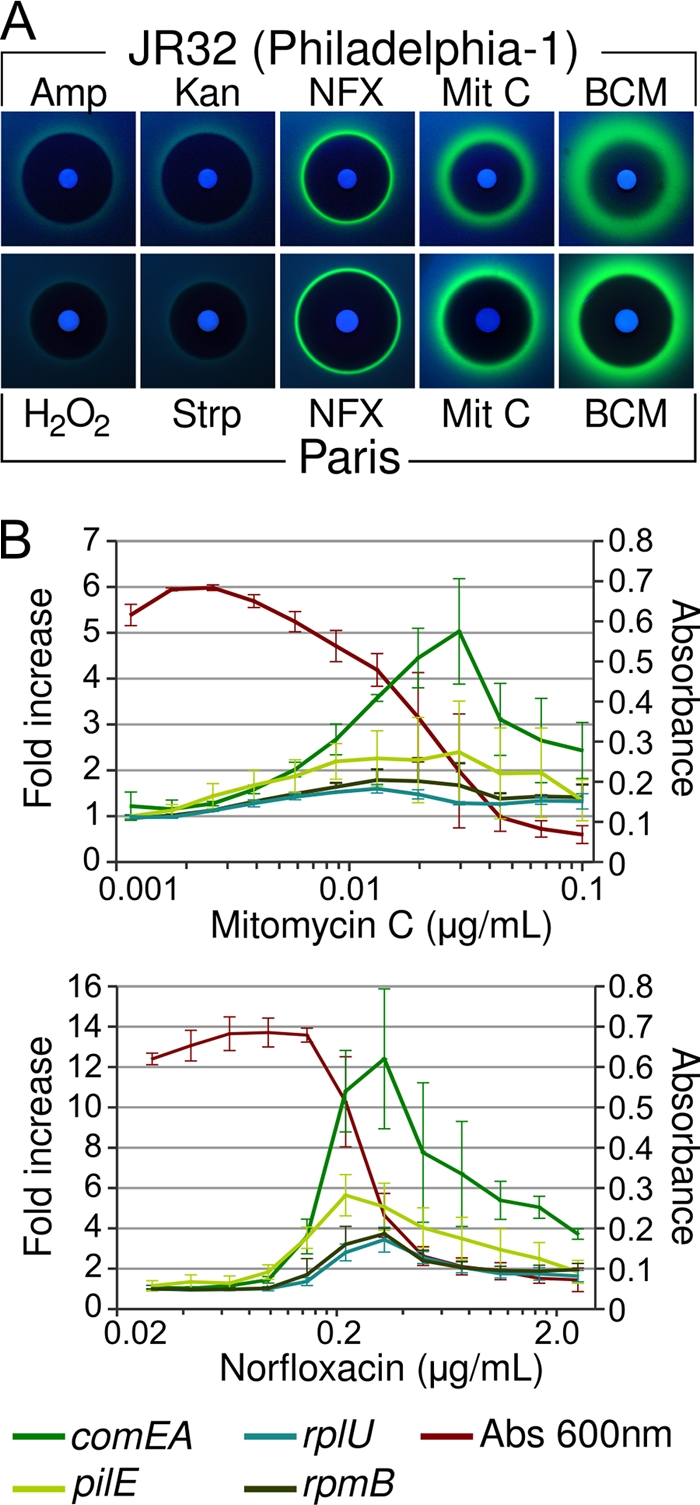
Expression of the competence gene comEA is induced by antibiotics and DNA-damaging agents. (A) L. pneumophila strains JR32 (a Philadelphia-1-derived strain) and Paris bearing the comEA-gfp reporter plasmid were grown on CYE plates with paper disks containing selected antibiotics and molecules. All shown molecules produce an inhibition zone (dark area around the disks). Mitomycin C (Mit C), norfloxacin (NFX), and bicyclomycin (BCM) but not hydrogen peroxide, streptomycin (Strp), and ampicillin (Amp) induce expression of comEA-gfp (green halo surrounding the inhibition zone). Photographs were taken with the same exposure time under long-UV light (365 nm). (B) Dose-response induction of comEA-gfp by norfloxacin and mitomycin C. L. pneumophila strain JR32 carrying plasmids with comEA-gfp, pilE-gfp, rplU-gfp, or rpmB-gfp fusions were grown to stationary phase in 96-well plates with increasing concentration of antibiotics. The maximum cell density (A600 [Abs 600 nm]) and maximum fold increase in gfp expression relative to cell density (GFP/Abs [A600]) are plotted as a function of antibiotic concentration. The maximum cell density displayed on the graphs corresponds to the strain carrying the comEA-gfp fusion. In both graphs, error bars represent standard deviations derived from three independent experiments.
Induction of comEA expression by antibiotics and DNA-damaging agents.
