Abstract
Lipopolysaccharide (LPS) is a major glycolipid present in the outer membrane (OM) of Gram-negative bacteria. The peculiar permeability barrier of the OM is due to the presence of LPS at the outer leaflet of this membrane that prevents many toxic compounds from entering the cell. In Escherichia coli LPS synthesized inside the cell is first translocated over the inner membrane (IM) by the essential MsbA flippase; then, seven essential Lpt proteins located in the IM (LptBCDF), in the periplasm (LptA), and in the OM (LptDE) are responsible for LPS transport across the periplasmic space and its assembly at the cell surface. The Lpt proteins constitute a transenvelope complex spanning IM and OM that appears to operate as a single device. We show here that in vivo LptA and LptC physically interact, forming a stable complex and, based on the analysis of loss-of-function mutations in LptC, we suggest that the C-terminal region of LptC is implicated in LptA binding. Moreover, we show that defects in Lpt components of either IM or OM result in LptA degradation; thus, LptA abundance in the cell appears to be a marker of properly bridged IM and OM. Collectively, our data support the recently proposed transenvelope model for LPS transport.
Lipopolysaccharide (LPS) is a complex glycolipid uniquely present in the outer layer of Gram-negative bacteria outer membrane (OM) (20, 21). LPS, also known as endotoxin, is one of the major virulence factors of Gram-negative bacteria and is responsible for the activation of the mammalian innate immune response (17). It consists of three distinct structural elements: lipid A (the hydrophobic moiety embedded in the OM), a core oligosaccharide, and the O antigen constituted of polysaccharide repeating units (21). LPS is essential in most Gram-negative bacteria, with the notable exception of Neisseria meningitidis (32). The lipid A-core moiety is synthesized in the cytoplasm and is flipped from the inner to the outer leaflet of the inner membrane (IM) by the essential ABC transporter MsbA (6, 19, 43). In bacterial strains producing the O antigen, ligation to the core oligosaccharide occurs at the periplasmic face of the IM, after MsbA-mediated translocation (21). Mature LPS, containing or not the O antigen, is then transported to the outer leaflet of the OM by a protein machine composed of seven recently discovered Lpt proteins (reviewed by Sperandeo et al. [28]) suggested to build up a complex (the Lpt complex) that spans the IM and OM. Indeed, these proteins are located at the IM (LptBCFG), in the periplasm (LptA), and at the OM (LptDE) (3, 23, 27, 29, 30, 33, 41). Genetic evidence suggests that the Lpt complex operates as a single device, since the depletion of any component leads to similar phenotypes, namely, failure to transport newly synthesized LPS to the cell surface and its accumulation at the outer leaflet of the IM (16, 23, 29). The LPS accumulating at the outer leaflet of the IM is decorated with colanic acid residues, and therefore this modification is diagnostic of defects in transport occurring downstream of the MsbA-mediated flipping of LPS to the periplasmic face of the IM (29).
Physical interaction between the different proteins of the machinery has been demonstrated for LptDE, which form a complex at the OM (41), and for the IM LptBCFG complex (18). LptD and LptE are responsible for the LPS assembly at the cell surface; LptE stabilizes LptD by interacting with its C-terminal domain, whereas LptE binds LPS, possibly serving as a substrate recognition site at the OM (5). LptC is an IM bitopic protein whose large soluble domain has a periplasmic localization (38). The crystal structure of LptC periplasmic domain has been recently solved and, like LptA, LptC has been shown to bind LPS in vitro (38). LptC physically interacts with the IM LptBFG proteins, and the LptBCFG complex is the IM ABC transporter that energizes the LPS transport (18). However, LptC seems not to be required for the ATPase activity of the transporter (18).
LptA expressed from an inducible promoter has a periplasmic localization and has been shown to bind both LPS and lipid A in vitro (27, 39). These data raise the possibility that LptA may act as a periplasmic chaperone for LPS transport across the periplasm. However, in N. meningitidis the LptA homologue was shown to be associated to the membrane fraction (2). Moreover, in the Escherichia coli LptA crystal structure obtained in the presence of LPS, the LptA monomers are packed as a linear filament (34), leading to the hypothesis that oligomers of LptA may be required to bridge the IM and the OM, thus facilitating LPS export. The observation that LPS is still transported to the OM in spheroplasts devoid of periplasmic content (35) is consistent with this idea. In line with these data it has been recently reported that all seven Lpt proteins physically interact and form a transenvelope complex spanning IM and OM (4).
In the present study we show that in vivo LptA and LptC physically interact and form a stable complex, suggesting that LptC may represent a docking site for LptA to the IM. Based on analyses of loss-of-function mutations in LptC, we predict that the C-terminal region of LptC is implicated in LptA binding and that LptC may form dimers. Finally, after analyzing the relative stability of the Lpt proteins when LptC, LptD, or LptE are either depleted or not functional, we suggest that LptA connects IM and OM via LptC and the LptDE complex and that LptA abundance in the cell may be used as a marker of properly bridged IM and OM.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids examined here are listed in Table 1. The oligonucleotide primers are listed in Table 2.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| AM604 | MC4100 ara+ | 41 |
| AM661 | AM604 ΔlptD::kan/araBp-lptD | 29 |
| AM689 | AM604 ΔlptE::kan Δ(λatt-lom)::blaaraBp-lptE | 41 |
| DH5α | Δ(argF-lac169) 80dlacZ58(M15) glnV44(AS) λ−rfbD1 gyrA96 recA1 endA1 spoT1 thi-1 hsdR17 | 11 |
| FL905 | AM604 Φ(kan araC araBp-lptC)1 | 29 |
| FL907 | AM604 Φ(kan araC araBp-lptA)1 | 29 |
| M15/pREP4 | F−lac thi mtl/pREP4 | Qiagen |
| Plasmids | ||
| pRSET | T7 promoter; Apr | Invitrogen |
| pET30b | T7 promoter; Kmr | Novagen |
| pET30-lptC-lk25-H | pET30b derivative, expresses a LptC-lk25-His6 version where lk is a 25-amino-acid linker region separating LptC residue 191 from the His6 affinity tag | This study |
| pQE30 | T5 promoter; Apr | Qiagen |
| pQEsH-lptC | pQE30 derivative, expresses His6-LptC24-191 | This study |
| pGS100 | pGZ119EH derivative, contains TIR sequence downstream of Ptac; Cmr | 30 |
| pGS103 | pGS100 Ptac-lptC | 30 |
| pGS103Δ177-191 | pGS103 Ptac-lptCΔ177-191 | This study |
| pGS103G56V | pGS103 Ptac-lptC(G56V) | This study |
| pGS103Y112S-G153R | pGS103 Ptac-lptC(Y112S-G153R) | This study |
| pGS103G153R | pGS103 Ptac-lptC(G153R) | This study |
| pGS108 | pGS100 Ptac-lptC-H | 30 |
| pGS108Δ177-191 | pGS108 Ptac-lptCΔ177-191-H | This study |
| pGS108G153R | pGS108 Ptac-lptC(G153R)-H | This study |
| pGS108G56V | pGS108 Ptac-lptC(G56V)-H | This study |
| pGS116 | pGS103Δ177-191 derivative expressing both the truncated LptCΔ177-191 and LptC-lk25 from pET30- lptC-lk25-H plasmid | This study |
Cmr, chloramphenicol resistance; Apr, ampicillin resistance; Kmr, kanamycin resistance.
TABLE 2.
Oligonucleotides
| Name | Sequence (5′-3′)a | Use and/or description |
|---|---|---|
| AP24 | cgactagtctagaTTAAGGCTGAGTTTGTTTG | Random mutagenesis with AP54; XbaI |
| AP54 | cgagaggaattcaccATGAGTAAAGCCAGACGTTGGG | pGS108Δ177-191 construction, with AP172 and random mutagenesis with AP24; EcoRI |
| AP149 | atatacatATGAGTAAAGCCAGACGTTG | pET30-lptC-H construction, with AP150; NdeI |
| AP150 | cgcgcaggtaccAGGCTGAGTTTGTTTGTTTTG | pET30-lptC-H construction, with AP149; KpnI |
| AP164 | GTCTATAACCCAGAAGTGGCACTAAGCTATCG | pGS108G56V construction, with AP165 |
| AP165 | CGATAGCTTAGTGCCACTTCTGGGTTATAGAC | pGS108G56V construction, with AP164 |
| AP166 | cgagatggatccATGGCCGAAAAAGACGATAC | pQEsH-lptC construction, with AP167; BamHI |
| AP167 | cgagatctgcagTTAAGGCTGAGTTTGTTTG | pQEsH-lptC construction, with AP166; PstI |
| AP168 | CTCGTCACGTTATACAGAACAACATTTAACTC | pGS108G153R and pGS103G153R construction, with AP169 |
| AP169 | GAGTTAAATGTTGTTCTGTATAACGTGACGAG | pGS108G153R and pGS103G153R construction, with AP168 |
| AP172 | gtgatcacatctagatcagtggtggtggtggtggtgTTCAATCAGCTCGGCGTTC | pGS108Δ177-191 construction, with AP54; insertion of C-terminal His6 tag into LptCΔ177-191; XbaI |
Uppercase letters, E. coli genomic sequence; underlined lowercase letters, restriction sites; boldface letters, codons mutated by site-directed mutagenesis.
