Abstract
The response regulator HrrA of the HrrSA two-component system (previously named CgtSR11) was recently found to be repressed by the global iron-dependent regulator DtxR in Corynebacterium glutamicum. Here, we provide evidence that HrrA mediates heme-dependent gene regulation in this nonpathogenic soil bacterium. Growth experiments and DNA microarray analysis revealed that C. glutamicum is able to use hemin as an alternative iron source and emphasize the involvement of the putative hemin ABC transporter HmuTUV and heme oxygenase (HmuO) in heme utilization. As a central part of this study, we investigated the regulon of the response regulator HrrA via comparative transcriptome analysis of an hrrA deletion mutant and C. glutamicum wild-type strain in combination with DNA-protein interaction studies with purified HrrA protein. Our data provide evidence for a heme-dependent transcriptional activation of heme oxygenase. Based on our results, it can be furthermore deduced that HrrA activates the expression of heme-containing components of the respiratory chain, namely, ctaD and the ctaE-qcrCAB operon encoding subunits I and III of cytochrome aa3 oxidase and three subunits of the cytochrome bc1 complex. In addition, HrrA was found to repress almost all genes involved in heme biosynthesis, including those for glutamyl-tRNA reductase (hemA), uroporphyrinogen decarboxylase (hemE), and ferrochelatase (hemH). Growth experiments with an hrrA deletion mutant showed that this strain is significantly impaired in heme utilization. In summary, our results provide evidence for a central role of the HrrSA system in the control of heme homeostasis in C. glutamicum.
Iron is a critical element for bacteria, being essential as a cofactor in a multitude of enzymes, poorly soluble in its oxidized form, and dangerous as ferrous iron by catalyzing the formation of reactive oxygen species. To avoid deprivation as well as high, toxic intracellular iron concentrations, organisms have evolved sophisticated regulatory systems. Previous studies demonstrated the transcriptional regulator DtxR as being the master regulator of iron-dependent gene expression in Corynebacterium glutamicum, a Gram-positive soil bacterium which is used industrially for large-scale amino acid production (13, 16). When the iron supply is sufficient, DtxR in complex with Fe2+ represses more than 50 genes, the majority of which are involved in iron acquisition, such as siderophore ABC transporters and siderophore binding proteins (8, 12, 49). Simultaneously, a number of genes are activated by DtxR (e.g., genes encoding the iron storage proteins ferritin and Dps). When iron becomes limiting, Fe2+ dissociates from DtxR and the protein loses its DNA-binding ability (14). Among the genes repressed by DtxR are also the putative hemin transport locus hmuTUV and the hmuO gene encoding heme oxygenase. Heme oxygenases are involved in the utilization of heme as an iron source by catalyzing the degradation of the tetrapyrrole ring to α-biliverdin, carbon monoxide, and free iron (40, 52). In Corynebacterium diphtheriae, the ABC transporter HmuTUV was shown to be required for the uptake of hemin, the oxidized form of heme as present in extracellular environments (1, 15, 26).
In C. glutamicum, four of the target genes repressed by DtxR under iron excess encode for themselves transcriptional regulators, which are the AraC-type regulator RipA, the ArsR-type regulators GlyR and Cg3082, and the response regulator HrrA, belonging to the HrrSA two-component system (previously named CgtSR11). Under iron limitation, when DtxR-mediated repression is relieved, RipA acts as a repressor of several genes encoding prominent iron-containing proteins (e.g., aconitase and succinate dehydrogenase) (50). GlyR was shown to activate expression of serine hydroxymethyltransferase; however, the link to iron metabolism is still not evident (41). The function of the two-component system HrrSA in C. glutamicum has not been studied yet. The genome of C. glutamicum encodes 13 two-component systems, some of which (MtrAB, PhoRS, and CitAB) have already been studied (10, 11, 25, 29, 37). Prototypical two-component systems consist of a response regulator and a cognate sensor histidine kinase; both proteins communicate via phosphorylation. Environmental signals influence the ability of the sensor protein to bring about the phosphorylation and dephosphorylation of the response regulator, which modulates gene expression (27, 45, 51).
