Abstract
Serratia sp. strain ATCC 39006 produces the red-pigmented antibiotic prodigiosin. Regulation of prodigiosin biosynthesis involves a complex hierarchy, with PigP a master transcriptional regulator of multiple genes involved in prodigiosin production. The focus of this study was a member of the PigP regulon, pigS, which encodes an ArsR/SmtB family transcriptional repressor. Mutations in pigS reduced production of prodigiosin by decreasing the transcription of the biosynthetic operon. The pigS gene is the first in a four-gene operon, which also encodes three membrane proteins (pmpABC) of the COG2391 (DUF395; YedE/YeeE) and COG0730 (DUF81; TauE/SafE) families that we propose constitute transport components for sulfur-containing compounds. We provide the first experimental evidence confirming the membrane localization of a COG2391 protein, that of PmpB. Divergently transcribed from pigS-pmpABC is a bicistronic operon (blhA-orfY), which encodes a metallo-β-lactamase and a coenzyme A-disulfide reductase containing a rhodanese homology domain, both of which may participate in reactions with sulfur-containing compounds. The overproduction of the BlhA and OrfY enzymes and the PmpABC membrane proteins differentially affected pigmentation. We have dissected the contributions of these various proteins and determined their importance in the control of prodigiosin production. PigS-mediated control of prodigiosin occurred via binding directly to a short inverted repeat sequence in the intergenic region overlapping the predicted −10 regions of both pigS and blhA promoters and repressing transcription. PigP was required for the activation of these promoters, but only in the absence of PigS-mediated repression.
Serratia sp. strain ATCC 39006 is a Gram-negative enteric bacterium originally isolated from a salt marsh in New Jersey, where it was associated with plant material (25). Serratia sp. ATCC 39006 produces several secondary metabolites, including a red antimicrobial pigment, prodigiosin (Pig), and a β-lactam antibiotic, carbapenem (Car) (6, 30). There is considerable interest in Pig and its derivatives due to their anticancer and immunosuppressive activities (34). The biosynthetic genes required for production of Pig and Car in Serratia sp. ATCC 39006 have been identified as pigA to -O (pigA-O) and carA to -H (carA-H), respectively (17, 35, 37), and several studies have revealed that biosynthesis is controlled via a complex hierarchical network of regulators (10-12, 14, 15, 29-31, 35, 36). Regulation includes a LuxIR-type quorum sensing (QS) system (SmaIR), which allows gene expression to be regulated in response to cell density via the production and detection of low-molecular-weight signal molecules called N-acyl homoserine lactones (AHLs) (11, 29, 31). The Serratia sp. ATCC 39006 QS system modulates secondary metabolism via the transcriptional regulation of four other regulators (CarR, an AHL-independent LuxR regulator; Rap, similar to RovA from Yersinia; PigR, an adenylate cyclase; and PigQ, a GacA response regulator) (11). Pig production in Serratia spp. can also be modulated by a number of environmental cues, including the availability of inorganic phosphate (15), carbon source (10), salt concentration, temperature, oxygen availability, and multiple metal ion concentrations (reviewed in reference 35).
Previously, PigP was identified as a master regulator of secondary metabolism in Serratia sp. ATCC 39006 (11). PigP is the founding member of a novel class of transcriptional regulators present in a restricted number of Enterobacteriaceae and has been shown to control secondary metabolism via transcriptional regulation of seven other regulators (CarR; PigQ; PigR; Rap; PigS, an ArsR family regulator; PigV, a homologue of YgfX from Escherichia coli; and PigX, a homologue of CsrD from Escherichi. coli containing a GGDEF/EAL domain) (11). Thus, the PigP regulon overlaps the QS regulon in Serratia sp. ATCC 39006.
Fineran et al. (11) identified pigS following a transposon mutagenesis screen due to the observed reduction in Pig production following disruption of pigS. In addition, expression of pigS was activated by the master regulator PigP (11). Sequencing revealed that pigS encoded a putative ArsR/SmtB family transcriptional regulator (11). ArsR/SmtB family regulators are metalloregulatory transcriptional repressors which can control the expression of genes linked to stress-inducing concentrations of heavy metals (5). Binding of ArsR/SmtB family regulators to metal ions, including Zn(II), Cd(II), Pb(II), Bi(II), Co(II), Ni(II) and Sb(II), has been observed (5). Upon metal binding, the DNA binding affinity of the protein is reduced, allowing derepression (5).
Here, we investigate the role of PigS in the regulation of Pig production. The pigS gene is the first gene in an operon encoding three putative membrane proteins of the COG2391 (clusters of orthologous groups) and COG0730 families, which is divergently transcribed from a second operon encoding BlhA, a metallo-β-lactamase superfamily protein, and OrfY, a coenzyme A-disulfide reductase containing a rhodanese homology domain (CoADR-RHD). Here we show that PigS binds directly to these divergent promoters and represses transcription and that in the absence of Pig, the master regulator, PigP, is required for activation. Currently, 30 to 40% of genes in newly sequenced bacteria have no known function (4, 13), and although challenging, it is clearly important to study these genes to understand bacterial physiology (26). We show that the balance between the pmpABC and blhA-orfY operons and their specific proteins is important for determining Pig levels and provide bioinformatic data that proteins of these families are functionally linked. In addition, we propose from in silico analyses that COG2391 proteins (including PmpAB), constitute transporters for sulfur-containing compounds and demonstrate that PmpB is localized to the cell membrane. To the best of our knowledge, this is the first experimental evidence confirming the membrane localization of any COG2391 protein.
MATERIALS AND METHODS
Bacterial strains, phage, and culture conditions.
Bacterial strains and plasmids are listed in Table 1. E. coli strains were grown at 37°C and Serratia sp. ATCC 39006 strains were grown at 30°C in LB medium (5 g liter−1 yeast extract, 10 g liter−1 Bacto tryptone, 5 g liter−1 NaCl) with shaking or on LB agar supplemented with 1.5% (wt/vol) agar. When required, medium was supplemented with antibiotics at the following concentrations; ampicillin (Ap), 100 mg ml−1; spectinomycin (Sp), 50 mg ml−1; tetracycline (Tc), 35 mg ml−1. The generalized transducing phage φOT8 (8) was used as described previously (31).
TABLE 1.
