Abstract
d-Alanine is a central component of the cell wall in most prokaryotes. d-Alanine synthesis in Escherichia coli is carried out by two different alanine racemases encoded by the alr and dadX genes. Deletion of alr and dadX from the E. coli genome results in a d-alanine auxotrophic phenotype. However, we have observed growth of prototrophic phenotypic revertants during routine culturing of a d-alanine auxotrophic strain. We present a detailed comparison of the proteome and transcriptome profiles of the d-alanine auxotroph and a prototrophic revertant strain. Most noticeably, a general upregulation of genes involved in methionine synthesis in the revertant strain was detected. The appearance of the revertant phenotype was genetically linked to point mutations in the methionine repressor gene (metJ). Our results reveal an alternative metabolic pathway which can supply essential d-alanine for peptidoglycan synthesis of alr- and dadX-deficient E. coli mutants and provide evidence for significant alanine racemase coactivity of the E. coli cystathionine beta-lyase (MetC).
Alanine racemases (EC 5.1.1.1) are unique prokaryotic enzymes that catalyze the reversible racemization of l- and d-alanine, the latter one being an essential component in the biosynthesis of the bacterial peptidoglycan of Gram-positive and Gram-negative bacteria (47). The bacteria investigated to date have been found to possess either one or two distinct alanine racemase genes. The alr gene encodes a constitutively expressed alanine racemase, which provides d-alanine for sufficient cross-linking of adjacent peptidoglycan strands in the cell wall. The second gene encodes the so-called catabolic alanine racemase, which is essential for l-alanine catabolism (24, 28, 41, 42, 48). In Escherichia coli, the alr-encoded alanine racemase is constitutively expressed, whereas the dadX-encoded enzyme is essential only for l-alanine catabolism, providing a substrate for a d-alanine-specific dehydrogenase encoded by the dadA gene (51). The dadX gene product provides a secondary source of d-alanine for cell wall biosynthesis.
d-Alanine auxotrophic E. coli, Bacillus subtilis, Corynebacterium glutamicum, Listeria monocytogenes, and Lactobacillus plantarum strains have been generated by inactivating genes encoding alanine racemases (15, 17, 24, 42, 43, 45). A strong selective pressure for maintenance of an alanine racemase (Dal)-encoding plasmid in a chromosomal dal mutant of Bacillus subtilis was observed upon growth on rich medium. In Lactobacillus plantarum, plasmids encoding alanine racemase (Alr) were efficiently selected in an alr-deficient Lactobacillus plantarum strain (5). In Listeria monocytogenes, two genes, an alanine racemase gene (dal) and a d-amino acid aminotransferase gene (dat), which control the synthesis of d-alanine, had to be inactivated in order to achieve complete d-alanine auxotrophy (46).
Under certain circumstances, the d-alanine auxotrophic phenotype was lost, indicating a redundancy of alanine racemase activity in bacteria. The d-alanine auxotrophic phenotype of a Bacillus subtilis dal mutant was lost when grown in minimal medium without l-alanine, suggesting that Bacillus subtilis possesses a second l-alanine-repressible alanine racemase. This finding was supported by the discovery of a second gene in the Bacillus subtilis genome sequence, which encodes a protein with high homology to alanine racemases (17). In E. coli only, the alr/dadX double knockout strain is dependent on d-alanine for growth (51). Removal of d-alanine from rich liquid medium during growth of an alanine racemase-deficient strain resulted in rapid cell lysis. However, this lysis was partially prevented when cells were grown in minimal medium (50). It was speculated that the protection could be from either osmotic protective pressure or derepression of a redundant alanine racemase, which however was not identified (50).
In this paper, we isolated and characterized E. coli BL21(DE3)ΔalrΔdadX mutants which were able to grow without d-alanine supplementation. Proteome and transcriptome analyses of one phenotypic revertant (PR) revealed that an alternative metabolic pathway involving the methionine regulon can be activated under certain growth conditions. Cystathionine beta-lyase (MetC), which provides homocysteine for the final steps in methionine synthesis, was shown to be the effector behind the d-alanine prototrophic phenotype.
MATERIALS AND METHODS
Replacement fragment construction.
The flanking regions of the alr, dadX, and metJ genes were PCR amplified using the primer pairs shown in Table 1. The resulting paired amplicons were then ligated simultaneously with a kanamycin resistance (Kmr) cassette containing an FLP recombination target (FRT) site (5′-GAAGTTCCTATTCTCTAGAAAGTATAGGAACTTC-3′) at both ends. For alr and dadX replacement, the upstream and downstream regions of the corresponding genes were amplified as the left arm and right arm, respectively, for homologous recombination. To replace metJ, the MetJ coding sequence and downstream region were amplified as the left arm and right arm, respectively.
TABLE 1.
Sequences of primers used in this study
| Primer | Oligonucleotide sequence (5′ to 3′)a | Enzyme | Ampliconb |
|---|---|---|---|
| FalrLA | acgcgtcgacGCGTTCCCGCGCACGCCGTA | SalI | alr upstream region |
| RalrLA | cccaagcttGCGGTTAATCACAACAGTTG | HindIII | |
| FalrRA | cccaagcttAACTTATTACGCGCCTGACT | HindIII | alr downstream region |
| RalrRA | cgggatccCCGGATGGTCCGCACCAAAC | BamHI | |
| FdadXLA | AACCGGAGACTGTCAGCTATTT | dadX upstream region | |
| RdadXLA | ACGGGTCATCTCGTTTCCTT | ||
| FdadXRA | catatgCGGTTGTGACGGTGTAACTTGTTGT | NdeI | dadX downstream region |
| RdadXRA | gagctcGGATCCAATTGGTCGGGAACGCGATCTTCCG | SacI | |
| FmetJL | gaagatctACGCGTCATGTGATGAAG | BglII | metJ coding regionc |
| RmetJL | cgccatatgTTAGTATTCCCACGTCTCCGG | NdeI | |
| FmetJR | cactgcagAGCAAAAAAGAGCGGCGCGG | PstI | metJ downstream region |
| RmetJR | cgagctcGCCGACTGCTTCAAAAACG | SacI |
Sequences complementary to the amplified regions are represented by uppercase letters, and newly added sequences are represented by lowercase letters. Incorporated cutting sites for restriction enzymes are underlined.
Flanking regions of individual genes for complementary recombination.
The metJ coding region is used as the left complementary region for recombination.
Gene disruption.