We further analyzed the dose response of comEA-gfp expression to the inducers (mitomycin C, norfloxacin, bicyclomycin, and hydroxyurea) in liquid media (Fig. 1B and see Fig. S2 in the supplemental material). Maximal induction of comEA-gfp occurs at concentrations that cause limited growth inhibition, suggesting that induction is stress related. In addition to the comEA-gfp fusions, we also tested the effect of norfloxacin and mitomycin C on additional gfp gene fusions (Fig. 1B). The rplU and rpmB gene are ribosomal protein-encoding genes whose expression is unchanged in constitutively competent mutants (6). The pilE gene is another gene upregulated during competence in L. pneumophila and may code for a putative type IV pilin similar to those commonly required for transformation in Gram-negative bacteria (6). Interestingly, rplU-gfp and rpmB-gfp are moderately induced by exposure to norfloxacin. This may be due to increased promoter activity induced by norfloxacin. Alternatively, this moderate induction may be the result of increased transcription caused by the altered topology of the reporter plasmid under these gyrase-inhibiting conditions. Importantly, the comEA-gfp gene fusion is more strongly upregulated than the control fusions (Fig. 1B). To a lesser extent, the pilE-gfp fusion is also induced by mitomycin C and norfloxacin. This is consistent with previous microarray data showing less induction of pilE compare to comEA in competent mutants (6).
All small molecules that induced expression of competence genes are known to cause a DNA replication stress. Mitomycin C is a DNA alkylation agent creating DNA lesions and arresting the DNA polymerase. Quinolones also block progression of DNA replication forks by inhibiting DNA gyrase, resulting in double-strand breaks (13). Hydroxyurea inhibits ribonucleotide reductase, lowers the pool of deoxynucleoside triphosphates (dNTPs), and slows down the DNA polymerase (27, 38). Bicyclomycin is a specific inhibitor of the Rho transcription terminator (26), which may increase the occurrence of R-loops and stalling of the replication fork, as in transcription termination-deficient mutants (20). In conclusion, induction of comEA-gfp appears to be a specific response to stress that causes stalling of the DNA replication fork and can lead to chromosome instability.
UV radiation caused the induction of comEA.
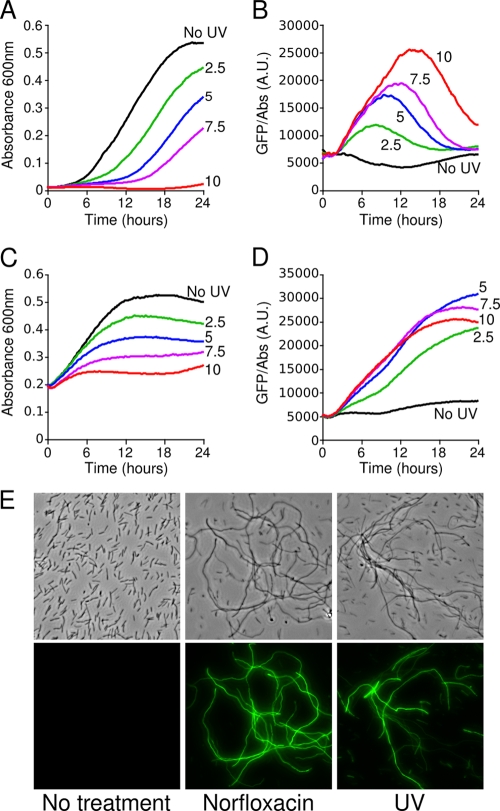
A nonchemical, well-characterized causal agent of DNA damage is exposure to UV radiation. UV radiation induces thymine dimers, which block the replication forks. L. pneumophila is a freshwater living organism and is likely to be exposed to UV radiation from natural sunlight. A Philadelphia-1-derived strain of L. pneumophila carrying the comEA-gfp fusion (JR32/pXDC91) was exposed to increasing doses of UV radiation (260 nm) and inoculated in rich medium at low or high cell densities. Bacterial growth and the expression of comEA-gfp were monitored every 10 min over a 24-h period (Fig. 2). The expression of comEA-gfp relative to the cell density is rather steady during bacterial growth in the absence of UV exposure. In contrast, exposure to UV radiation inhibited bacterial growth and induced the expression of comEA-gfp in a dose-dependent manner. The expression of comEA-gfp reaches a maximum when the growth of the bacterial population is the most affected and then decreases as the bacterial population returns to a fast growth rate. When UV-exposed L. pneumophila was inoculated at a higher density, comEA-gfp was also induced and remained at high levels, presumably because the culture could not reach exponential growth. Similar results were obtained with the Paris strain (data not shown). Observations by fluorescence microscopy confirmed a high induction of comEA-gfp by the quinolone norfloxacin, as well as by UV radiation, concomitant with cell filamentation (Fig. 2E). Most filamentous cells express elevated levels of comEA-gfp in response to norfloxacin or UV exposure (Fig. 2E). Filamentation is thought to be a consequence of cell division arrest and is a hallmark of a bacterial response to genotoxic stress. The checkpoint protein SulA inhibits cell division and is part of the SOS response (23). Since L. pneumophila lacks the sulA gene, filamentation may result here from the inhibition of cell division by an alternate mechanism.
