Abstract
The Lrp/AsnC family of transcription factors links gene regulation to metabolism in bacteria and archaea. Members of this family, collectively, respond to a wide range of amino acids as coregulators. In Escherichia coli, Lrp regulates over 200 genes directly and is well known to respond to leucine and, to a somewhat lesser extent, alanine. We focused on Lrp from Proteus mirabilis and E. coli, orthologs with 98% identity overall and identical helix-turn-helix motifs, for which a previous study nevertheless found functional differences. Sequence differences between these orthologs, within and adjacent to the amino acid-responsive RAM domain, led us to test for differential sensitivity to coregulatory amino acids. In the course of this investigation, we found, via in vivo reporter fusion assays and in vitro electrophoretic mobility shift experiments, that E. coli Lrp itself responded to a broader range of amino acids than was previously appreciated. In particular, for both the E. coli and P. mirabilis orthologs, Lrp responsiveness to methionine was similar in magnitude to that to leucine. Both Lrp orthologs are also fairly sensitive to Ile, His, and Thr. These observations suggest that Lrp ties gene expression in the Enterobacteriaceae rather extensively to physiological status, as reflected in amino acid pools. These findings also have substantial implications for attempts to model regulatory architecture from transcriptome measurements or to infer such architecture from genome sequences, and they suggest that even well-studied regulators deserve ongoing exploration.
The Leucine-responsive regulatory protein (Lrp)/AsnC family of transcription factors is broadly distributed and frequently ties bacterial metabolism to environmental signals, mediating transitions between “feast and famine” (11, 52). Family members are present in archaea as well as bacteria (12). Lrp/AsnC proteins include an N-terminal domain with a helix-turn-helix motif that interacts with DNA, as well as a C-terminal RAM (regulation of amino acid metabolism) domain that responds to amino acid coregulators (21, 25, 56, 63, 67). The C-terminal domain also mediates the formation of multimers (dimers, octamers, and hexadecamers), providing additional regulatory complexity (14, 16). Lrp from Escherichia coli (EcoLrp) is the most extensively studied protein in this family, and its structure has been determined (21) (see Fig. 1C). EcoLrp is a global regulatory protein that regulates ∼400 genes in E. coli, of which at least 130 involve direct interactions (17, 71). This regulation appears to help E. coli adapt between two major environments: “gut and gutter” (11).
FIG. 1.
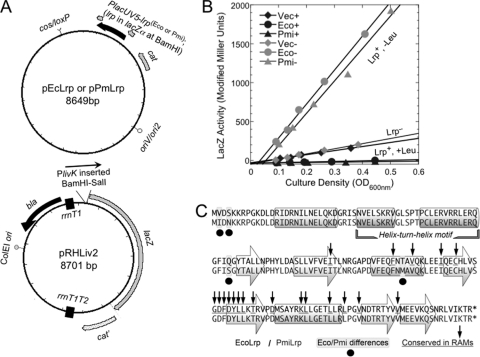
PlivK model system. (A) Plasmid diagrams. pEcLrp and pPmLrp are pCC1BAC-based plasmids containing (respectively) E. coli lrp or P. mirabilis lrp inserted into the BamHI site under the control of PlacUV5 and an artificial consensus ribosome-binding site. pRHLiv2 is derived from pKK223-3 (GenBank accession number M77749.1) and contains a PlivK-lacZ fusion transcriptionally isolated from the rest of the plasmid by strong bidirectional terminators (boxes). The cat gene of pRHLiv2 was inactivated to allow independent selection for each plasmid. (B) LacZ (β-galactosidase) activity is plotted versus culture optical density; a straight line indicates steady-state growth, and the slopes reflect relative levels of expression. In the key, “+” (black symbols) indicates the presence of 10 mM l-Leu in the MOPS-glucose medium, and “−” (gray symbols) indicates its absence. “Vec” is the pCC1BAC vector control (no Lrp), while “Eco” and “Pmi,” respectively, refer to EcoLrp and PmiLrp. (C) Sequences of Lrp from two members of the Enterobacteriaceae, Escherichia coli (EcoLrp; upper line) and Proteus mirabilis (PmiLrp; lower line), in single-letter amino acid code. The four sequence differences are indicated by black circles underneath. Indications of secondary structure (cylinders for helices and arrows for strands) are derived from the crystal structure of EcoLrp (21); the helix-turn-helix motif, primarily responsible for DNA sequence recognition, is shown. Positions of conserved residues from the RAM domain (regulation of amino acid metabolism (25)) are indicated by underlining of the EcoLrp sequence and vertical arrows.
Lrp orthologs from several genera are collectively responsive to a variety of amino acids, including Leu, Ala, Arg, Gln, His, Lys, Met, Phe, Pro, Thr, Trp, Tyr, and Val (though this has not yet been shown to involve direct effects in all cases). In contrast, as shown in Table 1, the well-studied E. coli Lrp ortholog (EcoLrp) has only been reported to respond to Leu (14-16, 28, 56, 65, 76) and Ala (6, 35, 43, 45, 79, 80). At least in the case of EcoLrp and Leu, their interaction modulates multimerization with associated effects on transcription (14, 16).
TABLE 1.
Coregulators of Lrp orthologs
| Species | Amino acida | Methodb | Reference(s)c |
|---|---|---|---|
| Escherichia coli | Leu | Indirect (in vivo) | e.g., 6, 38, 78 |
| Mutational analysis | 56 | ||
| EMSA | 7, 24, 80 | ||
| DNase I footprints | 41, 74 | ||
| Dynamic light scattering | 14, 16 | ||
| Ala | Indirect (in vivo) | 6, 35, 45 | |
| EMSA | 45, 80 | ||
| DNase I footprints | 80 | ||
| Butyrate | Indirect (in vivo) | 49 | |
| Actinobacillus pleuropneumoniae | Ile/Leu/Val mix | Indirect (in vivo) | 47 |
| Klebsiella aerogenes | Ala | Indirect (in vivo) | 33 |
| Mycobacterium tuberculosis | Phe, Tyr, Met, His, Lys, Arg, Pro, Thr, Gln | Crystallography | 67 |
| Neisseria meningitidis | Leu, Met | Crystallography | 63 |
| Proteus mirabilis | Leu | Indirect (in vivo) | 38 |
| Pseudomonas aeruginosa | d/l-Ala, Val | Indirect (in vivo) | 10 |
| Salmonella enterica serovar Typhimurium | Leu | Indirect (in vivo), EMSA, DNase I footprints | 4, 30, 42, 47 |
| Vibrio cholerae | Leu | Indirect (in vivo) | 38 |
l isomer, unless otherwise indicated.
“Indirect (in vivo)” refers to methods such as response of a lacZ reporter fusion when the amino acid is added to the growth medium.
Where many references report a particular type of observation, a representative subset is listed.