BL21(DE3) cells carrying a Red helper plasmid (pKD46) (E. coli Genetic Stock Center, Yale University) were grown in 50 ml of Luria-Bertani (LB) liquid containing ampicillin (100 μg/ml) at 30°C with 300 rpm of rotation until the optical density at 600 nm (OD600) reached 0.4. A total of 1 mM l-arabinose was then added to cell culture at 30°C for 1 h until the OD600 reached 0.6. The cells were made electrocompetent by being concentrated by 100-fold and being washed three times with ice-cold 10% glycerol. The replacement DNA fragments containing a Kmr gene flanked on both sides by homologous sequences of target genes were prepared by digestion of replacement vector with corresponding enzymes, gel purified, and suspended in water. Electroporation was done by using a Micropulser (Bio-Rad) with a 2.5-kV booster and 0.2-cm chamber using 40 μl of cells and 100 ng of replacement fragment, according to the manufacturer's instructions. The shocked cells were added to 1 ml super optimal broth with catabolite repression (SOC), incubated for 1 h at 37°C, and then spread onto LB agar containing kanamycin (50 μg/ml) and d-alanine (100 μM) to select Kmr transformants. Kmr colonies were screened by PCR with primers annealing to regions outside the mutated gene. The Kmr gene could be excised by introducing plasmid pCP20 encoding the FLP recombinase (E. coli Genetic Stock Center, Yale University) when necessary. Both plasmids pKD46 and pCP20 are thermosensitive for replication and were cured at 42°C (12, 30).
Isolation of revertants and phenotypic analysis.
The revertants were isolated by plating 50 μl BL21(DE3)ΔalrΔdadX cells growing in mid-log phase in LB liquid medium containing 100 μM d-alanine onto LB agar plates. The cell viability was evaluated by plating cells onto LB agar with or without supplementation of 100 μM d-alanine. For transmission electron microscopy (TEM), cells were harvested by low-speed centrifugation (1,000 × g, 5 min), washed twice in sodium cacodylate buffer (200 mM, pH 7.3), prefixed in 3% (wt/vol) glutaraldehyde, and fixed with 1% (wt/vol) osmium tetroxide (OsO4). After dehydration with increasing concentrations of ethanol, cells were embedded with EPOX 812 and polymerized at 60°C for 48 h. To improve contrast, the samples were stained with 4% (wt/vol) uranyl acetate and then with 0.4% (wt/vol) lead citrate. Ultrathin sections were deposited on a copper grid and viewed with a Hitachi H600 electron microscope at 80 kV using a 30-mm objective aperture.
Proteome analysis.
A single colony of strain BL21(DE3)ΔalrΔdadX or a phenotypic revertant (PR) strain named BL21(DE3)ΔalrΔdadXPR was inoculated in 3 ml LB medium with or without 100 μM d-alanine and grown overnight with constant aeration at 37°C. The next day, 50 ml LB medium with or without d-alanine was inoculated with 0.5 ml of the corresponding overnight cultures and grown at 37°C with constant aeration to an OD600 of 0.6. Cells were harvested by centrifugation at 3,000 × g for 10 min at 4°C. Pellets were washed gently on ice in phosphate-buffered saline (PBS) (50 mM, pH 7.2), centrifuged, and then stored at −80°C until used in proteome analysis or later microarray analysis.
Pellets corresponding to an amount with an OD600 of 19 were resuspended in lysis buffer consisting of 8 M urea, 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 2% (vol/vol) immobilized pH gradient (IPG) buffer (pH 3 to 10), and 1% (wt/vol) dithiothreitol (DTT) and sonicated on ice. Sonicated samples were centrifuged for 1 h at 40,000 × g to remove cell debris, and supernatants were stored at −80°C until used. Protein amounts were estimated using the 2-D Quant kit (product no. 80-6486-22; GE Healthcare), according to manufacturer's instructions.
IPG two-dimensional polyacrylamide gel electrophoresis (2-D PAGE) was used for proteome investigations. Nonlinear IPG 3-10 strips (24-cm; GE Healthcare) were used for isoelectric focusing with the Ettan IPGphor (GE Healthcare). The strips were rehydrated with 150 μg of protein in lysis buffer for 12 h at 30 V. The voltage was then increased as follows: 1 h at 100 V, 1 h at 250 V, 1 h at 500 V, 1 h at 1,000 V, 2 h at 3,000 V. Focusing was completed at 8,000 V (maximum 50-mA current, 20°C) until a total of ∼60,000 volt-hours were reached. After the first dimension was completed, the strips were equilibrated in a buffer containing 6 M urea, 1% (wt/vol) DTT, 50 mM Tris-HCl at pH 8.8, 30% (vol/vol) glycerol, 2% (wt/vol) sodium dodecyl sulfate (SDS), and bromphenol blue for 15 min. The strips were then equilibrated in the buffer in which DTT was replaced with 4% iodoacetamide for 15 min. The Ettan DALTsix electrophoresis unit (GE Healthcare) was used to separate proteins according to their molecular weight on 9 to 16% linear gradient polyacrylamide gels (25 by 20 by 0.1 cm) overnight at 100 V at 15°C.
After two-dimensional runs, E. coli proteins were visualized using a silver staining protocol according to Gharahdaghi et al. (19). Silver-stained gels were scanned at 300 dpi and analyzed using Image Master, version 5.0 (GE Healthcare). Protein spots, which were observed upregulated in the revertant strain on three independently run sets of gels, were excised and analyzed by mass spectrometry.
The general strategy for mass spectrometric analysis was performed essentially as previously reviewed (25). Excised protein spots were subjected to in-gel digestion using 12.5 ng/ml porcine trypsin (Promega) in 50 mM NH4CO3 buffer for enzymatic cleavage of the proteins, which have been reduced and alkylated with 10 mM DTT and 55 mM iodoacetamide, respectively. Tryptic peptides were desalted and concentrated by using reversed-phase microcolumns (Zip-Tip C18; Millipore), according to the manufacturer's instructions. Peptides were cocrystallized on matrix-assisted laser desorption ionization (MALDI) target plates using 4-hydroxy-α-cyanocinnamic acid by the dried-droplet method. Mass spectra were acquired on a prOTOF 2000 matrix-assisted laser desorption ionization-orthogonal time-of-flight (MALDI O-TOF) mass spectrometer interfaced with TOFworks software (Perkin Elmer/SCIEX). Proteins were identified using a threshold for peptide mass tolerance of 30 ppm by a submitting peptide mass peak list to database search (with species restrictions to E. coli) using the TOFworks software. A minimum of 25% sequence coverage for identifications was observed.
Microarray analysis.
Total RNA was isolated from the cell pellets prepared in the previous section using the protocol accompanying the RNeasy minikit (Qiagen), and cDNA synthesis and labeling were done as described in the Affymetrix GeneChip E. coli Genome Array technical manual. Affymetrix GeneChip E. coli genome arrays were used to analyze the complete E. coli transcriptome. Biotin-labeled fragmented cDNA (2.5 μg) was hybridized to E. coli genome arrays at 45°C for 16 h, as recommended in the GeneChip technical manual. The probed arrays were scanned at 570 nm at a resolution of 1.5 μm using Scanner3000 (Affymetrix). Gene expression levels were calculated with GCOS1.2 software. The mean hybridization signal of all probe sets on each array was scaled to a common value of 200. The Affymetrix detection algorithm was applied to determine whether a gene was expressed. The change between two experimental conditions (n-fold) was calculated by taking the mean of the log ratios of probe pair intensities across the two arrays. For each strain, three independent cultures were prepared, and the RNA was analyzed.
