Abstract
The fucose-/mannose-specific lectin LecB from Pseudomonas aeruginosa is transported to the outer membrane; however, the mechanism used is not known so far. Here, we report that LecB is present in the periplasm of P. aeruginosa in two variants of different sizes. Both were functional and could be purified by their affinity to mannose. The difference in size was shown by a specific enzyme assay to be a result of N glycosylation, and inactivation of the glycosylation sites was shown by site-directed mutagenesis. Furthermore, we demonstrate that this glycosylation is required for the transport of LecB.
Lectins are proteins of nonimmune origin that recognize and bind to specific carbohydrate structural epitopes without modifying them. This group of carbohydrate proteins function as central mediators of information transfer in biological systems and perform their duties by interacting with glycoproteins, glycolipids, and oligosaccharides (34). Lectins exist in a wide range of organisms, including viruses, bacteria, plants, and animals, and are believed to play a general and important role in cell-cell interactions (9). Many bacteria have an arsenal of different lectins for targeting glycosylated proteins of the host (21). One example, the lectin FimH at the top of type 1 pili from the uropathogenic Escherichia coli, recognizes terminally located d-mannose moieties on cell-bound glycoproteins to mediate adhesion between the bacterium and the urothelium (4, 20). Lectins are also of interest for medical and pharmaceutical applications, as exemplified by the galactoside-specific mistletoe lectin, which is widely used as a drug to support anticancer therapy (5).
Pseudomonas aeruginosa, an opportunistic pathogen associated with chronic airway infections, synthesizes two lectins, LecA and LecB (formerly also named PA-IL and PA-IIL) (11). Strains of P. aeruginosa which produce high levels of these virulence factors exhibit an increased virulence potential (12). Both lectins play a prominent role in human infection, since it was demonstrated that P. aeruginosa-induced otitis externa diffusa (46), as well as P. aeruginosa in respiratory tract infections (56) and cystic fibrosis patients (16), could successfully be treated by the application of a solution containing specific sugars. The sugar solutions presumably prevented lectin-mediated bacterial adhesion to the corresponding host cells. The expression of lectin genes in P. aeruginosa is coordinately regulated with certain other virulence factors and controlled via the quorum-sensing cascade and by the alternative sigma factor RpoS (60). Functional LecB consists of four 11.73-kDa subunits, each exhibiting high specificity for l-fucose and mannose and their derivates (10, 13), and the crystal structure was determined more recently (28, 32, 33). In cystic fibrosis, a characteristic increase of terminal fucosylation is found for airway epithelial glycoproteins, as well as a higher percentage of sialylated and sulfated oligosaccharides in Lewis A oligosaccharide side chains, which presumably represent preferential ligands for LecB (32), thereby contributing significantly to chronic respiratory Pseudomonas infections (38). Interestingly, LecA and LecB inhibit ciliary beating (31), hence inhibiting an important defense mechanism of the lung (2, 3). The subcellular localization of LecB has been an issue of discussion during recent years (14, 50). A LecB-deficient P. aeruginosa strain was impaired in biofilm formation, and LecB was shown to be located in the outer membrane, binding to yet-unidentified ligands on the surface of biofilm cells (50). This suggests that LecB may mediate the adhesion of P. aeruginosa cells to receptors that are located on its surface and facilitate biofilm formation, thereby promoting colonization of host tissues (50). Consequently, it was demonstrated that glycopeptide LecB-specific dendrimers can not only inhibit the formation of biofilms but also disperse them efficiently (22, 23). Although obviously residing in or at the outer membrane of P. aeruginosa, the secretion pathway used by LecB is not known so far. Type I secretion and Sec- or Tat-dependent translocation mechanisms appear not to be involved, since LecB does not contain any of the known signal peptides required for these pathways (30).
Here, we show that the LecB protein exists in two distinct forms which are present in the periplasmic space. Both the high-molecular-weight (HW LecB) and the low-molecular-weight (LW LecB) variant were functional in sugar binding and were purified from the periplasmic fraction of P. aeruginosa by affinity chromatography. Deglycosylation by N-glycosidase F treatment of HW LecB resulted in a decrease in the molecular mass of the lectin, showing that LecB is an N-glycosylated protein. Inactivation of the putative N glycosylation site resulted in mislocalization of this variant, showing that glycosylation is necessary for the proper secretion of LecB in P. aeruginosa.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. E. coli DH5α was used for cloning experiments, and E. coli S-17 for conjugal transfer. P. aeruginosa PAO1 was the wild-type strain, and LecB-mutant strain P. aeruginosa PATI2 was used as a host for the expression and subcellular localization of the nonglycosylated LecB variant.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype/phenotype | Reference or source |
|---|---|---|
| P. aeruginosa strains | ||
| PAO1 | Wild type | 19 |
| PATI2 | LecB mutant strain derived from PAO1; lecB::Gmr | 50 |
| E. coli strains | ||
| DH5α | supE44 Δ(lacZYA-argF)U169 (φ80dlacZΔM15) hsdR1 recA1 endA1 gyrA96 thi-1 relA1 | 61 |
| S17-1 | E. coli 294::[RP4-2 (Tc::Mu) (Km::Tn7)] pro res recA tra+ Tpr Smr | 42 |
| Plasmids | ||
| pBBC2 | pBBR1MCS containing a 398-bp XbaI/SacI fragment with lecB derived from pEC2 | 50 |
| pET22b | T7 expression vector for E. coli; PelB signal sequence, Apr | Novagen |
| pBBR1MCS | lacZα Cmrrep mob | 24 |
| pBBN22A | pBBR1MCS containing a 398-bp XbaI/SacI fragment bearing the lecB(N22A) genea | This study |
lecB(N22A) encodes LecB with an inactivated N glycosylation site.
Media and growth conditions.
Precultures for all experiments were grown overnight in 10 ml of LB medium at 37°C. Plasmid-carrying E. coli cells were selected with 100 μg ampicillin ml−1 and 50 μg chloramphenicol ml−1. In the case of plasmid- or cassette-carrying P. aeruginosa strains, 300 μg chloramphenicol ml−1 and 50 μg gentamicin ml−1 were added.