Bacteria were grown in LD medium (24). When required, 0.2% (wt/vol) l-arabinose (as an inducer of the araBp promoter), 0.1 or 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), 100 μg of ampicillin/ml, 25 μg of chloramphenicol/ml, and 25 μg of kanamycin/ml were added. Solid media were prepared as described above with 1% agar.
Plasmid construction.
Plasmid pGS108 expresses LptC with a C-terminal His6 tag (LptC-H) from the IPTG-inducible Ptac promoter (Table 1). Plasmids pGS108G56V, pGS103G153R, and pGS108G153R expressing LptCG56V-H, LptCG153R, and LptCG153R-H, respectively, were constructed by using a QuikChange site-directed mutagenesis kit (Stratagene). Codon 56 was changed from GGG to GTG by using the primer pair AP164-AP165, and codon 153 was changed from GGA to AGA by using the primer pair AP168-AP169 (Table 2). Plasmid pGS108Δ177-191 expressing LptCΔ177-191-H, truncated at residue 177, was constructed by PCR amplifying the lptC open reading frames from genomic MG1655 DNA with the primer pair AP54-AP172 (Table 2). The PCR product was EcoRI-XbaI digested and cloned into pGS108 cut with the same enzymes. The EcoRI-XbaI insert in pGS108Δ177-191, as well as the mutations in pGS108G56V, pGS108G153R, and pGS103G153R, was verified by sequencing.
Plasmid pQEsH-lptC expresses a soluble cytoplasmic version of LptC deprived of the transmembrane helix and with an N-terminal His6 affinity tag (sH-LptC) (Table 1). A 507-bp DNA fragment, encoding residues 24 to 191 of LptC, was amplified from MG1655 DNA with the primers AP166 and AP167 (Table 2) and cloned into the BamHI and PstI sites of pQE30 (Qiagen). The His6 affinity tag is separated from the first residue of LptC24-191 by two amino acids (G-S). The BamHI-PstI insert was verified by sequencing.
Plasmid pET30-lptC-lk25-H expresses a C-terminally His6-tagged version of LptC (LptC-lk25-H) with a 25-amino-acid linker region (GTDDDDKAMAISDPNSSSVDKLAAALE) containing the enterokinase recognition site (DDDDK) (Table 1). The lptC open reading frame was amplified from MG1655 DNA with the primers AP149 and AP150 (Table 2). The PCR product was NdeI-KpnI digested and cloned into the NdeI-KpnI sites of pET30b (Novagen). The NdeI-KpnI insert was verified by sequencing.
Plasmid pGS116 is a pGS103Δ177-191 derivative expressing LptC-lk25, together with LptCΔ177-191 from the IPTG-inducible Ptac promoter (Table 1). The XbaI-HindIII fragment of pET30-lptC-lk25-H, containing the ribosome binding site (RBS) region, the lptC gene without its stop codon and part of the plasmid multiple cloning site was inserted into the XbaI-HindIII sites of pGS103Δ177-191. The resulting plasmid expresses LptC (LptC-lk25) in frame with a 27-amino-acid coding sequence (GTDDDDKAMAISDPNSSSVDKLGCFGG). In this construct the His6 affinity tag is lost.
Random mutagenesis, screening, and genetic characterization of LptC mutants.
Random mutagenesis was performed by error-prone PCR using an unbalanced deoxynucleoside triphosphates concentration in the amplification reactions, as described previously (9). pGS103 expressing lptC from the Ptac promoter was used as a template (10 ng) in 50 μl with 50 pmol each of primers AP54 and AP24 (Table 2). The PCR conditions were as follows: 95°C denaturation for 2 min; 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; followed by a 5-min extension at 72°C. The products were gel purified, digested with XbaI and EcoRI, ligated into pGS100, and electroporated into FL905 cells. The transformants were plated onto LD agar with 0.2% arabinose and 25 μg of chloramphenicol/ml at 37°C. Single colonies were then inoculated into microtiter wells containing 100 μl of LD, serially diluted 10-fold, replica plated on LD-chloramphenicol agar plates supplemented with (permissive condition) or without (nonpermissive condition) 0.2% arabinose, and incubated overnight at 37°C. Plasmids unable to support FL905 growth in the absence of arabinose were identified and sequenced.
To test the effect of LptC mutant overexpression, serial dilutions of cultures grown overnight in LD-chloramphenicol were replica plated on LD-chloramphenicol agar plates supplemented or not with 0.5 mM IPTG.
In vivo cross-linking.
The cross-linking experiments were based on previously described methods (36) with some modifications. The cells were grown in 250 ml of LD medium supplemented with either chloramphenicol or ampicillin to an optical density at 600 nm (OD600) of 0.2, induced with 0.1 mM IPTG, and grown to an OD600 of ∼0.7. The cells were then harvested by centrifugation at 5,000 × g for 10 min. For treatment with DSP (di-thiobis[succinimidyl propionate]; Pierce), the cell pellet was washed with 25 ml of 20 mM potassium phosphate (pH 7.2) and 150 mM NaCl, resuspended in 25 ml of the same buffer, and then incubated for 15 min at 37°C. DSP dissolved in dimethyl sulfoxide was added to the cell suspension at a final concentration of 80 μg/ml, and the cells were incubated for 30 min at 37°C. The cross-linking reaction was quenched by the addition 1 M Tris-HCl (pH 7.4) to a final concentration of 20 mM, and the cells were harvested by centrifugation at 5,000 × g for 10 min.
For whole-cell extract analysis, the cells were grown in 50 ml of LD-chloramphenicol to an OD600 of 0.2, induced with 0.1 mM IPTG, and grown to an OD600 of ∼0.7. DSP treatment was performed as described above using two-thirds of each culture, whereas the reminder one-third was not treated. After the addition of 20 mM Tris-HCl (pH 7.4), the cells were centrifuged and washed twice in 20 mM potassium phosphate (pH 7.2) and 150 mM NaCl. The cell pellet was resuspended in 100 μl of sodium dodecyl sulfate (SDS) sample buffer containing or not containing 5% β-mercaptoethanol and boiled for 5 min.
Affinity purification.
Protein purification was performed as described previously (26). The cell pellets were resuspended in 4 ml of 50 mM potassium phosphate buffer (pH 8.0), 150 mM NaCl, 5 mM MgCl2, and 1% ZW3-14 (n-tetradecyl-N,N-dimethyl-3-ammonio-1-propanesulfonate) containing lysozyme (50 μg/ml), DNase I (50 μg/ml), and RNase I (50 μg/ml) and then lysed by shaking for 20 min at room temperature. To remove cell debris after lysis, the mixture was then centrifuged at 10,000 × g for 10 min. To the cleared lysate (whole-cell extract) 20 mM imidazole (pH 8.0) was added, and the final mixture was loaded onto a 0.4-ml Ni-NTA column. The column was first washed with 8 ml of 50 mM potassium phosphate buffer (pH 8.0), 300 mM NaCl, 20 mM imidazole, 0.1% Triton X-100, and 0.1% SDS and then eluted with 4 ml of 50 mM potassium phosphate buffer (pH 8.0), 300 mM NaCl, and 200 mM imidazole. The eluate was concentrated in an ultrafiltration device (Amicon Ultra [Millipore]; molecular weight cutoff, 10,000) by centrifugation at 5,000 × g. The concentrated sample was used for SDS-PAGE and Western blot analysis as described above.
Purification of sH-LptC.