Complementary expression plasmid construction.
Genomic DNA, isolated from the overnight culture of BL21(DE3) or BL21(DE3)ΔalrΔdadX, was used as the template for amplification of the full-length open reading frames of glyA, metB, metC, asd, ybdL, trpB, and malY. Primers with NdeI or XbaI at 5′ primer overhangs and BamHI at 3′ primer overhangs for each gene were designed to amplify the full-length genes. The gel-purified amplicons were ligated into pET11a expression vectors (Novagen), resulting in glyA, metB, metC, asd, ybdL, trpB, and malY expression constructs.
Western blotting.
Cells were collected by centrifugation and then resuspended in lysis buffer (20 mM Tris and 5 mM EDTA, pH 8.0). Cells were lysed by sonication and then centrifuged at 20,000 × g for 15 min to collect the soluble fraction. The soluble fraction was normalized to 10 OD units/ml and analyzed by immunoblotting using anti-E. coli MetC antibody (a polyclonal rabbit antibody developed against E. coli produced full-length MetC).
RESULTS
Phenotypic characterization of a BL21(DE3)ΔalrΔdadX strain and d-alanine prototrophic revertants.
To investigate whether a stable d-alanine auxotrophic E. coli strain can be made, an alr/dadX-deficient BL21(DE3) strain was obtained by sequential deletion of the alr and dadX genes using the λ Red recombinase-mediated recombination (12, 30).
The BL21(DE3)ΔalrΔdadX strain displayed a strict dependence on d-alanine on LB medium, indicating that disruption of both the alr and dadX genes resulted in a d-alanine auxotrophic phenotype (Table 2). However, upon characterization of the d-alanine dependence of the BL21(DE3)ΔalrΔdadX strain, prototrophic phenotypic revertants could be isolated from cells growing without an exogenous source of d-alanine (100 μM) on LB medium. If assuming that the plating efficiencies of BL21(DE3)ΔalrΔdadX and the BL21(DE3)ΔalrΔdadX phenotypic revertant are identical, we estimated the frequency of mutation to be 7 × 10−7. During the 16 h of cultivation on LB medium, there were at least 20 generations. Thus, the mutation rate is less than 5 × 10−8 per generation.
TABLE 2.
d-Alanine requirements for different alr/dadX-deficient strains
| Strain | Growth on selective LB mediuma |
|||
|---|---|---|---|---|
| Km+/d-ala− | Km+/d-ala+ | Km−/d-ala− | Km−/d-ala+ | |
| BL21(DE3)ΔalrΔdadX | N | N | N | V |
| BL21(DE3)ΔalrΔdadX metJ(R42C)-Kmr | V | V | V | V |
| BL21(DE3)ΔalrΔdadXPR | N | N | V | V |
| BL21(DE3)ΔalrΔdadXPRmetJ(R42C)-Kmr | V | V | V | V |
| BL21(DE3)ΔalrΔdadXPRmetJ-Kmr | N | V | N | V |
| BL21(DE3)ΔalrΔdadXΔmetC | N | N | N | V |
| BL21(DE3)ΔalrΔdadXPRΔmetC | N | N | N | V |
Km, kanamycin; d-ala, d-alanine; +, in the presence of reagent; −, in the absence of reagent; V, viable; N, not viable.
On rich liquid medium, the doubling time of the BL21(DE3)ΔalrΔdadX phenotypic revertant is almost the same as that of BL21(DE3)ΔalrΔdadX growing with 100 μM d-alanine supplementation in the medium for approximately 30 min. In order to explain the phenotype reversion of BL21(DE3)ΔalrΔdadX, the morphology of the BL21(DE3) wild type, BL21(DE3)ΔalrΔdadX, and one representative prototrophic phenotypic revertant, named BL21(DE3)ΔalrΔdadXPR, were examined by TEM. BL21(DE3)ΔalrΔdadX growing with an exogenous supply of d-alanine exhibits characteristic abnormalities, including increased roughness of the cell surface, collapse of the cell structure, and a ghost-like appearance in which the cells seem empty inside (Fig. 1). Thus, the d-alanine supplementation to the medium ensures the survival of BL21(DE3)ΔalrΔdadX but with visible impairments of cell wall integrity. In contrast, BL21(DE3)ΔalrΔdadXPR cells display morphology similar to that of wild-type cells, typical of this E. coli strain (Fig. 1). This indicates that d-alanine is still synthesized in the BL21(DE3)ΔalrΔdadX revertant despite the knockout of the two known E. coli alanine racemase genes.
FIG. 1.
Morphology analysis of BL21(DE3)ΔalrΔdadX and BL21(DE3)ΔalrΔdadXPR. Transmission electron microscopy shows the cell membrane structure of the BL21(DE3) cell growing on LB medium (A), the BL21(DE3)ΔalrΔdadX cell growing on LB medium supplemented with 0.1 mM d-alanine (B), and the BL21(DE3)ΔalrΔdadXPR cell growing on LB medium (C).
Comparative proteome and transcriptome analyses of BL21(DE3)ΔalrΔdadX and BL21(DE3)ΔalrΔdadXPR.
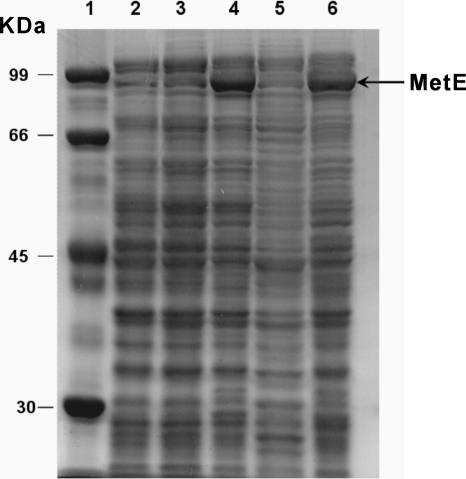
Of the 50 independently isolated phenotypic revertant colonies, 80% showed significant overexpression of E. coli MetE, as determined by SDS-PAGE and protein identification by MALDI peptide mass fingerprinting (Fig. 2, lane 4). To elucidate the molecular mechanism behind the d-alanine prototrophy of the phenotypic revertant, a comparative proteome investigation of BL21(DE3)ΔalrΔdadX and BL21(DE3)ΔalrΔdadXPR was performed (Fig. 3). By means of peptide mass fingerprinting (exemplified in Fig. 3B), the most significantly upregulated proteins in BL21(DE3)ΔalrΔdadXPR were identified as MetA, cystathionine gamma-synthetase (MetB), MetE, MetF, MetK, Asd, and serine hydroxymethyl transferase (GlyA). Thus, expression of enzymes involved in methionine synthesis is upregulated in BL21(DE3)ΔalrΔdadXPR (Fig. 3; Table 3). The function of these genes is depicted here (see Fig. 5), and methionine regulation in E. coli was reviewed in detail by Weissbach and Brot (49).