For subcellular localization experiments, P. aeruginosa PAO1 and P. aeruginosa PATI2 carrying the lecB expression plasmids were grown on nutrient broth (NB) agar plates at 37°C for 24 and 48 h, respectively. For purification of the LecB variants from the periplasm, P. aeruginosa PAO1 was grown for 48 h in NB medium.
Cell fractionation.
Cell fractionation was done as described by Tielker et al. (50). Bacterial cells (1.2 mg dry weight) were washed with 1 ml of 0.14 M NaCl and then centrifuged at 3,000 × g; the supernatant was sterile filtered and used to determine the content of LecB in the extracellular space. The cell pellet was suspended in 240 μl of 100 mM Tris-HCl (pH 8) containing 20% (wt/vol) sucrose. After the addition of 240 μl of the same buffer containing 5 mM EDTA and 20 μg lysozyme, the sample was incubated for 30 min at room temperature, spheroplasts were collected by centrifugation at 10,000 × g for 20 min, and the supernatant was used as the periplasmic fraction. Spheroplasts were disrupted by sonication (Sonifier W250; Branson) in 240 μl of 100 mM Tris-HCl (pH 8). After centrifugation for 5 min at 5,000 × g to remove intact cells and cell debris, the total membrane fraction was collected by centrifugation for 45 min at 13,000 × g and the supernatant was used as the cytoplasmic fraction. The total membrane fraction was suspended in 240 μl of 100 mM Tris-HCl (pH 8). Then, the proteins of the fractions were concentrated by precipitation with 10% (vol/vol) trichloroacetic acid. After the samples were washed using 80% (vol/vol) acetone, amounts equivalent to optical densities at 580 nm (OD580) of 0.5 and 1 per ml of each fraction were used for the Western blotting methods.
Purification of periplasmic LecB.
Functional LecB from the periplasm was purified from 500 mg of cells (dry weight) by its affinity toward mannose as described previously (51). Mannose was removed by repeated ultrafiltration (Vivaspin 6, 5-kDa cutoff; Vivascience, Hannover, Germany) and washed with 100 mM Tris-HCl (pH 8.0).
Separation of LecB variants.
LecB variants (1 mg/ml) were separated by high-performance liquid chromatography (HPLC), using an LC-10ai HPLC system (Shimadzu) with a TSKgel G2000SWXL column (Tosoh Bioscience). The column was equilibrated with 100 mM potassium phosphate buffer, pH 6.8. For calibration, thyroglobulin (670 kDa), IgG (150 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and uridine (0.244 kDa) were used.
SDS gel electrophoresis and Western blot analysis.
Protein samples were suspended in SDS-PAGE sample buffer, heated for 5 min at 98°C, and then loaded onto SDS-16% and SDS-12% polyacrylamide gels, followed by electrophoretic protein transfer to polyvinylidene difluoride (PVDF) membranes at 150 mA for 15 min and 300 mA for 20 min (Bio-Rad, Munich, Germany). The membranes were blocked overnight using 5% (wt/vol) skim milk in TBST (25 mM Tris-HCl, pH 8, 150 mM NaCl, 3 mM KCl, 0.2% [vol/vol] Tween 20) at 4°C. LecB, EstA, and DsbA were detected by incubating the membranes with specific polyclonal antibodies (50, 55, 59) at a dilution of 1:20,000, 1:85,000, or 1:100,000, respectively, in TBST, followed by an anti-rabbit IgG-horseradish peroxidase conjugate (Bio-Rad). The blots were developed with an ECL chemiluminescence kit (GE Healthcare).
Glucose-6-phosphate dehydrogenase assay.
Glucose-6-phosphate dehydrogenase was used as a cytoplasmic marker enzyme (8, 58). A stock solution of NADP (45 mM) and a stock solution of glucose-6-phosphate (110 mM) were diluted 1:100 in a buffer containing 55 mM Tris-HCl (pH 7.5) and 11 mM MgCl. Nine hundred-microliter volumes of this test solution were mixed with 100-μl samples from cytoplasm, periplasm, membrane, and supernatant, and the decrease in the optical density (DOD340/min) was monitored spectrophotometrically at 30°C for 90 s.
Protein identification by matrix-assisted laser desorption ionization mass spectrometry (MALDI MS).
Spots of interest were excised from polyacrylamide gels, digested overnight with chymotrypsin gold (Promega, United States), and eluted as described by Shevchenko et al. (41). The diluted proteins were desalted if necessary with a Ziptip C18 (Millipore, United States) and spotted on a prespotted AnchorChip (Bruker, Germany) with an HCCA (α-cyano-4-hydroxycinnamic acid) matrix. The masses of the peptides were determined with an Ultraflex III system (Bruker, Germany). Database search was carried out with MASCOT (Matrix Science).
Deglycosylation assay.
Glycosylation was shown as described earlier (47, 49), using N-glycosidase F (PNGase F; Roche, Penzberg, Germany) according to the manufacturer's manual. Ten micrograms of purified periplasmic LecB or 100 ng of purified HW LecB was incubated with 2 units of PNGase F in 50 mM sodium phosphate buffer (pH 7.5) for 16 h at 37°C. In the case of the time course experiment, samples containing 100 ng of HW LecB were taken off after 0, 6, 9, and 16 h. The samples were analyzed by SDS-PAGE and Western blot analysis using a LecB-specific antiserum. Samples treated identically but without the addition of PNGase F served as negative controls.
Inactivation of glycosylation site.
An overlap extension PCR method was used to introduce site-specific substitutions within the lecB open reading frame. Asparagine residues were mutated to alanine at position N22 using the oligonucleotide primers LIINdeI (5′-AAAACATATGGCAACACAAGGAGTGTTCAC-3′)/N22AUp (5′-GCCTTCGCCGCATCGTCCGGA-3′) and LIIBamHI (5′-AAAAGGATCCCTAGCCGAGCCAGTTGATC-3′)/N22ADWN (5′-TCCGGACGATGCGGCGAAGGC-3′) within the LecB coding sequence. Fragments were gel purified and subsequently used for overlap extension PCR with 5′ and 3′ flanking primers LIINdeI and LIIBamHI, respectively. The amplified DNA fragments were cloned into the NdeI/BamHI sites of pET22b, excised by digestion with SacI/XbaI, and ligated into pBBR1MCS which had been digested with the same enzymes. The resulting plasmid, pBBN22A, was introduced into P. aeruginosa PATI2 by conjugation.