Bacterial strains and phage used in this study
| Strain or plasmid | Genotype/phenotype | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F− φ80ΔdlacZM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 (rΚ− mΚ+) deoR thi-1 supE44 λ−gyrA96 relA1 | Gibco/BRL |
| SM10 λpir | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpir Kmr | 7 |
| Serratia | ||
| LacA (parental strain; WT) | Lac− derivative of Serratia sp. ATCC 39006, made by EMS mutagenesis | 31 |
| Mutants derived from LacA | ||
| 13S26L | pigP::mini-Tn5Sm/Sp pigS::mini-Tn5lacZ1 Spr Kmr | 11 |
| HSPIG26 | pigS::mini-Tn5lacZ1 Kmr | 11 |
| KFAAG6 | pigA::TnuidACm Cmr | N. Williamson, unpublished data |
| PIG13S | pigP::mini-Tn5Sm/Sp Spr | 11 |
| TG70 | pigS::mini-Tn5lacZ1 pigA::TnuidACm Kmr Cmr | This study |
| Plasmids | ||
| pQE-80L | Protein expression vector; Apr | Qiagen |
| pQE-80LoriT | pQE-80L containing oriT from RP4; Apr | J. Ramsay, unpublished data |
| pMAT29 | His6-PmpB expression vector, pQE-80LoriT based; Apr | This study |
| pTA25 | pigS promoter lacZ fusion (−86 to +44), Tcr | This study |
| pTA27 | Native PigS expression vector, pQE-80L based; Apr | This study |
| pTA132 | PmpABC expression vector, pQE-80LoriT based; Apr | This study |
| pTA133 | BlhAOrfY expression vector, pQE-80LoriT based; Apr | This study |
| pTA134 | PmpABC, BlhA-OrfY expression vector, pQE-80LoriT based; Apr | This study |
| pTA154 | BlhA expression vector, pQE-80LoriT based, Apr | This study |
| pTA155 | OrfY expression vector, pQE-80LoriT based; Apr | This study |
| pTA156 | PmpBC expression vector, pQE-80LoriT based; Apr | This study |
| pTA157 | PmpC expression vector, pQE-80LoriT based; Apr | This study |
| pTA159 | PmpAC expression vector, pQE-80LoriT based; Apr | This study |
| pTA160 | PmpA expression vector, pQE-80LoriT based; Apr | This study |
| pTA161 | PmpB expression vector, pQE-80LoriT based; Apr | This study |
| pTA162 | PmpAB expression vector, pQE-80LoriT based; Apr | This study |
| pTG7 | His6-PigS expression vector, pQE-80L based; Apr | This study |
| pTG41 | blhA promoter lacZ fusion (−151 to +18); Tcr | This study |
DNA manipulations and sequence analyses.
All molecular biology techniques, unless stated otherwise, were performed by standard methods (28). Sequence similarity searches were performed using BLAST (2), subcellular localization of proteins was predicted using PSORTb v3.0 (38), transmembrane spanning regions were detected using TMHMM v2.0 (21), and figures were generated using TOPO2 (www.sacs.ucsf.edu/TOPO2/). Functional linkages based on factors such as gene neighborhood, gene fusion, cooccurrence, and coexpression data were determined using STRING (18) set to COG-mode. Domain architectures were viewed using Pfam (http://pfam.sanger.ac.uk/), and some specific comparisons were made using MicrobesOnline (1). To determine the sequence of the pigS locus, a Serratia sp. ATCC 39006 cosmid library was constructed using the Epicentre Biotechnologies pWEB-TNC cosmid cloning kit, per the manufacturer's directions. A cosmid carrying the pigS locus was identified by PCR using primers PF64 (5′-GTACGAATTCAAAGGATCCCTATG-3′) and PF65 (5′-CTGACTGCAGCAATATTAATGGTC-3′), and primer walking was used to sequence the complete pigS locus.
Prodigiosin, β-galactosidase, and β-glucuronidase assays.
Prodigiosin production was assayed as described by Slater et al. (29). β-Galactosidase and β-glucuronidase activities were determined as described by Fineran et al. (11) and Gristwood et al. (15), respectively.
Mapping the transcriptional start site of pigS and RT-PCR.
The transcriptional start of pigS was determined by 5′ RACE (rapid amplification of cDNA ends) as described previously (12), except primers PF88 (5′-GAGTAAAACGAGGACTTTCGG-3′), PF87 (5′-CTAACTGCTGCGATAATGTTG-3′), and PF86 (5′-GTACTGCAGACGATCACTATTAGCC-3′) were used as specific primers. RNA for reverse transcriptase PCR (RT-PCR) was extracted from an early stationary-phase culture (10 h growth in LB medium) of Serratia strain LacA. A total of 250 ng of RNA was used for cDNA synthesis using Superscript II reverse transcriptase (Invitrogen) and random hexamer primers (Amersham). PCR was performed using primers PF64 and JCO24 (5′-TTTGGTACCCGAATTTGCGAACCCATTAG-3′), PF264 (5′-TTTGAATTCGAAATAGAAGGCGCAGGTC-3′) and PF265 (5′-CCACCTAAAGCTTCTGAGGG-3′), and PF266 (5′-TTTGAATTCCGCCAGTAAGGAGTTAACC-3′) and PF269 (5′-CGGACGCCATCTCGATATC-3′). For quantitative RT-PCR, RNA was extracted and cDNA synthesis was performed as described above using Serratia strains HSPIG26, PIG13S, and 13S26L. The quantitative PCRs (qPCRs) were performed using the Applied Biosystems SYBR green PCR mix and an Applied Biosystems Prism 7300. Primers OTG156 (5′-CAGCACCGTCAGTTACGTT-3′) and OTG157 (5′-GACCTGATCGCGTAAATAGTG-3′) were used to detect blhA mRNA, and primers 3916SF and 3916SR (36) were used to detect 16S rRNA. Cycle threshold (CT) values were calculated using ABI SDS software, and relative gene expression was calculated using 16S rRNA as the internal control and blhA mRNA/16S rRNA equal to a value of 1 in strain LacA.
Cellular localization experiments.
A construct that enabled expression of N-terminally His-tagged PmpB (pMAT29) was generated using primers MMO75 (5′-GATGGATCCATGAACTTACTTTTT-3′) and PF305 (5′-TTTAAGCTTGATTAATGCCTTTGTCTGG-3′). The PCR fragment was digested with EcoRI/HindIII and cloned into pQE-80LoriT, a derivative of pQE-80L containing the RP4 oriT. The plasmid generated (pMAT29) was conjugated into LacA using the E. coli donor strain SM10 λpir. LacA(pMAT29) was grown until an optical density at 600 nm (OD600) of 0.6 to 0.8 was reached, induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and incubated overnight. Cells were pelleted at 10,000 × g for 20 min, washed twice, and then resuspended in 50 mM potassium phosphate and lysed by several passages through a French press at 16,000 lb/in2. Cellular debris was removed by centrifugation at 27,000 × g for 15 min. The supernatant was subjected to centrifugation at 100,000 × g for 90 min. The supernatant (containing soluble fraction) was decanted, while the membrane pellet was washed twice in 50 mM potassium phosphate buffer and resuspended in 2 ml of potassium phosphate buffer. Protein quantification was determined using bicinchoninic acid (BCA) assays from Thermo Scientific, and fractions were kept on ice at 4°C before Western blot analysis using mouse monoclonal anti-His antibody (Sigma) and, as a secondary antibody, goat anti-mouse IgG-horseradish peroxidase (Santa Cruz). Bands were visualized on a ChemiDoc imaging system (Bio-Rad) using the SuperSignal West Pico chemiluminescent substrate kit (Pierce).
Plasmids for expression of pmpABC and blhA-orfY.