FIG. 2.
SDS-PAGE showing the effect of knock-in of metJ-Kmr and metJ(R42C)-Kmr. MetE of BL21(DE3)ΔalrΔdadXPR is downregulated after metJ-Kmr knock-in. Lane 1, molecular mass marker; lanes 2 to 6, BL21(DE3), BL21(DE3)ΔalrΔdadX, BL21(DE3)ΔalrΔdadXPR, BL21(DE3)ΔalrΔdadXPR with the metJ-Kmr knock-in, and BL21(DE3)ΔalrΔdadXPR with the metJ(R42C)-Kmr knock-in, respectively.
FIG. 3.
Comparative proteome analysis of BL21(DE3)ΔalrΔdadX and BL21(DE3)ΔalrΔdadXPR. (A) Silver-stained 2-D gel of BL21(DE3)ΔalrΔdadXPR. The identities of the marked spots are shown in Table 3. Solid-circles point at a degradation product of MetE. Enlargements a, c, e, and g (d-alanine auxotrophy) and b, d, f, and h (revertant strain) are exemplifying regions comprising identified proteins that were upregulated in the revertant strain compared to by d-alanine auxotrophy. Differently regulated spots are encircled. (a and b) GlyA and MetB. Solid circles indicates upregulated spots, which were identified as degradation products of MetE. (c and d) Two isoforms of MetF. (e and f) Two isoforms of Asd. (g and h) Two isoforms of full-length MetE. (B) Peptide mass spectrum of the P13 spot. Mass peaks matching MetB are indicated by numbered arrows corresponding to experimentally determined peptides, shown in the upper right corner. A total of 10 out of 11 peptides matched the theoretical tryptic peptides derived from the MetB sequence, with mass accuracy of less than 10 ppm. A total of 26% of the MetB sequence was covered by the peptides. “T” arrows point at peptides coming from the autodigestion of trypsin.
TABLE 3.
Upregulated genes in BL21(DE3)ΔalrΔdadXPR
| Gene | 2-D gel spot(s)c | Function | SLR (log2)a |
|
|---|---|---|---|---|
| Meanb | SD | |||
| asd | P9, P10 | Aspartate-semialdehyde dehydrogenase | 1.43 | 0.12 |
| folE | GTP cyclohydrolase I | 1.63 | 0.21 | |
| glyA | P3 | Serine hydroxymethyltransferase | ND | ND |
| metA | P11 | Homoserine transsuccinylase | 2.43 | 0.31 |
| metB | P13 | Cystathionine gamma-synthase | 2.20 | 0.56 |
| metC | Cystathionine beta-lyase (beta-cystathionase) | 2.27 | 0.40 | |
| metE | P1, P2, P15 | Tetrahydropteroyltriglutamate methyltransferase | 2.77 | 0.15 |
| metF | P6, P7 | 5,10-Methylenetetrahydrofolate reductase | 2.83 | 0.21 |
| metI | d- and l-Methionine transport protein (ABC superfamily, membrane) | 1.73 | 0.35 | |
| metK | P4 | S-Adenosylmethionine synthetase | ND | ND |
| metL | Aspartokinase II and homoserine dehydrogenase II | 2.27 | 0.75 | |
| metN | d- and l-Methionine transport protein (ABC superfamily, atp_bind) | 2.00 | 0.53 | |
| metQ | d-Methionine transport protein (ABC superfamily, peri_bind) | 1.57 | 0.32 | |
| rplJ | P14 | Ribosomal protein L10 | ND | ND |
| spf | Spot 42 RNA | 1.63 | 0.31 | |
| trpA | Tryptophan synthase, alpha protein | 1.23 | 0.23 | |
| trpB | Tryptophan synthase, beta protein | 1.33 | 0.21 | |
| trpC | N-(5-phosphoribosyl) anthranilate isomerase and indole-3-glycerolphosphate synthetase | 1.20 | 0.10 | |
| trpD | Anthranilate synthase component II, glutamine amido-transferase, and phosphoribosylanthranilate transferase | 1.60 | 0.10 | |
| trpE | Anthranilate synthase component I | 1.63 | 0.15 | |
| trpL | trp operon leader peptide | 1.47 | 0.32 | |
| ybdH | Putative oxidoreductase | 1.83 | 0.15 | |
| ybdL | Putative aminotransferase | 1.30 | 0.17 | |
| yfcF | orf, hypothetical protein | 1.53 | 0.35 | |
An SLR (signal log ratio) is applied to reflect the change (n-fold) between the BL21(DE3)ΔalrΔdadX cell and its revertant. The SLR was calculated by taking the ratio of the signal intensity (difference of the log2 value) between the two cells. Genes that showed at least a 2-fold increase in mRNA abundance relative to the control and had a present call by the Affymetrix algorithm were considered upregulated. ND, not determined.
An average value from three individual experiments.
“P” numbers denote proteins spots upregulated on 2-D gels and identified by peptide mass fingerprinting (Fig. 3B).
To verify whether any other genes are differentially expressed, the transcriptome profiles at exponential growth phases of BL21(DE3)ΔalrΔdadX and BL21(DE3)ΔalrΔdadXPR were compared. Three individual experiments were scaled to a common, global average expression level of 200 to correct for experimental variation. The gene was considered upregulated in BL21(DE3)ΔalrΔdadXPR only when it received an Affymetrix call of “present” in both tested [BL21(DE3)ΔalrΔdadXPR] and control [BL21(DE3)ΔalrΔdadX] arrays and showed a >2-fold change in gene expression with a P value of <0.05 (Student's t test) in all three individual experiments. Using this cutoff, we were able to identify 21 genes which were significantly upregulated in BL21(DE3)ΔalrΔdadXPR, including all methionine genes, which are responsible for the conversion of homoserine to l-methionine, and other genes associated with methionine metabolism (Table 3). MetL (aspartokinase II) is a bifunctional enzyme which, together with aspartate semialdehyde dehydrogenase (Asd), can convert l-aspartate to homoserine, the substrate of MetA. The metL and asd genes are repressed by excess l-methionine (44) and are upregulated in BL21(DE3)ΔalrΔdadXPR. Taken together with the observed upregulation of serine hydroxymethyl transferase (GlyA), which supplies 5,10-methylene-tetrahydrofolate for MetF (see Fig. 5) and GTP cyclohydrolase I (FolE), our results revealed that all genes responsible for linking l-aspartate, l-serine, and the folate pathway to the methionine pathway are upregulated in the BL21(DE3)ΔalrΔdadXPR strain. With the exception of MetK and GlyA, data obtained from microarray analysis verified the results from proteome analysis. Microarray data can be accessed through http://www.ebi.ac.uk/miamexpress/login.html under log-in name lshk_array_2005 and password lshk_array_2005.