Real-time PCR.
RNA isolation was performed with an RNeasy minikit (Qiagen, Germany) according to the protocol of the manufacturer. Afterwards, genomic DNA was digested by DNase (Promega, United States). Quantitative reverse transcription-PCR (qRT-PCR) was performed using the standard instrument settings on an ABI 7900HT instrument using TaqMan gene expression master mix. The expression levels were normalized to that of rpoD as a control housekeeping gene. Detection of lecB was achieved using a specific TaqMan gene expression assay (Applied Biosystems, Switzerland) (forward primer, GCACCAATAACGCCGTCATC; reverse primer, GCTGACCTGGACCTGTACCT; and assay, CAGGTGCTCAACTCC) and normalized to the level of the housekeeping gene rpoD (forward primer, ACGCGCGCATGCC; reverse primer, CTCGTCGGTCTCGTGGTT; and assay, TTCCTGCGCCTGTTCC).
RESULTS AND DISCUSSION
A variant of LecB with a greater molecular mass exists in the periplasm.
Different subcellular localization sites have been suggested for the mannose-/fucose-specific lectin LecB during recent years. A significant portion of the lectin has been found to reside in the cytoplasm (14), and in addition to the cytoplasmic localization of LecB, it has been shown that it is localized in the outer membrane of P. aeruginosa, most probably on the cell surface (50). Although this surface localization would perfectly explain the influence of LecB on biofilm formation (50), it is in fact not known at present how the protein traverses the cell envelope, since all secretion signals known in P. aeruginosa are missing (30, 50). Furthermore, LecB has been shown to influence type IV pilus formation and the extracellular activity of extracellular protease IV, suggesting that this protein may also affect the type II protein secretion of P. aeruginosa (44). Although the role LecB plays in these processes is completely unknown, this context suggests that LecB might exert its influence in the periplasm, which in turn would require a periplasmic localization of the protein itself.
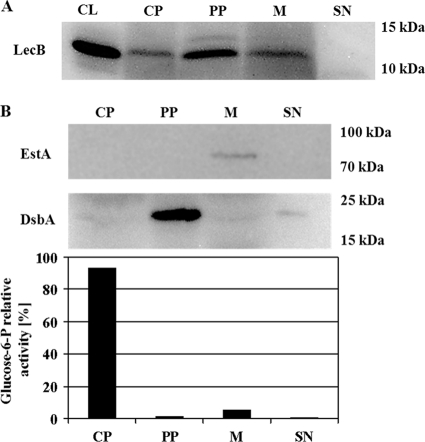
We proved this assumption when a signal became visible in Western blots in the periplasmic fraction of P. aeruginosa PAO1 cells grown on NB agar plates as saturated biofilms for 48 h at 37°C (Fig. 1A). Neither the outer membrane marker EstA (Fig. 1B) nor significant glucose-6-phosphate dehydrogenase activity was detected in the periplasm (Fig. 1B), indicating that detection of LecB in the periplasmic fraction was not the result of contamination with the outer membrane or cytoplasmic fraction. The purity of the periplasmic fraction was verified by demonstrating the exclusive presence of the periplasmic protein DsbA (55) in this fraction (Fig. 1B). Interestingly, a second protein band appeared which reacted specifically with the LecB antiserum and was exclusively present in the periplasmic fraction. This protein was of a slightly higher molecular weight than the LecB present in the other fractions (Fig. 1A). We thus named this variant high-molecular-weight LecB (HW LecB) to distinguish it from its low-molecular-weight variant (LW LecB) and asked whether this HW LecB variant was also functional in binding to mannose. This binding capability of HW LecB was assayed by purification of both variants from the periplasm by the same affinity chromatography which we established earlier (28, 51). Both variants were present in the purified lectin fraction in comparable amounts, showing that HW LecB also binds quantitatively to mannose and can be copurified with the low-molecular-weight variant from the periplasm (Fig. 2A). In an earlier study, cross-reactivity of the LecB antiserum with other P. aeruginosa proteins was excluded (50). Furthermore, both variants were analyzed by Western blotting (Fig. 2B) and were clearly identified as LecB protein by MALDI-TOF analysis (data not shown). The purity of the periplasmic fraction was verified by demonstrating the exclusive presence of the periplasmic protein DsbA (55) in this fraction (Fig. 2C).
FIG. 1.
Subcellular localization of LecB in P. aeruginosa PAO1 and controls to validate the cellular fractionation procedure. (A) Western blot analysis of cell fractions from P. aeruginosa PAO1. Cells were grown at 37°C on NB agar plates for 48 h, subcellular fractions were prepared as described in the text and separated by SDS-gel electrophoresis, and LecB was detected after transfer onto PVDF membranes using a LecB-specific antiserum. (B) Fractionation controls. The same cell fractions were analyzed using an EstA- and a DsbA-specific antiserum and by the distribution of relative glucose-6-phosphate dehydrogenase activities. The percentages of relative enzyme activities present in cytoplasm, periplasm, membrane, and culture supernatant are given. CL, cell lysate; CP, cytoplasm; PP, periplasm; M, membrane; SN, culture supernatant.
FIG. 2.
