Abstract
We observed movies of replisome trafficking during Streptomyces coelicolor growth. A replisome(s) in the spore served as a replication center(s) until hyphae reached a certain length, when a tip-proximal replisome formed and moved at a fixed distance behind the tip at a speed equivalent to the extension rate of the tip.
Members of the bacterial genus Streptomyces exhibit mycelial growth and sporulation that are reminiscent of those of filamentous fungi. Spores germinate to form germ tubes (7, 18), which extend to elongated hyphae through peptidoglycan incorporation at the hyphal tip (2, 4). Eventually, a mycelium is formed by branch emergence from the lateral hyphal walls. Incorporation of new cell wall material at the hyphal tip and branch points is, either directly or indirectly, dependent on the essential protein DivIVA (3). Typical cell division does not occur during vegetative growth, and elongated, multigenomic compartments are delimited by occasionally spaced septa. In contrast to most other bacteria, in this genus many genes required for cell division are inessential for vegetative growth and are required only during sporulation (14). The chromosomes in the vegetative hyphae seem to remain uncondensed and do not undergo typical segregation. Early studies using pulse-labeling revealed that hyphae did not show any region of preferential incorporation of the label and that replicating nucleoids were evenly distributed along the hyphae (12, 13). This indicates that DNA replication does not depend on nucleoid location; the corollary of this is that mechanisms must exist to allow chromosomes to populate the extending hypha. However, Yang and Losick (19) were unable to find any evidence that DNA replication activity was concentrated at the apex. Further studies using a functional DnaN-enhanced green fluorescent protein (EGFP) fusion demonstrated that DNA replication takes place in both apical and subapical compartments of Streptomyces coelicolor vegetative hyphae (17). Moreover, replication is asynchronous, and only selected chromosomes undergo replication at any given time (17). Recently, a new method for monitoring Streptomyces hyphal growth in real time using time-lapse microscopy was established by Jyothikumar et al. (11). It showed that, following germination, hyphal extension occurred at about 20 μm h−1 when grown on mannitol-supplemented minimal medium at 30°C. Here, we present the results obtained by application of this method (11) to study replisome dynamics in vegetative mycelium. Captured images were processed using IPlabs 3.7 image processing software (BD Biosciences Bioimaging, Rockville, MD). Eleven Z sections (step size, 0.5 μm) of both phase-contrast and fluorescent images were captured at each time point at 10-min intervals and used to render three-dimensional images. DnaN-EGFP in S. coelicolor J3337 was visualized with a fluorescein isothiocyanate (FITC) filter and an exposure time of 100 ms.
DNA replication is limited to the spore until after significant hyphal extension.
An earlier study of S. coelicolor J3337 spores showed that two or three replisomes could be observed in germinating spores (17). In order to determine the sequence of events during germination, we performed time-lapse microscopy at 10-min intervals and generated movies of germinating S. coelicolor J3337 spores (n = 20) (11). As reported previously, the earliest observable event during spore germination was spore swelling (7, 18), which occurred before the appearance of a visible replisome (Fig. 1 A; see also Movie S1A in the supplemental material). Replisomes separated into two distinct foci in germinating spores (Fig. 1A; see also Movie S1A); this occurred either before or just after the emergence of a germ tube. It is not possible to say whether the separation of the original DnaN-EGFP focus represented two independent replication factories replicating two independent chromosomes or two sister replisomes positioned at separated replication forks of the same chromosome, as in Escherichia coli (16), although the subsequent reunification of the foci as a single spot suggests the latter. Following the appearance of a germ tube, further separation of foci was observed into three or more spots (data not shown) before a tip-proximal replisome appeared 2.50 (±1.05) μm from the tip when the germ tube was 7.56 (±2.47) μm in length (Fig. 1A; see also Movie S1A in the supplemental material). A schematic representation of replisome behavior during germination is shown in Fig. 1B.
FIG. 1.