Vectors were created to allow expression of the pmpABC and blhA-orfY operons as outlined below. The pmpABC and blhA-orfY operons were amplified by PCR, using primer pair PF264 and PF265 and primer pair PF266 and PF267 (5′-TTTAAGCTTCTCGAGTCAGGCGGACAACGTC-3′), respectively. PCR fragments were cloned into pQE-80LoriT, giving plasmids pTA132 (pmpABC) and pTA133 (blhA-orfY). A plasmid (pTA134) for overexpression of both pmpABC and blhA-orfY operons from divergent IPTG-inducible promoters was made by cloning the blhA-orfY region from pTA133 on XhoI sites into pTA132 cut with XhoI. A plasmid expressing blhA (pTA154) was constructed by digesting pTA133 with EcoRI and SphI and ligating the fragment containing blhA into pQE-80LoriT. An OrfY expression plasmid (pTA155) was generated by PCR using primers PF302 (5′-TTTGAATTCAAAAAAGGAGTTTGCTGTG-3′) and PF267 and ligating the EcoRI/HindIII-digested product into pQE-80LoriT. A series of plasmids with different combinations of pmpABC were generated. Plasmid pTA132 (pmpABC) was digested with BamHI/HindIII, which removed pmpBC, blunted with Klenow fragment, and religated, giving plasmid pTA160 (pmpA). The pmpB and pmpAB genes were amplified using primer MMO67 (5′-GATGAATTCAGGAGGACAGGGATGAACTTACTTTTT-3′) or PF264 and PF305 (5′-TTTAAGCTTGATTAATGCCTTTGTCTGG-3′), and the EcoRI/HindIII-digested products were ligated into pQE-80LoriT, giving plasmids pTA161 (pmpB) and pTA162 (pmpAB), respectively. Next, pmpBC and pmpC were amplified with primer MMO67 or MMO69 (5′-GATGAATTCAGGAGGACAGGGATGTTCATTTCATTA-3′) and primer PF265, and the products were ligated into pQE-80LoriT, resulting in plasmids pTA156 and pTA157, respectively. To generate a plasmid expressing PmpAC, pTA132 was digested with BamHI/SphI, blunted with Klenow fragment, and religated to yield plasmid pTA159. Plasmids were conjugated into LacA and HSPIG26 using E. coli SM10 λpir. Where indicated, expression of these plasmids was induced with 0.1 mM IPTG.
Construction of pigS and blhA promoter lacZ fusions and assay conditions.
The pigS and blhA promoters were amplified using primers PF58 (5′-GCATGAATTCGGTTAACTCCTTAC-3′) and PF59 (5′-GCACAAGCTTAGGGATCCTTTC-3′) and primers OTG173 (5′-ATGTGAATTCGCTAATCGTCGCGTTATCC-3′) and OTG172 (5′-ATGTAAGCTTCTCGATATGTAGTGTCATGG-3′), respectively. The resulting PCR products were cloned into EcoRI/HindIII-digested pRW50 (10, 22), giving plasmids pTA25 and pTG41, and promoter expression was determined as described previously (10).
Native and His-tagged PigS expression plasmids.
Constructs that enabled expression of native, untagged PigS, and hexahistidine-tagged PigS (His6-PigS; PigS protein with the N-terminal extension MRGSHHHHHHGS) were created as outlined below. The pigS gene was amplified by PCR, using primers PF64 and PF65 for native PigS or OTG3 (5′-ATAGGCATGCGATAACGCGACGATTAGCC-3′) and OTG4 (5′-ATAGAAGCTTTTAATGGTCATGACCTGCG-3′) for His6-PigS. The resulting PCR fragments were cloned into pQE-80L, yielding plasmids pTA27 and pTG7, respectively. Expression of plasmid pTG7 in both E. coli and Serratia sp. ATCC 39006 was induced with 1 mM IPTG.
Purification of His6-PigS.
A 500-ml culture of strain HSPIG26 carrying pTG7 was grown to an OD600 of 0.6 to 0.8, induced with 1 mM IPTG, and incubated overnight at 16°C. Cells were harvested by centrifugation at 4°C, and His6-PigS was purified using Ni-nitriloacetic acid (Qiagen) as described previously (14). Purified His6-PigS was dialyzed overnight at 4°C against 2 liters of storage buffer (250 mM NaCl, 20 mM Tris-HCl [pH 7.9], 1 mM EDTA, 1 mM dithiothreitol [DTT], 50% [wt/vol] glycerol) and stored at −20°C.
EMSAs.
The pigS-blhA intergenic region was PCR amplified from LacA genomic DNA using primers PF58 and OTG42 (5′-AGGGATCCTTTCAAACTAAG-3′), resulting in a 130-bp DNA fragment (fragment A). Truncated fragments of the pigS-blhA intergenic region were obtained by PCR amplification from LacA genomic DNA using forward primers OTG41 (5′-CTATTGCCTTAAACAA-3′), OTG43 (5′-CAAGACTTGAATTAAATATAC-3′), OTG44 (5′-ATATTATATTATATAACATTG-3′), and OTG68 (5′-ATAACATTGTAAAGTGTTTTGTTG-3′), and the reverse OTG42 to give 105-bp (fragment B), 84-bp (fragment C), 64-bp (fragment D), and 52-bp (fragment E) DNA fragments, respectively. DNA fragments were 3′-end labeled with DIG-11-ddUTP using terminal transferase, and electrophoretic mobility shift assay (EMSA) reactions were carried out according to the DIG gel shift kit protocol (Roche). Each 20-μl reaction mixture contained the indicated amount of His6-PigS and 1.5 nM DIG-labeled DNA in binding buffer [20 mM HEPES, pH 7.6, 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM DTT, 0.02% (wt/vol) Tween, and 30 mM KCl], 5 μg ml−1 poly-l-lysine, and 50 μg ml−1 poly[d(I-C)]. Where indicated, reaction mixtures also contained a 50-fold excess (75 nM) of unlabeled competitor DNA. The pigS-blhA fragment A was used as a specific competitor, and a promoter DNA fragment from Pectobacterium carotovorum was used as a nonspecific competitor (kindly donated by Tom Burr). Polyacrylamide gel electrophoresis, electroblotting, and chemiluminescent detection were carried out according to the DIG gel shift kit protocol (Roche).
RESULTS
PigS is a transcriptional regulator of Pig biosynthesis.
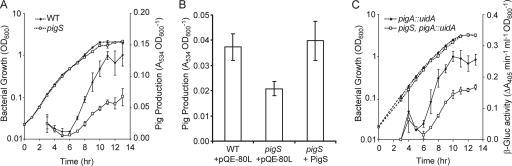
It was unknown to what extent the ArsR/SmtB family regulator PigS affected prodigiosin production and whether this was due to transcriptional control of pigA-O. In a pigS mutant strain, Pig production was reduced by approximately 50% throughout growth compared with Pig production by the WT (throughout this study, WT refers to the LacA parental strain) (Fig. 1 A). There was no effect on Car or AHL production in the pigS mutant strain compared with the WT (data not shown). Pig was restored to WT levels following complementation of the pigS mutation in trans (Fig. 1B), confirming that the absence of pigS was responsible for the observed phenotype. Expression of a chromosomal pigA::uidA transcriptional fusion was reduced 1.5-fold throughout growth in a pigS mutant background compared with its expression in the parental strain (Fig. 1C). Therefore, the phenotypic effects on Pig production following mutation of pigS were occurring, at least in part, at the level of transcription of the Pig biosynthetic operon, consistent with PigS being a predicted transcriptional regulator.
FIG. 1.
Pig production and pigA-O transcription are reduced in a pigS mutant. (A) Pig production by the WT (diamonds) or a pigS mutant strain (HSPIG26) (unfilled squares) throughout growth. (B) Pig production by the WT and a pigS mutant (HSPIG26) in the presence of empty vector control pQE-80L and Pig production by a pigS mutant (HSPIG26) in the presence of pTA27, encoding native PigS at late exponential phase (9 h), are shown. (C) β-Glucuronidase activity was measured from a chromosomal pigA::uidA fusion in an otherwise WT background (KFAAG6) (diamonds) or in a pigS mutant background (TG70) (unfilled squares) throughout growth. Solid lines represent either Pig or β-glucuronidase assays, and dashed lines represent bacterial growth. Data shown are the means ± SD of the results of three independent experiments.
Sequence analysis of the pigS locus.