It would be very useful for bioinformatic inference of cell physiology (or for understanding its evolution) to assume conserved regulatory properties for conserved regulatory proteins. However, the appropriate limits for such extrapolation between species have not yet been well defined, and orthologous regulators do not always play the same roles (31, 34, 39, 40, 59). In a previous study, we found distinct regulatory differences between Lrp orthologs from Vibrio cholerae, Proteus mirabilis, and E. coli, even when they were expressed in the same background from the same expression sequences (38). These differences were seen despite the orthologs' very high sequence identity and completely conserved helix-turn-helix motifs. One of the few sequence differences between these orthologs lay within a region involved in coregulator interactions (25, 56) (see Fig. 1C). We accordingly explored the possibility that differences within the coregulator binding domains could help explain differences in Lrp regulatory behavior. While no substantial differences were observed, we found that both Lrp orthologs responded to a wider range of amino acids than had been previously reported.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Most bacterial strains used in this study were derivatives of the E. coli W3110-based BE10.2 background (46) and contained the pCC1BAC-based plasmid pVec (vector control), pEcLrp (E. coli lrp), or pPmLrp (P. mirabilis lrp), together with the compatible reporter plasmid pRHLiv2 (PlivK-lacZ) or pPM2005 (PgltB-lacZ) (38) (see Fig. 1A). The lrp-bearing plasmids contain the respective lrp open reading frames (ORFs), with a consensus Shine-Dalgarno ribosome binding site, downstream of the vector's PlacUV5 promoter. The reporter plasmids are derived from pBH403, which in turn is derived from pKK223-3 (GenBank accession number M77749.1). E. coli strain SPB107 carries two chromosomal fusions: PlivK-lacZ as the result of a λplacMu insertion and PlacUV5-lrp from background strain AAEC546 (7, 8). Cells were grown in baffled flasks with gyratory shaking at 37°C except in the 20-amino-acid screening experiment, in which they were grown at 37°C in 5-ml capped polypropylene culture tubes on a rotator. These cultures were grown in morpholinopropane sulfonate (MOPS) glucose minimal medium (50) from Teknova (Hollister, CA). Background expression from PlacUV5 is sufficient (38), so no isopropyl-β-d-thiogalactopyranoside (IPTG) inducer was used. For protein purification, cells were grown with aeration in STG medium (LB containing 0.2% glycerol and 50 mM potassium phosphate at pH 7.4) (46). Antibiotics were used where indicated: ampicillin (100 μg/ml), tetracycline (10 μg/ml), and chloramphenicol (15 μg/ml). Media were supplemented with amino acids as indicated at 10 mM, except for tryptophan (5 mM). Overnight cultures were inoculated from M9-glucose agar plates (66), which had been streaked the previous day from frozen stocks. These starter cultures were serially diluted just after inoculation to ensure that exponential-phase cells could be used as the main culture inoculum the next day. Main culture inoculation involved 1:50 dilution into fresh medium of overnight cultures having an optical density at 600 nm (OD600) of 0.4 to 0.8.
β-Galactosidase assays.
Experimental cultures were inoculated (1:250) from overnight ones. Between OD600 values of 0.1 and 0.8, 1-ml culture samples were collected and lysed by vortex mixing for 30 s with 50 μl of chloroform and 25 μl of 10% (wt:vol) SDS. To determine β-galactosidase levels, the rate of o-nitrophenyl-β-d-galactopyranoside (ONPG) hydrolysis was plotted against culture absorbance and fitted by linear regression to yield β-galactosidase activity (38, 57). When plotting against culture absorbance, this term is excluded from the calculation of β-galactosidase activity, which is thus expressed in “modified Miller units.” The original unit description is in reference 48.
Western blot analysis.
Equal volumes of cell cultures were centrifuged at 13,000 (13K) × g for 2 min, and pellets were suspended in SDS buffer and boiled for 10 min. Protein concentrations were determined by the Lowry-based RC DC protocol (Bio-Rad, Hercules, CA). Equal amounts of protein were loaded on 10% polyacrylamide gels, electrophoresed at 110 V in 1× Tris-glycine SDS buffer (66), and electroblotted to polyvinylidene difluoride (PVDF) using an Xcell apparatus (Invitrogen, Carlsbad, CA). The blotted membrane was blocked with 5% powdered milk in PBST (137 mM sodium chloride, 2 mM potassium chloride, 10 mM dibasic sodium phosphate, 1.7 mM monobasic potassium phosphate, 0.05% Tween 20, pH 7.4) and probed with a 1:10,000 dilution of rabbit anti-EcoLrp polyclonal antiserum and a 1:25,000 dilution of horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (a gift of Darren Sledjeski). Detection made use of ECLplus reagents (GE Health Science, Piscataway, NY) per the manufacturer's instructions. Protein bands were visualized on an UltraLum 16vS imaging system (Omega, Claremont, CA), and densitometry was performed using the ImageJ software program (62).
Protein purification.
Native Lrp protein was purified as previously reported (46). In short, E. coli JWD3-1 cells were grown in 500 ml STG medium and induced with 0.5 mM IPTG when the culture reached an OD600 of 1.0 to 1.5. Cells were grown for 2 h postinduction and were then pelleted and frozen until purification. For purification, cells were sonicated in TG10ED buffer (3 ml/g cells; 10 mM Tris [pH 8.0], 10% glycerol, 0.1 mM EDTA, 0.2 M NaCl, and 0.1 mM dithiothreitol [DTT]) with 100 μl 1.14 M phenylmethylsulfonyl fluoride (PMSF) per 500 ml cells grown. Sonication was carried out in a cup horn probe (Ultrasonics, Plainview, NY) at maximum power for five rounds of 1-min duration, each separated by 2 min on ice. The lysate was centrifuged for 30 min at 15K × g, and the resulting supernatant was loaded onto a 1- by 12-cm BioRex70 cation exchange column (Bio-Rad, Hercules, CA) equilibrated with TG10ED. Proteins were eluted with a 0.2 to 1.0 M NaCl gradient, and fractions were analyzed by examining stained SDS polyacrylamide gels for the Lrp 18.9-kDa band. Fractions containing Lrp were pooled and concentrated with VivaSpin concentrators having a 10,000-molecular-weight cutoff (Sartorius Stedim, Dusseldorf, Germany). Concentrated Lrp fractions were then loaded onto a 1- by 28-cm Superose12 column (GE Healthcare, Uppsala, Sweden) equilibrated with TG10ED buffer. Fractions containing highly purified Lrp were concentrated and dialyzed into MES buffer (10 mM N-morpholinoethane sulfonate, pH 6.25, 0.1 mM EDTA, and 0.2 M KCl). For stability of Lrp in sensitive assays of binding or multimerization, we have found that transfer to the MES buffer must occur within 96 h of lysis.
Electrophoretic mobility shift assays.
DNA fragments encompassing the livK promoter (PlivK) were amplified using the following primers: forward, 5′-TACTCCGGCC AGATCTTTTGCA; reverse, 5′-TAGTCAAAAA TCCCCATTCGTGA. There are two starting points for livK transcription, separated by 24 bp (1); the PlivK fragment ranged from −305 (relative to P1) through +170 (relative to P2). PgltB DNA was obtained using the following primers forward, 5′-CGAGGGATCC GGTACCGCGG TCTAGATACC GTCACGGTTA GGGCAG; reverse, 5′-CGCCGTCGAC TCGCCCCCTT GTTGTCCTTT. Thus, relative to the starting point of transcription (53), the PgltB fragment ranged from −573 to +107.
Purified Lrp was mixed with 23 nM DNA in a solution containing 40 mM Tris (pH 7.4), 60 mM KCl, 0.1 mM EDTA, 5% glycerol, 80 mM NaCl, and 1 mM DTT, with a given l-amino acid at the concentration indicated above. The samples were incubated at 23°C for 20 min prior to the addition of 1 μl Novex high-density Tris-borate-EDTA (TBE) sample buffer (Invitrogen, Carlsbad, CA) and immediately loaded onto a 1.5-mm 4% acrylamide TBE gel in an Xcell apparatus (Invitrogen). Samples were electrophoresed at 110 V until they entered the gel and then resolved at 80 V at room temperature. The gel was stained with 0.5 μg/ml ethidium bromide and visualized with an UltraLum imager (Omega, Claremont, CA). Densitometry was performed with ImageJ (62).
RESULTS
PlivK as a sensitive measure of Lrp response to coregulators.
To investigate potential functional differences between Lrp orthologs, we used the E. coli livKHMGF promoter (PlivK) fused to the reporter gene lacZ. PlivK was chosen because it is both activated by Lrp in the absence of leucine and repressed by Lrp in the presence of leucine (7, 28, 38), thus giving a wide dynamic range. Lrp orthologs from E. coli and P. mirabilis were introduced into an lrp-Tn10 strain of E. coli on the plasmids pEcLrp and pPmLrp (38) (Fig. 1 A). These plasmids, based on the low-copy pCC1BAC vector (75), have identical expression sequences upstream of both orthologs and have been used in previous studies (38). The compatible reporter plasmid carries a PlivK-lacZ transcriptional fusion, isolated from the rest of the plasmid by flanking terminators. As shown in Fig. 1B, Lrp from E. coli or P. mirabilis has essentially identical effects on PlivK-lacZ, with ∼8× activation in the absence of Leu and repression to nearly background levels in the presence of Leu.
Several amino acids besides leucine elicit Lrp-dependent differential expression of PlivK.