M15/pREP4 carrying the plasmid pQEsH-lptC was grown at 30°C in LD containing kanamycin (25 μg/ml) and ampicillin (100 μg/ml) for 18 h. This culture was diluted 1:100 in fresh medium and grown until mid-logarithmic phase (OD600, ∼0.6). The expression of sH-LptC was induced overnight at 20°C by adding IPTG to a final concentration of 0.5 mM. Cells were then harvested by centrifugation (5,000 × g, 10 min). The cell pellet was resuspended in buffer A (50 mM NaH2PO4 [pH 8.0] containing 300 mM NaCl, 10 mM imidazole, and 10% glycerol), followed by incubation for 30 min at 4°C with shaking in the presence of lysozyme (0.2 mg/ml), DNase (100 μg/ml), 10 mM MgCl2, and 1 mM phenylmethylsulfonyl fluoride. After 10 cycles of sonication (10-s pulses), the unbroken cells were removed by centrifugation (39,000 × g, 30 min). The soluble sH-LptC protein was purified from the supernatant by using Ni-NTA agarose (Qiagen). The column was washed with 10 column volumes of 4% buffer B (50 mM NaH2PO4 [pH 8.0] containing 300 mM NaCl, 500 mM imidazole, and 10% glycerol) in buffer A. The protein was eluted by using a stepwise gradient obtained by mixing buffer B with buffer A in 5 steps (10, 20, 50, 70, and 100% buffer B). At each step, 1 column volume was flowed through the column. Elution fractions were monitored by 12.5% polyacrylamide SDS-PAGE. The pooled fractions containing purified protein were dialyzed against 100 volumes of buffer C (50 mM NaH2PO4 [pH 8.0], 100 mM NaCl) for gel filtration analysis, and 100 volumes of 10 mM ammonium acetate pH 7.5 for mass spectrometry. Protein concentrations were determined by using a Coomassie (Bradford) assay kit (Pierce) with bovine serum albumin as the standard.
Mass spectrometry.
Electrospray ionization-mass spectrometry (ESI-MS) experiments were performed on a hybrid quadrupole-time-of-flight mass spectrometer (QSTAR Elite; Applied Biosystems, Foster City, CA) equipped with a nano-ESI sample source. Metal-coated borosilicate capillaries (Proxeon, Odense, Denmark), with medium-length emitter tips (1-μm internal diameter), were used to infuse the samples. The instrument was calibrated using the renin inhibitor (1,757.9 Da) (Applied Biosystems) and its fragment (109.07 Da) as standards. Spectra were acquired in the 500 to 5,000 m/z range, with accumulation times of 1 s, an ion-spray voltage of 1,300 V, a declustering potential of 40 V, and an instrument interface at room temperature. Spectra were averaged over a period of at least 3 min.
Analytical gel filtration chromatography.
Size-exclusion chromatography was performed on an ÄKTA purifier liquid-chromatography system (GE Healthcare, Amersham Place, Little Chalfont, United Kingdom), using a prepacked, Superdex 75HR column (30 by 1 cm; GE Healthcare, Amersham Place, Little Chalfont, United Kingdom). Chromatography was carried out at room temperature in 50 mM NaH2PO4 (pH 8.0)-100 mM NaCl at a flow rate of 0.5 ml/min and monitored by the eluate absorbance at 280 nm. The calibration curve was constructed by using the following standards (0.5 mg/ml): transferrin (81,000 Da), chicken ovalbumin (43,000 Da), chymotrypsin (21,500 Da), bovine cytochrome c (12,200 Da), and aprotinin (6,500 Da) (Sigma Aldrich, St. Louis, MO). LptC was injected at a concentration of 1 mg/ml.
Determination of LptA, LptC, and LptE levels.
LptA, LptC, and LptE levels were assessed in FL905 and its derivatives expressing mutant LptC or in AM661 or AM689 strains by Western blot analysis with polyclonal antibody raised in mouse or rabbit against peptides (LptA) or whole proteins (LptE, LptC, and AcrB). Bacterial cultures grown at 37°C in LD supplemented with 0.2% arabinose and 25 μg of chloramphenicol/ml when required were harvested by centrifugation after they had reached an OD600 of 0.2, washed in LD, and diluted to OD600s of 0.01 (AM689), 0.004 (AM661), 0.0004 (AM604, FL905/pGS103Δ177-191, FL905/pGS103, FL905, FL905/pGS103G56V, and FL905/pGS108G153R) in fresh media with or without 0.2% arabinose and with 25 μg of chloramphenicol/ml. Growth was monitored by measuring the OD600. Samples for protein analysis were centrifuged (16,000 × g, 5 min), and pellets were resuspended in a volume (in ml) of SDS sample buffer equal to one-eighth of the total OD of the sample. The samples were boiled for 5 min, and equal volumes (25 μl) were analyzed by SDS-12.5% PAGE. Proteins were transferred onto nitrocellulose membranes (GE Healthcare), and Western blot analysis was performed as previously described (27). Polyclonal sera raised against LptA (GenScript Corp.) were used as primary antibody at a dilution of 1:2,000, whereas polyclonal sera against LptC, LptE (kindly provided by D. Kahne), and AcrB (kindly provided by K. M. Pos) were used at a dilution of 1:5,000. As secondary antibodies, sheep anti-mouse immunoglobulin G-horseradish peroxidase (HRP) conjugate (GE Healthcare) and donkey anti-rabbit immunoglobulin G-HRP conjugate (GE Healthcare) were used at dilutions of 1:5,000.
Analyses of LptC-H mutant stability.
The expression of LptC was induced with 0.1 mM IPTG at an OD600 of 0.3 to 0.4 in cultures grown in LD-chloramphenicol, and 1-ml samples were taken immediately before and 15, 30, and 60 min after induction. Samples preparation and Western blotting were performed as described above, and wild-type and mutant versions of LptC-H were visualized by using HisProbe-HRP (Pierce) according to the manufacturer's instructions.
Total LPS extraction and analysis.
LPS extraction from AM604 and FL905 transformed or not with plasmids carrying wild-type or mutant LptC was performed as previously described. Samples of a total OD600 of 2 were taken, and LPS was extracted from cell pellets by a mini phenol-water extraction technique (22). Briefly, the cells were resuspended in water and pelleted (5 min, 10,000 × g) to remove the exopolysaccharides and then resuspended in 0.3 ml of potassium phosphate buffer (pH 7.0) and thoroughly vortexed; 0.3 ml of phenol equilibrated with 0.1 M Tris-HCl (pH 5.5) was added, and the suspension was vortexed. The tubes were placed in a 65°C heating block for 15 min with thorough vortexing every 5 min and then cooled on ice. After centrifugation (10,000 × g, 5 min), the water phase was removed, dialyzed (molecular mass cutoff, 2,000 to 4,000 Da) against phosphate buffer (pH 7.0), and lyophilized. The lyophilized material was then dissolved in 30 μl of water. LPS was separated by N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine-SDS-PAGE (31) and visualized by silver staining according to the Hitchcock and Brown method as described previously (27).
RESULTS
LptC stably associates with LptA.
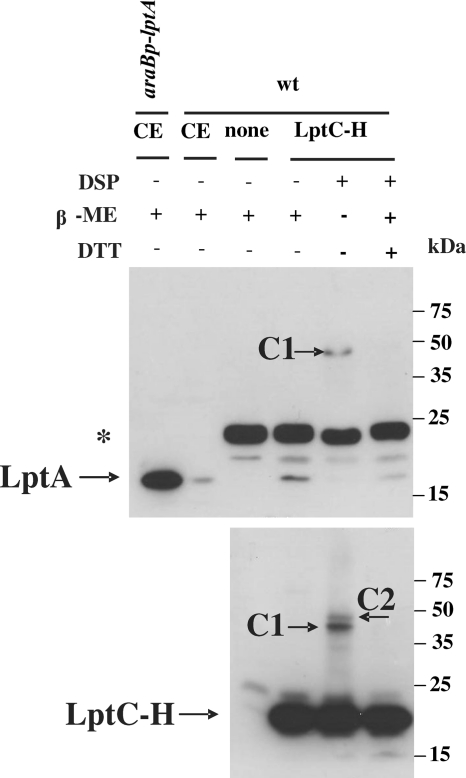
Previous work by our and other laboratories (27, 39) suggested that LptA expressed from an inducible promoter has a periplasmic localization. However, evidence of direct physical interaction between the seven Lpt proteins has been recently reported and LptA has been shown to associate with both IM and OM (4). Since the bitopic IM LptC protein possesses a large C-terminal periplasmic domain (E26-P191) (38), we hypothesized that LptA binding to the IM could be mediated by LptC. To address this issue, we probed the interaction between LptA and LptC by affinity purification followed by immunoblotting. A C-terminal His tagged version of LptC (LptC-H) overexpressed from plasmid pGS108 in the wild-type strain AM604 was used as bait in copurification experiments. AM604 harboring the pRSET vector, which allows the basal expression of the His tag alone, was used as a negative control. To detect possible weak or transient interactions, in vivo cross-linking using di-thiobis(succinimidyl propionate) (DSP) (42) was also performed. As shown in Fig. 1, LptA copurified with LptC-H even when DSP was not added to the cells overexpressing LptC-H, suggesting a stable LptA-LptC interaction. When samples treated with DSP were not reverted by a reducing agent, high-molecular-weight bands appeared (Fig. 1, upper and lower panel, bands C1 and C2). C1 may correspond to an LptA-LptC complex, whereas the C2 band that appears only in the lower panel might correspond to an LptC-LptC complex, as inferred by the molecular weight. These data suggest that LptA and LptC interact and form a stable complex.