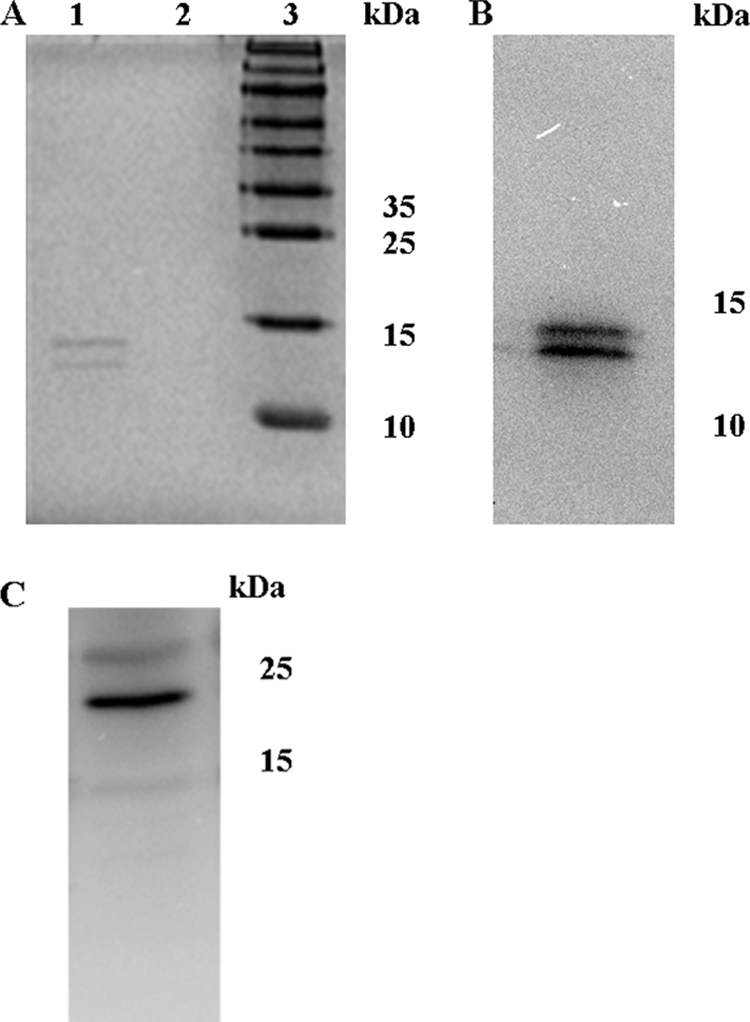
Two distinct forms of the LecB protein are present in the periplasm. Proteins from the periplasmic fraction were loaded on a mannose agarose column to isolate proteins with affinity toward mannose. (A) SDS-PAGE analysis of the eluate fraction after affinity chromatography and a washing step (lane 1, eluate fraction; lane 2, wash fraction; lane 3, molecular mass marker). (B) Identification of the two forms as LecB by Western blot analysis. (C) Western blot of the periplasmic fraction using a DsbA-specific antiserum.
Periplasmic HW LecB is an N-glycosylated protein.
Posttranslational modifications of proteins can significantly increase their apparent molecular weight (47, 53). Glycosylation is a very common modification in eukaryotic cells, in which proteins targeted for extracytoplasmic localization are labeled by covalent attachment of sugars to the protein (54). Glycan residues can be attached either on hydroxyl groups of serine and threonine (O glycosylation) or amino groups of asparagine (N glycosylation) (26, 48). In addition to functions in targeting, protein glycosylation is important for stabilization of proteins (43).
In bacteria, glycosylation has been described in principle, but so far, the number of examples of glycosylation of prokaryotic proteins is rather limited. In P. aeruginosa, glycosylation has been described for the pilin and flagellin subunits of type IV pili and flagella (7, 40, 53). However, these proteins are O glycosylated, whereas N glycosylation has not been described so far for this bacterium. Moreover, the studies that showed O glycosylation have been performed with P. aeruginosa strains other than the prototype strain P. aeruginosa PAO1. Nevertheless, using the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/) for the detection of glycosylation sites, we have identified residue N22 within the LecB sequence as representing a putative N glycosylation site (NXS/T) (data not shown). Based on the assumption that LecB might in fact be N glycosylated, we then tested whether glycosylation of LecB was responsible for the increased molecular weight of the periplasmic variant observed in P. aeruginosa PAO1 by using N-glycosidase F from Flavobacterium meningosepticum (Roche, Penzberg, Germany), which is known to specifically remove N glycosylation from proteins (49). After appropriate incubation of the LecB variants purified from the periplasm with the enzyme, the HW variant completely disappeared (Fig. 3A, lane 3) in comparison with the control sample incubated identically but without enzyme (Fig. 3A, lane 2). In contrast, LW LecB remained unaffected by the N-glycosidase and appeared with the same electrophoretic mobility as the deglycosylated form. Separation of HW LecB from LW LecB using size exclusion chromatography and subsequent incubation of HW LecB with N-glycosidase F for 16 h at 37°C resulted again in degradation of the HW variant to the lower-molecular-weight species (Fig. 3B, lane 5, and C), whereas identical incubation without N-glycosidase F had no effect (Fig. 3B, lane 4). These results strongly indicate that HW LecB is in fact an N-glycosylated protein.
FIG. 3.