FIG. 2.
UV-irradiated L. pneumophila expresses elevated levels of the competence gene comEA. The JR32 strain bearing the comEA-gfp reporter plasmid was exposed to increasing doses of 254-nm UV radiation (0, 2.5, 5.0, 7.5, and 10 J/m2) and then inoculated into rich medium in microtiter plates at low (A600 = 0.02, equivalent to OD600 = 0.1 [A and B]) or high (A600 = 0.2, equivalent to OD600 = 0.1 [C and D]) cell densities. Bacterial growth (A600 [A and C]) and comEA-gfp expression relative to cell density (GFP/OD, arbitrary units [B and D]) were measured every 10 min over a 24-h period. (E) Microscopic observation of individual L. pneumophila cells after exposure to UV radiation and norfloxacin. The JR32 strain bearing the comEA-gfp reporter plasmid was treated with a single dose of UV radiation (7.5 J/m2) or with a chronic exposure to norfloxacin. After a 24-h growth period, the bacteria were observed using white-light microscopy (top row) and fluorescence microscopy (bottom row). All photographs were taken with the same exposure time.
Induction of competence by genotoxic agents is RecA independent.
The RecA recombinase is a key component of the bacterial response to DNA damage. It is involved in the regulation of the transcriptional activity that follows sensing of the DNA damages, processing of the damages, and repair of the damage-induced stalled replication forks (24, 29). We assumed that RecA might play similar roles in L. pneumophila and would thus be required for the induction of competence in response to genotoxic stress. As expected, a L. pneumophila recA deletion mutant shows high sensitivity to UV radiation (Fig. 3 A), confirming the requirement of RecA for DNA repair. We then analyzed comEA expression in response to UV radiation and norfloxacin exposure by Northern blotting. As predicted by the comEA-gfp reporter, the steady-state levels of the comEA transcript increased with exposure to UV radiation and norfloxacin in the wild-type strain (Fig. 3B and C). Since RecA is required for the transcriptional response to DNA damage, we tested whether comEA induction still occurred in a recA mutant. Unexpectedly, increased expression of comEA is also observed in the recA mutant strain (Fig. 3B and C). Presumably because the recA mutant is unable to repair and remove the DNA lesions, comEA induction occurs in the recA mutant at lower doses of UV radiation and norfloxacin than in the wild-type strain. The finding that comEA is induced by genotoxic stresses in a RecA-independent manner indicates that competence development is independent of an SOS response. Interestingly, in L. pneumophila, RecA is not induced after UV stress and mitomycin C treatment (30, 42), suggesting that L. pneumophila may lack a prototypic SOS response.
FIG. 3.
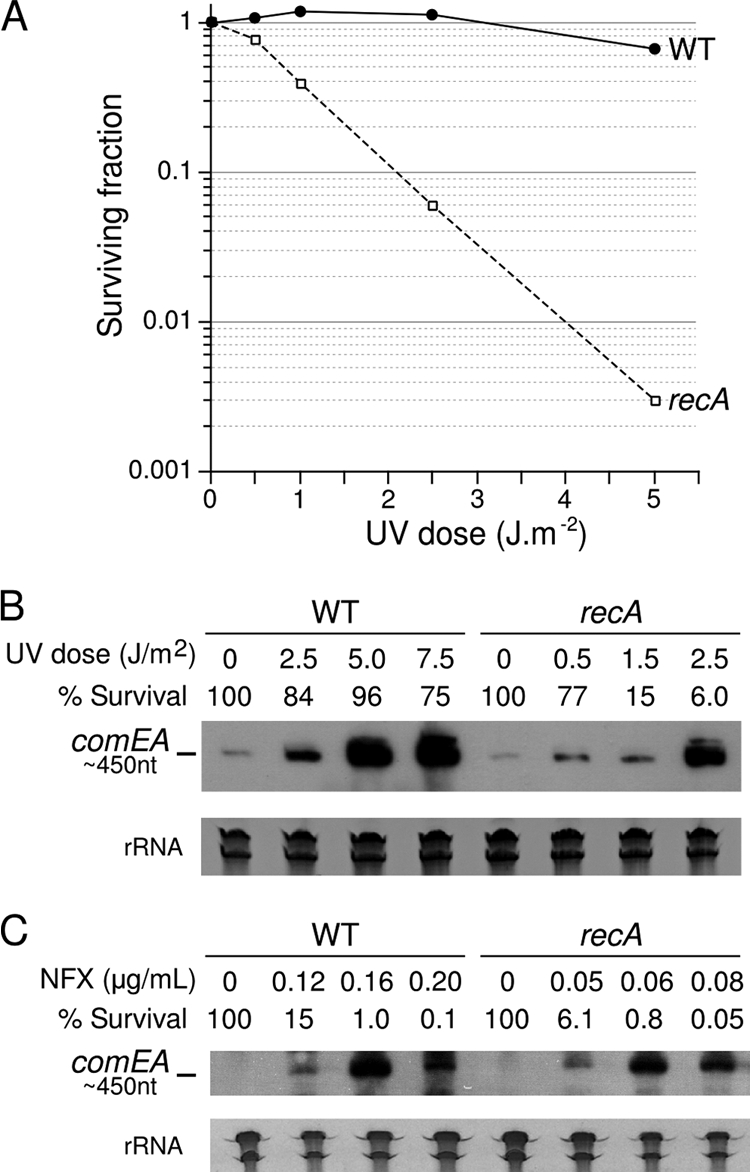
RecA is required for survival after UV exposure but is dispensable for the induction of elevated comEA levels after UV irradiation or under chronic exposure to norfloxacin. (A) The L. pneumophila strain Paris and the recA mutant were exposed to increasing doses of UV radiation and inoculated in liquid medium for overnight recovery. The cultures were plated to determine the survival by counting CFU. (B and C) Northern blot analysis of the comEA transcript in wild-type L. pneumophila strain (Paris) and recA mutant after exposure to increasing doses of UV radiation (A) and chronic exposure to increasing doses of norfloxacin (B). The survival of the treated cells is expressed as a percentage of the untreated population. Lower doses of UV radiation and norfloxacin were used with the recA mutant. Ethidium bromide staining of rRNA was used to ensure that equal amounts of total RNA were loaded into each lane.