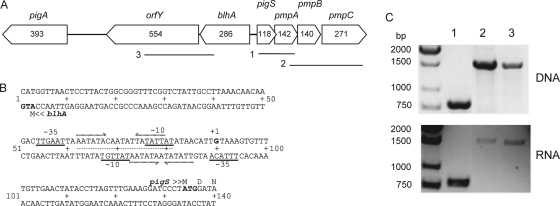
Sequencing of the region surrounding pigS revealed five additional genes predicted to be in two operons (Fig. 2 A) that were closely linked to the pigA-O biosynthetic cluster; the translational start of pigA is located 3.6 kb upstream of the start of pigS. The pmpA and pmpB (putative membrane protein) genes encode putative COG2391 membrane proteins (also known as the YeeE/YedE or DUF395; domain of unknown function). The pmpC gene encodes a COG0730 (TauE/SafE or DUF81) (20, 33) putative membrane protein. For consistency, we will refer to COG numbers throughout this article. The blhA and orfY genes, divergently transcribed from pigS, are predicted to encode two cytoplasmic proteins, a metallo-β-lactamase superfamily protein (COG0491) and a coenzyme A-disulfide reductase containing a rhodanese homology domain (CoADR-RHD; COG0446 and COG0607).
FIG. 2.
The pigS locus. (A) The Serratia sp. ATCC 39006 pigS locus consists of two divergent operons, bearing six genes. Numbers shown represent the lengths in amino acids of the putative protein products. Solid lines indicate products generated by RT-PCR, as described in Materials and Methods and shown in panel C. (B) The pigS-blhA intergenic region. The pigS transcriptional start site is denoted +1, and potential −10 and −35 sites of the pigS and blhA promoters are underlined. Inverted repeats are indicated by dashed arrows, and a dotted line indicates a conserved BigR box consensus sequence (3). (C) RT-PCRs were performed to determine the operonic nature of blhA-orfY and pigS-pmpABC. RT-PCR products were generated with the following primer combinations: 1, PF64 and JCO24; 2, PF264 and PF265; and 3, PF266 and PF269. The locations of the PCR products are shown in panel A.
blhA-orfY and pigS-pmpABC are divergently transcribed operons.
The pigS transcriptional start site was mapped, a single transcriptional start site (+1) was identified, and located upstream of this are putative −10 and −35 regions, based on the E. coli σ70 consensus sequences (16). Putative −10 and −35 regions were also predicted for the blhA promoter. However, attempts to map the +1 start site were unsuccessful (data not shown). Two inverted repeat sequences were identified within the pigS-blhA intergenic region; these were predicted to be regulatory protein binding sites (Fig. 2B). RT-PCR was performed to determine whether the six genes in the pigS locus were transcribed as two operons. RT-PCR of regions spanning the gene junctions generated products of the predicted size (Fig. 2C) with no products in the control lacking RT (−RT control) (data not shown). These results indicated that the blhA-orfY genes are bicistronic and that the pigS-pmpABC genes comprise a four-gene operon.
Expression of pigS-pmpABC and blhA-orfY is controlled by PigP and PigS.
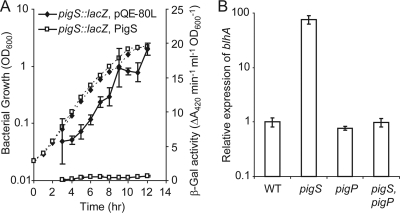
The transcriptional organization, and the similarity of PigS to ArsR family transcriptional repressors, suggested that PigS might control the expression of the pigs-pmpABC and blhA-orfY operons. Expression of a chromosomal pigS::lacZ transcriptional fusion was measured throughout growth in the presence of plasmid-encoded PigS or an empty vector control. β-Galactosidase activity was reduced by PigS, indicating that PigS is negatively autoregulatory and represses expression of pigS-pmpABC (Fig. 3 A). In contrast, it was previously shown that the master regulator PigP activates pigS-pmpABC transcription (11). Quantitative RT-PCR was performed to investigate whether PigS and PigP also regulate transcription from the divergent blhA promoter (Fig. 3B). Expression of blhA was increased 75-fold in a pigS mutant compared with the WT, indicating that PigS represses transcription of blhA-orfY from the blhA promoter. Mutation of pigP returned blhA transcripts to WT levels in the pigS mutant background but had no effect in a WT background. These data indicate that blhA-orfY expression is activated by PigP but only when there is also derepression in the absence of the repressor, PigS. While the effect of either protein might be direct or indirect based on these data, we will show later that the PigS effects are direct.
FIG. 3.
PigS autoregulates and, along with PigP, controls expression of blhA. (A) β-Galactosidase activity was measured from a chromosomal pigS::lacZ fusion (HSPIG26) in the presence of an empty vector control, pQE-80L (diamonds), or pTA27, encoding native PigS (unfilled squares), throughout growth. Solid lines represent β-galactosidase assays, and dashed lines represent bacterial growth. (B) Quantitative RT-PCR was used to measure blhA transcript levels, relative to the WT (LacA), in a pigS mutant (HSPIG26), a pigP mutant (PIG13S), and a pigS pigP double mutant (13S26L). Data shown are the means ± SD of the results of three independent experiments.
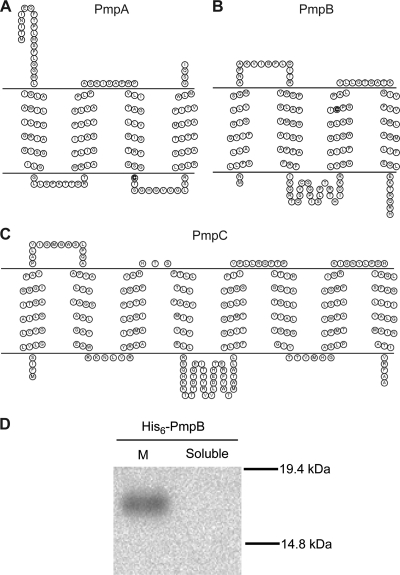
PmpA, PmpB, and PmpC are predicted to be inner membrane proteins involved in the transport of sulfur-containing compounds.
The PigS- and PigP-mediated control of pigS-pmpABC prompted further analysis of PmpABC. PmpA and PmpB are members of the uncharacterized COG2391 family of putative inner membrane proteins with a conserved cysteine and four transmembrane helices (Fig. 4 A and B). PmpC is a member of the COG0730 family of proteins, which Weinitschke et al. (33) proposed to be novel permeases for the transport of anions (typically sulfur containing) across cytoplasmic membranes. Proteins of this group have been implicated in the uptake of 4-toluenesulfonate (19, 23), sulfate, and sulfite (27) and in the export of sulfite (33) and sulfoacetate (20). PmpC was predicted to contain eight transmembrane domains (Fig. 4C). Since there is no experimental evidence demonstrating the membrane localization of COG2391 or COG0730 proteins, we performed cellular fractionation experiments to localize PmpABC. Unfortunately, despite testing multiple induction conditions, we could not express N-terminally His-tagged or C-terminally FLAG-tagged PmpA or PmpC (data not shown). However, His6-PmpB was overexpressed in the WT and, following cellular fractionation, was detected by Western blotting in the membrane fraction yet was absent from the soluble fraction (Fig. 4D).
FIG. 4.
PmpABC are predicted to localize to the membrane. Topology mapping of PmpA (A), PmpB (B), and PmpC (C) was performed using TMHMM v2.0 (21), and figures were generated using TOPO2. The highly conserved cysteine residues in PmpA and PmpB are shown in bold. (D) Cellular localization of N-terminally His-tagged PmpB (His6-PmpB) expressed in LacA was performed as described in Materials and Methods. Fifty micrograms of total protein from the membrane fraction (M) or soluble fraction was analyzed by Western blotting to detect the presence of His6-PmpB.