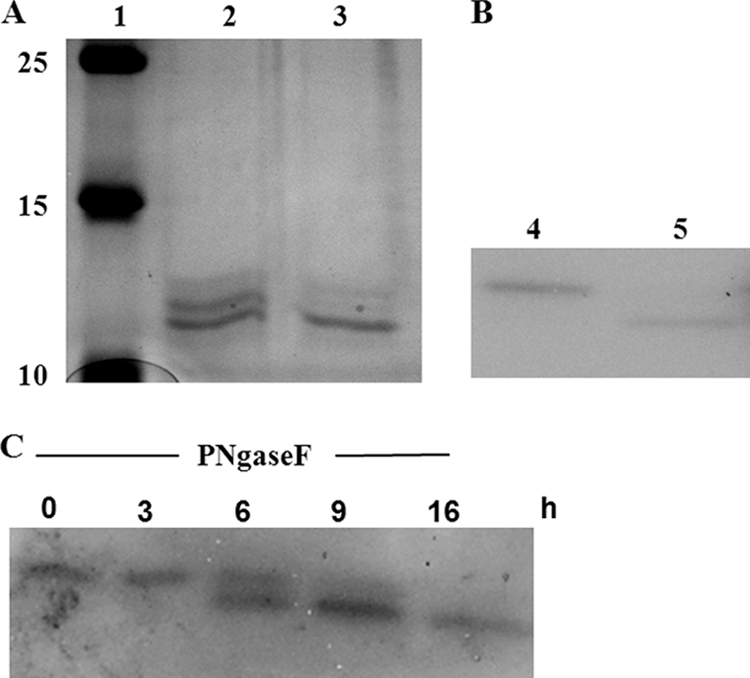
Deglycosylation of the high-molecular-weight variant of LecB (HW LecB). The affinity-purified periplasmic LecB variants (A) and size exclusion chromatography-separated HW LecB (B) were exposed to N-glycosidase F from Flavobacterium meningosepticum (PNGase F; Roche). Amounts of 10 μg of purified periplasmic LecB and 100 ng of separated HW LecB were incubated with 2 U of PNGase F or without any enzyme in 50 mM sodium-phosphate buffer (pH 7.5) in a final volume of 10 μl for 16 h at 37°C. Lanes: 1, molecular weight marker (in thousands); 2 and 4, periplasmic LecB (2) and HW LecB (4) without the addition of PNGase F; 3 and 5, periplasmic LecB (3) and HW LecB (5) with PNGase F treatment. (C) Time course of the deglycosylation of HW LecB. HW LecB was incubated with PNGase F for 16 h at 37°C. After incubation periods of 0, 3, 6, 9, and 16 h, samples that each contained 100 ng of HW LecB were taken off. Samples were analyzed by SDS-PAGE and Western blotting using a LecB-specific antiserum.
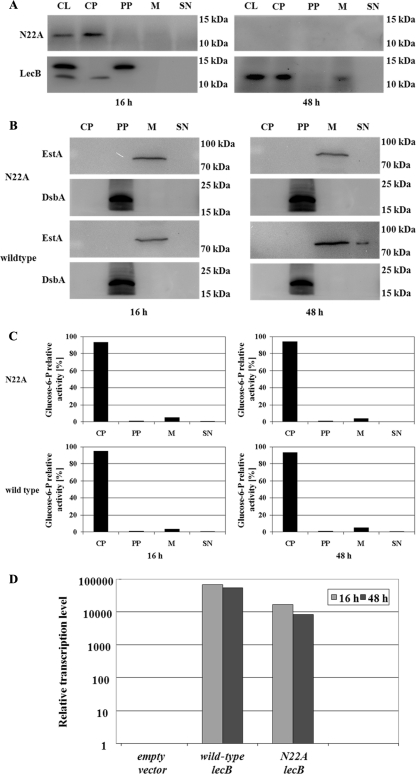
To confirm these results and to examine the biological impact of possible N glycosylation on LecB localization, the amino acid asparagine N22 was mutagenized to alanine to inactivate the putative N glycosylation site. Subsequently, the variant gene was cloned into the expression vector pBBR1MCS. Using this vector, lecB is under transcriptional control of the constitutive lac promoter, which not only uncouples lecB expression from its native regulation but also results in a moderate overexpression. Under these conditions, LecB transport is not affected and the protein can perfectly reach the outer membrane (50). For subcellular localization of the LecB variant with the N22A mutation, the variant gene was expressed in comparison to the wild-type gene in the background of the LecB-deficient strain P. aeruginosa PATI2. The cells were grown as an unsaturated biofilm on the surface of NB agar plates at 37°C. This method has been shown to result in the formation of unsaturated biofilms (45) of the type that is also found in the lungs of CF patients suffering from a P. aeruginosa infection (29).
After growth periods of 16 and 48 h, cells were fractionated and analyzed to compare the presence of the mutant LecB and the wild type. Detection of EstA, DsbA, and glucose-6P-dehydrogenase in the membrane, periplasmic (Fig. 4B), or cytoplasmic fraction (Fig. 4C), respectively, served as controls to validate the cellular fractionation procedure. Both strains produced LecB in the cytoplasm after 16 h of growth (Fig. 4A), but in contrast to the wild-type lectin, the mutated LecB accumulated in the cytoplasm and both HW and LW LecB were absent from the periplasm (Fig. 4A), demonstrating that the integrity of this N glycosylation site is required for proper transport of LecB to its final destination in the membrane. After a growth period of 48 h (Fig. 4A), LecB was present in the cytoplasm and the membrane fraction in the strain harboring the wild-type gene, as described in an earlier study (50), whereas the mutated LecB completely disappeared from the cells. Surprisingly, both molecular-weight forms of wild-type LecB were not seen in the periplasm after a growth period of 48 h (Fig. 4A), whereas they were present in the P. aeruginosa wild-type strain PAO1, as shown in Fig. 1A. The major difference between the two experiments is the fact that in the experiment whose results are shown in Fig. 1A, wild-type cells were used, whereas in the experiment whose results are shown in Fig. 4A, the strain used was the lecB-negative P. aeruginosa strain PATI2 carrying pBBR1MCS with the wild-type lecB gene under the control of the lac promoter provided by the vector. Due to this, the expression of LecB was increased and uncoupled from its native regulation. In cells grown for only 16 h, both forms of LecB are present in the cell lysate fraction. The HW LecB form is also present, in a remarkably large amount, in the periplasmic fraction in this growth period.
FIG. 4.