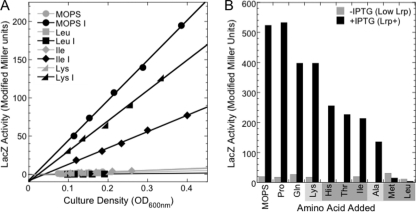
Lrp is well known to respond to leucine (hence its name as the Leucine-responsive regulatory protein). However, other amino acids are also known to affect Lrp behavior, even in E. coli, yet are rarely considered (Table 1 and references therein). We examined the effects of all 20 amino acids on PlivK-lacZ to functionally compare the E. coli and P. mirabilis orthologs of Lrp (EcoLrp and PmiLrp). For this screening, we grew triplicate cultures of E. coli carrying PlivK-lacZ on one plasmid and producing either EcoLrp or PmiLrp from a compatible plasmid (Fig. 1A) in capped tubes rotated at 37°C. The cultures were grown in MOPS minimal glucose medium supplemented with one amino acid at a concentration of 10 mM (except for Trp, which was added at 5 mM). Samples were collected when the OD600 was between 0.4 and 0.8, and β-galactosidase levels were measured (see Fig. S1 in the supplemental material). The amino acids could be divided into four classes. The first three classes are amino acids having little effect (<25% reduction in expression relative to no added amino acids), those having intermediate effects (25 to 75%), and those with strong effects (>75%). The fourth class had toxic or strong effects on growth. Most interesting are the strongly effective amino acids, which included Leu (as expected) but also Met, His, Thr, and Ile, which have not been reported to affect EcoLrp. The known EcoLrp coregulator Ala (6, 43, 45, 79, 80) had only intermediate effects in this assay. The fourth class of amino acids included Ser, Val, and Cys, which were therefore excluded from further studies with the exception of Ser, since it appeared to have Lrp ortholog-specific effects. The growth effects of these three amino acids were expected based on the Val sensitivity of E. coli K-12 (2, 20), the conditional auxotrophy for Ile in the presence of excess Ser (19), and the inhibition of threonine deaminase by Cys (29).
Use of a plasmid-independent method to assess EcoLrp-dependent coregulator effects.
To test further the apparent breadth of Lrp-coregulator interactions revealed by our in vivo and in vitro studies, we used a strain system independent of that used for Fig. S1 in the supplemental material. This alternative system has a chromosomal PlivK-lacZ fusion (due to λplacMu integration) in an independent host background, E. coli strain AAEC546 (which carries a chromosomal Plac-lrp fusion) (7, 8). This strain was grown in the absence of IPTG, which yields Lrp levels undetectable via Western blotting, or in the presence of IPTG, which yields ∼0.35 ng Lrp per ml of cell extract (equivalent to an intracellular monomer concentration of ∼1.5 mM) (9). LacZ activity was assessed for cultures grown with or without IPTG in well-aerated MOPS glucose medium having no added amino acids or with 10 mM Leu, Met, Ala, Gln, His, Ile, Lys, Pro, or Thr. The results are shown in Fig. 2 and were consistent with Leu, Met, Ile, His, and Thr acting as strong coregulators of Lrp. In particular, Met and Leu were associated with actual repression of PlivK (less LacZ activity than in the “Lrp−” cells), while Ile, His, and Thr antagonized activation (more LacZ activity than in the Lrp− cells but less than when no amino acid was added). Additionally, Ala showed strong antagonism of Lrp-dependent activation in this experimental system, consistent with previous results involving other promoters (6, 18, 45, 80). Lys and Gln yielded modest antagonism of activation, while Pro had little effect.
FIG. 2.
In vivo coregulator assays using a chromosomal reporter. (A) LacZ assays were carried out with E. coli strain SPB107, which carries a chromosomal Plac-lrp fusion and a PlivK-lacZ fusion. LacZ activity is plotted against culture density (OD600). The cultures were grown in MOPS-glucose medium, ±10 mM each of the indicated amino acids and ±0.4 mM IPTG. R values for least-squares fit were all >0.9. Cultures lacking IPTG (low Lrp) but containing His, Thr, or Ala grew poorly and were not assayed. (B) Bar chart of the slopes resulting from the analysis in panel A. Slopes from experiments in the absence of IPTG (low Lrp) are shown as gray bars, while black bars are from cultures containing IPTG (Lrp+). Shading below the graph relates to shading in Fig. S1 in the supplemental material, with light gray indicating amino acids that had an intermediate effect in that experiment and dark gray indicating a strong coregulator.
Further analysis of strong coregulatory effects on Lrp at PlivK.
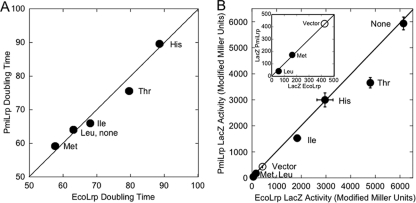
The most striking finding, from Fig. S1 in the supplemental material and from Fig. 2, is that amino acids besides Leu strongly affected control of the PlivK promoter by both EcoLrp and PmiLrp. Accordingly, we repeated in more detail the Fig. S1 experiment for the class of strong coregulators (Leu, Ile, Met, His, and Thr), with two refinements. First, we grew larger cultures producing EcoLrp or PmiLrp in well-aerated flasks. When β-galactosidase activity is plotted versus culture density (as in Fig. 1B and 2A), linearity indicates relatively steady-state growth, and the slopes very accurately reflect relative levels of lacZ expression. The resulting LacZ slopes in the presence of EcoLrp were plotted against those in the presence of PmiLrp in a correlogram (Fig. 3 B). With respect to both growth rates (Fig. 3A) and PlivK-lacZ expression, the points all fall on or very close to the correlogram lines, indicating very similar effects for both Lrp orthologs. As shown in Fig. 3A, some of the effects of Thr and His (but not of Ile or Met) may result from their ortholog-independent slowing of growth. Comparison to results in the absence of Lrp (vector control) reveals that Met and Leu are associated with actual repression (Fig. 3B, inset), while Ile, Thr, and His reduce the extent of activation but do not cause repression.
FIG. 3.
Correlograms for the strong coregulator amino acids of Lrp. (A) Growth rates of EcoLrp cells versus PmiLrp cells bearing PlivK-lacZ. The strong coregulators were present at 10 mM. (B) LacZ activities of EcoLrp versus PmiLrp. Means of triplicate cultures are shown, along with standard errors; where error bars are not visible, they were smaller than the symbols. Points below the vector control (open circle) suggest repression, while points above vector but below “None” (no amino acid added) suggest decreased activation. The inset shows the apparent repression on an expanded scale.
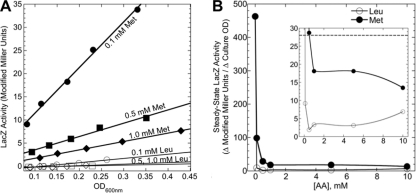
The second refinement of our strong-coregulator analysis was determining the dose-response analysis for Met (and, for comparison, Leu). Up to this point, we had been using relatively high coregulator concentrations (10 mM). For Leu, this is common and is justified in that naturally occurring shifts from minimal to richer environments would lead to transient intracellular spikes of Leu due to the fully induced high-affinity uptake system from branched-chain amino acids (of which LivK happens to be a component). However, for Met, it is unclear whether 10 mM is a physiologically relevant concentration. Accordingly, we repeated the experiment shown in Fig. 2 over a range of Leu and Met concentrations, from 0.1 to 10 mM. Figure 4 A shows that the cultures gave linear LacZ activity versus culture density relationships (all tested cultures gave linear results; a subset is shown for clarity). Figure 4B shows that even 0.1 mM Met gave substantial repression. The extrapolated amino acid concentrations giving 50% reductions in expression were 0.06 mM for Met and 0.05 mM for Leu. For comparison, 1.0 mM Ile had no detectible effect (not shown).
FIG. 4.
Dose-response analysis of corepressors Leu and Met effects on PlivK. (A) LacZ assays were carried out with E. coli strain SPB107, which carries a chromosomal Plac-lrp fusion and a PlivK-lacZ fusion. LacZ activity is plotted against culture density (OD600). The cultures were grown in MOPS-glucose medium, with a 0 to 10 mM concentration of either l-leucine or l-methionine, and 0.4 mM IPTG (to induce expression of EcoLrp). R values for least-squares fit were all >0.9. (B) The slopes from panel A are plotted versus the concentration of Leu or Met. The inset has an expanded y axis scale, and the dotted line indicates the slope value from a parallel culture grown without IPTG (very low [Lrp]) and with no added amino acids.