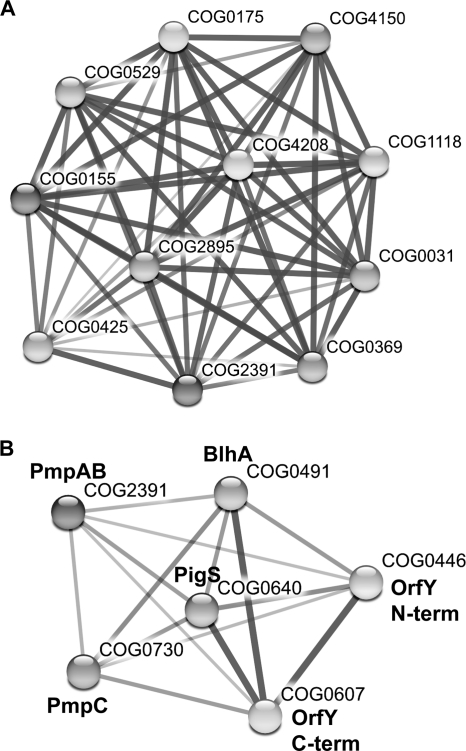
To gain insight into the possible role of COG2391 (PmpAB) proteins, STRING (18) was used in COG-mode. STRING identifies putative functional linkages between proteins by weighting and integrating information about factors such as their gene neighborhood, gene fusion, cooccurrence, and coexpression. Functional relationships were predicted, and the top 10 are shown in Fig. 5 A. Associated functions include SirA, involved in disulfide bond formation, and other proteins involving sulfur-containing compounds (for details, see Fig. 5A). Interestingly, COG2391 (PmpAB) domains are also found fused as a single protein to SirA (COG0425) or COG0607 (C-terminal of OrfY) domains in some species. Therefore, we propose that PmpAB are involved in the transport of sulfur-containing molecules and may be linked to proteins involved in sulfur transfer reactions (see below).
FIG. 5.
PmpAB proteins are associated with proteins involved in sulfur metabolism, and members of the pigS-pmpABC and blhA-orfY system are frequently associated. (A) Phylogenomic profile of functions related to COG2391 (PmpAB; DUF395) shows possible interactions generated in STRING (high confidence, 10 top interactors shown). Shown are COG0425 (SirA), a predicted regulator of disulfide bond formation; COG2895, GTPase-sulfate adenylate transferase subunit 1; COG4208, ABC-type sulfate transport system, permease component; COG0031, cysteine synthase; COG0175, 3-phosphoadenosine 5-phosphosulfate sulfotransferase; COG0529, adenylylsulfate kinase and related kinases; COG1118, ABC-type sulfate/molybdate transport systems, ATPase component; COG0155, sulfite reductase, beta subunit (hemoprotein); COG4150, ABC-type sulfate transport system, periplasmic component; and COG0369, sulfite reductase, alpha subunit (flavoprotein). (B) Putative functional interactions between all six domain members of the blhA-orfY and pigS-pmpABC operons. In both panels A and B, the thickness of the lines relates to the degree of confidence in an association.
BlhA and OrfY are predicted to function in sulfur transfer reactions with PmpABC.
The coregulation of pmpABC and blhA-orfY indicated that they might be involved in similar processes. Insight into the role of OrfY comes from the recent crystal structure of the dimeric Bacillus anthracis protein BaCoADR-RHD (32). Like OrfY, BaCoADR-RHD contains both a coenzyme A disulfide reductase (COG0446) and a rhodanese homology domain (COG0607). The physiological role of BaCoADR-RHD is unclear but via an interesting mechanism is proposed to reduce polysulfide (S52−) to HS− using flavin adenine dinucleotide (FAD) as a cofactor and reducing equivalents from NADH. We predict that OrfY is involved in disulfide reduction, but the substrate is unknown. BlhA is a member of the metallo-β-lactamase superfamily and is related to the ubiquitous COG0491 family of Zn-dependent hydrolases. COG0491 proteins include β-lactamases, thiolesterases, lactonases, and glyoxalases, but their diverse functions make it difficult to accurately predict the function of BlhA. Gene neighborhood analyses (STRING [18]) using COG0491 (BlhA) demonstrated that four of the eight most commonly linked domains belonged to COG0730 (PmpC), COG0446 and COG0607 (both OrfY), and PigS (COG0640), and similar results were obtained using either domain of OrfY as the query (data not shown). Interestingly, there are examples of gene fusions between COG0491 (BlhA) and COG0446 and/or COG0607 (OrfY) domains, and COG0607 (OrfY C terminus) is also found fused to COG0640 (PigS). To investigate the associations of all domains in the divergent pigS-pmpABC and blhA-orfY operons, STRING was used (COG queries) to predict functional linkages between these six different domains (Fig. 5B). Our data presented suggest that BlhA and OrfY are acting in similar processes related to sulfur transfer or disulfide reduction reactions that are linked to the PmpABC transport components of sulfur-containing molecules.
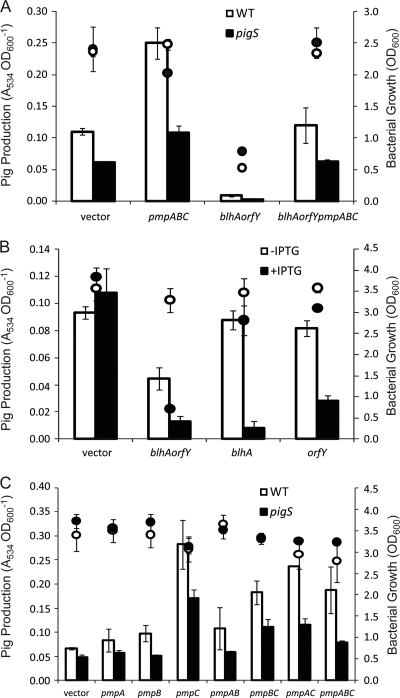
Overexpression of blhA-orfY, relative to pmpABC levels, decreases Pig production.
We hypothesized that the decrease in Pig production in the pigS mutant was the result of increased expression of blhA-orfY in the absence of the PigS repressor. Furthermore, due to the transposon insertion within pigS, it was likely that pmpABC would no longer be under correct PigS-mediated control. To test this model, and the proposed functional linkage of both divergent operons, we measured the effect of overexpression of blhA-orfY (pTA133), pmpABC (pTA132), or both the blhA-orfY and pmpABC operons (pTA134) on Pig production in a WT or pigS mutant background. Overexpression of blhA-orfY in the WT or pigS mutant resulted in reduced Pig production and growth inhibition (Fig. 6 A). Overexpression of pmpABC in the WT or pigS mutant resulted in increased Pig production (Fig. 6A). However, the concomitant overexpression of blhA-orfY and pmpABC restored Pig production, and growth, to levels comparable to those of the parental strains, indicating that the balance between expression levels of these operons is important for Pig production and for avoiding growth inhibition (Fig. 6A).
FIG. 6.