Secretion defect of the nonglycosylated LecB variant in the LecB-deficient strain P. aeruginosa PATI2, and controls to validate the cellular fractionation procedure. The asparagine residue (N) of the putative N glycosylation site (NXS/T) was replaced by alanine (A) by site-directed mutagenesis. The variant gene was cloned into pBBR1MCS under transcriptional control of the lac promoter and expressed in the LecB-deficient background of strain P. aeruginosa PATI2. (A) Subcellular localization of wild-type and mutant LecB (N22A mutant) in P. aeruginosa biofilm cells. Localization was determined by isolation of cellular compartments after growth periods of 16 and 48 h at 37°C on NB agar plates and subsequent Western immunoblotting using a LecB-specific antiserum. (B and C) Validation of cellular fractionation procedure was performed by immunoblotting using an antiserum raised against the periplasmic protein DsbA and the outer membrane protein EstA (B) and by distribution of the activity of the cytoplasmic glucose-6-phosphate dehydrogenase (C). The percentages of total enzyme activities present in cytoplasm, periplasm, membrane, and culture supernatant are given. (D) Relative transcription levels of the lecB variant gene lecB(N22A) in P. aeruginosa cells. RNA was isolated from the lecB-deficient P. aeruginosa strain PATI2 containing the plasmid pBBN22A [lecB(N22A)] encoding mutant LecB after growth periods of 16 and 48 h. As a positive control, mRNA isolated from the lecB-deficient P. aeruginosa strain PATI2 containing the lecB expression plasmid pBBC2 (wild-type lecB) was used. Detection of lecB and lecB(N22A) was achieved by using a specific TaqMan gene expression assay (Applied Biosystems, Switzerland). The transcription levels of the genes were normalized to that of the housekeeping gene rpoD and related to the lecB transcription level of the lecB-deficient strain PATI2 containing the empty vector pBBR1MCS. All values for the transcription levels investigated in this study represent means of at least three measurements; standard deviations were less than 5%. CL, cell lysate; CP, cytoplasm; PP, periplasm; M, membrane; SN, culture supernatant.
Given the hypothesis that glycosylation of LecB is a transitory event and the protein needs to be deglycosylated when it reaches the membrane destination, this requires an appropriate enzyme activity, which may be absent in this growth period or may be not present in sufficient amounts. This may lead to accumulation of the HW LecB form in the periplasm, consistent with the finding that no (deglycosylated) LecB is in the membrane fraction. The disappearance of mutated LecB from the cells suggests that a mechanism exists in P. aeruginosa by which the aberrant protein was efficiently removed from the cell, although it was similarly expressed on the transcriptional level at both sampling times, as was verified by RT-PCR, shown quantitatively in Fig. 4D. This indicates that the proper production of LecB depends on an intact N glycosylation site at this position and again supports the idea that as a consequence of not being glycosylated, the LecB variant N22 is degraded upon production in the living cell. N glycosylation of proteins has been shown to be important for protein stability in vivo (37). The influence of N glycosylation on the stability of the proteinase cathepsin E has been demonstrated via a similar site-directed mutagenesis analysis that showed that the removal of putative N glycosylation sites resulted in severely reduced stability of the protein (62).
We have shown here that a high-molecular-weight variant of LecB from P. aeruginosa is present in the periplasmic space and that it can be transformed into the smaller variant that is also present in other subcellular fractions by the activity of a commercial N-glycosidase F. Inactivation of the predicted N glycosylation site asparagine 22 resulted in accumulation of the protein in the cytoplasm. N glycosylation of LecB appears to be essential for the production of LecB and its transport to the outer membrane in P. aeruginosa. Although it remains to be further analyzed whether outer membrane LecB is completely deglycosylated after secretion is completed, this suggests that glycosylation is a novel transitory event during secretion. The transport mechanism used to direct LecB to the membrane is currently not known; however, our results show for the first time that a periplasmic intermediate exists and that the transport process thus is a two-step transport mechanism (15, 36, 52). The subunits of type IV pili and flagella have been the only described glycosylated proteins in P. aeruginosa. However, these proteins stay glycosylated after secretion (7, 40, 53), whereas LecB appears to be deglycosylated when it finally resides in the membrane. Some genes involved in this O glycosylation process have been identified (27). Interestingly, type IV pili and flagella, like the lectin, represent important adhesive surface structures which contribute to the virulence of P. aeruginosa (6, 35), which may imply that glycosylation might be a common feature of adhesins in this pathogenic bacterium. Furthermore, several other pathogenic bacteria are known to contain glycosylated adhesive proteins, among them Neisseria gonorrhoeae and Helicobacter pylori (17, 39), with glycosylation-defective mutants showing attenuated virulence-associated properties (1). Interestingly, N-linked glycans were found to be attached via the eukaryotic consensus sequence (NXS/T) on proteins in the intestinal Gram-negative pathogen Campylobacter jejuni (18, 25). The pathway responsible for N glycosylation in this bacterium appears to act at the cytoplasmic membrane, and key enzymes involved are glycosyltransferases of the so-called pgl gene cluster (1, 63). The genome sequence of P. aeruginosa PAO1 was analyzed for the presence of orthologues of these proteins from Campylobacter jejuni using the NBCI BLAST/blastp suite (www.ncbi.nlm.nih.gov/BLAST/) (data not shown). P. aeruginosa does not contain a complete cluster with similarity to the pgl gene cluster, and thus, the N glycosylation pathway from P. aeruginosa appears to be distinct from the pathway of C. jejuni or may at least be differently organized. Interestingly, the genes PA1385 and pslA encode proteins with strong similarity to the glycosyltransferases PglA and PglC, respectively, which are involved in the assembly of the heptasaccharide on a lipid carrier (1), and the P. aeruginosa protein MsbA shows a strong similarity to the flippase MsbA, which mediates the transport of the heptasaccharide from the cytoplasm to the periplasm (57). It will be interesting to further characterize the role of these candidates to elucidate the mechanism of N glycosylation in P. aeruginosa. Moreover, it will be of importance to analyze whether glycosylation is limited to adhesins or is also present in other proteins of P. aeruginosa.
Footnotes
Published ahead of print on 7 January 2011.
REFERENCES
- 1.Abu-Qarn, M., J. Eichler, and N. Sharon. 2008. Not just for Eukarya anymore: protein glycosylation in Bacteria and Archaea. Curr. Opin. Struct. Biol. 18:544-550. [DOI] [PubMed] [Google Scholar]
- 2.Adam, E. C., B. S. Mitchell, D. U. Schumacher, G. Grant, and U. Schumacher. 1997. P. aeruginosa PA-II lectin stops human ciliary beating: therapeutic implications of fucose. Am. J. Respir. Crit. Care Med. 155:2102-2104. [DOI] [PubMed] [Google Scholar]
- 3.Adam, E. C., D. U. Schumacher, and U. Schumacher. 1997. Cilia from a cystic fibrosis patient react to cilitoxic Pseudomonas aeruginosa II lectin in a similar manner to normal control cilia—a case report. J. Laryngol. Otol. 111:760-762. [DOI] [PubMed] [Google Scholar]
- 4.Beachey, E. H. 1980. Bacterial adherence. Chapman and Hall, London, England.