Competence-stimulating genotoxic stresses do not induce adaptive mutagenesis.
The molecules and conditions found to induce comEA are known inducers of the bacterial SOS response. The SOS response is a damage-induced response with two components: (i) induced activity of DNA repair and (ii) a transient and genome-wide hypermutable state. The LexA repressor is the master regulator of the SOS response (3). RecA-stimulated self-degradation of LexA alleviates the repression of genes involved in DNA repair and error-prone polymerases responsible for the hypermutable state (17). A pan-genome search for proteins with a LexA-like architecture, i.e., an N-terminal helix-turn-helix DNA-binding domain and a C-terminal S24 peptidase domain, identified five potential repressors (see Fig. S4 in the supplemental material). All five LexA-like encoding genes we found in L. pneumophila genomes are located in the vicinity of phage-related genes or mobile genetic elements. LexA homologs can often be confused with other RecA-interacting proteins, including the phage repressors of the cI family that also share the LexA-like architecture. All of the identified LexA-like proteins in L. pneumophila appear to be more closely related to cI phage repressors rather than to LexA orthologs (see Fig. S4 in the supplemental material). Hence, the absence of a definite LexA homolog suggests that L. pneumophila lacks a canonical SOS response. However, the L. pneumophila genome shows genes coding the SOS-inducible DNA polymerases IV and V, and a hypermutable state may therefore exist in L. pneumophila. Expression of this SOS phenotypic response may be experimentally checked by testing the induction of a hypermutator phenotype under conditions known to induce the SOS response. The frequency of mutations in the rpoB gene conferring rifampin resistance is often used as a readout of the genome-wide hypermutable state. The mutations in rpoB that confer rifampin resistance in L. pneumophila are similar to those in E. coli (33). L. pneumophila was exposed to UV radiation or to the quinolone nalidixic acid and plated on medium containing rifampin to determine the frequency of rifampin-resistant mutants (RifR). Exposure of L. pneumophila to increasing dose of UV resulted in a steady but moderate increase in the RifR mutation rates. The maximum increase of ∼7-fold is much lower than the >200-fold increase observed for E. coli under the same conditions (Table 2). In addition, the increase in mutation rate appears to be proportional to the UV dose, suggesting that mutagenesis is a consequence of direct damages or error-prone repair rather than induction of a hypermutable state. Quinolones also induce the SOS response but, unlike UV radiation, they do not chemically alter the DNA. Exposure of L. pneumophila to nalidixic acid resulted in a 2- to 3-fold increase of mutagenesis (Table 2). Our results suggest the absence of a stress-induced hypermutable state in L. pneumophila. Considering the absence of both an inducible hypermutable state and a LexA homolog, we propose that L. pneumophila lacks a prototypic SOS response.
TABLE 2.
Mutation frequency to rifampin resistance after UV irradiation and chronic exposure to the quinolone nalidixic acida
| Treatment | Avg ± SD |
||
|---|---|---|---|
| % Survival | Frequency (10−8) | Fold increase for L. pneumophila and E. colib | |
| UV (J/m2) | |||
| 0 | 100 | 4.1 ± 1.0 | 1 |
| 5 | 96 ± 21 | 10.8 ± 3.9 | 2.6 ± 1.1 (92) |
| 7.5 | 77 ± 22 | 14.9 ± 6.9 | 3.6 ± 1.8 (178) |
| 10 | 23 ± 13 | 30.1 ± 6.4 | 7.3 ± 2.9 (223) |
| Nalidixic acid (μg/ml) | |||
| 0 | 100 | 5.8 ± 1.0 | 1 |
| 0.16 | 27 ± 11 | 10.8 ± 3.1 | 1.9 ± 0.8 (ND) |
| 0.24 | 0.8 ± 0.7 | 14.6 ± 7.9 | 2.6 ± 1.6 (ND) |
Values are averages from three independent experiments (n = 3). ND, not determined.