Overexpression of PmpABC and/or BlhA-OrfY. (A) Pig production by WT (white bars) or pigS mutant strains (HSPIG26) (black bars) after 12 h of growth carrying pQE-80LoriT (vector), pTA132 (pmpABC), pTA133 (blhA-orfY), and pTA134 (pmpABC blhA-orfY). Bacterial growth is indicated by white (WT) or black (pigS mutant) circles. (B) Pig production by WT carrying pQE-80LoriT (vector), pTA133 (blhAorfY), pTA154 (blhA), and pTA155 (orfY) after 12 h of growth. Cultures were grown in the absence of IPTG induction (white bars) or with 0.1 mM IPTG (black bars). Bacterial growth is indicated by white (−IPTG) or black (+IPTG) circles. (C) Pig production by WT (white bars) or pigS mutant strains (HSPIG26) (black bars) carrying pQE-80LoriT (vector), pTA160 (pmpA), pTA161 (pmpB), pTA157 (pmpC), pTA162 (pmpAB), pTA156 (pmpBC), pTA159 (pmpAC), or pTA132 (pmpABC) after 12 h of growth. Bacterial growth is indicated by white (WT) or black (pigS mutant) circles. Except where indicated, cultures were induced with 0.1 mM IPTG. Data shown are the means ± SD of the results of three independent experiments.
To assess the role of BlhA and OrfY further, the effect of overexpressing blhA or orfY on Pig production was measured in a WT background. As overexpression of blhA-orfY in the WT or pigS mutant had been shown to result in a growth defect (Fig. 6A), we also assessed the impact of leaky blhA and orfY expression from the pQE-80L promoter in the absence of IPTG induction. Leaky expression of blhA-orfY resulted in reduced Pig production without growth inhibition, but leaky expression of either blhA or orfY did not reduce Pig levels or growth (Fig. 6B). However, following IPTG induction, overexpression of BlhA or OrfY alone caused reduced Pig production but did not inhibit growth. This demonstrates that both BlhA and OrfY affect Pig levels and that when overexpressed together they are detrimental to growth.
Similarly, the effect of pmpABC on Pig production was investigated in more detail. Overexpression of pmpA or pmpB, or pmpAB, did not significantly affect Pig production in either the WT or pigS mutant (Fig. 6C). Overexpression of pmpC in the WT or pigS mutant caused an increase in Pig production greater than that caused by overexpression of the entire pmpABC operon. Overexpression of pmpBC or pmpAC partially attenuated the hyperpigmentation observed following overexpression of pmpC alone (Fig. 6C). These data suggest that PmpC is responsible for the increased Pig levels following overexpression of the pmpABC operon and that PmpA and PmpB partially limit or oppose the phenotypic effect of PmpC.
PigS regulates transcription via direct binding to the pigS-blhA intergenic region.
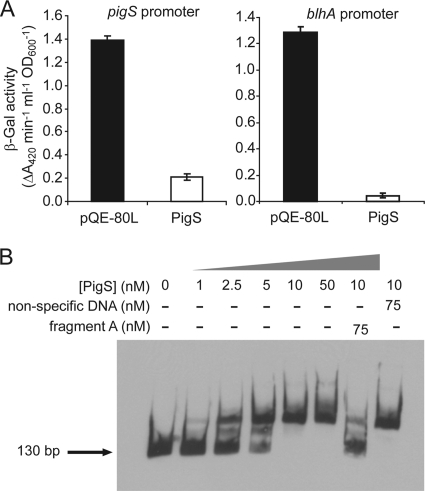
The regulation and overexpression data above indicated that PigS was a repressor of the divergent blhA-orfY and pmpABC operons, the products of which affect Pig production. To characterize PigS repression further in a heterologous host, β-galactosidase production from E. coli strains carrying vectors with either the pigS or blhA promoter fused to a promoterless lacZ (pTA25 or pTG41) was measured in the presence or absence of PigS (pTA27 and pQE-80L). In both cases, promoter activity was decreased by PigS (Fig. 7 A), indicating that PigS represses transcription, possibly via direct binding to the pigS-blhA intergenic region without requiring additional Serratia sp. ATCC 39006 proteins (e.g., PigP). Since PigS is an ArsR family transcriptional regulator, this reporter system was used to investigate whether PigS-mediated repression was allosterically inhibited by the presence of metal ions [Fe(III), Cd(III), Zn(II), Ni(II), Mn(II), Mg(II), Cu(II), Li(I), Ag(I), K(I)]. However, derepression was not observed, providing no evidence that these molecules act as PigS ligands (data not shown).
FIG. 7.
PigS represses the pigS and blhA promoters by direct binding to the pigS-blhA intergenic region. (A) β-Galactosidase activity was measured in E. coli carrying pTA25 or pTG41 (pigS or blhA promoters upstream of a promoterless lacZ gene) and either an empty vector control (pQE-80L) (black bar) or pTA27, encoding native PigS (white bar). Data shown are the means ± SD of the results of three independent experiments. (B) Binding of His6-PigS to the pigS-blhA intergenic region (fragment A) was assessed using EMSAs. Each lane contains 1.5 nM DIG-labeled fragment A DNA and the indicated amount of His6-PigS. Competition experiments were carried out in which a 50-fold excess (75 nM) of either unlabeled fragment A or a nonspecific fragment was added.
To test direct binding by PigS, a 130-bp DNA fragment containing the complete pigS-blhA intergenic region was combined with increasing concentrations of N-terminally hexahistidine-tagged PigS (His6-PigS) and assessed for binding using EMSAs. There was a decrease in mobility of the 130-bp fragment with increasing concentrations of His6-PigS, the majority of which was shifted by 10 nM His6-PigS (Fig. 7B). Binding was outcompeted by a 50-fold excess of unlabeled pigS-blhA intergenic region but not by a nonspecific control, demonstrating that binding is specific.
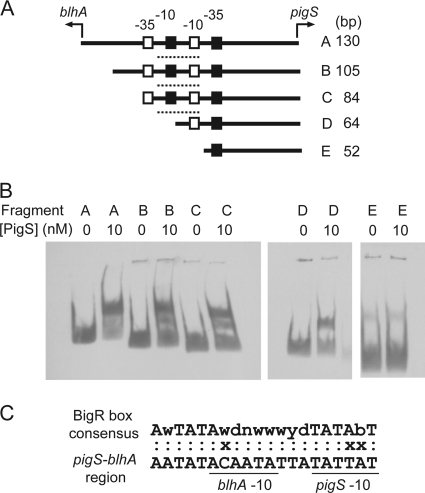
To more precisely identify the binding site(s) of His6-PigS, truncated fragments of the pigS-blhA intergenic region (Fig. 8 A) were used in an EMSA (Fig. 8B). His6-PigS bound to the 130-, 105-, 84-, and 64-bp fragments (fragments A, B, C, and D) but did not bind to the 52-bp fragment (fragment E). This implied that the His6-PigS binding site lies within the region overlapping the pigS −10 region sequence. Using EMSAs, it was determined that there was no specific binding by His6-PigS to the pigA promoter nor did PigS affect expression of the pigA promoter cloned in E. coli, indicating that PigS does not directly regulate pigA-O transcription (data not shown).
FIG. 8.
PigS binds directly to the pigS-blhA intergenic region. (A) The pigS-blhA intergenic region (fragment A) and truncated fragments (fragments B to E) were generated by PCR as described in Materials and Methods. The predicted pigS and blhA −10 and −35 sequences are represented by unfilled and filled boxes, respectively. A dashed line indicates the location of the putative BigR box consensus sequence (3). (B) Binding of His6-PigS to truncated fragments of the pigS-blhA intergenic region was assessed using EMSAs. Each lane contains a 1.5 nM concentration of the appropriate fragment of DIG-labeled DNA and either 0 or 10 nM His6-PigS, as indicated in the figure. (C) The predicted PigS binding region shows similarity (17/20 nucleotides) to the BigR box consensus sequence, which has 100% conserved bases represented by capital letters (3). The blhA and pigS −10 sequences are underlined. An “x” indicates nucleotides that do not match the BigR consensus; a colon indicate nucleotides that match.