FIG. 1.
LptA interacts with LptC in vitro. Affinity chromatography experiments were performed in the presence or absence of DSP in AM604/pGS108 overexpressing LptC-H (wt/LptC-H) and AM604/ pRSET (wt/none) expressing the His tag element only, as a negative control. LptA and LptC were detected in Ni-NTA column-enriched fractions by Western blot analysis with anti-LptA antibody and HisProbe-HRP, respectively. LptA detected in crude cell extract (CE) of AM604 (wt) and of FL907 mutant overexpressing LptA (araBp-lptA) was used as a marker. Samples were treated with dithiothreitol (DTT) and/or β-mercaptoethanol (β-ME) to revert the cross-linking. Equal amounts of protein (2 μg) were loaded into each lane. C1 and C2, high-molecular-weight complexes. The SlyD protein of 24 kDa cross-reacting with anti-LptA antibodies is labeled (*).
Isolation of inactive lptC mutant alleles.
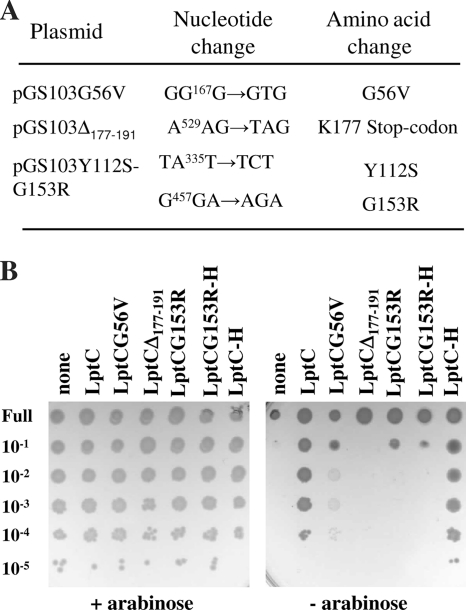
To better characterize LptA-LptC interaction and to define the molecular role of LptC in LPS transport, we searched for point mutations that inactivate LptC function. Random mutations were introduced by error-prone PCR into lptC carried by pGS103 (Table 1), and the mutagenized plasmids were tested for complementation of LptC+-depleted cells, as described in Materials and Methods. Briefly, LptC+ depletion strain FL905 was transformed with the mutagenized plasmids in the presence of arabinose, and plasmids unable to support FL905 growth in the absence of arabinose were isolated. Of 1,664 transformants analyzed, we obtained 21 clones unable to fully complement FL905 in the nonpermissive conditions. Most noncomplementing clones harbored multiple mutations, as assessed by sequencing the mutant alleles. Nevertheless, three plasmids harbored only one or two mutations in LptC (Fig. 2A), namely, G56V, K177Stop (which generates a truncated protein lacking the C-terminal 15 amino acids [Δ177-191]), and Y112S-G153R (a double substitution mutant in the C-terminal region). FL905/pGS103Δ177-191 and FL905/pGS103Y112S-G153R growth was completely inhibited on agar medium in the absence of arabinose, whereas FL905/pGS103G56V growth was severely impaired in the absence of arabinose and formed “dust-like colonies” in this condition (Fig. 2B).
FIG. 2.
Mutations affecting LptC function. (A) Mutations in the lptC gene in the pGS103-derived plasmids. Nucleotide coordinate numbers are based on the designation of the start codon of lptC as 1. The amino acid numbers are based on the predicted sequence of LptC that is 191 amino acids in length. (B) Plating efficiency of FL905 (araBp-lptC) transformed with plasmid pGS103 carrying the wild-type protein (LptC and LptC-H, respectively), or plasmids carrying the LptC mutated proteins LptCG56V, LptCΔ177-191, LptCG153R, LptCG153R-H, or plasmid pGS100 (none), in agar plate containing chloramphenicol supplemented (+) or not (−) with arabinose. Serial dilutions are given on the left side of the panel.
The G153R substitution was also found in other multiple mutants isolated in our screening associated with different amino acid substitutions, suggesting that G153R could be the residue responsible of the observed phenotype. This was confirmed by the fact that both pGS103G153R and pGS108G153R, which expresses a His6-tagged LptCG153R mutant protein generated by site-directed mutagenesis (LptCG153R-H) (Table 1), were unable to complement FL905 (Fig. 2B). In all of the experiments described below, FL905/pGS108G153R was used. The mutations described above fall in conserved regions of the protein (see, for example, the sequence alignments of LptC orthologues from proteobacteria in reference 38).
Stability of the LptC mutant proteins.
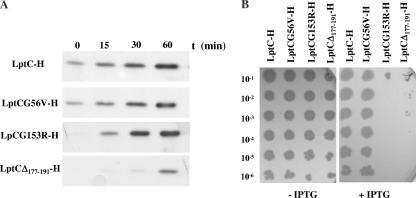
To assay the stability of LptC mutant proteins, we examined the level of ectopically expressed LptC-H (which can be distinguished from the endogenous wild-type protein) and its mutant derivatives in the wild-type strain AM604 upon induction with IPTG, assuming that the level of de novo-synthesized proteins correlates with their stability. It should be noted that in these conditions the chromosomal wild-type copy of lptC is expressed from its natural promoter. Detection of the proteins using HisProbe-HRP revealed that the level of LptCG56V-H was comparable to that of LptC-H and only the basal level (before IPTG induction) of LptCG153R-H was affected, suggesting that this protein is only slightly unstable (Fig. 3A). On the contrary, the level of the truncated LptCΔ177-191-H protein was severely reduced under these conditions, indicating that the truncated protein is intrinsically unstable.
FIG. 3.
LptC mutant stability in vivo and effects of their overexpression. (A) LptC mutant stability. AM604 cells harboring pGS108 (LptC-H), pGS108G56V (LptCG56V-H), pGS108G153R (LptCG153R-H), and pGS108Δ177-191 (LptCΔ177-191-H) were grown to the early logarithmic phase. Expression of LptC-H or its mutant forms were induced by the addition of 0.1 mM IPTG. Samples for protein analysis were obtained 0, 15, 30, and 60 min postinduction and analyzed using HisProbe-HRP. Equal amount of cells (0.12 OD600 units) was loaded into each lane. (B) Overexpression of LptCΔ177-191 and LptCG153R leads to cell lethality. Serial dilutions (indicated on the left) of overnight cultures of AM604 carrying the plasmids pGS108, pGS108G56V, pGS108Δ177-191, and pGS108G153R were replica plated onto agar plate supplemented (+) or not (−) with 0.5 mM IPTG and incubated overnight.
We then tested the effect of LptC mutants overexpression on E. coli growth by plating wild-type strain AM604 harboring pGS108, pGS108G56V, pGS108Δ177-191, and pGS108G153R on agar medium in the absence or in the presence of IPTG. Overexpression of LptCG56V did not impair growth; on the contrary, we found that overexpression of Δ177-191 and G153R LptC-H mutants abolished AM604 growth (Fig. 3B). Since LptC is part of a multiprotein complex these data suggest that either the mutants may titrate a wild-type interacting Lpt factor(s) or that their toxicity may be due to the overexpression of a misfolded protein.
The C-terminal region of LptC is required for LptA binding.
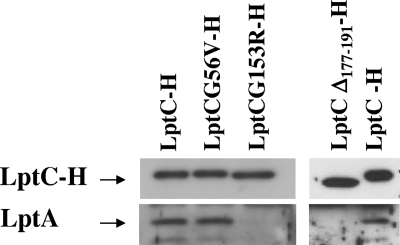
To test whether the G56V, Δ177-191, and G153R mutations could affect LptC interaction with LptA, we compared the wild-type and the mutant proteins for their ability to copurify LptA. Whole-cell extracts of the wild-type strain AM604 overexpressing LptC-H or the mutant His-tagged derivatives (G56V, Δ177-191, and G153R; Table 1) were subjected to affinity purification and LptA and LptC-H detected by Western blotting with anti-LptA antibodies and HisProbe-HRP, respectively. As shown in Fig. 4, LptCG56V-H retained the ability to copurify LptA, whereas the mutations in the LptC C-terminal region (G153R and Δ177-191) severely impaired LptA-LptC complex formation.
FIG. 4.