The two-component system HrrSA of C. diphtheriae, which shows the highest sequence identity to C. glutamicum HrrSA (sensor kinases, 56%; response regulators, 86%), was shown to be involved in the heme-dependent activation of hmuO and also acts as repressor of hemA encoding glutamyl-tRNA reductase, a heme biosynthesis enzyme (5). A second two-component system involved in heme-dependent expression of hmuO in C. diphtheriae is the ChrSA system, consisting of the response regulator ChrA and the sensor kinase ChrS (4, 5, 39). Recent studies of ChrS signal sensing postulated a mechanism by which autophosphorylation of the conserved histidine residue of ChrS is triggered by the direct interaction of heme with the N-terminal sensor domain of ChrS (6, 20). The mechanism of HrrS activation has not been studied yet.
In this study, we show via a combination of comparative transcriptomics with DNA-protein interaction studies that the response regulator HrrA of C. glutamicum on one hand activates expression of genes coding for heme oxygenase and heme-containing components of the respiratory chain and on the other hand represses transcription of operons encoding enzymes involved in heme biosynthesis. These results present comprehensive insights into the HrrA regulon and provide evidence for a global function of the HrrSA two-component system in the control of heme homeostasis in C. glutamicum.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains and plasmids used or constructed in this work are listed in Table 1. For growth experiments, 5 ml of brain heart infusion (BHI) medium (Difco Laboratories, Detroit, MI) was inoculated with C. glutamicum colonies from a fresh BHIS agar (BHI agar with 0.5 M sorbitol) plate and incubated overnight at 30°C and 170 rpm. This preculture was used to inoculate the main culture consisting of 50 ml CGXII minimal medium (21) with 4% (wt/vol) glucose, 250 μM ferrous iron chelator 2,2′-dipyridyl, and either 2.5 μM FeSO4 or 2.5 μM hemin (Sigma-Aldrich) to an optical density at 600 nm (OD600) of about 1. The trace element solution and the iron source were added after autoclaving. A 1 mM hemin stock solution was prepared in 100 mM KOH and stored at 4°C. For growth on plates, C. glutamicum strains grown in BHIS preculture were adjusted to an OD600 of about 1, and serial dilutions (100 to 10−7) were spotted (5 μl each) on CGXII minimal medium plates, which were prepared as described for liquid cultures with an additional 1.5% (wt/vol) agar. For DNA microarray analysis, cells were harvested in the exponential growth phase at an OD600 of 5 to 6. Escherichia coli DH5α or BL21(DE3) cells were grown aerobically in LB medium on a rotary shaker (150 rpm) or on LB agar plates at 37°C (36). When appropriate, the media contained kanamycin (25 μg ml−1 for C. glutamicum or 50 μg ml−1 for E. coli) or ampicillin (100 μg ml−1 for E. coli).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| C. glutamicum | ||
| ATCC 13032 | Biotin-auxotrophic wild type | 23 |
| 13032ΔhrrSA mutant | In-frame deletion of the genes cg3247 and cg3248 | 25 |
| 13032ΔhrrA mutant | In-frame deletion of cg3247 | This study |
| 13032Δhmu mutant | In-frame deletion of the hmu operon (cg0466-cg0469) | This study |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (f80lacZDM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen |
| BL21(DE3) | F-ompT hsdSB(rB- mB-) gal dcm (DE3) | 47 |
| Plasmids | ||
| pK19mobsacB | Kanr; vector for allelic exchange in C. glutamicum (pK18 oriVE.coli sacB lacZα) | 38 |
| pK19mobsacB-ΔhrrA | Kanr; pK19mobsacB derivative containing an overlap extension PCR product covering the up- and downstream regions of hrrA (cg3247) | This study |
| pK19mobsacB-Δhmu | Kanr; pK19mobsacB derivative containing a overlap extension PCR product covering the up- and downstream regions of the hmu operon (htaA hmuT hmuU hmuV cg0466-cg0469) | This study |
| pK19mobsacB-ΔhmuO | Kanr; pK19mobsacB derivative containing a overlap extension PCR product covering the up- and downstream regions of hmuO (cg2445) | This study |
| pMal-c | Ampr PtaclacIq ColE1 oriV; E. coli expression vector for overproduction of E. coli MBP (MalE) fusion proteins without signal peptide | New England Biolabs |
| pMBP-HrrSΔ1-248 | Ampr; pMal-c derivative for overproduction of the HrrS kinase domain (residues 249-487) fused to the C terminus of E. coli MBP | This study |
| pET28b | Kanr; vector for overexpression of genes in E. coli, adding an N-terminal or a C-terminal hexahistidine tag to the synthesized protein (pBR322 oriVE. coli PT7 lacI) | Novagen |
| pET28b-hrrA | Kanr pET28b-Streptag derivative for overproduction of HrrA (Cg3247) with an N-terminal hexahistidine tag | This study |
Recombinant DNA work.