Lrp levels are not substantially altered by the strong coregulators.
If the addition of amino acids to the media resulted in altered amounts of Lrp, this might explain differences in Plivk-lacZ expression. This is especially a concern since Plrp responds to the nutrient environment and ppGpp: Lrp levels are lower in rich media (13, 37), so individual amino acids might reduce the Lrp concentration. In addition, the lrp gene is autogenously regulated by Lrp (72), and coregulators can normally affect Lrp levels in that way. However, in these experiments the lrp genes were controlled by PlacUV5 (see Methods); this promoter is not controlled by Lrp and is relatively insensitive to ppGpp (60, 61). Nevertheless, to determine the levels of Lrp, we collected samples from mid-exponential cultures (grown as in the experiment described in Fig. 3) and performed Western blot analysis with a polyclonal anti-Lrp antiserum. Lrp levels were affected minimally or not at all by the presence of the strong Lrp coregulators (Fig. 5). Leu, Met, Ile, and Thr appeared to increase Lrp levels only slightly (25 to 45%), while His and Arg had no effect. Furthermore, these differences do not correspond to the effects on PlivK-lacZ expression shown in Fig. 3B.
FIG. 5.
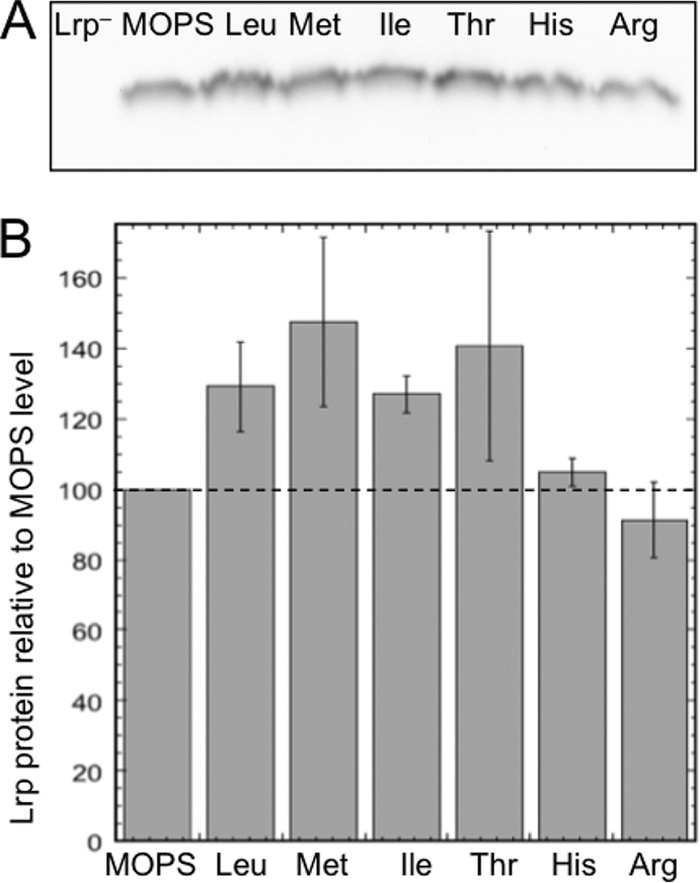
Western blot analysis of intracellular EcoLrp levels in the presence of the strong coregulator amino acids. Cells were grown under the same conditions as those used for the LacZ assays, and samples were collected in mid-exponential phase at a culture OD600 of ∼0.5. Equal amounts of protein were loaded on SDS gels, blotted, and probed with polyclonal anti-Lrp antiserum as described in Materials and Methods. Arg, which has no apparent effect on PlivK-lacZ expression, was included as a control. (A) Image of one of the triplicate blots. (B) Images such as those shown in panel A were quantified using ImageJ, and values were normalized to those from cells grown in the absence of amino acids. Error bars indicate standard errors from the three experiments.
Effects of strong coregulators on Lrp binding to target promoters.
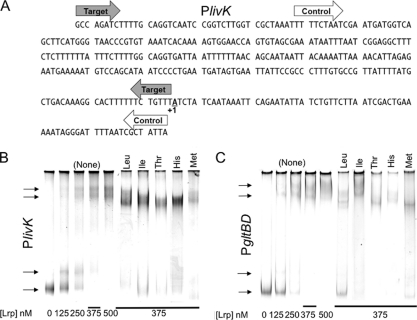
To help determine if the amino acids in the strong coregulator class directly interact with Lrp, we performed electrophoretic mobility shift assays (EMSAs) in which 10 mM amino acid was added to the binding reaction. Since no difference in PlivK-lacZ expression was seen between EcoLrp and PmiLrp for this class of amino acids (Fig. 3B), these experiments used only EcoLrp. The respective amino acids were also cast into the gels and included in the loading buffer at 10 mM to maintain their concentrations during electrophoresis. We examined effects on two promoters. PlivK was used to allow comparison to our other results (Fig. 6 A and B). Binding of Lrp to PlivK had not been demonstrated previously via EMSA, though chromatin immunoprecipitation analyses have detected it (17; A. Khodursky, personal communication). Figure 6A shows the 298-bp target DNA and an overlapping 326-bp specificity control that did not shift at any tested Lrp concentration (not shown). Since leucine switches Lrp between activating and repressing this PlivK, we suspected that the coregulators might not greatly affect binding per se. In fact, only minor effects were seen, though some weakening of EcoLrp binding to PlivK was seen with all five amino acids (Leu, Ile, Met, His, and Thr), as reflected in the rate of disappearance of unshifted promoter DNA with increasing Lrp concentrations (see Fig. S2A and B in the supplemental material).
FIG. 6.
Mobility shift assays with EcoLrp and strong coregulator amino acids. Two promoter targets were used (both at 23 nM): PlivK (A and B) and PgltBD (C). The indicated amino acids were included in the loading buffer and the gel to maintain their concentrations during electrophoresis. Concentrations of the Lrp protein used, calculated as the monomer, were 0, 125, 250, 375, and 500 nM. The 500-nM concentration corresponds to 250 nM as dimers, 62 nM as octamers, and 31 nM as hexadecamers. (A) Sequence of PlivK, showing the transcription start (+1) and boundaries of the EMSA target (gray arrows) or the overlapping specificity control (white arrows). The control fragment did not shift at any of the Lrp concentrations (not shown). (B and C) Negative image of ethidium-stained gel photographed under UV. A composite image is shown, with the first five lanes showing EcoLrp titration in the absence of amino acids, while the remainder are the 375 nM EcoLrp lanes from five separate gels with the indicated amino acids cast into the gel. Arrows indicate observed DNA bands in the absence of amino acids. The PlivK target is shown in panel B, and the PgltB target is shown in panel C.
We also tested PgltBD DNA (Fig. 6C). Lrp activates PgltBD, and Leu reduces Lrp binding, though the in vivo effects of Leu on gltBD transcription are minimal, possibly due to increased activation efficiency (9, 24, 74). Lrp binds between −140 and −260 bp relative to the start of transcription (74), and the DNA fragment used here encompassed that full range. Only His and Leu detectably weakened DNA binding by EcoLrp, as judged by residual unshifted DNA (see Fig. S2C and D in the supplemental material). The Leu results are consistent with those in an earlier study (24); our results are smaller in magnitude, probably reflecting our use of a lower coregulator concentration (10 mM versus 30 mM). However, differences were observed in the distribution of shifted bands, indicating that some amino acids affected EcoLrp-DNA complexes, even if they only minimally affected the fraction of unbound DNA (Fig. 6C). Lrp alone yielded two distinct shifted bands. Leu resulted in a triplet of shifted bands. In the presence of Met or Thr, Lrp yielded only one shifted band, apparently corresponding to the middle band of the triplet seen in the presence of Leu. Since these differences were not seen with PlivK DNA or in the absence of Lrp, they do not appear to be electrophoretic artifacts due to the presence of the various amino acids. These banding differences thus tend to support a direct interaction between the strong coregulator class of amino acids and Lrp.
Some coregulators may have differential effects on PlivK regulation by the E. coli and P. mirabilis Lrp orthologs.