We also observed upregulation of the MetD methionine transporters (MetI, MetN, and MetQ), which actively transport l-methionine or d-methionine across the cell membrane (26) and are regulated by MetJ (27, 53). We did not observe upregulation of MetH neither at the protein level nor at the transcriptional level in our study. MetH is the only Met gene that does not contain MetJ recognition sequences (Met box) in its promoter and is regulated indirectly by a complex interplay of MetR and vitamin B12 (6, 32).
The methionine aminotransferase YbdL (16) is upregulated in BL21(DE3)ΔalrΔdadXPR. This confirms a previous prediction of MetJ binding sites being located in its promoter (27). A hypothetical protein, YbdH, was also upregulated, and its coding gene is located next to ybdL in the E. coli genome but transcribed in the opposite direction.
Besides general upregulation of genes involved in methionine metabolism, all genes carried by the trpEDCBA operon involved in the conversion of chorismate to l-tryptophan were found upregulated. This indicates that two major metabolic pathways are affected in BL21(DE3)ΔalrΔdadXPR.
We specifically evaluated the expression level of MalY, a regulator of the maltose operon, as it was previously shown to have alanine racemase activity in Treponema denticola (4). When 2-D gels of BL21(DE3)ΔalrΔdadX and BL21(DE3)ΔalrΔdadXPR were compared, MalY was not found upregulated in BL21(DE3)ΔalrΔdadXPR (data not shown), as observed also by transcriptome analysis.
Mutations in metJ or the general derepression of Met genes is directly responsible for alanine racemase-independent survival of the BL21(DE3)ΔalrΔdadX strain.
The proteome and transcriptome data suggest that the phenotype of BL21(DE3)ΔalrΔdadXPR is caused by mutations in either an enhancer or a repressor of Met genes, resulting in upregulation of all Met genes during normal growth with sufficient methionine. MetR and MetJ are the two general regulators involved in the regulation of genes in the methionine synthesis pathway. All known Met genes, except metH but including the metR transcriptional activator, are repressed by MetJ and its cofactor S-adenosylmethionine (SAM), the product of MetK (20-23). Thus, the metR, metJ, and metK genes were sequenced from independently isolated phenotypic revertants, and their sequences were compared to the wild-type sequence from the d-alanine auxotrophic strain. We sequenced the metJ gene of 7 phenotypic revertants which overexpressed MetE and found that all of them had mutations in the metJ gene. The BL21(DE3)ΔalrΔdadXPR strain had a single point mutation, which introduced an R42C mutation in the MetJ protein. Five other phenotypic revertants had single point mutations, resulting in a MetJ protein with the following mutations: A12T, G15S, L36F, H50N, and A60T. The last revertant had a 2-base-pair insertion, which results in an N-terminally truncated MetJ protein comprising 45 amino acids. No mutation in metR or metK was identified.
To obtain direct evidence for a linkage between mutations in MetJ and d-alanine prototrophy, a replacement of the metJ(R42C) gene in the BL21(DE3)ΔalrΔdadXPR genome by wild-type metJ or the metJ(R42C) gene was carried out. A selection marker, the Kmr cassette, was introduced into the genome together with the target gene by gene replacement. To rule out the possibility that the effect introduced by the metJ-Kmr gene is due to any polar effect caused by insertion of the Kmr cassette right after the metJ loci, the metJ(R42C)-Kmr gene was applied as a control. The d-alanine auxotrophy was completely restored in BL21(DE3)ΔalrΔdadXPR by introducing the metJ-Kmr gene but not the metJ(R42C)-Kmr gene (Table 2). In agreement with the phenotype change, MetE expression was normalized by metJ-Kmr but not metJ(R42C)-Kmr (Fig. 2), which verified that the mutated metJ(R42C) protein is not fully functional.
To rule out the possibility that mutations in other genes besides metJ contribute to the phenotypes observed, the metJ(R42C)-Kmr gene was introduced into the BL21(DE3)ΔalrΔdadX genome to replace the wild-type metJ gene. The BL21(DE3)ΔalrΔdadX strain carrying metJ(R42C)-Kmr could grow on LB medium without d-alanine supplementation (Table 2). To verify that upregulation of the methionine pathway genes can directly compensate for the absence of the DadX and Alr alanine racemases, the growth of the d-alanine auxotroph was analyzed on l-methionine-deficient LB medium. Upon plating of the BL21(DE3)ΔalrΔdadX strain on l-methionine-deficient LB medium, supplementation of d-alanine was no longer required for growth (data not shown). With an increase of the l-methionine concentration in medium, the growth rate of BL21(DE3)ΔalrΔdadX decreased. When the l-methionine concentration reached 1 mM, complete growth inhibition of BL21(DE3)ΔalrΔdadX (but not that of the phenotypic revertant strain) was observed. This shows that the d-alanine auxotrophic strain becomes prototrophic when l-methionine is depleted from the growth medium. Thus, all together these data suggested that E. coli possesses another l-methionine-repressible alanine racemase.
Cystathionine beta-lyase (MetC) alone can complement the d-alanine requirement of the BL21(DE3)ΔalrΔdadX strain.
To elucidate whether a single MetJ-regulated gene can encode a protein with previously unknown alanine racemase activity, which could alone complement the alr/dadX deletion, candidates were chosen from the identified upregulated proteins. As alanine racemases belong to the large family of PLP (pyridoxal 5′-phosphate) cofactor enzymes, it was anticipated that the target protein would also belong to this family. From the entire E. coli proteome, 56 proteins were predicted to belong to this category of enzymes using the TagIdent tool (18), of which several were found to be upregulated in BL21(DE3)ΔalrΔdadXPR. Besides GlyA, which was previously reported to possess limited alanine racemase activity (10, 11, 37), MetB, MetC, Asd, TrpB, and YbdL were also annotated as PLP cofactor-requiring enzymes. Plasmids expressing these genes under the control of the T7 promoter were transformed into BL21(DE3)ΔalrΔdadX. The transformants were plated on LB medium with or without 0.1 mM d-alanine supplementation. Following overnight culturing at 37°C, only the MetC-expressing plasmid was able to completely reverse the d-alanine auxotrophic phenotype of BL21(DE3)ΔalrΔdadX (Fig. 4).
FIG. 4.