Standard methods like PCR, restriction, or ligation were carried out according to established protocols (36). E. coli was transformed by the RbCl method (19). DNA sequencing was performed by Agowa (Berlin, Germany). The oligonucleotides were synthesized by Eurofins MWG Operon (Ebersfeld, Germany) and are listed in Table S1 in the supplemental material.
In-frame deletion mutants of the genes hrrA (cg3247) and hmuO (cg2445), as well as the hmu operon (genes htaA [cg0466], hmuT [cg0467], hmuU [cg0468], and hmuV [cg0469]), were constructed via the two-step homologous recombination procedure as described previously (31). Here, the procedure will be exemplified for hrrA. The deletion methods used for hmuO and the hmu operon were performed comparably; the same oligonucleotide nomenclature was used. Briefly, the corresponding up- and downstream regions were amplified with the oligonucleotide pairs DhrrA-1/DhrrA-2 and DhrrA-3/DhrrA-4. The resulting PCR products served as the template for an overlap extension PCR with the oligonucleotides DhrrA-1 and DhrrA-4. The resulting PCR products of ca. 1 kb were digested with XmaI and XbaI and cloned into the vector pK19mobsacB. The pK19mobsacB derivatives pK19mobsacB-ΔhrrA, pK19mobsacB-Δhmu, and pK19mobsacB-ΔhmuO were then suitable for performing an allelic exchange by homologous recombination in the chromosome of C. glutamicum ATCC 13032 (38), resulting in the respective C. glutamicum ΔhrrA, Δhmu, and ΔhmuO mutant strains.
For overproduction and purification of HrrA with an N-terminal hexahistidine tag, the hrrA coding region was cloned into the expression vector pET28b (Novagen). The gene hrrA (cg3247) was amplified with the oligonucleotides hrrA-for and hrrA-rev, thereby introducing restriction sites for NdeI and XhoI. The HrrA protein encoded by the resulting plasmid, pET28b-hrrA, contains 20 additional amino acids (MGSSHHHHHHSSGLVPRGSH) at the N terminus. For overproduction, the plasmid was transferred into E. coli BL21(DE3).
For phosphorylation studies with the sensor kinase HrrS, the DNA region encoding the kinase domain (amino acid residues 249 to 487) was amplified with the oligonucleotides HrrS_K_for and HrrS_K_rev, thereby introducing EcoRI and HindIII restriction sites. Subsequently, the fragment was cloned into the expression vector pMal-c, resulting in the plasmid pMBP-HrrSΔ1-248, which enables the expression of the HrrS kinase domain fused to the C terminus of maltose binding protein (MBP) lacking its signal peptide.
Global gene expression analysis.