The results shown in Fig. S1 in the supplemental material suggest that some amino acids, having more limited overall effects than the strong coregulators described above, appeared to differentially affect EcoLrp and PmiLrp control of PlivK (Gln, Pro, and Ser). Since such differences could have important physiological implications, we again repeated the experiment analyzed in Fig. S1 in more detail, growing larger cultures in well-aerated flasks and plotting β-galactosidase activity versus culture density (as in Fig. 1B). The resulting LacZ slopes in the presence of EcoLrp were plotted against those in the presence of PmiLrp in a correlogram (Fig. 7 B). Compared to the vector control (Lrp−), both Lrp orthologs substantially activated PlivK in the absence of exogenous amino acids, as expected (Fig. 7B, “None”). Ile and Lys, included as controls because they showed no differential effects between EcoLrp and PmiLrp in Fig. S1, again yielded minimal (Lys) to no (Ile) differences between the Lrp orthologs (Fig. 7B).
FIG. 7.
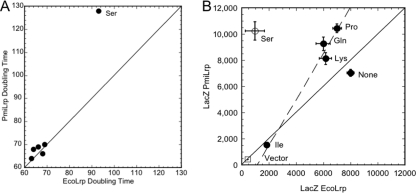
Correlogram of effects of PmiLrp versus those of EcoLrp on PlivK-lacZ in the presence of selected amino acids. MOPS-glucose cultures with plasmids as in Fig. 1A included no added amino acids (“None”) or 10 mM Ile (strongly depressive group), Lys (moderately depressive), Gln and Pro (weakly depressive), and Ser (like Pro, appearing in Fig. 2 to have differential effects on EcoLrp and PmiLrp). (A) Growth rates. Only Ser yielded a vastly different doubling time. (B) LacZ activity slopes from experiments such as those shown in Fig. 1B, plotted for EcoLrp versus PmiLrp (Lrp− vector control is plotted on both axes as an open square). The solid line indicates expected results if EcoLrp and PmiLrp had identical effects; the dashed line fits the data (excluding the vector control, no amino acid addition, and Ser). Means for triplicate cultures are shown, along with standard errors; where error bars are not visible, they were smaller than the symbols.
In contrast to both Ile and Lys, in the experiment shown in Fig. S1, Ser, Pro, and Gln had differential effects on PlivK-lacZ activities depending on the Lrp ortholog present. Pro (and perhaps Gln) again yielded this effect in the experiment shown in Fig. 7B. The responses to Lys, Gln, and Pro are all consistent with EcoLrp having ∼70% of the activity of PmiLrp on PlivK-lacZ expression (dotted line in Fig. 7B). This differential effect between PmiLrp and EcoLrp is amino acid specific, since it was not seen in the absence of exogenous amino acids (“None”) or with the strongly effective amino acid class (see previous sections). However, the results for Gln and Pro in Fig. 7B differ from those shown in Fig. 2, where Gln and Pro yielded higher PlivK-lacZ expression with EcoLrp than with PmiLrp. Among possible explanations of this difference is the lower-aeration conditions used for Fig. 2 (capped test tubes versus shaken flasks). Nevertheless, the apparent sensitivity to growth parameters demands caution in interpreting these results.
Serine showed the strongest ortholog-specific effects (Fig. 7B). Relative to the absence of exogenous amino acids, Ser increased PlivK-lacZ activity by ∼50% when PmiLrp was present but decreased it by nearly 90% when EcoLrp was present. This effect might be explained indirectly, at least in part, by the parallel effects on growth rate in each case (Fig. 7A). (Some batches of Ser have very recently been reported to contain an E. coli growth inhibitor of unknown nature [58]; if this is in any way involved in our observation, it would still represent a major difference between strains differing only in the Lrp ortholog produced.) While our LacZ-versus-culture-density analyses (e.g., Fig. 1B) account for growth rate differences per se, they do not correct for growth-rate-dependent differences in the gene expression machinery (e.g., concentrations of RNA polymerase and ribosomes) (22, 36). Whether these differential effects of Ser are direct or not, they nonetheless represent a surprisingly large difference between cells differing only in having Lrp orthologs that differ at just 2% of their residues (Fig. 1C). Again, however, it is noteworthy that in the experiment shown in Fig. S1 in the supplemental material, the differential pattern was reversed, with higher expression of PlivK-lacZ in the presence of EcoLrp than in that of PmiLrp.
We next carried out EMSAs to determine whether the effects of Pro and Gln on EcoLrp and PmiLrp are direct. Based on the results shown in Fig. S1 in the supplemental material, we tested Asn, since it had also yielded mildly differential effects between Lrp orthologs. Fig. S3A shows typical results from experiments in which the respective amino acids were included in the loading buffer and incorporated into the gel such that coregulator levels were maintained at 10 mM during electrophoresis. As with the strong coregulators, only modest effects on binding were seen. Fig. S3B shows the densitometry results of the unshifted bands, normalized (for each Lrp protein) to the results when no amino acids were added. This normalization corrects for possible differences in the fraction of active protein in each of the two preparations and for different intrinsic affinities for PlivK. The results suggest that Gln stimulates binding of both Lrp orthologs to PlivK (more-rapid disappearance of the unshifted band), while Asn and Pro stimulate PmiLrp binding but not that of EcoLrp.
DISCUSSION
Global regulators are key to understanding the regulatory architecture of bacterial cells or inferring it from genome sequences. The transcriptional regulators of E. coli and probably of most bacteria follow a power law distribution with respect to the number of promoters controlled: the top seven together control about half of all E. coli genes (44). As one of these top seven global regulators, Lrp plays a central role in E. coli cell physiology (71), and we have studied it as a representative of this class of proteins (7, 9, 24, 38, 54, 55, 71, 74).
Inferring regulation via extrapolation from genome sequences is particularly important for bacteria that cannot yet be grown easily in the laboratory, when microarray and other transcriptional data may not be available. Such approaches assume a fairly complete understanding of the conserved regulators (e.g., see reference 5). However, in a previous study, when we used microarray analysis to compare E. coli strains differing only in producing the native EcoLrp or the orthologous PmiLrp from P. mirabilis, substantial differences in gene expression patterns were seen (38). This was striking, given that EcoLrp and PmiLrp differ at only 4/164 amino acid positions (Fig. 1C), and it led us to explore here whether Lrp interactions with coregulators might explain part of the unexpected behavior.
We have found, through effects on both in vivo reporter fusions and in vitro DNA-binding assays, that a wider range of amino acids appear to affect EcoLrp and PmiLrp than had previously been appreciated. In this respect, EcoLrp and PmiLrp appear to resemble some other Lrp orthologs (Table 1).
We used PlivKHMGF (PlivK) as our primary model of an Lrp-regulated promoter. It is activated by Lrp (7, 32, 71), and in the presence of exogenous Leu it is repressed by Lrp (7, 17, 71). This promoter thus provides a relatively sensitive readout of Lrp-dependent interactions with the coregulator Leu. Global analyses suggest that PlivK also binds Crp (27) and Ihf (26). Effects of Crp are unlikely in our experiments, since the cultures were all grown with glucose as the carbon source (68-70). Similarly, except for the experiments involving Ser (Fig. 7A) and possibly His and Thr (Fig. 3A), cultures were in unrestricted logarithmic growth, so Ihf levels should have been fairly constant (3, 73). Excluding Glu, the tested amino acids gave remarkably constant expression of PlivK-lacZ in Lrp− cultures (white bars in Fig. S1 in the supplemental material and gray bars in Fig. 2B). In contrast, in the presence of Lrp, the coregulators had a range of transcriptional effects on PlivK (Fig. 8).
FIG. 8.
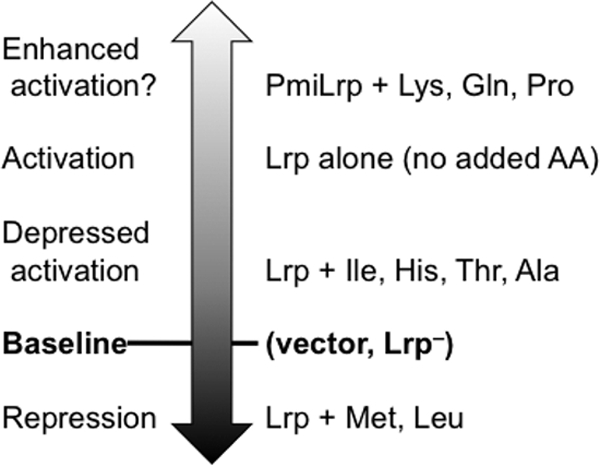
Summary of coregulator effects on Lrp regulation of PlivK. The relative level of expression is on a unitless vertical scale. Repression is indicated by levels lower than the Lrp− vector control baseline. Depressed or enhanced activation is inferred relative to the expression level in Lrp+ cells with no added amino acids.