The values for E. coli are given in parentheses.
UV radiation induces genetic exchange in stressed L. pneumophila populations.
Full induction of competence by a genotoxic stress should allow L. pneumophila to take up and integrate an exogenously added transforming DNA fragment. In order to directly assess the transformability of L. pneumophila under genotoxic stress, the L. pneumophila strain Paris was exposed to increasing doses of UV radiation, and free transforming DNA was added to the culture. UV exposure increased the number of transformants in the bacterial population by nearly 4 orders of magnitude (Fig. 4 A). No transformants were obtained in a strain lacking the comEC gene encoding the transmembrane DNA channel of the DNA uptake machinery (12). Both Paris and Philadelphia-1 isolates could be transformed with linear DNA after UV exposure; however, no transformants could be obtained with the laboratory strain JR32 (derived from Philadelphia-1). The reason for this is currently unknown, but it may be linked to the mutations and/or deletions acquired by JR32 during the derivation process from the original Philadelphia-1 isolate (39). The high transformability of UV-exposed L. pneumophila suggests that genetic exchange could occur within a UV-stressed population. To test for genetic exchange between bacteria within a stressed population, two L. pneumophila Paris strains carrying distinct selectable chromosomal markers were mixed, exposed to UV, and cocultured in liquid media. In the absence of UV exposure, no L. pneumophila isolate carrying both markers could be obtained. However, a recombinant progeny carrying the two markers was detected at a frequency of ∼10−5 after exposure to UV radiation (Fig. 4B). The addition of DNase I to the culture media strongly reduced the size of the recombinant population, suggesting that the entire recombinant population resulted from transformation by released DNA. Indeed, exposure to UV radiation may cause the release of DNA from heavily damaged cells, which can subsequently be imported by the surviving cells. These results reveal that following UV stress, L. pneumophila is prone to acquire genetic information from the environment and from its siblings, a form of parasexual behavior that can promote the transmission of genes that confer a selective advantage within a population.
FIG. 4.
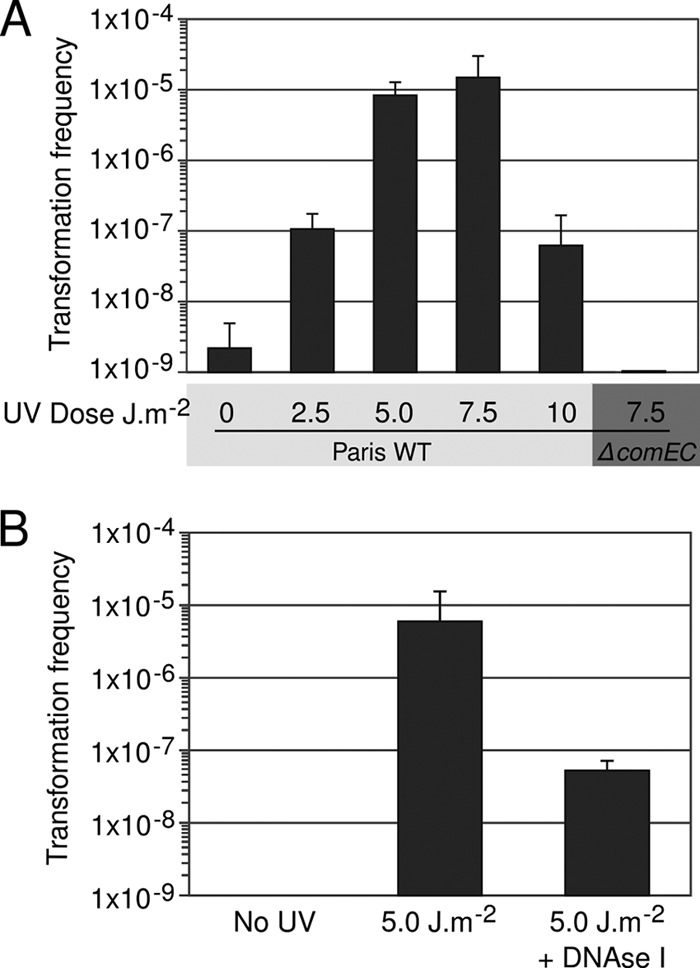
Transformability of UV-irradiated L. pneumophila cells. (A) Transformation with exogenous DNA. Wild-type L. pneumophila strain (Paris WT) and transformation-deficient mutant comEC (ΔcomEC) were exposed to various doses of UV light and inoculated into rich liquid medium in the presence of 1 μg of DNA conferring kanamycin resistance. The cultures were then plated on selective and nonselective media. Transformation frequency represents the number of CFU on selective plates containing kanamycin versus the number of CFU on nonselective plates. (B) Genetic exchange within a stressed population. Two L. pneumophila strains carrying either a gentamicin or a kanamycin chromosomal marker were mixed, exposed to a single dose of UV irradiation (5.0 J/m2), and inoculated in liquid media with or without DNase I (no exogenous DNA added). The transformation frequency represents the number of CFU on selective plates containing kanamycin and gentamicin versus the number of CFU on nonselective plates. In both graphs, error bars represent the standard deviations derived from three independent experiments.