DISCUSSION
In this study, we set out to determine the role of the ArsR family protein PigS, a member of the PigP regulon, in the control of Pig production in Serratia. PigS repressed the level of Pig at least partly via the transcription of the biosynthetic genes pigA-O. However, PigS did not directly activate, or bind near, the pigA promoter. Despite the lack of direct regulation of pigA-O, PigS was shown to represses transcription from two divergent operons which drive the expression of six genes (blhA, orfY, pigS, pmpA, pmpB, and pmpC) via direct binding to the blhA-pigS intergenic region. Since pigS is within one of these operons, it is autoregulated. In addition, PigP, the founding member of a novel class of transcriptional regulators (11), activates transcription from the blhA and pigS promoters in the absence of repression by PigS. It is currently unknown whether PigP acts directly at the pigS and blhA promoters.
The genomic context of pigS shares some similarity with the bigR locus from the plant pathogen Xylella fastidiosa 9a5c and other members of the Rhizobiales (3). Barbosa and Benedetti show that in X. fastidiosa, the ArsR/SmtB family regulator, BigR, controls transcription of a five-gene operon, encoding a metallo-β-lactamase superfamily protein (COG0491-COG3453 fusion), BigR, two COG2391 (PmpAB) proteins, and a COG0730 (PmpC) protein (3). However, the bigR locus does not encode an orthologue of OrfY. This operon was implicated in biofilm growth in X. fastidiosa and in Agrobacterium tumefaciens. BigR bound directly to a 20-bp region spanning the blh −10 sequence, and a BigR box consensus sequence was proposed (3). PigS bound the pigS-blhA intergenic region near a BigR boxlike sequence (17/20 matches to BigR box consensus), which overlaps the predicted pigS and blhA −10 sequences (Fig. 8C). However, PigS was still able to bind to the 64-bp fragment D, in which 8 nucleotides of the putative BigR-like box had been removed, indicating that either a BigR-like half-site is sufficient to bind PigS or that the BigR box is not required for binding. We suggest that in the absence of a specific inducer molecule, PigS represses blhA and pigS via direct binding to the intergenic region, potentially via the BigR-like box, and occludes RNA polymerase. When present, an inducer may bind PigS, causing disassociation from the target DNA and allowing transcriptional activation by PigP. Consistent with the findings for BigR (3), we could not identify a metal as the inducer of PigS.
We demonstrated that the effect of PigS on Pig biosynthesis was mediated via BlhA, OrfY, and PmpABC. Expression of either or both the metallo-β-lactamase BlhA and the coenzyme A disulfide reductase with a rhodanese homology domain, OrfY, decreased Pig production. In contrast, expression of the putative transporters PmpABC increased Pig levels. These effects were alleviated by the concomitant overexpression of both operons, suggesting that the balance between the expression of PmpABC and BlhA and OrfY is important. However, the increased Pig levels resulting from pmpABC overexpression were mediated via PmpC, with PmpA and PmpB inhibiting the phenotypic effect of PigC. These data support the suggestion that the complete pigS locus is not only coordinately regulated (by PigS and PigP) but that these genes represent a complete functional system.
Despite numerous COG2391 (PmpAB) proteins in bacteria, archaea, and eukaryotes, to the best of our knowledge, there is no experimental evidence suggesting their function. We demonstrated that PmpB was localized to the cell membrane and provide the first experimental data supporting the prediction that COG2391 proteins are integral membrane proteins. Based on functional partnership predictions, we propose a possible role of COG2391 proteins (and PmpAB) as transporters of sulfur-containing compounds. Interestingly, COG2391 proteins contain a highly conserved cysteine (Fig. 4), which might enable disulfide bond formation or the generation of a reactive persulfide that could participate in S transfer reactions (24). In E. coli, cobalt stress was recently shown to disrupt Fe/S biogenesis and decrease yeeE (COG2391) by ∼2.6-fold (9). Furthermore, yeeE was activated by IscR, the master regulator of Fe/S biogenesis in E. coli.
COG0730 (PmpC) proteins are present in bacteria, archaea, and eukaryotes, and a number of studies indicate that COG0730 proteins may import or export sulfur-containing molecules in bacteria (19, 20, 23, 27, 33). Despite the genetic linkage between PmpAB and PmpC (and their COG groups), and the possibility that they are involved in transport of S compounds, COG2391 and COG0730 are not always found together. This suggests that these proteins can act together in a process in some organisms but perform functionally separable tasks. Indeed, our expression data show that PmpC (COG0730) acts independently but that PmpA and PmpB (COG2391) can temper the effects of PmpC. What is transported via this system awaits identification, but it, or a product of BlhA and/or OrfY, may be the inducer(s) of PigS, enabling expression of these operons under appropriate conditions. Of interest are two highly conserved cysteines present in the PigS/BigR subfamily of ArsR regulators (3), which we predict could be involved in modulating its DNA-binding activity.
Experimental and bioinformatic data demonstrate that OrfY and BlhA are linked to a process similar to that of PmpABC with regard to Pig production. It is likely that OrfY is involved in disulfide reduction reactions and the mechanism is understood (32) but the substrate is unknown. The evidence suggests that BlhA is acting in concert with OrfY, but the details are unclear due to the diverse roles of COG0491 (BlhA) proteins. BlhA is unlikely to act as an AHL lactonase since levels of lactone were unaffected in the BlhA-overexpressing pigS mutant (data not shown). Likewise, a β-lactamase function of the related BigR system from X. fastidiosa and A. tumefaciens could not be demonstrated (3).
Due to the challenges involved in deciphering the function of conserved uncharacterized proteins, there is still much to learn about the role of the pigS locus and related genes in other species. Based on the current data, we propose one possible model in which the enzymes OrfY and BlhA generate a S-containing product (z), which is currently unidentified. Product z may indirectly inhibit pigA-O transcription and hence Pig levels. PmpC acts as a transporter, exporting z and maintaining appropriate intracellular levels. PmpAB partially inhibit or modulate this PmpC-mediated effect, possibly by also acting as transporters. Mutation of pigS, or overexpression of blhA-orfY, increases levels of z within the cell, causing reduced Pig production. The concomitant overexpression of pmpABC allows excess z to be exported, and thus appropriate intracellular z levels, and Pig, are restored. Overexpression of PmpABC results in reduced intracellular z levels, and hence Pig levels are increased.
In conclusion, we have characterized in detail an ArsR family transcriptional regulator which modulates expression of the prodigiosin antibiotic genes via the control of divergent operons that are predicted to constitute a system involved in the transport and modification of a S-containing compound(s). In addition, we have provided experimental and bioinformatic evidence that PmpB (COG2391) is membrane associated and have provided a prediction on the function of this uncharacterized family of proteins in the transport of sulfur-containing molecules. This study represents a step forward in understanding the role of COG2391 and COG0730 proteins and their association with metallo-β-lactamase (COG0491) and CoADR-RHD (COG0446 and COG0607) proteins. These components are important in the control of prodigiosin antibiotic production in Serratia sp. ATCC 39006, presumably by functioning in the transport and modification of sulfur-containing molecules. The clear phenotypes, and their newly hypothesized roles, will provide a highly tractable system to further unravel the function of these poorly characterized proteins and their regulation by PigS.
Acknowledgments
We thank Josh Ramsay for plasmid pQE-80LoriT and Neil Williamson for strain KFAAG6.
This work was supported by the BBSRC, United Kingdom, the University of Otago, and the Marsden Fund, Royal Society of New Zealand. M.B.M. was supported by a Bright Futures Top Achiever doctoral scholarship from the Tertiary Education Commission of New Zealand.