Possible basis for varied effects of different coregulators.
The additional coregulator amino acids have varied effects on the EcoLrp regulatory pattern of PlivK. Similar to a model for Lrp suggested earlier (74), coregulator binding may reduce Lrp affinity to DNA but at the same time increase the ability of the remaining bound Lrp to activate the promoter. According to this view, the net result would depend on the affinity of the given target DNA sequence and the relative effects on Lrp DNA affinity and activation efficiency. Decreased binding and increased activation efficiency may both result from the Lrp octamer-hexadecamer equilibrium, which is driven toward the smaller form by (at least) Leu (14, 16). In being able to switch between activation and repression upon coregulator binding, AraC at ParaBAD resembles EcoLrp or PmiLrp at PlivK (23, 64). Thus, AraC-arabinose control may serve as a useful model for understanding Lrp-amino acid regulation. It is not yet clear how any Lrp protein activates transcription.
Another possible basis for regulatory flexibility in Lrp proteins may involve coregulator binding. It is interesting that the amino acid-sensing RAM domain of Lrp (25) contains one of the four differences between EcoLrp and PmiLrp (Fig. 1C). MtbLrp structures of cocrystals indicate the simultaneous binding of multiple amino acids per monomer; significantly, this involves two distinct binding pockets (67). Different amino acids were selectively bound at the two sites. Occupancy of one of these sites, analogous to the Leu-binding site in EcoLrp (21), caused a conformational change that seemed likely to influence multimerization, while occupancy of the other site had distinct effects that seemed more likely to alter interactions with RNA polymerase or other transcription factors.
Met acts as an Lrp corepressor at PlivK.
Leu is an Lrp corepressor of the promoter for livKHMGF, an operon specifying a high-affinity uptake system for branched-chain amino acids (7, 28, 38). In this study, Met was found to have strong effects on Lrp-dependent regulation of PlivK, similar to those shown by Leu (Fig. 2 and 4; see also Fig. S1 in the supplemental material). While Lrp coregulator promiscuity has been seen in other species (Table 1), this effect of Met on the Escherichia and Proteus Lrp orthologs has not been reported previously and has significant physiological implications. In Neisseria meningitidis, Met-Lrp interactions appear to adapt metabolism to nutrient-poor environments (63), and for Mycobacterium tuberculosis, Met was shown to bind Lrp (67). Since Lrp activates PlivK while Lrp·Leu or Lrp·Met represses it, one would expect Lrp to bind PlivK whether or not the coregulator is present. In fact, EMSA revealed that Met, like Leu, only mildly affects binding of EcoLrp or PmiLrp to PlivK (Fig. 6B; see also Fig. S2A and B), so presumably Met and Leu alter the protein-DNA complex such that it interferes with transcription. It is noteworthy that, at least in E. coli, the AdoMet synthetase gene (metK) is repressed by Lrp and induced by Leu (51; see also Fig. S3 of reference 17), and the same pattern was observed for the Met transporter genes metNIQ (17). Our results suggest that the MetNIQ and MetK substrate Met probably also acts as a metK inducer via Lrp.
Modulating Lrp activation of PlivK.
Of the amino acids tested in depth (Fig. 2 to 4 and 7), only Met and Leu reduced PlivK-lacZ expression to less than that seen in Lrp+ cells when no amino acids were added (Fig. 3B, inset). The other amino acids having substantial effects all appeared to modulate the extent of activation, in that PlivK-lacZ expression levels were higher than those in the Lrp− vector control but were either similar to or lower than those in Lrp+ cells with no added amino acids (Fig. 8). The results for some amino acids were consistent across all of our experiments (Table 2). This group included the two amino acids yielding strong repression (Leu and Met) and five others substantially decreasing the extent of activation (Ala, Ser, His, Thr, and Ile). The other amino acids (Gln, Pro, and Lys) had complex in vivo effects that were sensitive to the plasmid versus chromosomal location of the genes, the culture conditions (such as the level of aeration), or both. Accordingly, we will not discuss them further.
TABLE 2.
Relative amino acid effects on EcoLrp control of PlivK-lacZ
| Amino acid added | Relative expression of PlivK-lacZb |
||
|---|---|---|---|
| Fig. S1 | Fig. 2 | Fig. 3 or 7 | |
| MOPSa | (1.00) | (1.00) | (1.00) |
| Gln | 1.04 | 0.76 | 1.25 |
| Pro | 0.78 | 1.02 | 1.47 |
| Ala | 0.54 | 0.26 | ND |
| Ser | 0.53 | NDc | 0.20 |
| Lys | 0.40 | 0.76 | 1.29 |
| Hisd | 0.24 | 0.49 | 0.48 |
| Thrd | 0.15 | 0.43 | 0.78 |
| Iled | 0.12 | 0.41 | 0.38, 0.30 |
| Leud | 0.03 | 0.01 | 0.01 |
| Metd | 0.004 | 0.03 | 0.03 |
MOPS alone; no amino acid was added.
Data for EcoLrp in indicated figures, divided by the value with no added amino acids. The experiment for Fig. S1 used triplicate culture tube-grown cells with a single-time assay using a plasmid-based system; that for Fig. 2 used shaken flasks with multiple samples and linear fit versus culture density and with chromosomal genes; those for Fig. 3 and 7 used shaken flasks and linear fit and a plasmid-based system. Fig. S1 is found in the supplemental material.
ND, not determined.
Qualitatively consistent results in three independent in vivo analyses.
Ile, His, and Thr substantially reduced the extent of PlivK activation relative to that when no amino acids were added (Fig. 3B; see also Fig. S1 in the supplemental material) and did so equally with EcoLrp and PmiLrp. Mycobacterium tuberculosis Lrp binds His and (with substantially lower affinity) Thr (67). In vivo studies indicate that Val affects Pseudomonas aeruginosa Lrp regulation (10); given the roles of Leu, Ile, and Met, we suspect Val might also interact with EcoLrp, but we did not test this due to growth inhibition. Decreased activation could result from reduced Lrp binding, reduced activation efficiency, or both. The EMSA results suggest that these three amino acids affect PlivK binding at least as much as Leu does (Fig. 6B; see also Fig. S2A and B), though His is the only one that has as great an effect as Leu on PgltBD binding (Fig. 6C; see also Fig. S2C and D). However, these are not strong effects, suggesting that conformational changes in the Lrp-DNA complex are primarily responsible for the reduced PlivK transcription. With respect to the regulatory logic of these amino acids serving as coregulators, global analysis suggests that EcoLrp regulates the HisJQMP uptake system, in addition to the Thr transporters YgjU, TdcC, and SdaC (17).
Our observations, revealing unexpected coregulator breadth for one of the global regulators at the apex of the gammaproteobacterial “operating system” (44, 77), have major physiological consequences. They also reveal that extrapolatory inferences of regulatory architecture may require a greater depth of knowledge of the transcription factors than has been assumed to date.
Supplementary Material
Acknowledgments
We thank Robert Lintner and Pankaj Mishra for generating some of the plasmids used, Darren Sledjeski for providing antiserum, Arkady Khodursky for sharing unpublished results, and J. David Dignam for advice and use of HPLC equipment. We thank Ivana de la Serna, R. Mark Wooten, Isabel Novella, Sean Corum, and the anonymous reviewers for advice and comments on the manuscript.
This work was supported by funds from NIH grant R01 AI54716 and a Research Challenge award from the University of Toledo to R.M.B. B.R.H. was also supported in part by a graduate fellowship from the University of Toledo Health Science Campus.