Effect of lptC mutations on LptC-LptA interaction. Whole-cell extracts from AM604 cells transformed with plasmids pGS108 carrying the wild-type protein (LptC-H) or the LptC-mutated proteins LptCG56V-H, LptCG153R-H, and LptCΔ177-191-H were subjected to affinity chromatography. Equal amounts (2.5 μg) of Ni-NTA column-enriched LptC-H and its mutant versions were separated by SDS-12.5% PAGE and analyzed by Western blotting with anti-LptA antibody and HisProbe-HRP, respectively.
Effects of different lptC alleles on Lpt complex and LPS transport.
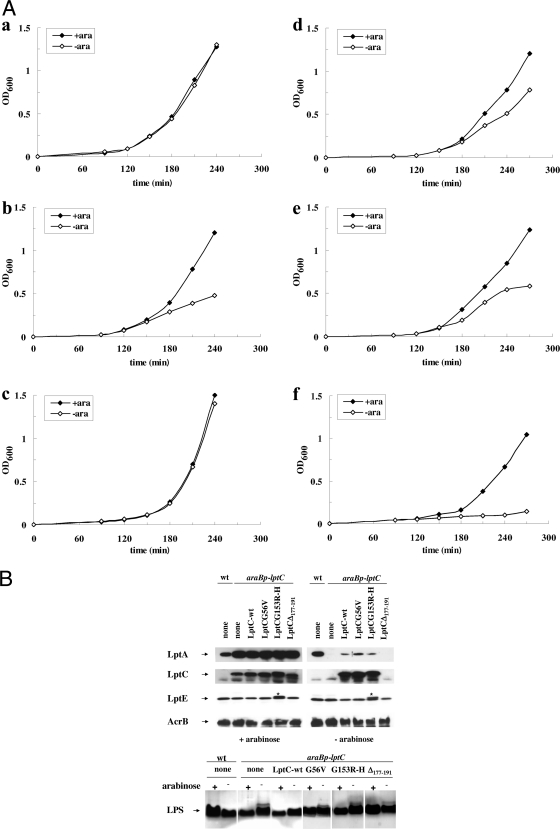
To gain better insights into the molecular role of LptC in the Lpt complex, we tested the effects of the three lptC mutant alleles on the steady-state level of LptA and LptE, the latter as a representative of the LptDE OM complex, and on LPS transport. The LptC+ depletion strain FL905 and its derivatives harboring pGS103, pGS103G56V, pGS103Δ177-191, and pGS108G153R were grown to the exponential phase and shifted into a medium lacking arabinose (nonpermissive condition) to deplete the chromosomally encoded LptC wild type, while allowing expression of the mutant proteins. Samples were then taken from cultures grown in the presence or absence of arabinose for 240 min (AM604, FL905, and FL905/pGS103) or 270 min (FL905/pGS103G56V, FL905/pGS103Δ177-191, and FL905/pGS108G153R) after the shift to nonpermissive conditions (Fig. 5A) and analyzed by Western blotting with anti-LptA, anti-LptC, and anti-LptE antibodies. The level of the IM protein AcrB was used as a sample loading control.
FIG. 5.
Effect of lptC mutations on Lpt proteins complex and LPS transport. (A) Growth curves of AM604 (wt; a), FL905 (araBp-lptC; b) strains and FL905 carrying plasmids expressing LptC (c), LptCG56V (d), LptCG153R-H (e), and LptCΔ177-191 (f). Cells growing exponentially in LD containing arabinose were harvested, washed, and subcultured in arabinose-supplemented (⧫) or arabinose-free (⋄) medium. Growth was monitored by measuring the OD600. Samples for protein and LPS analyses were taken from cells grown in the presence (+ara) or absence (−ara) of arabinose at 240 (wt, FL905, and FL905/pGS103) or 270 min (FL905/pGS103G56V, FL905/pGS108G153R, and FL905/pGS103Δ177-191) after the shift into fresh medium. (B) Steady-state levels of LptA, LptC, and LptE and LPS profile. Protein samples were subjected to Western blot analysis with anti-LptA, anti-LptC, anti-LptE, and anti-AcrB (as a loading control) antibodies. Anti-LptE antibodies cross-react with LptC-H His tag (*). Total LPS was extracted from mutant cells grown in the same conditions and at the same time points of protein analysis. LPS extracted from cultures with a total OD600 of 2 were separated by 18% Tricine-SDS-PAGE and silver stained. Equal amounts of cells, based on the OD measurement (0.2 OD600 units for protein analysis, 0.4 for LPS analysis), were loaded into each lane.
As shown in Fig. 5B (upper part), the level of physiologically expressed LptC seemed very low since the protein was undetectable in the wild-type strain with our antibody preparation. However, LptC was detected when ectopically expressed from a plasmid or from the araBp promoter (Fig. 5B). In FL905 cells expressing LptCG56V and LptCG153R, the level of the mutant proteins was comparable under permissive and nonpermissive conditions, whereas LptCΔ177-191 was undetectable in the noncomplemented strain (see below).
In the LptC+ depletion strain FL905 grown in the presence of arabinose, lptA is expressed from the upstream araBp promoter, and the level of LptA is higher than in the wild-type AM604 strain, where the protein is expressed from its natural promoter (29). The level of LptA in LptC+-depleted cells expressing LptCG153R-H was similar to that observed in the positive control (FL905 complemented by wild-type LptC), whereas LptA appeared slightly more abundant in LptC+-depleted cells expressing LptCG56V. On the contrary, in noncomplemented LptC+-depleted cells and in cells ectopically expressing LptCΔ177-191 LptA was undetectable. It thus appears that the absence of LptC protein caused by either depletion (noncomplemented LptC+-depleted cells) or mutation (LptC+-depleted cells expressing LptCΔ177-191) induces LptA destabilization. The abundance of LptE did not substantially change upon depletion of LptC with or without overexpression of any mutant LptC, indicating that the steady-state level of the OM component LptE was not affected by LptC depletion or mutations (Fig. 5B, upper part). The steady-state level of LptE thus served in these experiments also as a sample loading control.
Depletion of any Lpt protein leads to the production of LPS decorated by colanic acid; this phenotype is diagnostic of defects in LPS transport occurring downstream of the MsbA-mediated flipping of lipid A-core to the periplasmic face of the IM (29). We therefore analyzed the LPS profile in lptC mutant strains. The total LPS was extracted from nondepleted and LptC+-depleted FL905 complemented with wild-type and mutant lptC alleles, and the LPS profiles were analyzed as described previously (29). As shown in Fig. 5B (lower part), LPS decorated with colanic acid could be detected in LptC+-depleted FL905 complemented by each of the mutant alleles but not by wild-type lptC, indicating that each of the above LptC mutations impair LPS transport.
Evidence for LptC oligomerization in vivo.
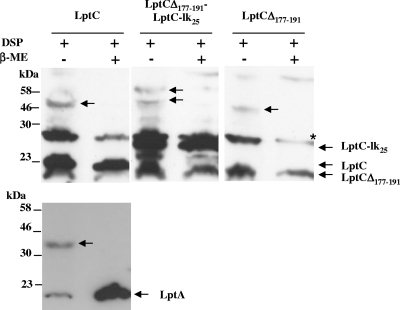
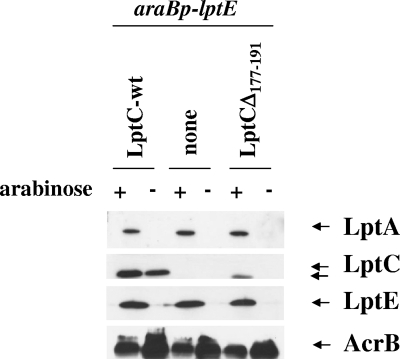
As noted above, LptCΔ177-191 mutant protein was not detectable upon depletion of the chromosomally encoded LptC+, whereas, when LptC+ was coexpressed from the araBp promoter, the LptCΔ177-191 level was higher (notice that the wild-type and truncated proteins can be distinguished by their different molecular weights; Fig. 5B, upper part). On the contrary, the LptCG56V and LptCG153R mutant proteins remained abundant upon depletion of wild-type LptC. LptCΔ177-191 is an intrinsically unstable protein (Fig. 3A) and appears to be stabilized by overexpression of LptC+, which is consistent with the idea that LptC might interact with itself to form a dimer (see also Fig. 1) or a multimer. An alternative explanation is that upon depletion envelope stress response is triggered and periplasmic proteases are induced that might degrade the unstable LptC truncated protein. To probe LptC oligomerization in vivo, we performed in vivo cross-linking experiments using the wild-type strain AM604 transformed with different LptC constructs. In DSP-treated AM604 cells expressing wild-type LptC a band of ∼46 kDa was visible consistent with the formation of an LptC-LptC complex (Fig. 6, upper panel). The 46-kDa band disappeared when the DSP cross-linker was reverted with a reducing agent. When samples were analyzed by Western blotting with anti-LptA antibodies, a band of ∼43 kDa was visible that possibly corresponded to an LptA-LptC complex (Fig. 6, lower panel). In DSP-treated AM604 cells expressing both LptCΔ177-191 and an LptC version carrying an additional 27 amino acids at the C-terminal end (LptC-lk25), two high-molecular-mass bands with an apparent masses of approximately 46 and 52 kDa appeared, possibly corresponding to LptC-lk25-LptCΔ177-191 and LptC-lk25-LptC-lk25 complexes. In DSP-treated cells expressing LptCΔ177-191 the level of the truncated protein was low compared to the LptC wild type and LptC-lk25, a finding consistent with the notion that the truncated protein is intrinsically unstable. However, an high-molecular-mass band of ∼44 kDa appeared, suggesting that even the truncated protein is able to dimerize (Fig. 6, upper panel). These data suggest that LptC can form dimers in vivo. However, this experiment does not prove that dimerization with wild-type LptC protects the truncated LptCΔ177-191 protein version from degradation in the LptC+ depletion strain FL905. Therefore, we examined the LptC levels in the lptE depletion strain AM689 grown under permissive and nonpermissive conditions and expressing either the wild-type or the truncated LptCΔ177-191 proteins. As shown in Fig. 7, LptCΔ177-191 was undetectable when the lptE depletion strain was grown in the absence of arabinose, an observation in line with the idea that LptCΔ177-191 is degraded by proteases induced under stress conditions.