Footnotes
Published ahead of print on 23 December 2010.
REFERENCES
- 1.Alm, E. J., et al. 2005. The MicrobesOnline Web site for comparative genomics. Genome Res. 15:1015-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbosa, R. L., and C. E. Benedetti. 2007. BigR, a transcriptional repressor from plant-associated bacteria, regulates an operon implicated in biofilm growth. J. Bacteriol. 189:6185-6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Busenlehner, L. S., M. A. Pennella, and D. P. Giedroc. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27:131-143. [DOI] [PubMed] [Google Scholar]
- 6.Bycroft, B. W., C. Maslen, S. J. Box, A. Brown, and J. W. Tyler. 1987. The isolation and characterisation of (3R,5R)- and (3S,5R)-carbapenem-3-carboxylic acid from Serratia and Erwinia species and their putative biosynthetic role. J. Chem. Soc. Chem. Commun. (Camb.) 21:1623-1625. [Google Scholar]
- 7.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, T. J., et al. 2010. Characterization of a broad-host-range flagellum-dependent phage that mediates high-efficiency generalized transduction in, and between, Serratia and Pantoea. Microbiology 156:240-247. [DOI] [PubMed] [Google Scholar]
- 9.Fantino, J. R., B. Py, M. Fontecave, and F. Barras. 2010. A genetic analysis of the response of Escherichia coli to cobalt stress. Environ. Microbiol. 12:2846-2857. [DOI] [PubMed] [Google Scholar]
- 10.Fineran, P. C., L. Everson, H. Slater, and G. P. Salmond. 2005. A GntR family transcriptional regulator (PigT) controls gluconate-mediated repression and defines a new, independent pathway for regulation of the tripyrrole antibiotic, prodigiosin, in Serratia. Microbiology 151:3833-3845. [DOI] [PubMed] [Google Scholar]
- 11.Fineran, P. C., H. Slater, L. Everson, K. Hughes, and G. P. Salmond. 2005. Biosynthesis of tripyrrole and beta-lactam secondary metabolites in Serratia: integration of quorum sensing with multiple new regulatory components in the control of prodigiosin and carbapenem antibiotic production. Mol. Microbiol. 56:1495-1517. [DOI] [PubMed] [Google Scholar]
- 12.Fineran, P. C., N. R. Williamson, K. S. Lilley, and G. P. Salmond. 2007. Virulence and prodigiosin antibiotic biosynthesis in Serratia are regulated pleiotropically by the GGDEF/EAL domain protein, PigX. J. Bacteriol. 189:7653-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galperin, M. Y., and E. V. Koonin. 2004. ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res. 32:5452-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gristwood, T., P. C. Fineran, L. Everson, and G. P. Salmond. 2008. PigZ, a TetR/AcrR family repressor, modulates secondary metabolism via the expression of a putative four-component resistance-nodulation-cell-division efflux pump, ZrpADBC, in Serratia sp. ATCC 39006. Mol. Microbiol. 69:418-435. [DOI] [PubMed] [Google Scholar]
- 15.Gristwood, T., P. C. Fineran, L. Everson, N. R. Williamson, and G. P. Salmond. 2009. The PhoBR two-component system regulates antibiotic biosynthesis in Serratia in response to phosphate. BMC Microbiol. 9:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harley, C. B., and R. P. Reynolds. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res. 15:2343-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris, A. K., et al. 2004. The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology 150:3547-3560. [DOI] [PubMed] [Google Scholar]
- 18.Jensen, L. J., et al. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37:D412-D416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Junker, F., and A. M. Cook. 1997. Conjugative plasmids and the degradation of arylsulfonates in Comamonas testosteroni. Appl. Environ. Microbiol. 63:2403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krejcik, Z., et al. 2008. Sulfoacetate released during the assimilation of taurine-nitrogen by Neptuniibacter caesariensis: purification of sulfoacetaldehyde dehydrogenase. Arch. Microbiol. 190:159-168. [DOI] [PubMed] [Google Scholar]
- 21.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 22.Lodge, J., J. Fear, S. Busby, P. Gunasekaran, and N. R. Kamini. 1992. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol. Lett. 74:271-276. [DOI] [PubMed] [Google Scholar]
- 23.Mampel, J., et al. 2004. A novel outer-membrane anion channel (porin) as part of a putatively two-component transport system for 4-toluenesulphonate in Comamonas testosteroni T-2. Biochem. J. 383:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller, E. G. 2006. Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nat. Chem. Biol. 2:185-194. [DOI] [PubMed] [Google Scholar]
- 25.Parker, W. L., et al. 1982. SQ 27,860, a simple carbapenem produced by species of Serratia and Erwinia. J. Antibiot. (Tokyo). 35:653-660. [DOI] [PubMed] [Google Scholar]
- 26.Roberts, R. J. 2004. Identifying protein function— call for community action. PLoS Biol. 2:E42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rückert, C., et al. 2005. Functional genomics and expression analysis of the Corynebacterium glutamicum fpr2-cysIXHDNYZ gene cluster involved in assimilatory sulphate reduction. BMC Genomics 6:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York, NY.
- 29.Slater, H., M. Crow, L. Everson, and G. P. Salmond. 2003. Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and -independent pathways. Mol. Microbiol. 47:303-320. [DOI] [PubMed] [Google Scholar]
- 30.Thomson, N. R., et al. 1997. The rap and hor proteins of Erwinia, Serratia and Yersinia: a novel subgroup in a growing superfamily of proteins regulating diverse physiological processes in bacterial pathogens. Mol. Microbiol. 26:531-544. [DOI] [PubMed] [Google Scholar]
- 31.Thomson, N. R., M. A. Crow, S. J. McGowan, A. Cox, and G. P. Salmond. 2000. Biosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing control. Mol. Microbiol. 36:539-556. [DOI] [PubMed] [Google Scholar]
- 32.Wallen, J. R., et al. 2009. Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking. Biochemistry 48:9650-9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinitschke, S., K. Denger, A. M. Cook, and T. H. Smits. 2007. The DUF81 protein TauE in Cupriavidus necator H16, a sulfite exporter in the metabolism of C2 sulfonates. Microbiology 153:3055-3060. [DOI] [PubMed] [Google Scholar]
- 34.Williamson, N. R., et al. 2007. Anticancer and immunosuppressive properties of bacterial prodiginines. Future Microbiol. 2:605-618. [DOI] [PubMed] [Google Scholar]
- 35.Williamson, N. R., P. C. Fineran, F. J. Leeper, and G. P. Salmond. 2006. The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Microbiol. 4:887-899. [DOI] [PubMed] [Google Scholar]
- 36.Williamson, N. R., P. C. Fineran, W. Ogawa, L. R. Woodley, and G. P. Salmond. 2008. Integrated regulation involving quorum sensing, a two-component system, a GGDEF/EAL domain protein and a post-transcriptional regulator controls swarming and RhlA-dependent surfactant biosynthesis in Serratia. Environ. Microbiol. 10:1202-1217. [DOI] [PubMed] [Google Scholar]
- 37.Williamson, N. R., et al. 2005. Biosynthesis of the red antibiotic, prodigiosin, in Serratia: identification of a novel 2-methyl-3-n-amyl-pyrrole (MAP) assembly pathway, definition of the terminal condensing enzyme, and implications for undecylprodigiosin biosynthesis in Streptomyces. Mol. Microbiol. 56:971-989. [DOI] [PubMed] [Google Scholar]
- 38.Yu, N. Y., et al. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]