(A and B)BL21(DE3)ΔalrΔdadX cells were transformed with expression plasmids encoding different potential alanine racemases. Cells were grown on LB medium supplemented with 100 μM d-alanine (A) or without d-alanine (B) but with a 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) induction of pET11a (1), alr (2), asd (3), glyA (4), metB (5), metC (6), trpB (7), and ybdL (8). MetC has in vivo alanine racemase activity, which results in the phenotypic conversion of BL21(DE3)ΔalrΔdadX from a d-alanine auxotroph to a d-alanine prototroph. (C) Western blotting using anti-E. coli MetC polyclonal antibody. Lanes 1 to 3, 5, 10, and 20 ng of purified recombinant MetC, respectively; lanes 4 to 6, cell extracts from BL21(DE3)ΔalrΔdadXPR, BL21(DE3)ΔalrΔdadX, and BL21(DE3)ΔalrΔdadXPRΔmetC, respectively. Note: the band above the MetC band in the lysates is due to an unspecific detection by the polyclonal antibody.
When the metC gene was deleted from the BL21(DE3)ΔalrΔdadXPR genome, the strain could no longer grow on LB plates without d-alanine (Table 2). This confirms that MetC is the effector behind the d-alanine prototrophic phenotype. Furthermore, Western blots of cells lysates using anti-MetC polyclonal antibody demonstrated that MetC is highly upregulated in d-alanine prototrophic BL21(DE3)ΔalrΔdadXPR but not in BL21(DE3)ΔalrΔdadX [or metC-deficient BL21(DE3)ΔalrΔdadXPR] (Fig. 4).
DISCUSSION
The genes involved in the biosynthesis of l-methionine from homoserine and tetrahydrofolate (Fig. 5) are regulated by the following: (i) repression of the Met genes by the MetJ aporepressor and its corepressor, S-adenosylmethionine (SAM), when E. coli is cultivated in the presence of methionine; (ii) activation of specific Met genes mediated by the MetR transcriptional activator and its coactivator, homocysteine; and iii) repression of specific Met genes by vitamin B12. The MetJ aporepressor dimerizes upon binding to its target DNA sequences (34, 38, 39) in a fashion that is greatly facilitated by the cobinding of SAM, the product of MetK (35, 36). Mutations in either metJ or metK may result in similar phenotypes, which show constitutive expression of the Met genes even in the presence of excess methionine, as observed for the BL21(DE3)ΔalrΔdadX phenotypic revertant. The regulatory nucleotide regions controlling transcription of metA, metB, metC, metF, metE, and metK and the genes from the metD operon all share a tandemly repeated 8-base-pair sequence, the so-called “Met box” (3, 13). In this study, all the genes involved in the conversion of l-aspartate or l-serine to l-methionine and its catabolite, SAM, were found upregulated in the BL21(DE3)ΔalrΔdadXPR strain (Fig. 5), as described in a similar study of a metJ knockout strain (29). The dependency of the methionine regulon for disruption of the d-alanine deficiency of the BL21(DE3)ΔalrΔdadX strain was directly supported by our observations of the effect of the l-methionine concentration on growth. In minimal medium, the BL21(DE3)ΔalrΔdadX and BL21(DE3)ΔalrΔdadXPR strains grew equally well at concentrations of up to 100 μM l-methionine, but only the BL21(DE3)ΔalrΔdadXPR strain was able to grow at concentrations of 1 mM l-methionine or more. The rich LB medium used in our investigations contains a concentration of free l-methionine of approximately 1 mM. At zero l-methionine or low l-methionine concentrations, the derepression of the Met genes is sufficient to complement the d-alanine auxotrophy of the BL21(DE3)ΔalrΔdadX strain, whereas the BL21(DE3)ΔalrΔdadXPR strain is much less sensitive to different intracellular amounts of l-methionine due to an impaired MetJ repressor.
FIG. 5.
Schematic representation of the main conclusions of this paper. Methionine pathway showing the relationship between the Met repressor (MetJ) and MetJ responsive genes and the novel link to d-alanine and peptidoglycan synthesis via MetC. Stippled arrows indicate activator action, and solid lines indicate repressor action.
E. coli strains containing single point mutations in the metJ gene can display a significant increase in the level of intracellular l-methionine and activity of key enzymes of the methionine pathway (1, 8, 9, 31). The position of the single MetJ mutations differed considerably among the seven investigated BL21(DE3)ΔalrΔdadX phenotypic revertants but may all affect the interaction of MetJ with the Met box, MetJ dimerization, and/or the important protein-protein interactions with the corepressor SAM, as judged from analysis of the structure of the MetJ operator complex (40).
The knock-in of the wild-type metJ gene but not of the metJ(R42C) gene restores d-alanine auxotrophy in the BL21(DE3)ΔalrΔdadXPR strain, showing that the MetJ mutation is directly responsible for the BL21(DE3)ΔalrΔdadXPR phenotype. 2-D gel analysis indicates that MetE is unstable or rapidly degraded by the cell (Fig. 3A), which might explain the absence of overexpressed MetE for some of the BL21(DE3)ΔalrΔdadX phenotypic revertant isolates. Certain BL21(DE3)ΔalrΔdadX phenotypic revertant isolates may be able to circumvent d-alanine auxotrophy through upregulation of proteins with alanine racemase coactivity, other than MetC (e.g., MalY). However, our data suggest that the primary mechanism for development of the revertant phenotype is a strong selectional pressure toward inactivation or functional impairment of MetJ.
Alanine racemases belong to the diverse group of PLP (vitamin B6)-dependent enzymes (11, 33), catalyzing the formation of an aldimine intermediate, which is the basis for decarboxylation, transamination, β-elimination, and racemization reactions. Different PLP enzymes may share the same common fold comprising PLP and similar catalytic mechanisms despite low sequence similarity (14). We therefore evaluated upregulated candidates, which belonged to this group, for alanine racemase activity.
Serine hydroxymethyl transferase (GlyA) transaminates both l-alanine and d-alanine and also catalyzes an alanine racemase reaction (10, 11, 37). However, the reported GlyA racemase coactivity is not sufficient to fully compensate for the alr/dadX deletions in our study.
Cystathionine gamma-synthetase (MetB) catalyzes the formation of cystathionine, which is subsequently used by MetC for the formation of homocysteine (Fig. 5). MetB and MetC are evolutionarily related enzymes, which share 36% sequence similarity (3) and belong to the same branch of PLP-dependent enzymes (2). Only the plasmid expressing MetC could abolish the d-alanine auxotrophy of the BL21(DE3)ΔalrΔdadX strain. We observed a strong upregulation of MetC in BL21(DE3)ΔalrΔdadXPR, and the d-alanine prototrophy could be abolished by removal of metC from the BL21(DE3)ΔalrΔdadXPR genome. We have also found that purified recombinant E. coli MetC exhibits in vitro alanine racemase activity (our unpublished results).
Taken together, we therefore conclude that MetC, due to significant alanine racemase coactivity, is the effector protein behind the d-alanine prototrophic revertant phenotype. To our knowledge, this is the first time that a protein directly involved in the synthesis of l-methionine has been shown to have alanine racemase activity.