Footnotes
Published ahead of print on 17 December 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adams, M. D., et al. 1990. Nucleotide sequence and genetic characterization reveal six essential genes for the LIV-I and LS transport systems of Escherichia coli. J. Biol. Chem. 265:11436-11443. [PubMed] [Google Scholar]
- 2.Ambartsoumian, G., R. D'Ari, R. T. Lin, and E. B. Newman. 1994. Altered amino acid metabolism in lrp mutants of Escherichia coli K12 and their derivatives. Microbiology 140(Pt. 7):1737-1744. [DOI] [PubMed] [Google Scholar]
- 3.Aviv, M., H. Giladi, G. Schreiber, A. B. Oppenheim, and G. Glaser. 1994. Expression of the genes coding for the Escherichia coli integration host factor are controlled by growth phase, rpoS, ppGpp and by autoregulation. Mol. Microbiol. 14:1021-1031. [DOI] [PubMed] [Google Scholar]
- 4.Baek, C. H., S. Wang, K. L. Roland, and R. Curtiss III. 2009. Leucine-responsive regulatory protein (Lrp) acts as a virulence repressor in Salmonella enterica serovar Typhimurium. J. Bacteriol. 191:1278-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumbach, J., S. Rahmann, and A. Tauch. 2009. Reliable transfer of transcriptional gene regulatory networks between taxonomically related organisms. BMC Syst. Biol. 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berthiaume, F., et al. 2004. Influence of L-leucine and L-alanine on Lrp regulation of foo, coding for F1651, a Pap homologue. J. Bacteriol. 186:8537-8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhagwat, S. P., M. R. Rice, R. G. Matthews, and R. M. Blumenthal. 1997. Use of an inducible regulatory protein to identify members of a regulon: application to the regulon controlled by the leucine-responsive regulatory protein (Lrp) in Escherichia coli. J. Bacteriol. 179:6254-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blomfield, I. C., P. J. Calie, K. J. Eberhardt, M. S. McClain, and B. I. Eisenstein. 1993. Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J. Bacteriol. 175:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borst, D. W., R. M. Blumenthal, and R. G. Matthews. 1996. Use of an in vivo titration method to study a global regulator: effect of varying Lrp levels on expression of gltBDF in Escherichia coli. J. Bacteriol. 178:6904-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boulette, M. L., et al. 2009. Characterization of alanine catabolism in Pseudomonas aeruginosa and its importance for proliferation in vivo. J. Bacteriol. 191:6329-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvo, J. M., and R. G. Matthews. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58:466-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charlier, D., M. Roovers, T. L. Thia-Toong, V. Durbecq, and N. Glansdorff. 1997. Cloning and identification of the Sulfolobus solfataricus lrp gene encoding an archaeal homologue of the eubacterial leucine-responsive global transcriptional regulator Lrp. Gene 201:63-68. [DOI] [PubMed] [Google Scholar]
- 13.Chen, C. F., et al. 1997. Metabolic regulation of lrp gene expression in Escherichia coli K-12. Microbiology 143(Pt. 6):2079-2084. [DOI] [PubMed] [Google Scholar]
- 14.Chen, S., and J. M. Calvo. 2002. Leucine-induced dissociation of Escherichia coli Lrp hexadecamers to octamers. J. Mol. Biol. 318:1031-1042. [DOI] [PubMed] [Google Scholar]
- 15.Chen, S., Z. Hao, E. Bieniek, and J. M. Calvo. 2001. Modulation of Lrp action in Escherichia coli by leucine: effects on non-specific binding of Lrp to DNA. J. Mol. Biol. 314:1067-1075. [DOI] [PubMed] [Google Scholar]
- 16.Chen, S., M. H. Rosner, and J. M. Calvo. 2001. Leucine-regulated self-association of leucine-responsive regulatory protein (Lrp) from Escherichia coli. J. Mol. Biol. 312:625-635. [DOI] [PubMed] [Google Scholar]
- 17.Cho, B. K., C. L. Barrett, E. M. Knight, Y. S. Park, and B. O. Palsson. 2008. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105:19462-19467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crost, C., et al. 2004. Influence of environmental cues on transcriptional regulation of foo and clp coding for F165(1) and CS31A adhesins in Escherichia coli. Res. Microbiol. 155:475-482. [DOI] [PubMed] [Google Scholar]
- 19.Daniel, J., and A. Danchin. 1979. Involvement of cyclic AMP and its receptor protein in the sensitivity of Escherichia coli K 12 toward serine: excretion of 2-ketobutyrate, a precursor of isoleucine. Mol. Gen. Genet. 176:343-350. [DOI] [PubMed] [Google Scholar]
- 20.De Felice, M., et al. 1977. Growth inhibition of Escherichia coli K-12 by L-valine: a consequence of a regulatory pattern. Mol. Gen. Genet. 156:1-7. [DOI] [PubMed] [Google Scholar]
- 21.de los Rios, S., and J. J. Perona. 2007. Structure of the Escherichia coli leucine-responsive regulatory protein Lrp reveals a novel octameric assembly. J. Mol. Biol. 366:1589-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennis, P. P., and H. Bremer. 1974. Macromolecular composition during steady-state growth of Escherichia coli B/r. J. Bacteriol. 119:270-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dirla, S., J. Y. Chien, and R. Schleif. 2009. Constitutive mutations in the Escherichia coli AraC protein. J. Bacteriol. 191:2668-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ernsting, B. R., J. W. Denninger, R. M. Blumenthal, and R. G. Matthews. 1993. Regulation of the gltBDF operon of Escherichia coli: how is a leucine-insensitive operon regulated by the leucine-responsive regulatory protein? J. Bacteriol. 175:7160-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ettema, T. J., A. B. Brinkman, T. H. Tani, J. B. Rafferty, and J. Van Der Oost. 2002. A novel ligand-binding domain involved in regulation of amino acid metabolism in prokaryotes. J. Biol. Chem. 277:37464-37468. [DOI] [PubMed] [Google Scholar]
- 26.Grainger, D. C., D. Hurd, M. D. Goldberg, and S. J. Busby. 2006. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 34:4642-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grainger, D. C., D. Hurd, M. Harrison, J. Holdstock, and S. J. Busby. 2005. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. U. S. A. 102:17693-17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haney, S. A., J. V. Platko, D. L. Oxender, and J. M. Calvo. 1992. Lrp, a leucine-responsive protein, regulates branched-chain amino acid transport genes in Escherichia coli. J. Bacteriol. 174:108-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris, C. L. 1981. Cysteine and growth inhibition of Escherichia coli: threonine deaminase as the target enzyme. J. Bacteriol. 145:1031-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hecht, K., S. Zhang, T. Klopotowski, and G. F. Ames. 1996. D-histidine utilization in Salmonella typhimurium is controlled by the leucine-responsive regulatory protein (Lrp). J. Bacteriol. 178:327-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hershberg, R., and H. Margalit. 2006. Co-evolution of transcription factors and their targets depends on mode of regulation. Genome Biol. 7:R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung, S. P., P. Baldi, and G. W. Hatfield. 2002. Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J. Biol. Chem. 277:40309-40323. [DOI] [PubMed] [Google Scholar]
- 33.Janes, B. K., and R. A. Bender. 1999. Two roles for the leucine-responsive regulatory protein in expression of the alanine catabolic operon (dadAB) in Klebsiella aerogenes. J. Bacteriol. 181:1054-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janga, S. C., and E. Perez-Rueda. 2009. Plasticity of transcriptional machinery in bacteria is increased by the repertoire of regulatory families. Comput. Biol. Chem. 33:261-268. [DOI] [PubMed] [Google Scholar]
- 35.Kim, S. H., B. L. Schneider, and L. Reitzer. 2010. Genetics and regulation of the major enzymes of alanine synthesis in Escherichia coli. J. Bacteriol. 192:5304-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klumpp, S., Z. Zhang, and T. Hwa. 2009. Growth rate-dependent global effects on gene expression in bacteria. Cell 139:1366-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landgraf, J. R., J. Wu, and J. M. Calvo. 1996. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J. Bacteriol. 178:6930-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lintner, R. E., et al. 2008. Limited functional conservation of a global regulator among related bacterial genera: Lrp in Escherichia, Proteus and Vibrio. BMC Microbiol. 8:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lozada-Chavez, I., S. C. Janga, and J. Collado-Vides. 2006. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 34:3434-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madan Babu, M., S. A. Teichmann, and L. Aravind. 2006. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 358:614-633. [DOI] [PubMed] [Google Scholar]