DNA microarray analysis was used to compare the genome-wide mRNA levels of C. glutamicum grown on heme versus FeSO4 as the iron source. In a second experiment, the transcriptomes of the C. glutamicum wild type and the ΔhrrA mutant were compared. For RNA preparation, the strains were first cultivated overnight in CGXII minimal medium containing 4% (wt/vol) glucose and 1 μM FeSO4 as the iron source. For the main culture, cells from 1 μM FeSO4 precultures were used to inoculate CGXII minimal medium with 4% (wt/vol) glucose and either 2.5 μM FeSO4 or 2.5 μM hemin as the iron source. At an OD600 of 5 to 6, 20 ml of the cultures was poured into ice-containing tubes precooled to −20°C and cells were harvested by centrifugation (4 min, 4,200 × g, 4°C). The cell pellets were directly used for RNA isolation as described before (29) or stored at −20°C. All DNA microarray analyses were performed with custom-made DNA microarrays based on 70-mer oligonucleotides obtained from Operon Biotechnologies. Three independent biological replicates were performed. The experimental details for handling these microarrays were described before (18).
Overproduction and purification of HrrA.
E. coli BL21(DE3) carrying the plasmid pET28b-hrrA was grown in 200 ml LB medium with 50 μg/ml kanamycin at 37°C. Expression was induced at an OD600 of ∼0.7 with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and incubation was continued at 30°C. Four hours after induction, cells were harvested by centrifugation and stored at −20°C. For cell extract preparation, cells were resuspended in 3 ml of TNI-5 buffer (20 mM Tris-HCl, pH 7.9, 300 mM NaCl, 5 mM imidazole) containing Complete protease inhibitor cocktail (Roche Diagnostics). The cells were disrupted by being passed three times through a French pressure cell (SLM Aminco, Spectronic Instruments, Rochester, NY) at 207 MPa followed by centrifugation (20 min, 5,000 × g, 4°C). The supernatant was subjected to ultracentrifugation (1 h, 150,000 × g, 4°C). HrrA present in the supernatant was purified by nickel-activated nitrilotriacetic acid-agarose (Novagen). After the column had been washed with TNI-20 buffer (containing 20 mM imidazole), HrrA protein was eluted with TNI-200 buffer (containing 200 mM imidazole). Fractions containing HrrA were pooled, and the elution buffer was exchanged against bandshift buffer (20 mM Tris-HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 5% [vol/vol] glycerol, 0.5 mM EDTA).
Overproduction and purification of MBP-HrrSΔ1-248.
For phosphorylation studies, MBP-HrrSΔ1-248 was overproduced in E. coli BL21(DE3). Cultivation, cell disruption, and the preparation of the soluble supernatant were performed as described above for HrrA. MBP-HrrSΔ1-248 present in the supernatant after ultracentrifugation was purified by affinity chromatography on amylose resin (New England BioLabs) equilibrated with TNM buffer (50 mM Tris-HCl, pH 8.0, 200 mM NaCl, 5 mM MgCl2). After being washed with 15 column volumes of TNM buffer, MBP-HrrSΔ1-248 was eluted with 3 column volumes of TNM buffer containing 10 mM maltose. Fractions containing MBP-HrrSΔ1-248 were pooled, the buffer was exchanged against bandshift buffer, and the purified protein was kept at 4°C and immediately used for phosphorylation studies. After storage for about 2 days, activity was significantly reduced.
In vitro phosphorylation assays.
To determine its autophosphorylation activity, 12 μM MBP-HrrSΔ1-248 was incubated with 0.25 μM [γ-33P]ATP (10 mCi/ml; GE Healthcare) and 80 μM nonradioactive ATP. The assay mixture was incubated at room temperature, and at different time points aliquots were removed, mixed with an equal volume of 2× SDS loading buffer (124 mM Tris-HCl, pH 6.8, 20% [vol/vol] glycerol, 4.6% [wt/vol] SDS, 1.4 M β-mercaptoethanol, 0.01% [wt/vol] bromophenol blue), and kept on ice. For analysis of phosphorylation of HrrA by MBP-HrrSΔ1-248, a 2-fold molar excess of purified HrrA was added to MBP-HrrSΔ1-248 that had been preincubated for 20 min with [γ-33P]ATP, and the mixture was incubated for another 60 min. At different time points, samples were taken (taking into consideration the reduced MBP-HrrSΔ1-248 concentration) and handled as described above. Subsequently, without prior heating, the samples were subjected to 12% SDS-PAGE. The dried gel was analyzed with a BAS-1800 phosphorimager (Fujifilm).