FIG. 6.
LptC dimerization in vivo. Wild-type strain AM604 cells transformed with different LptC constructs were treated with the cross-linking DSP agent in vivo. Cell extracts were prepared and analyzed by Western blotting with anti-LptC and anti-LptA antibodies. Samples were treated with β-mercaptoethanol (β-ME) to revert the cross-linking. Equal amount of cells, (1.6 OD600 units) was loaded into each lane. An asterisk (*) labels a band that cross-reacts with the anti-LptC antibody.
FIG. 7.
Steady-state levels of LptA and LptC upon LptE depletion. Protein samples from AM689 cells transformed with various plasmids and grown in the presence (+) or absence (−) of arabinose 180 min after a shift in the nonpermissive condition were analyzed by Western blotting with anti-LptA, anti-LptC, and anti-LptE antibodies. Equal amounts of cells (0.2 OD600 units) were loaded into each lane. AcrB was used as a loading control.
Purified LptC is a dimer in solution.
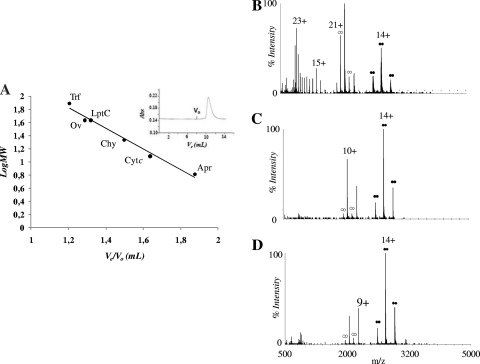
In order to probe the oligomeric state of LptC in vitro, the apparent molecular weight of the pure protein was tested by size-exclusion chromatography and ESI-MS under nondenaturing conditions. To do this, a preparation of the soluble version of LptC with an N-terminal His6 tag and lacking the first 23 amino acids of the transmembrane domain was analyzed (sH-LptC [see Materials and Methods]) by size-exclusion chromatography. The protein eluted as a single, slightly asymmetric peak on a Superdex75 column (Fig. 8A, inset). The LptC elution volume corresponded to an estimated molecular mass of ∼43,000 Da, relative to globular calibrants in the range 6,500 to 81,000 Da (Fig. 8A), suggesting a dimeric assembly. This conclusion was confirmed by nano-ESI-MS, which provides complementary information by direct detection and weighing of protein noncovalent complexes (13, 14). In Fig. 8B, C, and D, the spectra of pure sH-LptC preparations in the presence of ammonium acetate, which is known to favor detection of protein complexes by mass spectrometry, are reported. At 5 μM protein and 50 mM ammonium acetate (Fig. 8B), the spectrum shows signals of LptC monomers and dimers. Mass deconvolution gives a value of 20,536.8 (±0.25) Da for the monomer and 41,073.9 (±1.02) Da for the dimer which are in close agreement with the values calculated from the amino acid sequence (20,535.85 Da for the monomer and 41,071.7 Da for the dimer). Both species are characterized by conformational heterogeneity, as indicated by multimodal charge-state distributions (15), with maxima at 10+, 15+, and 23+ for the monomer and at 14+ and 21+ for the dimer. A 10-fold increase in the ionic strength results in a simpler spectrum, with only one monomeric and one dimeric species centered, respectively, on the 10+ and 14+ ions (Fig. 8C). This result suggests that the conformational heterogeneity observed at a lower ionic strength might reflect partial protein unfolding induced by the experimental conditions used. In Fig. 8D, the effect of protein concentration on the spectrum at high ionic strength is shown. The relative intensity of the monomer decreases as the protein concentration is increased from 5 to 20 μM, a finding consistent with a monomer-dimer equilibrium in the original liquid sample. The calculated relative amount of the dimer is ∼70%. These data provide direct evidence of a predominant dimeric state of LptC in solution. However, it should be underscored that the apparent dissociation constant (in the micromolar range, according to these results) could be affected by the peculiar conditions imposed by electrospray. Overall, these data suggest that the LptC dimerizes and that the soluble portion of the protein is sufficient to promote dimerization.
FIG. 8.
LptC dimerization in vitro. (A) Apparent molecular weight (MW) of LptC as determined by gel filtration. The relative elution volume of sH-LptC in 50 mM sodium phosphate (pH 8.0)-100 mM NaCl was compared to that of globular standards. The chromatogram is shown in the inset. Trf, transferrin; Ov, ovalbumin; Chy, chymotrypsin; Cytc, cytochrome c; Apr, aprotinin; Ve, elution volume; Vo, void volume; Abs, absorbance at 280 nm. (B, C, and D) Quaternary structure of LptC by mass spectrometry. The nano-ESI-MS spectra of 5 μM LptC in 50 mM ammonium acetate (pH 7.5) (B), 5 μM LptC in 500 mM ammonium acetate (pH 7.5) (C), and 20 μM LptC in 500 mM ammonium acetate (pH 7.5) (D) are shown. The main charge state of each peak envelope is labeled by the corresponding number of charges. Dimer-specific peaks are labeled by double circles. Solid symbols correspond to compact dimers, and open symbols correspond to less compact dimers.
Effect of LptD and LptE depletion on Lpt complex assembly.
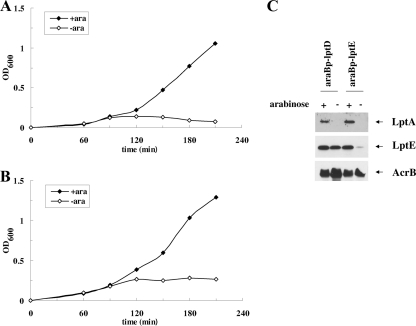
The experiments described above showed that the steady-state level of LptA is affected in the absence of wild-type LptC or by the Δ177-191 mutation, which severely impairs LptCΔ177-191 stability. It is possible that the absence of a proper IM docking structure for LptA results in LptA degradation in vivo. Therefore, the LptA level in the cell could be diagnostic of the properly bridged IM and OM. We thus tested whether the absence of the OM LptDE complex could also exert a similar effect on LptA stability. The AM661 and AM689 strains, in which the LptD and LptE expression is driven by the inducible araBp promoter (29), were grown under permissive conditions to exponential phase and then shifted to media lacking arabinose to deplete LptD and LptE, respectively. Samples for protein analyses were taken from cultures grown in the presence or in the absence of arabinose 210 min after the shift to nonpermissive conditions (Fig. 9A) and then processed for Western blot analysis with anti-LptA, anti-LptE, and anti-AcrB antibodies. As shown in Fig. 9B, a decreased steady-state level of LptA is observed upon LptE and LptD depletion. Overall, our data suggest that when the Lpt machinery is not functional for lack of either IM or OM components, LptA is destabilized, probably because it is not properly assembled in the Lpt complex.
FIG. 9.
LptA level upon LptD and LptE depletion. (A and B) Growth curves of AM661 (araBp-lptD [A]) and AM689 (araBp-lptE [B]). Cells growing exponentially in LD containing arabinose were harvested, washed, and subcultured in arabinose-supplemented (⧫) or arabinose-free (⋄) medium. Growth was monitored by measuring the OD600. (C) Steady-state levels of LptA. Samples for protein analysis taken from cells grown in the presence (+) or absence (−) of arabinose at 210 min after the shift into nonpermissive condition were analyzed by Western blotting with anti-LptA and anti-LptE antibodies. Equal amounts of cells (0.2 OD600 units) were loaded into each lane. AcrB was used as the loading control.