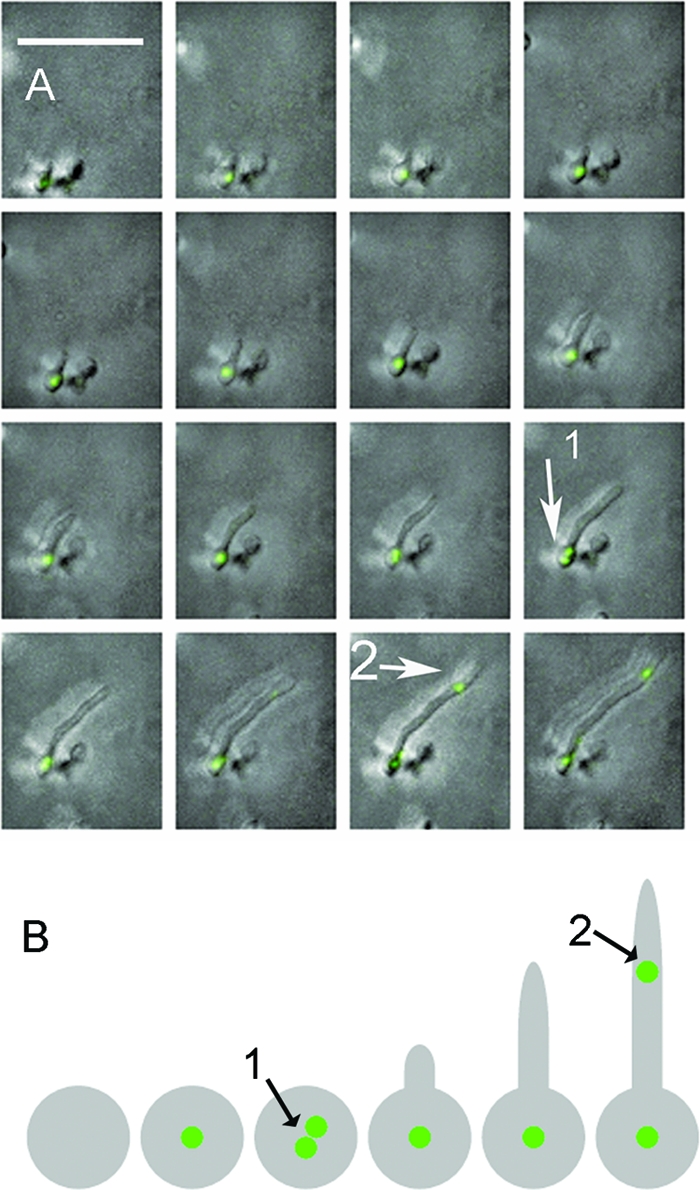
Replisome separation during S. coelicolor spore germination. Time-lapse movies of replisome formation in germinating spores of S. coelicolor J3337 were generated after image capture at 10-min intervals; representative images of germination (A) and a schematic depiction of replisome behavior during S. coelicolor spore germination (B) that display replisome focus separation in the spore (arrow 1) and appearance of another replisome toward the hyphal tip (arrow 2) are shown. Experimental conditions were exactly as described previously (11). The movie is Movie S1A in the supplemental material.
Replisomes follow tips at a speed equivalent to the rate of hyphal extension.
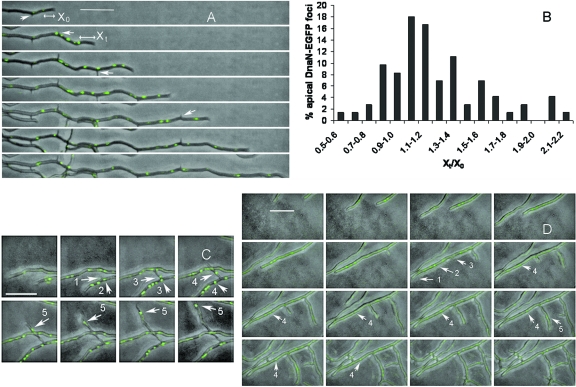
Movies of growing substrate hyphae of S. coelicolor J3337 were also generated. Both apical and subapical replisomes followed the extending tip at equivalent speeds (Fig. 2 A; see also Movie S2A in the supplemental material). Movies were broken into separate frames (n = 73) to enable measurements to be made of tip extension, tip-to-replisome distance, and replisome-to-replisome distance. The rate of tip extension was 16.52 (±5.68) μm h−1; this is a significantly lower (P < 1 × 10−5; Student's t test, assuming unequal variance) rate than previously reported for S. coelicolor K113 (6, 11), and presumably this is due to some impairment of DnaN efficiency when fused to EGFP. In parallel, we also measured the rate of progression of the apical replisome at 16.30 (±10.25) μm h−1; Student's t test showed that there was no significant difference between replisome progression and tip extension (P < 0.87) and indicates that there may be an association between tip extension and replisome progression similar to nuclear trafficking in filamentous fungi (5). Apical replisomes were always positioned some distance, on average 5.32 (±2.00) μm, from the tip, and at whatever distance the initial replisome was first observed behind the extending tip (Fig. 2A, X0), the replisome tended to remain at that distance behind the tip through subsequent time frames (Xt), so that Xt/X0 ≈ 1(Fig. 2A and B). Taken together, these data support a model where apical replisomes are positioned at a landmark, some distance from the tip of a young germ tube, and are somehow tied to that position as the tip extends. Tip-distal replisomes also followed the extending tip at an equivalent speed and were separated by an average distance of 4.01 (±2.39) μm. For analysis, hyphae were divided into regions and the mean distance between replisomes calculated in each region (region 1, tip to branch 1, 3.98 μm; region 2, branch 1 to branch 2, 3.38 μm; region 3, branch 2 to branch 3, 4.04 μm; region 4, branch 3 to branch 4, 3.62 μm; region 5, branch 4 to branch 5, 4.07 μm), showing that the distance between replisomes did not increase with increasing distance from the hyphal tip. These data are therefore consistent with earlier reports that DNA replication takes place throughout the length of the hyphae and is not restricted to the tip (12, 13, 19). Clearly, a number of candidate proteins, such as FilP (1), DivIVA (3, 8), ParA (10), or ParB (9), exist that might play a role in associating replisomes, possibly via the apical chromosome, with the tip. However, the mechanism by which this is accomplished and how this effect is exerted over 5 μm from the tip is unknown. Trafficking of replisomes along hyphae suggests that nucleoids flow through the extending hypha toward, but never reaching, the extending tip. It also remains to be seen how the distribution of the apical nucleoid is accomplished to allow nucleoid population of the extending hypha, as well as the deposition of subapical nucleoids for replication in tip-distal regions.
FIG. 2.
Replisome trafficking during S. coelicolor hyphal growth. Time-lapse movies of hyphal growth (Movies S2A, S2C, and S2D in the supplemental material) were generated after image capture at 10-min intervals. Horizontal white bars represent 10 μm. (A) Trafficking of replisomes during hyphal growth. White arrows indicate abortive branches that formed but did not extend; X0 is the distance of the apical replisome from the tip in the first time frame, and Xt is the distance of the apical replisome from the tip at subsequent time frames. (B) Histogram showing distribution Xt/X0 of apical replisomes from each frame of all movies (n = 73); when Xt/X0 ≈ 1, the distance from the tip to the apical replisome was the same as at the first time point at which it was measured. (C) Time-lapse mosaic showing trafficking of replisome foci into an extending branch; branch emergence (arrow 1), replisome located at the branch point (arrow 2), movement of two replisomes along primary hypha and branch (arrows 3, 4, and 5). (D) Arrest of a primary hypha (arrow 1) and replisomes (arrow 2) following branch emergence (arrow 3). Replisomes in primary hypha subsequently faded (arrow 4), while trafficking replisome foci were diverted into the branch (arrow 5). Experimental conditions were exactly as described previously (11).
Replisome absence prevents tip extension in emerging branches.
Previous studies with S. coelicolor K113 showed that branches often formed but did not extend (11); we also made this observation with S. coelicolor J3337. It was evident that these abortive branches did not receive a replisome (Fig. 2A, white arrows; see also Movie S2A in the supplemental material) and were bypassed by replisomes in the primary hypha. However, when a replisome (EGFP foci) was assembled in a stalled hypha, the branch resumed extending (Fig. 2C; see also Movie S2C in the supplemental material). This effect was also observed in primary hyphae (Fig. 1; see also Movie S2D in the supplemental material), where diversion of replisomes into a branch (i.e., away from the tip of the primary hypha) was accompanied by arrest in growth of the primary hypha and an equivalent cessation of replisome trafficking. Earlier studies demonstrated an association of DNA replication with peptidoglycan synthesis in Streptomyces antibioticus (15); consequently, it is possible that S. coelicolor hyphae require a replicating nucleoid close to the hyphal tip in order for cell wall extension to take place.
Conclusions.