- 41.Marasco, R., et al. 1994. In vivo footprinting analysis of Lrp binding to the ilvIH promoter region of Escherichia coli. J. Bacteriol. 176:5197-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall, D. G., B. J. Sheehan, and C. J. Dorman. 1999. A role for the leucine-responsive regulatory protein and integration host factor in the regulation of the Salmonella plasmid virulence (spv) locus in Salmonella typhimurium. Mol. Microbiol. 34:134-145. [DOI] [PubMed] [Google Scholar]
- 43.Martin, C. 1996. The clp (CS31A) operon is negatively controlled by Lrp, ClpB, and L-alanine at the transcriptional level. Mol. Microbiol. 21:281-292. [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Antonio, A., and J. Collado-Vides. 2003. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 6:482-489. [DOI] [PubMed] [Google Scholar]
- 45.Mathew, E., J. Zhi, and M. Freundlich. 1996. Lrp is a direct repressor of the dad operon in Escherichia coli. J. Bacteriol. 178:7234-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matthews, R. G., Y. Cui, D. Friedberg, and J. M. Calvo. 2000. Wild-type and hexahistidine-tagged derivatives of leucine-responsive regulatory protein from Escherichia coli. Methods Enzymol. 324:322-329. [DOI] [PubMed] [Google Scholar]
- 47.McFarland, K. A., and C. J. Dorman. 2008. Autoregulated expression of the gene coding for the leucine-responsive protein, Lrp, a global regulator in Salmonella enterica serovar Typhimurium. Microbiology 154:2008-2016. [DOI] [PubMed] [Google Scholar]
- 48.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 49.Nakanishi, N., et al. 2009. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology 155:521-530. [DOI] [PubMed] [Google Scholar]
- 50.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newman, E. B., et al. 1998. Lack of S-adenosylmethionine results in a cell division defect in Escherichia coli. J. Bacteriol. 180:3614-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newman, E. B., R. D'Ari, and R. T. Lin. 1992. The leucine-Lrp regulon in E. coli: a global response in search of a raison d'etre. Cell 68:617-619. [DOI] [PubMed] [Google Scholar]
- 53.Oliver, G., et al. 1987. Determination of the nucleotide sequence for the glutamate synthase structural genes of Escherichia coli K-12. Gene 60:1-11. [DOI] [PubMed] [Google Scholar]
- 54.Paul, L., R. M. Blumenthal, and R. G. Matthews. 2001. Activation from a distance: roles of Lrp and integration host factor in transcriptional activation of gltBDF. J. Bacteriol. 183:3910-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paul, L., P. K. Mishra, R. M. Blumenthal, and R. G. Matthews. 2007. Integration of regulatory signals through involvement of multiple global regulators: control of the Escherichia coli gltBDF operon by Lrp, IHF, Crp, and ArgR. BMC Microbiol. 7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Platko, J. V., and J. M. Calvo. 1993. Mutations affecting the ability of Escherichia coli Lrp to bind DNA, activate transcription, or respond to leucine. J. Bacteriol. 175:1110-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Platko, J. V., D. A. Willins, and J. M. Calvo. 1990. The ilvIH operon of Escherichia coli is positively regulated. J. Bacteriol. 172:4563-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Potrykus, K., H. Murphy, N. Philippe, and M. Cashel. 2010. ppGpp is the major source of growth rate control in E. coli. Environ. Microbiol. [Epub ahead of print.] doi: 10.1111/j.1462-2920.2010.02357.x. [DOI] [PMC free article] [PubMed]
- 59.Price, M. N., P. S. Dehal, and A. P. Arkin. 2007. Orthologous transcription factors in bacteria have different functions and regulate different genes. PLoS Comput. Biol. 3:1739-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Primakoff, P. 1981. In vivo role of the relA+ gene in regulation of the lac operon. J. Bacteriol. 145:410-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Primakoff, P., and S. W. Artz. 1979. Positive control of lac operon expression in vitro by guanosine 5′-diphosphate 3′-diphosphate. Proc. Natl. Acad. Sci. U. S. A. 76:1726-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rasband, W. 2010. ImageJ. Documentation. http://rsb.info.nih.gov/ij/docs/index.html.
- 63.Ren, J., et al. 2007. The structure and transcriptional analysis of a global regulator from Neisseria meningitidis. J. Biol. Chem. 282:14655-14664. [DOI] [PubMed] [Google Scholar]
- 64.Rodgers, M. E., N. D. Holder, S. Dirla, and R. Schleif. 2009. Functional modes of the regulatory arm of AraC. Proteins 74:81-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roesch, P. L., and I. C. Blomfield. 1998. Leucine alters the interaction of the leucine-responsive regulatory protein (Lrp) with the fim switch to stimulate site-specific recombination in Escherichia coli. Mol. Microbiol. 27:751-761. [DOI] [PubMed] [Google Scholar]
- 66.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 67.Shrivastava, T., and R. Ramachandran. 2007. Mechanistic insights from the crystal structures of a feast/famine regulatory protein from Mycobacterium tuberculosis H37Rv. Nucleic Acids Res. 35:7324-7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tagami, H., and H. Aiba. 1995. Role of CRP in transcription activation at Escherichia coli lac promoter: CRP is dispensable after the formation of open complex. Nucleic Acids Res. 23:599-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tagami, H., T. Inada, T. Kunimura, and H. Aiba. 1995. Glucose lowers CRP* levels resulting in repression of the lac operon in cells lacking cAMP. Mol. Microbiol. 17:251-258. [DOI] [PubMed] [Google Scholar]
- 70.Takahashi, H., T. Inada, P. Postma, and H. Aiba. 1998. CRP down-regulates adenylate cyclase activity by reducing the level of phosphorylated IIA(Glc), the glucose-specific phosphotransferase protein, in Escherichia coli. Mol. Gen. Genet. 259:317-326. [DOI] [PubMed] [Google Scholar]
- 71.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. U. S. A. 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, Q., J. Wu, D. Friedberg, J. Plakto, and J. M. Calvo. 1994. Regulation of the Escherichia coli lrp gene. J. Bacteriol. 176:1831-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weglenska, A., B. Jacob, and A. Sirko. 1996. Transcriptional pattern of Escherichia coli ihfB (himD) gene expression. Gene 181:85-88. [DOI] [PubMed] [Google Scholar]
- 74.Wiese, D. E., II, B. R. Ernsting, R. M. Blumenthal, and R. G. Matthews. 1997. A nucleoprotein activation complex between the leucine-responsive regulatory protein and DNA upstream of the gltBDF operon in Escherichia coli. J. Mol. Biol. 270:152-168. [DOI] [PubMed] [Google Scholar]
- 75.Wild, J., and W. Szybalski. 2004. Copy-control pBAC/oriV vectors for genomic cloning. Methods Mol. Biol. 267:145-154. [DOI] [PubMed] [Google Scholar]
- 76.Willins, D. A., C. W. Ryan, J. V. Platko, and J. M. Calvo. 1991. Characterization of Lrp, an Escherichia coli regulatory protein that mediates a global response to leucine. J. Biol. Chem. 266:10768-10774. [PubMed] [Google Scholar]
- 77.Yan, K. K., G. Fang, N. Bhardwaj, R. P. Alexander, and M. Gerstein. 2010. Comparing genomes to computer operating systems in terms of the topology and evolution of their regulatory control networks. Proc. Natl. Acad. Sci. U. S. A. 107:9186-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang, L., R. T. Lin, and E. B. Newman. 2002. Structure of the Lrp-regulated serA promoter of Escherichia coli K-12. Mol. Microbiol. 43:323-333. [DOI] [PubMed] [Google Scholar]
- 79.Zhi, J., E. Mathew, and M. Freundlich. 1998. In vitro and in vivo characterization of three major dadAX promoters in Escherichia coli that are regulated by cyclic AMP-CRP and Lrp. Mol. Gen. Genet. 258:442-447. [DOI] [PubMed] [Google Scholar]
- 80.Zhi, J., E. Mathew, and M. Freundlich. 1999. Lrp binds to two regions in the dadAX promoter region of Escherichia coli to repress and activate transcription directly. Mol. Microbiol. 32:29-40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.