DISCUSSION
The results presented here establish a link between cellular pathways for sensing chromosome instability and for actively importing exogenous DNA. We initially hypothesized that induction of competence by genotoxic agents was a direct consequence of DNA damage and that sensors of DNA damage would be required for competence development. However, UV radiation and norfloxacin still induce competence in a recA mutant (Fig. 3). Similarly, norfloxacin induces comEA in addB, recO, recN, uvrA, and rpoS mutants (data not shown). Therefore, the induction of competence by genotoxic agents may not be a direct consequence of DNA damage. Indeed, bleomycin, a natural antibiotic that induces extensive DNA strand breaks does not induce competence (see Table S1 in the supplemental material). Also, fluoroquinolones induce competence under specific conditions. High doses of fluoroquinolone induce irreversible DNA strand breaks, whereas low concentration produce reversible “frozen” gyrase intermediates which block the DNA polymerase (13). High doses of fluoroquinolone (10 times higher than the 50% inhibitory concentration [IC50]) fail to induce comEA expression during either chronic or temporary exposure (see Fig. S2 in the supplemental material and data not shown). Rather, fluoroquinolones induce competence at concentrations near or below the IC50 (Fig. 1B and see Fig. S2 in the supplemental material), suggesting that competence expression is here the consequence of replication fork stalling. Consistent with this hypothesis, we found that comEA is induced by hydroxyurea (see Fig. S2), a small molecule inhibitor of class I ribonucleotide reductase that depletes the dNTP pool and is commonly used to study DNA damage-independent replication fork stalling (27, 38). We currently favor the hypothesis that stalling of the DNA replication fork is the primary signal leading to competence development.
While investigating the possible involvement of the SOS response in the induction of competence, several lines of evidence suggested the lack of this prototypic regulatory network in L. pneumophila. First, in L. pneumophila, RecA is not induced following UV stress and mitomycin C treatment (30, 42). Second, we could identify no definite homolog of the SOS response master regulator LexA. Third, exposure to UV radiation did not induce a mutagenic state. This may not be an uncommon situation since many other bacterial pathogens lack the LexA-encoding gene (2). In many respects, L. pneumophila is reminiscent of the human pathogen Streptococcus pneumoniae. Like L. pneumophila, S. pneumoniae lacks an SOS response (19), and fluoroquinolones and mitomycin C have been shown to induce competence in this organism (36). In this bacterium, induction of competence had been previously proposed to be a general response to stress (10). Although it is not known whether UV radiation induces competence in S. pneumoniae, competence development induced by DNA-damaging antibiotics indicated that it is an alternative response to the SOS response (36). The induction of competence genes by DNA damage has also been recently reported in Helicobacter pylori (11). The DNA damage response of H. pylori is distinct from the SOS response and includes competence genes. Although this bacterium is constitutively competent under laboratory conditions, exposure to the fluoroquinolone ciprofloxacin further increased transformation frequencies. The striking similarities in the behavior of these three distant organisms suggest that the induction of competence by genotoxic stresses may not be an unusual situation in the bacterial world.
We cannot exclude that other signals may also trigger competence in L. pneumophila. However, we report here for the first time that UV radiation can induce bacterial competence and should now be considered as a potential signal of competence development in other bacteria. Like most living organisms in their natural habitat, L. pneumophila is exposed to sunlight-emitted UV radiation, and the induction of competence may have been initially evolved to respond to DNA-damaging radiation. It may be fortuitous that the same response is induced by some antibiotics because, under specific conditions, they produce DNA alterations structurally similar to those produced by UV radiation.
The induction of competence under genotoxic conditions raises the question of the benefit of such response for bacteria. Of what use is active DNA import to bacterial cells harboring a compromised chromosome? Natural transformation has previously been proposed to be a mechanism of DNA repair where the imported DNA would serve as a template for recombinational repair of the recipient's damaged chromosome (22, 32, 44). Experimental evidence supporting this hypothesis is limited, and its interpretation has been questioned (14). This “DNA import for repair” hypothesis has also been criticized on the grounds that UV radiation does not induce or enhance the competence of the SOS-proficient bacteria Bacillus subtilis and Haemophilus influenzae (37). Although competence is indeed induced by genotoxic stress in L. pneumophila and S. pneumoniae, this finding does not necessarily imply that the actively imported DNA is being used for recombinational repair. If it were, a transformation-deficient mutant should be more sensitive to UV radiation. We tested this prediction and did not observe a statistically different survival of an L. pneumophila comEC mutant compared to the wild-type strain (data not shown). Therefore, the “DNA import for repair” hypothesis remains controversial and needs to be investigated further, eventually under growth conditions of higher cell density such as in a biofilm community or, for L. pneumophila, in a confined intracellular environment.