- 5.Beuth, J., B. Stoffel, H. L. Ko, J. Jeljaszewicz, and G. Pulverer. 1995. Mistellektin-1: neue therapeutische Perspektiven in der Onkologie. Onkologie 18:36-40. [Google Scholar]
- 6.Burrows, L. L. 2005. Weapons of mass retraction. Mol. Microbiol. 57:878-888. [DOI] [PubMed] [Google Scholar]
- 7.Castric, P., F. J. Cassels, and R. W. Carlson. 2001. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J. Biol. Chem. 276:26479-26485. [DOI] [PubMed] [Google Scholar]
- 8.de Smet, M. J., J. Kingma, and B. Witholt. 1978. The effect of toluene on the structure and permeability of the outer and cytoplasmic membranes of Escherichia coli. Biochim. Biophys. Acta 506:64-80. [DOI] [PubMed] [Google Scholar]
- 9.Gabius, H. J., S. Andre, H. Kaltner, and H. C. Siebert. 2002. The sugar code: functional lectinomics. Biochim. Biophys. Acta 1572:165-177. [DOI] [PubMed] [Google Scholar]
- 10.Garber, N. C., U. Guempel, N. Gilboa-Garber, and R. J. Doyle. 1987. Specificity of the fucose-binding lectin of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 48:331-334. [Google Scholar]
- 11.Gilboa-Garber, N. 1982. Pseudomonas aeruginosa lectins. Methods Enzymol. 83:378-385. [DOI] [PubMed] [Google Scholar]
- 12.Gilboa-Garber, N., and N. Garber. 1992. Microbial lectins, p. 541-591. In H. J. Allen and E. C. Kisailus (ed.), Glycoconjugates: composition, structure and function. M. Dekker, Inc., New York, NY.
- 13.Gilboa-Garber, N., D. J. Katcoff, and N. C. Garber. 2000. Identification and characterization of Pseudomonas aeruginosa PA-IIL lectin gene and protein compared to PA-IL. FEMS Immunol. Med. Microbiol. 29:53-57. [DOI] [PubMed] [Google Scholar]
- 14.Glick, J., and N. Garber. 1983. The intracellular localization of Pseudomonas aeruginosa lectins. J. Gen. Microbiol. 129:3085-3090. [DOI] [PubMed] [Google Scholar]
- 15.Hardie, K. R., S. Pommier, and S. Wilhelm. 2009. The secreted proteins of Pseudomonas aeruginosa: their export machineries, and how they contribute to pathogenesis, p. 451-458. In K. Wooldridge (ed.), Bacterial secreted proteins: secretory mechanisms and role in pathogenesis. Caister Academic Press, Norwich, United Kingdom.
- 16.Hauber, H. P., et al. 2008. Inhalation with fucose and galactose for treatment of Pseudomonas aeruginosa in cystic fibrosis patients. Int. J. Med. Sci. 5:371-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hegge, F. T., et al. 2004. Unique modifications with phosphocholine and phosphoethanolamine define alternate antigenic forms of Neisseria gonorrhoeae type IV pili. Proc. Natl. Acad. Sci. U. S. A. 101:10798-10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hitchen, P. G., and A. Dell. 2006. Bacterial glycoproteomics. Microbiology 152:1575-1580. [DOI] [PubMed] [Google Scholar]
- 19.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung, C. S., et al. 2002. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol. Microbiol. 44:903-915. [DOI] [PubMed] [Google Scholar]
- 21.Imberty, A., E. P. Mitchell, and M. Wimmerová. 2005. Structural basis of high-affinity glycan recognition by bacterial and fungal lectins. Curr. Opin. Struct. Biol. 15:525-534. [DOI] [PubMed] [Google Scholar]
- 22.Johansson, E. M., et al. 2008. Inhibition and dispersion of Pseudomonas aeruginosa biofilms by glycopeptide dendrimers targeting the fucose-specific lectin LecB. Chem. Biol. 15:1249-1257. [DOI] [PubMed] [Google Scholar]
- 23.Kolomiets, E., et al. 2009. Glycopeptide dendrimers with high affinity for the fucose-binding lectin LecB from Pseudomonas aeruginosa. Chem. Med. Chem. 4:562-569. [DOI] [PubMed] [Google Scholar]
- 24.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800-802. [PubMed] [Google Scholar]
- 25.Kowarik, M., et al. 2006. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 25:1957-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lengeler, K. B., D. Tielker, and J. F. Ernst. 2008. Protein-O-mannosyltransferases in virulence and development. Cell Mol. Life Sci. 65:528-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindhout, T., P. C. Lau, D. Brewer, and J. S. Lam. 2009. Truncation in the core oligosaccharide of lipopolysaccharide affects flagella-mediated motility in Pseudomonas aeruginosa PAO1 via modulation of cell surface attachment. Microbiology 155:3449-3460. [DOI] [PubMed] [Google Scholar]
- 28.Loris, R., D. Tielker, K.-E.Jaeger, and L. Wyns. 2003. Structural basis of carbohydrate recognition by the lectin LecB from Pseudomonas aeruginosa. J. Mol. Biol. 331:861-870. [DOI] [PubMed] [Google Scholar]
- 29.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma, Q., et al. 2003. Protein secretion systems of Pseudomonas aeruginosa and P. fluorescens. Biochim. Biophys. Acta 1611:223-233. [DOI] [PubMed] [Google Scholar]
- 31.Mewe, M., et al. 2005. Pseudomonas aeruginosa lectins I and II and their interaction with human airway cilia. J. Laryngol. Otol. 119:595-599. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell, E., et al. 2002. Structural basis for oligosaccharide-mediated adhesion of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Nat. Struct. Biol. 9:918-921. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell, E. P., et al. 2005. High affinity fucose binding of Pseudomonas aeruginosa lectin PA-IIL: 1.0 Å resolution crystal structure of the complex combined with thermodynamics and computational chemistry approaches. Proteins 58:735-746. [DOI] [PubMed] [Google Scholar]
- 34.Nilsson, C. L. 2007. Lectins: analytical tools from nature, p. 1-13. In C. L. Nilsson (ed.), Lectins: analytical technologies. Elsevier, Oxford, United Kingdom.