DISCUSSION
LptA interaction with IM and OM Lpt components.
The Lpt proteins constitute a machinery for LPS transport to the cell surface. Genetic and biochemical evidence indicates that the components of the machinery work in a concerted way. In fact, upon depletion of any of the seven Lpt proteins, the LPS assembly pathway is blocked in nearly the same fashion (23, 29); moreover, it has been recently shown that the seven Lpt proteins physically interact and constitute a transenvelope complex connecting IM and OM (4). However, little is known about how specific Lpt factors interact with each other. We show here by copurification experiments that in vivo LptA binding to IM occurs through binding to LptC, although we cannot rule out the possibility that LptA may also interact with other IM components such as LptFG, which possess periplasmic loops (23).
LptC depletion results in a significant decrease of LptA level. Similarly, the LptA level in the cells decreases upon LptD or LptE depletion, strongly suggesting that LptA also interacts with the LptDE complex at the OM and that in a non-properly assembled Lpt complex LptA may be degraded. It is likely that LptC and the LptDE complex play a structural role and prevent LptA degradation. Instability and/or degradation of components of protein complexes when proven or proposed interacting partners are either depleted or not functional is not unusual and provides indirect evidence of physical and functional interaction. The loss of any protein of the PilMNOP IM complex required for biogenesis of type IV pili in Pseudomonas aeruginosa results in instability of the other interacting partners (1); similarly, EspL and EspM, which belong to the type II secretion pathway required for cholera toxin secretion in Vibrio cholerae, have been shown to participate in mutual stabilization by protecting each other from proteolysis (25). Finally, Chng and coworkers demonstrated that LptD can only be overexpressed with LptE and that purified LptE is resistant to proteolytic degradation when in complex with LptD, thus indicating a structural role of LptE in stabilizing the folded state or facilitating the folding and assembly of LptD (5).
Interaction of LptA with LptC and the LptDE complex supports the model proposed by Kahne and coworkers (4), which posits that LptA interacts with both the IM and the OM. Consistent with this model are previous findings showing that in the crystals obtained in the presence of LPS, the LptA monomers are packed as a linear filament (34), suggesting that LptA oligomerization is functional to bridging the IM and OM. Since LptA consists of a conserved domain also found in the N-terminal periplasmic domain of LptD, we suggest that the latter protein may be the docking site for LptA at the OM (2, 30) and thus that LptA is anchored to the IM and OM through LptC and LptD, respectively.
LptC structure-function relationship.
The analysis of loss-of-function mutations in lptC allowed us to further support the structural role of LptC-LptA interaction in preventing LptA degradation and to define functional regions in the LptC protein. The LptCG56V substitution falls in a stretch of relatively conserved residues and, based on the recently reported crystal structure, this residue is contained in one of the two disordered regions of the protein (residues 24 to 58) where no electron density was observed (38). The G56V amino acid substitution does not abolish interaction with LptA and in cells expressing LptCG56V the LptA level seems even higher than that observed in cells expressing wild-type LptC. However, LptCG56V impairs LPS transport, as demonstrated by the decreased viability and the appearance of LPS decorated with colanic acid in cells expressing the mutant protein only. These data suggest that this mutation does not affect IM and OM bridging by the Lpt complex, although the complex is not functional. Since LptC has been shown to bind LPS in vitro (38), it may be possible that the G56V mutation impairs such binding. An alternative hypothesis can be that during LPS transport LptC may undergo conformational changes, as suggested by the recently reported crystal structure analysis (38), that could be compromised by the G56V mutation.
The LptCG153R mutant protein displays a reduced affinity to LptA since it is not able to copurify LptA. As for LptCG56, G153 also falls in a stretch of conserved residues (38). The inability of LptCG153R to copurify LptA suggests that the binding determinants are located in the C-terminal region of the protein. In cells expressing LptC+ or LptCG153R, the LptA level is comparable, suggesting that the residual binding activity of LptCG153R mutant protein, overexpressed from a plasmid, is sufficient to prevent LptA degradation. Nevertheless, LptC+-depleted FL905 cells expressing LptCG153R are not viable (Fig. 2B) and accumulate colanic acid decorated LPS (Fig. 5B), which indicates that transport is impaired.
LptCΔ177-191 is highly unstable (Fig. 3A) and fails to interact with LptA (Fig. 4), and thus the assembly of the Lpt complex is compromised. Interestingly, the C-terminal deletion of LptCΔ177-191 removes the second disordered region of the protein (residues 185 to 191) (38). It is well established that unfolded or only partly folded proteins in their native states fold into an ordered structure on binding a partner molecule/protein (7, 8). A well-studied example is the binding of colicins to disordered regions of their OM receptors as a key step in their translocation into the target cell (12, 37). Therefore, the intrinsically disordered region in the C-terminal end of LptC (residues 185 to 191) might be reorganized and folded upon binding to LptA.
We show here that in the Lpt machinery, LptC interacts with LptA. The recently solved crystal structure of LptC reveals a striking structural similarity to LptA despite the fact that the two proteins do not share sequence similarity (38). LptC is also part of the LptBCFG complex (18). A possible candidate for interaction in this complex is LptF since its periplasmic loop displays a structural similarity to LptC (http://zhang.bioinformatics.ku.edu/I-TASSER/). We provide evidence here that LptC, when overexpressed, is also able to dimerize in vivo (Fig. 6A). In addition to the in vivo data, size-exclusion chromatography and ESI-MS experiments showed that a soluble LptC version missing the N-terminal transmembrane domain (sH-LptC) forms dimers in vitro whose abundance increases with increasing protein concentration and ionic strength. In a very recent study it was reported that LptC exists as a monomer in solution (38). It is possible that this discrepancy is due to the different approaches and experimental conditions used. However, we suggest that the dimeric form is not the physiological preferred state in wild-type cells and that in the Lpt complex a single LptC molecule is present; our data suggest that LptC overexpression in vivo may shift the equilibrium between heterodimerization (interaction with LptA and/or LptF in the LptBFG complex) and homodimerization. It has been shown that the relative stoichiometry of the IM PilM/N/O/P proteins is crucial in promoting pilus assembly in P. aeruginosa. When overexpressed, PilO may form homodimers. However, the heterodimer PilN/O is the physiologically preferred state, occurring when PilO is expressed from the chromosome (1). The subunit ratio of the LptBCFG complex (LptB/LptF/LptG/LptC) has been proposed to be 2:1:1:1 (predicted molecular mass of 157.2 kDa) (18). However, the molecular mass of the LptBCFG complex determined by size-exclusion chromatography was ∼330 kDa (18), which would be consistent with the ability of LptC to interact with itself, thus resulting in dimerization of LptBCFG complex.
As discussed above, the LptCΔ177-191 truncated protein is intrinsically unstable, since it may expose unfolded regions and become a protease substrate (40). Alternatively, LptCΔ177-191 could not associate with the LptBFG complex and might thus be degraded in the periplasm. In fact, it has been reported that unbalanced expression of LptB in an LptFG wild-type background results in the loss of IM association and the degradation of soluble LptB molecules in the cytoplasm (18). LptC could also be also stabilized by the interaction with LptA, the level of which does indeed increase when the LptC+ depletion strain FL905 is grown with arabinose. A similar codependence has been observed for the IM divisome subcomplex FtsB/FtsL/FtsQ, where it has been shown that FtsL and FtsB are codependent for stabilization (10). However, our data (Fig. 7) suggest that the stability of LptC does not depend on LptA. In fact, upon LptE depletion LptA is undetectable since the OM docking site is missing, but the LptC level is not affected.
In conclusion, we provide evidence here that depletion of LptC, LptD, LptE, or mutations in LptC abolishing its function break the Lpt machinery by impairing either its assembly (LptCΔ177-191) or functionality (LptCG153R and LptCG56V). The lack of LptC (because the protein is depleted or mutated) or LptDE removes the IM and OM LptA docking sites, and the LptA level in the cell decreases. Based on these data, we suggest that the steady-state level of LptA is controlled at the protein stability level by the assembly of the Lpt complex since the inability of the protein to properly interact with IM and OM docking sites results in its degradation. Therefore, the LptA level could be used as a marker of properly bridged IM and OM components. Our genetic and biochemical data confirm that LptC plays a key role in docking LptA and other Lpt factors at the IM, in addition to a direct role in LPS transport, since the protein has also been shown to bind LPS (38). Overall, the data presented here strongly support the transenvelope complex model for LPS transport.