It is intriguing that competence induction by genotoxic stress occurs in two such distantly related bacteria which both lack an SOS system with its mutagenic properties. Stress calls for adjustment, and the transient, mutagenic activity of SOS-dependent error-prone polymerases provides genetic diversity and most likely has adaptive properties (18). Supporting the idea that increased mutagenesis may be important for adaptive evolution, error-prone DNA polymerases enhance long-term survival and evolutionary fitness (28, 45). Transformation is a well-accepted mechanism for acquisition of genetic diversity and exploration of the fitness landscape. We hypothesize that DNA damage-induced competence has evolved to provide genetic diversity in organisms unable to access it via the transient hypermutagenic state linked to the SOS response. In addition to L. pneumophila and S. pneumoniae, other important pathogens that lack LexA (2) may respond to genotoxic stress and antibiotics by expressing competence. This raises the concern that antibiotics such as fluoroquinolones may be responsible for increased rates of horizontal gene transfer.
Supplementary Material
Acknowledgments
We are grateful to Robert Washburn and Max Gottesman for providing bicyclomycin and for helpful discussions. We thank the two anonymous reviewers for their constructive comments and detailed corrections. We also thank Jean-Pierre Claverys for encouraging and stimulating discussions.
This study was supported by PHS grants AI23549 and AI064481 (awarded to H.A.S.) from the National Institute of Allergy and Infectious Diseases. Support was also provided to X.C. by the Fondation pour la Recherche Médicale.
Footnotes
Published ahead of print on 17 December 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Almahmoud, I., E. Kay, D. Schneider, and M. Maurin. 2009. Mutational paths toward increased fluoroquinolone resistance in Legionella pneumophila. J. Antimicrob. Chemother. 64:284-293. [DOI] [PubMed] [Google Scholar]
- 2.Ambur, O. H., et al. 2009. Genome dynamics in major bacterial pathogens. FEMS Microbiol. Rev. 33:453-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butala, M., D. Zgur-Bertok, and S. J. W. Busby. 2009. The bacterial LexA transcriptional repressor. Cell. Mol. Life Sci. 66:82-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cazalet, C., et al. 2008. Multigenome analysis identifies a worldwide distributed epidemic Legionella pneumophila clone that emerged within a highly diverse species. Genome Res. 18:431-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cazalet, C., et al. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165-1173. [DOI] [PubMed] [Google Scholar]
- 6.Charpentier, X., S. P. Faucher, S. Kalachikov, and H. A. Shuman. 2008. Loss of RNase R induces competence development in Legionella pneumophila. J. Bacteriol. 190:8126-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, I., and E. C. Gotschlich. 2001. ComE, a competence protein from Neisseria gonorrhoeae with DNA-binding activity. J. Bacteriol. 183:3160-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chien, M., et al. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966-1968. [DOI] [PubMed] [Google Scholar]
- 9.Claverys, J., B. Martin, and P. Polard. 2009. The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol. Rev. 33:643-656. [DOI] [PubMed] [Google Scholar]
- 10.Claverys, J., M. Prudhomme, and B. Martin. 2006. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 60:451-475. [DOI] [PubMed] [Google Scholar]
- 10a.de Felipe, K. S., et al. 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 187:7716-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorer, M. S., J. Fero, and N. R. Salama. 2010. DNA damage triggers genetic exchange in Helicobacter pylori. PLoS Pathog. 6:e1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draskovic, I., and D. Dubnau. 2005. Biogenesis of a putative channel protein, ComEC, required for DNA uptake: membrane topology, oligomerization and formation of disulfide bonds. Mol. Microbiol. 55:881-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drlica, K., M. Malik, R. J. Kerns, and X. Zhao. 2008. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 52:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Franco, I. S., H. A. Shuman, and X. Charpentier. 2009. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell. Microbiol. 11:1435-1443. [DOI] [PubMed] [Google Scholar]
- 17.Friedberg, E. C., et al. 2005. DNA repair and mutagenesis, 2nd ed. ASM Press, Washington, DC.