- 35.Ramphal, R., and S. K. Arora. 2001. Recognition of mucin components by Pseudomonas aeruginosa. Glycoconj. J. 18:709-713. [DOI] [PubMed] [Google Scholar]
- 36.Rosenau, F., and K. Jaeger. 2000. Bacterial lipases from Pseudomonas: regulation of gene expression and mechanisms of secretion. Biochimie 82:1023-1032. [DOI] [PubMed] [Google Scholar]
- 37.Rudd, P. M., T. Elliott, P. Cresswell, I. A. Wilson, and R. A. Dwek. 2001. Glycosylation and the immune system. Science 291:2370-2376. [DOI] [PubMed] [Google Scholar]
- 38.Scanlin, T. F., and M. C. Glick. 2001. Glycosylation and the cystic fibrosis transmembrane conductance regulator. Respir. Res. 2:276-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schirm, M., et al. 2004. Structural, genetic, and functional characterization of the flagellin glycosylation process in Helicobacter pylori. J. Bacteriol. 186:6721-6727. [DOI] [PubMed] [Google Scholar]
- 40.Schirm, M., et al. 2004. Structural and genetic characterization of glycosylation of type a flagellin in Pseudomonas aeruginosa. J. Bacteriol. 186:2523-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 42.Simon, R., M. O'Connell, M. Labes, and A. Puhler. 1986. Plasmid vectors for the genetic analysis and manipulation of Rhizobia and other gram-negative bacteria. Methods Enzymol. 118:640-659. [DOI] [PubMed] [Google Scholar]
- 43.Skropeta, D. 2009. The effect of individual N-glycans on enzyme activity. Bioorg. Med. Chem. 17:2645-2653. [DOI] [PubMed] [Google Scholar]
- 44.Sonawane, A., J. Jyot, and R. Ramphal. 2006. Pseudomonas aeruginosa LecB is involved in pilus biogenesis and protease IV activity but not in adhesion to respiratory mucins. Infect. Immun. 74:7035-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinberger, R. E., A. R. Allen, H. G. Hansa, and P. A. Holden. 2002. Elongation correlates with nutrient deprivation in Pseudomonas aeruginosa-unsaturates biofilms. Microb. Ecol. 43:416-423. [DOI] [PubMed] [Google Scholar]
- 46.Steuer, M. K., et al. 1993. Hemmung der bakteriellen Adhäsion durch Lektinblockade bei durch Pseudomonas aeruginosa induzierter Otitis externa im Vergleich zur lokalen Therapie mit Antibiotika. Otorhinolaryngol. Nova 3:19-25. [Google Scholar]
- 47.Swanson, A. F., and C. C. Kuo. 1991. Evidence that the major outer membrane protein of Chlamydia trachomatis is glycosylated. Infect. Immun. 59:2120-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szymanski, C. M., and B. W. Wren. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3:225-237. [DOI] [PubMed] [Google Scholar]
- 49.Tarentino, A. L., C. M. Gómez, and T. H. Plummer, Jr. 1985. Deglycosylation of asparagine-linked glycans by peptide: N-glycosidase F. Biochemistry 24:4665-4671. [DOI] [PubMed] [Google Scholar]
- 50.Tielker, D., et al. 2005. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 151:1313-1323. [DOI] [PubMed] [Google Scholar]
- 51.Tielker, D., F. Rosenau, K.-M. Bartels, T. Rosenbaum, and K.-E. Jaeger. 2006. Lectin-based affinity tag for one-step protein purification. Biotechniques 41:327-332. [DOI] [PubMed] [Google Scholar]
- 52.Tommassen, J., A. Filloux, M. Bally, M. Murgier, and A. Lazdunski. 1992. Protein secretion in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 9:73-90. [DOI] [PubMed] [Google Scholar]
- 53.Totten, P. A., and S. Lory. 1990. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J. Bacteriol. 172:7188-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ungar, D. 2009. Golgi linked protein glycosylation and associated diseases. Semin. Cell Dev. Biol. 20:762-769. [DOI] [PubMed] [Google Scholar]
- 55.Urban, A., M. Leipelt, T. Eggert, and K.-E. Jaeger. 2001. DsbA and DsbC affect extracellular enzyme formation in Pseudomonas aeruginosa. J. Bacteriol. 183:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Bismarck, P., R. Schneppenheim, and U. Schumacher. 2001. Successful treatment of Pseudomonas aeruginosa respiratory tract infection with a sugar solution—a case report on a lectin based therapeutic principle. Klin. Padiatr. 213:285-287. [DOI] [PubMed] [Google Scholar]
- 57.Weerepana, E., and B. Imperiali. 2006. Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems. Glycobiology 16:91-101. [DOI] [PubMed] [Google Scholar]
- 58.Wilhelm, S., J. Tommassen, and K.-E. Jaeger. 1999. A novel lipolytic enzyme located in the outer membrane of Pseudomonas aeruginosa. J. Bacteriol. 181:6977-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilhelm, S., A. Gdynia, P. Tielen, F. Rosenau, and K.-E. Jaeger. 2007. The autotransporter esterase EstA of Pseudomonas aeruginosa is required for rhamnolipid production, cell motility, and biofilm formation. J. Bacteriol. 189:6977-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winzer, K., et al. 2000. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J. Bacteriol. 182:6401-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woodcock, D. M., et al. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yasuda, Y., et al. 1999. Role of N-glycosylation in cathepsin E. A comparative study of cathepsin E with distinct N-linked oligosaccharides and its nonglycosylated mutant. Eur. J. Biochem. 266:383-391. [DOI] [PubMed] [Google Scholar]
- 63.Young, N. M., et al. 2002. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277:42530-42539. [DOI] [PubMed] [Google Scholar]