Gel shift assays.
For testing the binding of HrrA to putative target promoters, purified protein (0 to 380 nM) was mixed with DNA fragments (500 bp; final concentration, 15 nM) in a total volume of 20 μl. The binding buffer contained 20 mM Tris-HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 5% (vol/vol) glycerol, 0.5 mM EDTA, and 0.005% (wt/vol) Triton X-100. Approximately 15 nM (100 ng/lane, 500-bp fragments) promoter regions of putative nontarget genes (cytP, cg0645, and pck) was used as negative controls. For phosphorylation of HrrA, the protein had been incubated for 60 min with MBP-HrrSΔ1-248 (half the concentration of HrrA) and 5 mM ATP before the DNA fragments were added. The reaction mixture was incubated at room temperature for 20 min and then loaded onto a 10% nondenaturing polyacrylamide gel. Electrophoresis was performed at room temperature and 170 V with 1× TBE (89 mM Tris base, 89 mM boric acid, 2 mM EDTA) as the electrophoresis buffer. The gels were subsequently stained with Sybr green I (Sigma-Aldrich). All PCR products used in the gel shift assays were purified with the Qiagen PCR purification kit.
Microarray data accession number.
Processed and normalized data as well as experimental details conformed to the MIAME guidelines (9) and were stored in the in-house microarray database (35) for further analysis and in the Gene Expression Omnibus (GEO) repository under accession no. GSE26122.
RESULTS
The C. glutamicum HrrSA (CgtSR11) two-component system—genomic organization and sequence similarities.
The derepression of cgtR11 (cg3247) encoding the response regulator CgtR11 under iron limitation suggested that the CgtSR11 system (here renamed HrrSA) might be involved in some aspect related to iron acquisition (12, 49). A search for orthologous proteins of CgtR11 and CgtS11 revealed a high similarity to the two-component systems HrrSA and ChrSA of C. diphtheriae. CgtS11 shows 56% and 30% sequence identity to HrrS and ChrS, respectively, and CgtR11 shows 86% and 50% sequence identity to HrrA and ChrA, respectively. Both two-component systems of C. diphtheriae were recently shown to be involved in the heme-dependent activation of hmuO expression and repression of hemA transcription (5). Due to the high sequence similarity to HrrSA and ChrSA, an involvement of the C. glutamicum CgtSR11 system in the control of genes required for heme utilization appeared likely. To stay consistent with the described orthologous system of C. diphtheriae, we decided to rename CgtSR11 as “HrrSA” (heme-responsive regulator sensor and activator).
The gene hrrS (cg3248) encoding the sensor kinase HrrS is located upstream of and codirectional with hrrA (cg3247) coding for the response regulator HrrA. The two genes are separated by an intergenic region of 98 bp. We previously showed that hrrA is repressed by the iron-dependent regulator DtxR under conditions of sufficient iron supply and that purified DtxR binds to a DNA fragment covering the intergenic region (49). The most likely DtxR binding site (ATGAGTAAGGCTAGACTAA) is centered 97 bp upstream of the start codon of hrrA and partially overlaps with the coding region of hrrS. Thus, only expression of hrrA, but not that of hrrS, is regulated by DtxR in response to iron availability.
Heme utilization of C. glutamicum.