- 18.Galhardo, R. S., P. J. Hastings, and S. M. Rosenberg. 2007. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 42:399-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasc, A. M., N. Sicard, J. P. Claverys, and A. M. Sicard. 1980. Lack of SOS repair in Streptococcus pneumoniae. Mutat. Res. 70:157-165. [DOI] [PubMed] [Google Scholar]
- 20.Harinarayanan, R., and J. Gowrishankar. 2003. Host factor titration by chromosomal R-loops as a mechanism for runaway plasmid replication in transcription termination-defective mutants of Escherichia coli. J. Mol. Biol. 332:31-46. [DOI] [PubMed] [Google Scholar]
- 21.Herriott, R. M., E. M. Meyer, and M. Vogt. 1970. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J. Bacteriol. 101:517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoelzer, M. A., and R. E. Michod. 1991. DNA repair and the evolution of transformation in Bacillus subtilis. III. Sex with damaged DNA. Genetics 128:215-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huisman, O., R. D'Ari, and S. Gottesman. 1984. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc. Natl. Acad. Sci. U. S. A. 81:4490-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janion, C. 2008. Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. Int. J. Biol. Sci. 4:338-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnsborg, O., V. Eldholm, and L. S. Håvarstein. 2007. Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 158:767-778. [DOI] [PubMed] [Google Scholar]
- 26.Kohn, H., and W. Widger. 2005. The molecular basis for the mode of action of bicyclomycin. Curr. Drug Targets Infect. Disord. 5:273-295. [DOI] [PubMed] [Google Scholar]
- 27.Krakoff, I. H., N. C. Brown, and P. Reichard. 1968. Inhibition of ribonucleoside diphosphate reductase by hydroxyurea. Cancer Res. 28:1559-1565. [PubMed] [Google Scholar]
- 28.Loh, E., J. J. Salk, and L. A. Loeb. 2010. Optimization of DNA polymerase mutation rates during bacterial evolution. Proc. Natl. Acad. Sci. U. S. A. 107:1154-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lusetti, S. L., and M. M. Cox. 2002. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 71:71-100. [DOI] [PubMed] [Google Scholar]
- 30.Marra, A., and H. A. Shuman. 1992. Genetics of Legionella pneumophila virulence. Annu. Rev. Genet. 26:51-69. [DOI] [PubMed] [Google Scholar]
- 31.Meibom, K. L., M. Blokesch, N. A. Dolganov, C. Wu, and G. K. Schoolnik. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824-1827. [DOI] [PubMed] [Google Scholar]
- 32.Michod, R. E., M. F. Wojciechowski, and M. A. Hoelzer. 1988. DNA repair and the evolution of transformation in the bacterium Bacillus subtilis. Genetics 118:31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen, K., P. Hindersson, N. Hoiby, and J. M. Bangsborg. 2000. Sequencing of the rpoB gene in Legionella pneumophila and characterization of mutations associated with rifampin resistance in the Legionellaceae. Antimicrob. Agents Chemother. 44:2679-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ninio, S., J. Celli, and C. R. Roy. 2009. A Legionella pneumophila effector protein encoded in a region of genomic plasticity binds to Dot/Icm-modified vacuoles. PLoS Pathog. 5:e1000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Provvedi, R., and D. Dubnau. 1999. ComEA is a DNA receptor for transformation of competent Bacillus subtilis. Mol. Microbiol. 31:271-280. [DOI] [PubMed] [Google Scholar]
- 36.Prudhomme, M., L. Attaiech, G. Sanchez, B. Martin, and J. Claverys. 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313:89-92. [DOI] [PubMed] [Google Scholar]
- 37.Redfield, R. J. 1993. Evolution of natural transformation: testing the DNA repair hypothesis in Bacillus subtilis and Haemophilus influenzae. Genetics 133:755-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenkranz, H. S., A. J. Garro, J. A. Levy, and H. S. Carr. 1966. Studies with hydroxyurea. I. The reversible inhibition of bacterial DNA synthesis and the effect of hydroxyurea on the bactericidal action of streptomycin. Biochim. Biophys. Acta 114:501-515. [PubMed] [Google Scholar]
- 39.Samrakandi, M. M., S. L. G. Cirillo, D. A. Ridenour, L. E. Bermudez, and J. D. Cirillo. 2002. Genetic and phenotypic differences between Legionella pneumophila strains. J. Clin. Microbiol. 40:1352-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sexton, J. A., and J. P. Vogel. 2004. Regulation of hypercompetence in Legionella pneumophila. J. Bacteriol. 186:3814-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stone, B. J., and Y. A. Kwaik. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szeto, L. 1992. The function and regulation of the Legionella pneumophila major secretory protein, and RecA protein. Ph.D. thesis. Columbia University, New York, NY.
- 43.Wiater, L. A., A. B. Sadosky, and H. A. Shuman. 1994. Mutagenesis of Legionella pneumophila using Tn903 dlllacZ: identification of a growth-phase-regulated pigmentation gene. Mol. Microbiol. 11:641-653. [DOI] [PubMed] [Google Scholar]
- 44.Wojciechowski, M. F., M. A. Hoelzer, and R. E. Michod. 1989. DNA repair and the evolution of transformation in Bacillus subtilis. II. Role of inducible repair. Genetics 121:411-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeiser, B., E. D. Pepper, M. F. Goodman, and S. E. Finkel. 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. U. S. A. 99:8737-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zusman, T., E. Degtyar, and G. Segal. 2008. Identification of a hypervariable region containing new Legionella pneumophila Icm/Dot translocated substrates by using the conserved icmQ regulatory signature. Infect. Immun. 76:4581-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.