It was reported that E. coli MalY, a regulator of the maltose operon, can abolish the methionine requirement of a MetC mutant. This proves that both MetC and MalY have cystathionine beta-lyase activity (52) and thus potentially similar catalytic mechanisms. Furthermore, it was recently reported that cystalysin from Treponema denticola possesses alanine racemase activity (4, 7). E. coli MalY shows 51% sequence similarity to Treponema denticola cystalysin, and we found that a plasmid expressing E. coli MalY was indeed able to complement the alr/dadX deletion (data not shown). However, there is no significant sequence similarity between E. coli MalY (or Treponema denticola cystalysin) and E. coli MetC, which clearly illustrates the broad functional overlap among different PLP-dependent enzymes. In contrast to metC, malY is not regulated by MetJ, and both proteome and microarray analyses from this study rule out the possibility that MalY is causing the d-alanine prototrophic phenotype of the BL21(DE3)ΔalrΔdadXPR strain. Thus, our findings show that two E. coli cystathionine beta-lyases connected to very different metabolic pathways can each catalyze the racemization between l-alanine and d-alanine.
Acknowledgments
We thank Baoping Wang for continuous advice and lab technicians Malene Dahl, Qinfen Wang, and Junhua Wang for excellent technical assistance.
Footnotes
Published ahead of print on 30 December 2010.
REFERENCES
- 1.Ahmed, A. 1973. Mechanism of repression of methionine biosynthesis in Escherichia coli. I. The role of methionine, S-adenosylmethionine and methionyl-transfer ribonucleic acid in repression. Mol. Gen. Genet. 123:299-324. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, F. W., E. Sandmeier, P. K. Mehta, and P. Christen. 1994. Evolutionary relationships among pyridoxal-5′-phosphate-dependent enzymes. Regio-specific alpha, beta and gamma families. Eur. J. Biochem. 219:953-960. [DOI] [PubMed] [Google Scholar]
- 3.Belfaiza, J., et al. 1986. Evolution in biosynthetic pathways: two enzymes catalyzing consecutive steps in methionine biosynthesis originate from a common ancestor and possess a similar regulatory region. Proc. Natl. Acad. Sci. U. S. A. 83:867-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertoldi, M., B. Cellini, A. Paiardini, M. Di Salvo, and V. C. Borri. 2003. Treponema denticola cystalysin exhibits significant alanine racemase activity accompanied by transamination: mechanistic implications. Biochem. J. 371:473-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bron, P. A., et al. 2002. Use of the alr gene as a food-grade selection marker in lactic acid bacteria. Appl. Environ. Microbiol. 68:5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, X. Y., et al. 1989. Methionine synthesis in Escherichia coli: effect of the MetR protein on metE and metH expression. Proc. Natl. Acad. Sci. U. S. A. 86:4407-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cellini, B., M. Bertoldi, A. Paiardini, S. D'Aguanno, and C. B. Voltattorni. 2004. Site-directed mutagenesis provides insight into racemization and transamination of alanine catalyzed by Treponema denticola cystalysin. J. Biol. Chem. 279:36898-36905. [DOI] [PubMed] [Google Scholar]
- 8.Clandinin, M. T., and A. Ahmed. 1973. Mechanism of repression of methionine biosynthesis in Escherichia coli. II. The effect of metJ mutations on the free amino acid pool. Mol. Gen. Genet. 123:325-331. [DOI] [PubMed] [Google Scholar]
- 9.Collier, C. D., and J. R. Johnson. 1990. The Escherichia coli K-12 metJ193 allele contains a point mutation which alters the hydrophobic pocket responsible for in vitro binding of S-adenosylmethionine: effects on cell growth and induction of met regulon expression. J. Bacteriol. 172:3918-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contestabile, R., et al. 2000. Role of tyrosine 65 in the mechanism of serine hydroxymethyltransferase. Biochemistry 39:7492-7500. [DOI] [PubMed] [Google Scholar]
- 11.Contestabile, R., et al. 2001. l-Threonine aldolase, serine hydroxymethyltransferase and fungal alanine racemase. A subgroup of strictly related enzymes specialized for different functions. Eur. J. Biochem. 268:6508-6525. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson, B. E., and I. Saint-Girons. 1989. The Escherichia coli regulatory protein MetJ binds to a tandemly repeated 8 bp palindrome. Mol. Microbiol. 3:1639-1648. [DOI] [PubMed] [Google Scholar]
- 14.Denessiouk, K. A., A. I. Denesyuk, J. V. Lehtonen, T. Korpela, and M. S. Johnson. 1999. Common structural elements in the architecture of the cofactor-binding domains in unrelated families of pyridoxal phosphate-dependent enzymes. Proteins 35:250-261. [DOI] [PubMed] [Google Scholar]
- 15.Diderichsen, B. 1986. A genetics system for stabilization of cloned genes in Bacillus subtilis, p. 35-46. In A. Gansean and J. A. Hoch (ed.), Bacillus molecular genetics and biotechnology applications. Academic Press, New York, NY.
- 16.Dolzan, M., et al. 2004. Crystal structure and reactivity of YbdL from Escherichia coli identify a methionine aminotransferase function. FEBS Lett. 571:141-146. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari, E., D. J. Henner, and M. Y. Yang. 1985. Isolation of an alanine racemase gene from Bacillus subtilis and its use for palsmid maintenance in B. subtilis. Biotechnology (NY) 3:1003-1007. [Google Scholar]
- 18.Gasteiger, E., et al. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ.