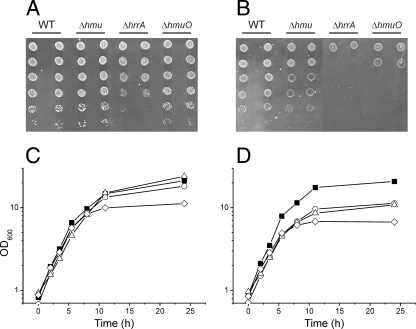
In the first series of experiments, we tested whether the nonpathogenic organism C. glutamicum is able to use heme as an iron source. The type strain ATCC 13032 was cultivated on CGXII agar plates (Fig. 1A and B) or in liquid culture with CGXII minimal medium (Fig. 1C and D) containing either 2.5 μM FeSO4 or 2.5 μM hemin as the iron source. In all experiments, 250 μM 2,2′-dipyridyl was added as a chelator of ferrous iron to ensure iron limitation of the cells. As shown in Fig. 1C and D, the liquid cultures grew with almost identical growth rates up to an OD600 of about 20. As high concentrations of hemin (>5 μM) are toxic for C. glutamicum, cells were generally grown under iron limitation to compare different iron sources. Under nonlimiting conditions (36 μM FeSO4), a typical C. glutamicum wild-type culture reaches a final OD600 of about 60. Further experiments could show that C. glutamicum is also able to extract heme iron from heme proteins such as hemoglobin (data not shown). In control experiments with CGXII medium lacking an iron source, cells stopped growing after approximately 1 to 2 duplications at an OD600 of about 3. This clearly demonstrated that growth on hemin (Fig. 1D) is not just a result of consumption of stored iron from the FeSO4 preculture but that C. glutamicum is able to extract heme iron when hemin is added as the sole iron source.
FIG. 1.
Growth of the C. glutamicum ATCC 13032 wild-type (WT) strain and Δhmu, ΔhrrA, and ΔhmuO deletion mutants on agar plates (A and B) or in liquid culture (C and D). For panels A and B, serial dilutions of the indicated strains were spotted on CGXII minimal medium plates containing either 2.5 μM FeSO4 (A) or 2.5 μM hemin (B) as the iron source. The plates were incubated at 30°C for 48 h. For panels C and D, the C. glutamicum wild-type strain (filled squares) and ΔhrrA (triangles), ΔhmuO (diamonds), and Δhmu (circles) mutants were cultivated in CGXII minimal medium with 4% (wt/vol) glucose and either 2.5 μM FeSO4 (C) or 2.5 μM hemin (D) as the iron source. When CGXII minimal medium without an iron source was inoculated with wild-type cells, growth stopped at an OD600 of about 3 (data not shown).
Influence of heme on global gene expression.
To identify genes of C. glutamicum necessary for heme utilization, we compared the transcriptomes of wild-type cells grown with either heme or FeSO4 as the iron source. Table 2 shows a selection of genes upregulated in heme-grown cells (also see Table S2 in the supplemental material). Among the genes showing the highest upregulation on heme was the putative hemin transport operon cg0466-cg0469, including the putative hemin ABC transporter hmuTUV and the gene encoding the secreted hemin-binding protein HtaA. The genes htaB (cg0470) and htaC (cg0471), located downstream of the hmu operon, and htaD (cg3156) encode putative secreted hemin-binding proteins and also showed a significant upregulation in heme-grown cells. The mRNA level of hmuO encoding heme oxygenase, an enzyme necessary for the release of iron from the tetrapyrrol ring, was increased more than 10-fold on heme. In recent studies, the ABC transporter HrtAB was shown to be required for dealing with toxic heme concentrations and a role in heme efflux was postulated in several Gram-positive species (2). The orthologous genes hrtA and hrtB of C. glutamicum showed a high upregulation in heme-grown cells, consistent with a similar function of this system in C. glutamicum. The only downregulated gene having a function related to heme metabolism is hemH encoding ferrochelatase, a protein involved in protoheme IX biosynthesis. Other genes of heme biosynthesis enzymes show no significantly changed mRNA ratio.
TABLE 2.