Replisomes of S. coelicolor appear prior to germ tube emergence before separating to form distinct foci. Apical replisomes form in the locality of, but not directly at, the hyphal tip and follow behind the hyphal tip at a speed equivalent to the rate of hyphal extension. Replisomes also form in tip-distal regions and track along the hypha behind the extending tip. Branch extension is associated with the presence of a replisome in the newly formed branch.
Supplementary Material
Acknowledgments
This work was supported by U.K. BBSRC grant BB/D521657/l and by the Polish Ministry of Science and Higher Education (grant N301 2035 33). J.Z.-C. gratefully acknowledges financial support received from the Foundation for Polish Science (MISTRZ Program).
Footnotes
Published ahead of print on 30 December 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bagchi, S., H. Tomenius, L. M. Belova, and N. Ausmees. 2008. Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces. Mol. Microbiol. 70:1037-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767-776. [DOI] [PubMed] [Google Scholar]
- 3.Flardh, K. 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 49:1523-1536. [DOI] [PubMed] [Google Scholar]
- 4.Flardh, K., and M. J. Buttner. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7:36-49. [DOI] [PubMed] [Google Scholar]
- 5.Freitag, M., P. C. Hickey, N. B. Raju, E. U. Selker, and N. D. Read. 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41:897-910. [DOI] [PubMed] [Google Scholar]
- 6.Grantcharova, N., W. Ubhayasekera, S. L. Mowbray, J. R. McCormick, and L. Flardh. 2003. A missense mutation in ftsZ differentially affects vegetative and developmentally controlled cell division in Streptomyces coelicolor A3(2). Mol. Microbiol. 47:645-656. [DOI] [PubMed] [Google Scholar]
- 7.Hardisson, C., M. B. Manzanal, J. A. Salas, and J. E. Suarez. 1978. Fine structure, physiology and biochemistry of arthrospore germination in Streptomyces antibioticus. J. Gen. Microbiol. 105:203-214. [DOI] [PubMed] [Google Scholar]
- 8.Hempel, A. M., S.-B. Wang, M. Letek, J. A. Gil, and K. Flardh. 2008. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J. Bacteriol. 190:7579-7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jakimowicz, D., K. Chater, and J. Zakrzewska-Czerwinska. 2002. The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the linear chromosome. Mol. Microbiol. 45:1365-1377. [DOI] [PubMed] [Google Scholar]
- 10.Jakimowicz, D., P. Zydek, A. Kois, J. Zakrzewska-Czerwinska, and K. F. Chater. 2007. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol. Microbiol. 65:625-641. [DOI] [PubMed] [Google Scholar]
- 11.Jyothikumar, V., E. J. Tilley, R. Wali, and P. R. Herron. 2008. Time-lapse microscopy of Streptomyces coelicolor growth and sporulation. Appl. Environ. Microbiol. 74:6774-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kummer, C., and S. Kretschmer. 1986. DNA replication behaviour of individual nucleoids of two Streptomyces strains. J. Basic Microbiol. 26:219-223. [Google Scholar]
- 13.Kummer, C., and S. Kretschmer. 1986. DNA replication is not restricted to specific regions in young vegetative Streptomyces mycelia. J. Basic Microbiol. 26:27-31. [DOI] [PubMed] [Google Scholar]
- 14.McCormick, J. R. 2009. Cell division is dispensable but not irrelevant in Streptomyces. Curr. Opin. Microbiol. 12:689-698. [DOI] [PubMed] [Google Scholar]
- 15.Miguelez, E. M., C. Martin, M. B. Manzanal, and C. Hardisson. 1992. Growth and morphogenesis in Streptomyces. FEMS Microbiol. Lett. 100:351-359. [DOI] [PubMed] [Google Scholar]
- 16.Reyes-Lamothe, R., C. Possoz, O. Danilova, and D. J. Sherratt. 2008. Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133:90-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruban-Osmialowska, B., D. Jakimowicz, A. Smulczyk-Krawczyszyn, K. F. Chater, and J. Zakrzewska-Czerwinska. 2006. Replisome localization in vegetative and aerial hyphae of Streptomyces coelicolor. J. Bacteriol. 188:7311-7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharples, G. P., and S. T. William. 1976. Fine structure of spore germination in actinomycetes. J. Gen. Microbiol. 96:323-332. [Google Scholar]
- 19.Yang, M. C., and R. Losick. 2001. Cytological evidence for association of the ends of the linear chromosome in Streptomyces coelicolor. J. Bacteriol. 183:5180-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.