- 19.Gharahdaghi, F., C. R. Weinberg, D. A. Meagher, B. S. Imai, and S. M. Mische. 1999. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis 20:601-605. [DOI] [PubMed] [Google Scholar]
- 20.Greene, R. C. 1996. Biosynthesis of methionine, p. 542-560. In F. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 21.Greene, R. C., J. S. Hunter, and E. H. Coch. 1973. Properties of metK mutants of Escherichia coli K-12. J. Bacteriol. 115:57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greene, R. C., C. H. Su, and C. T. Holloway. 1970. S-Adenosylmethionine synthetase deficient mutants of Escherichia coli K-12 with impaired control of methionine biosynthesis. Biochem. Biophys. Res. Commun. 38:1120-1126. [DOI] [PubMed] [Google Scholar]
- 23.Hobson, A. C., and D. A. Smith. 1973. S-Adenosylmethionine synthetase in methionine regulatory mutants of Salmonella typhimurium. Mol. Gen. Genet. 126:7-18. [DOI] [PubMed] [Google Scholar]
- 24.Hols, P., et al. 1997. The alanine racemase gene is essential for growth of Lactobacillus plantarum. J. Bacteriol. 179:3804-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen, O. N., M. R. Larsen, and P. Roepstorff. 1998. Mass spectrometric identification and microcharacterization of proteins from electrophoretic gels: strategies and applications. Proteins 2(Suppl.):74-89. [DOI] [PubMed] [Google Scholar]
- 26.Kadner, R. J. 1975. Regulation of methionine transport activity in Escherichia coli. J. Bacteriol. 122:110-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, R., T. W. Blackwell, and D. J. States. 2001. Conformational model for binding site recognition by the E. coli MetJ transcription factor. Bioinformatics 17:622-633. [DOI] [PubMed] [Google Scholar]
- 28.Lobocka, M., J. Hennig, J. Wild, and T. Klopotowski. 1994. Organization and expression of the Escherichia coli K-12 dad operon encoding the smaller subunit of d-amino acid dehydrogenase and the catabolic alanine racemase. J. Bacteriol. 176:1500-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marincs, F., I. W. Manfield, J. A. Stead, K. J. McDowall, and P. G. Stockley. 2006. Transcript analysis reveals an extended regulon and the importance of protein-protein co-operativity for the Escherichia coli methionine repressor. Biochem. J. 396:227-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy, K. C., K. G. Campellone, and A. R. Poteete. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321-330. [DOI] [PubMed] [Google Scholar]
- 31.Nakamori, S., S. Kobayashi, T. Nishimura, and H. Takagi. 1999. Mechanism of l-methionine overproduction by Escherichia coli: the replacement of Ser-54 by Asn in the MetJ protein causes the derepression of l-methionine biosynthetic enzymes. Appl. Microbiol. Biotechnol. 52:179-185. [DOI] [PubMed] [Google Scholar]
- 32.Old, I. G., D. Margarita, R. E. Glass, and I. Saint-Girons. 1990. Nucleotide sequence of the metH gene of Escherichia coli K12 and comparison with that of Salmonella typhimurium LT2. Gene 87:15-21. [DOI] [PubMed] [Google Scholar]
- 33.Paiardini, A., R. Contestabile, S. D'Aguanno, S. Pascarella, and F. Bossa. 2003. Threonine aldolase and alanine racemase: novel examples of convergent evolution in the superfamily of vitamin B6-dependent enzymes. Biochim. Biophys. Acta 1647:214-219. [DOI] [PubMed] [Google Scholar]
- 34.Phillips, S. E., et al. 1989. Cooperative tandem binding of met repressor of Escherichia coli. Nature 341:711-715. [DOI] [PubMed] [Google Scholar]
- 35.Saint-Girons, I., et al. 1986. Interactions of the Escherichia coli methionine repressor with the metF operator and with its corepressor, S-adenosylmethionine. J. Biol. Chem. 261:10936-10940. [PubMed] [Google Scholar]
- 36.Shoeman, R., et al. 1985. Regulation of methionine synthesis in Escherichia coli: effect of metJ gene product and S-adenosylmethionine on the in vitro expression of the metB, metL and metJ genes. Biochem. Biophys. Res. Commun. 133:731-739. [DOI] [PubMed] [Google Scholar]
- 37.Shostak, K., and V. Schirch. 1988. Serine hydroxymethyltransferase: mechanism of the racemization and transamination of D- and L-alanine. Biochemistry 27:8007-8014. [DOI] [PubMed] [Google Scholar]
- 38.Smith, A. A., and R. C. Greene. 1984. Cloning of the methionine regulatory gene, metJ, of Escherichia coli K12 and identification of its product. J. Biol. Chem. 259:14279-14281. [PubMed] [Google Scholar]
- 39.Smith, A. A., R. C. Greene, T. W. Kirby, and B. R. Hindenach. 1985. Isolation and characterization of the product of the methionine-regulatory gene metJ of Escherichia coli K-12. Proc. Natl. Acad. Sci. U. S. A. 82:6104-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somers, W. S., and S. E. Phillips. 1992. Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands. Nature 359:387-393. [DOI] [PubMed] [Google Scholar]
- 41.Strych, U., H. C. Huang, K. L. Krause, and M. J. Benedik. 2000. Characterization of the alanine racemases from Pseudomonas aeruginosa PAO1. Curr. Microbiol. 41:290-294. [DOI] [PubMed] [Google Scholar]
- 42.Strych, U., R. L. Penland, M. Jimenez, K. L. Krause, and M. J. Benedik. 2001. Characterization of the alanine racemases from two mycobacteria. FEMS Microbiol. Lett. 196:93-98. [DOI] [PubMed] [Google Scholar]
- 43.Tauch, A., S. Gotker, A. Puhler, J. Kalinowski, and G. Thierbach. 2002. The alanine racemase gene alr is an alternative to antibiotic resistance genes in cloning systems for industrial Corynebacterium glutamicum strains. J. Biotechnol. 99:79-91. [DOI] [PubMed] [Google Scholar]
- 44.Theze, J., D. Margarita, G. N. Cohen, F. Borne, and J. C. Patte. 1974. Mapping of the structural genes of the three aspartokinases and of the two homoserine dehydrogenases of Escherichia coli K-12. J. Bacteriol. 117:133-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, R. J., H. G. Bouwer, D. A. Portnoy, and F. R. Frankel. 1998. Pathogenicity and immunogenicity of a Listeria monocytogenes strain that requires D-alanine for growth. Infect. Immun. 66:3552-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verch, T., Z. K. Pan, and Y. Paterson. 2004. Listeria monocytogenes-based antibiotic resistance gene-free antigen delivery system applicable to other bacterial vectors and DNA vaccines. Infect. Immun. 72:6418-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wasserman, S. A., E. Daub, P. Grisafi, D. Botstein, and C. T. Walsh. 1984. Catabolic alanine racemase from Salmonella typhimurium: DNA sequence, enzyme purification, and characterization. Biochemistry 23:5182-5187. [DOI] [PubMed] [Google Scholar]
- 48.Wasserman, S. A., C. T. Walsh, and D. Botstein. 1983. Two alanine racemase genes in Salmonella typhimurium that differ in structure and function. J. Bacteriol. 153:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weissbach, H., and N. Brot. 1991. Regulation of methionine synthesis in Escherichia coli. Mol. Microbiol. 5:1593-1597. [DOI] [PubMed] [Google Scholar]
- 50.Wijsman, H. J. 1972. The characterization of an alanine racemase mutant of Escherichia coli. Genet. Res. 20:269-277. [DOI] [PubMed] [Google Scholar]
- 51.Wild, J., J. Hennig, M. Lobocka, W. Walczak, and T. Klopotowski. 1985. Identification of the dadX gene coding for the predominant isozyme of alanine racemase in Escherichia coli K12. Mol. Gen. Genet. 198:315-322. [DOI] [PubMed] [Google Scholar]
- 52.Zdych, E., R. Peist, J. Reidl, and W. Boos. 1995. MalY of Escherichia coli is an enzyme with the activity of a beta C-S lyase (cystathionase). J. Bacteriol. 177:5035-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, Z., et al. 2003. A transporter of Escherichia coli specific for L- and D-methionine is the prototype for a new family within the ABC superfamily. Arch. Microbiol. 180:88-100. [DOI] [PubMed] [Google Scholar]