Acknowledgments
We are grateful to Daniel Kahne for providing the LptC and LptD antibodies. We thank K. M. Pos for providing the AcrB antibodies. We also thank Chiara Raimondi for excellent technical assistance.
This study was supported in part by MIUR (Ministero dell'Istruzione Università e Ricerca) grant PRIN 200824M2HX (A.P.) and Regione Lombardia Cooperazione Scientifica e Tecnologica Internazionale grant 16876 SAL-18 (A.P.).
Footnotes
Published ahead of print on 17 December 2010.
REFERENCES
- 1.Ayers, M., et al. 2009. PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J. Mol. Biol. 394:128-142. [DOI] [PubMed] [Google Scholar]
- 2.Bos, M. P., V. Robert, and J. Tommassen. 2007. Biogenesis of the Gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61:191-214. [DOI] [PubMed] [Google Scholar]
- 3.Bos, M. P., B. Tefsen, J. Geurtsen, and J. Tommassen. 2004. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl. Acad. Sci. U. S. A. 101:9417-9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chng, S. S., L. S. Gronenberg, and D. Kahne. 2010. Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry 49:4565-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chng, S. S., N. Ruiz, G. Chimalakonda, T. J. Silhavy, and D. Kahne. 2010. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc. Natl. Acad. Sci. U. S. A. 107:5363-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doerrler, W. T., H. S. Gibbons, and C. R. Raetz. 2004. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 279:45102-45109. [DOI] [PubMed] [Google Scholar]
- 7.Dyson, H. J., and P. E. Wright. 2002. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 12:54-60. [DOI] [PubMed] [Google Scholar]
- 8.Dyson, H. J., and P. E. Wright. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell. Biol. 6:197-208. [DOI] [PubMed] [Google Scholar]
- 9.Eom, G. T., J. K. Song, J. H. Ahn, Y. S. Seo, and J. S. Rhee. 2005. Enhancement of the efficiency of secretion of heterologous lipase in Escherichia coli by directed evolution of the ABC transporter system. Appl. Environ. Microbiol. 71:3468-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez, M. D., and J. Beckwith. 2009. Divisome under construction: distinct domains of the small membrane protein FtsB are necessary for interaction with multiple cell division proteins. J. Bacteriol. 191:2815-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 12.Hecht, O., H. Ridley, J. H. Lakey, and G. R. Moore. 2009. A common interaction for the entry of colicin N and filamentous phage into Escherichia coli. J. Mol. Biol. 388:880-893. [DOI] [PubMed] [Google Scholar]
- 13.Heck, A. J., and R. H. Van Den Heuvel. 2004. Investigation of intact protein complexes by mass spectrometry. Mass Spectrom. Rev. 23:368-389. [DOI] [PubMed] [Google Scholar]
- 14.Invernizzi, G., A. Natalello, M. Samalikova, and R. Grandori. 2007. Protein-protein and protein-ligand interactions studied by electrospray-ionization mass spectrometry. Protein Pept. Lett. 14:894-902. [DOI] [PubMed] [Google Scholar]
- 15.Kaltashov, I. A., and R. R. Abzalimov. 2008. Do ionic charges in ESI MS provide useful information on macromolecular structure? J. Am. Soc. Mass Spectrom. 19:1239-1246. [DOI] [PubMed] [Google Scholar]
- 16.Ma, B., C. M. Reynolds, and C. R. Raetz. 2008. Periplasmic orientation of nascent lipid A in the inner membrane of an Escherichia coli LptA mutant. Proc. Natl. Acad. Sci. U. S. A. 105:13823-13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyake, K. 2004. Innate recognition of lipopolysaccharide by Toll-like receptor 4-MD-2. Trends Microbiol. 12:186-192. [DOI] [PubMed] [Google Scholar]
- 18.Narita, S., and H. Tokuda. 2009. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 583:2160-2164. [DOI] [PubMed] [Google Scholar]
- 19.Polissi, A., and C. Georgopoulos. 1996. Mutational analysis and properties of the msbA gene of Escherichia coli, coding for an essential ABC family transporter. Mol. Microbiol. 20:1221-1233. [DOI] [PubMed] [Google Scholar]
- 20.Raetz, C. R., et al. 2009. Discovery of new biosynthetic pathways: the lipid A story. J. Lipid Res. 50(Suppl.):S103-S108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reuhs, B. L., R. W. Carlson, and J. S. Kim. 1993. Rhizobium fredii and Rhizobium meliloti produce 3-deoxy-d-manno-2-octulosonic acid-containing polysaccharides that are structurally analogous to group II K antigens (capsular polysaccharides) found in Escherichia coli. J. Bacteriol. 175:3570-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz, N., L. S. Gronenberg, D. Kahne, and T. J. Silhavy. 2008. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105:5537-5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabbattini, P., F. Forti, D. Ghisotti, and G. Dehò. 1995. Control of transcription termination by an RNA factor in bacteriophage P4 immunity: identification of the target sites. J. Bacteriol. 177:1425-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandkvist, M., L. P. Hough, M. M. Bagdasarian, and M. Bagdasarian. 1999. Direct interaction of the EpsL and EpsM proteins of the general secretion apparatus in Vibrio cholerae. J. Bacteriol. 181:3129-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sklar, J. G., T. Wu, D. Kahne, and T. J. Silhavy. 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 21:2473-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sperandeo, P., et al. 2007. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J. Bacteriol. 189:244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sperandeo, P., G. Dehò, and A. Polissi. 2009. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim. Biophys. Acta 1791:594-602. [DOI] [PubMed] [Google Scholar]
- 29.Sperandeo, P., et al. 2008. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J. Bacteriol. 190:4460-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sperandeo, P., C. Pozzi, G. Dehò, and A. Polissi. 2006. Non-essential KDO biosynthesis and new essential cell envelope biogenesis genes in the Escherichia coli yrbG-yhbG locus. Res. Microbiol. 157:547-558. [DOI] [PubMed] [Google Scholar]
- 31.Sprott, G. D., G. Ferrante, and I. Ekiel. 1994. Tetraether lipids of Methanospirillum hungatei with head groups consisting of phospho-N,N-dimethylaminopentanetetrol, phospho-N,N,N-trimethylaminopentanetetrol, and carbohydrates. Biochim. Biophys. Acta 1214:234-242. [DOI] [PubMed] [Google Scholar]
- 32.Steeghs, L., et al. 1998. Meningitis bacterium is viable without endotoxin. Nature 392:449-450. [DOI] [PubMed] [Google Scholar]
- 33.Stenberg, F., et al. 2005. Protein complexes of the Escherichia coli cell envelope. J. Biol. Chem. 280:34409-34419. [DOI] [PubMed] [Google Scholar]
- 34.Suits, M. D., P. Sperandeo, G. Dehò, A. Polissi, and Z. Jia. 2008. Novel structure of the conserved Gram-negative lipopolysaccharide transport protein A and mutagenesis analysis. J. Mol. Biol. 380:476-488. [DOI] [PubMed] [Google Scholar]
- 35.Tefsen, B., J. Geurtsen, F. Beckers, J. Tommassen, and H. de Cock. 2005. Lipopolysaccharide transport to the bacterial outer membrane in spheroplasts. J. Biol. Chem. 280:4504-4509. [DOI] [PubMed] [Google Scholar]
- 36.Thanabalu, T., E. Koronakis, C. Hughes, and V. Koronakis. 1998. Substrate-induced assembly of a contiguous channel for protein export from Escherichia coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 17:6487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tozawa, K., et al. 2005. Clusters in an intrinsically disordered protein create a protein-binding site: the TolB-binding region of colicin E9. Biochemistry 44:11496-11507. [DOI] [PubMed] [Google Scholar]
- 38.Tran, A. X., C. Dong, and C. Whitfield. 2010. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J. Biol. Chem. 285:33529-33539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran, A. X., M. S. Trent, and C. Whitfield. 2008. The LptA protein of Escherichia coli is a periplasmic lipid A-binding protein involved in the lipopolysaccharide export pathway. J. Biol. Chem. 283:20342-20349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickner, S., M. R. Maurizi, and S. Gottesman. 1999. Posttranslational quality control: folding, refolding, and degrading proteins. Science 286:1888-1893. [DOI] [PubMed] [Google Scholar]
- 41.Wu, T., et al. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 103:11754-11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zgurskaya, H. I., and H. Nikaido. 2000. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J. Bacteriol. 182:4264-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, Z., K. A. White, A. Polissi, C. Georgopoulos, and C. R. Raetz. 1998. Function of Escherichia coli msbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 273:12466-12475. [DOI] [PubMed] [Google Scholar]