Genes differentially expressed in the C. glutamicum wild type grown with heme instead of iron sulfate as the sole iron sourcea
| Gene ID | Gene name | Annotation | Heme/ Fe ratiob |
|---|---|---|---|
| cg0465 | Hypothetical protein | 8.01 | |
| cg0466 | htaA | Secreted heme transport-associated protein | 8.66 |
| cg0467 | hmuT | Hemin-binding periplasmic protein precursor | 7.67 |
| cg0468 | hmuU | Hemin transport system, permease protein | 6.43 |
| cg0469 | hmuV | Hemin transport system, ATP-binding protein | 5.55 |
| cg0470 | htaB | Secreted heme transport-associated protein | 12.37 |
| cg0471 | htaC | Secreted heme transport-associated protein | 5.71 |
| cg1734 | hemH | Ferrochelatase precursor | 0.52 |
| cg2202 | hrtB | ABC-type transport system, permease component | 3.45 |
| cg2204 | hrtA | ABC-type transport system, ATPase component | 4.97 |
| cg2444 | Hypothetical protein | 3.15 | |
| cg2445 | hmuO | Heme oxygenase | 10.88 |
| cg3156 | htaD | Secreted heme transport-associated protein | 18.24 |
This table shows genes with a putative heme-related function, such as heme uptake, utilization, or biosynthesis. A list of all genes with a ≥2-fold-altered mRNA level is provided in Table S2 in the supplemental material.
Average mRNA ratio of genes showing a ≥2-fold-altered mRNA level (heme/Fe) of C. glutamicum cells grown in CGXII minimal medium with 2.5 μM hemin or 2.5 μM FeSO4 as the iron source. The mRNA ratios represent average values from three independent DNA microarray experiments (P ≤ 0.05).
Role of hmu genes for heme utilization.
Deletion of the hmu operon (cg0466-cg0469; htaA, hmuT, hmuV, and hmuU) encoding a putative hemin ABC transporter resulted in a slight but significant growth defect when hemin was supplied as the iron source (Fig. 1B and D). Both the growth rate and the final OD600 were reduced compared to those of the wild type in heme-containing media. A mutant with deletion of hmuO encoding heme oxygenase was significantly impaired when grown on hemin (Fig. 1B and D). These findings support a role for the Hmu ABC transporter in heme uptake and highlight the importance of heme oxygenase in recycling of heme iron, which apparently is also of relevance in FeSO4-grown cells facing iron starvation. A C. glutamicum mutant lacking htaB and htaC in addition to the hmu operon showed the same growth phenotype as the Δhmu mutant (data not shown).
Growth phenotype of an hrrA deletion mutant.
In order to assess the function of the response regulator HrrA in iron-dependent gene regulation, an hrrA deletion mutant was constructed. In liquid culture, this ΔhrrA mutant showed a similar growth phenotype to the Δhmu mutant (Fig. 1C): i.e., a reduced growth rate and final OD600 in heme-containing medium. However, when spotted on hemin plates, the ΔhrrA mutant showed an even stronger growth delay than the mutant lacking heme oxygenase and was also impaired when FeSO4 was added as the iron source (Fig. 1A and B). These results suggest a key function of HrrA in the control of heme utilization and metabolism.
Transcriptome analysis of a ΔhrrA mutant.
To identify the regulon of the response regulator HrrA, the transcriptomes of the ΔhrrA mutant and the wild type grown in CGXII minimal medium with either FeSO4 or hemin were compared. Genes showing a more than 2-fold-altered mRNA level in the ΔhrrA mutant in FeSO4- or heme-grown cells are summarized in Table S3 in the supplemental material. In Table 3, all genes with a putative function in heme metabolism are shown. Generally, the microarray data indicated that the ΔhrrA mutant shows a stronger iron starvation response than the wild type when cultivated with heme as the iron source. Several genes belonging to the DtxR regulon showed higher mRNA levels in the mutant strain, whereas genes of the RipA regulon coding for iron-containing proteins (e.g., succinate dehydrogenase or nitrate reductase) showed lower mRNA levels in the ΔhrrA mutant (see Table S3 in the supplemental material). This is most likely a result of the downregulation of genes involved in heme utilization, such as heme oxygenase. The corresponding hmuO gene, despite being a target of DtxR repression, showed a 20-fold-decreased mRNA level in the ΔhrrA mutant compared to the wild type when the cells were grown with heme as the iron source, whereas the hmuO mRNA level was 10-fold decreased on FeSO4. Similar to hmuO, the mRNA levels of the hmu operon, htaA, and htaD, all of which are targets of DtxR, also are lowered in the ΔhrrA mutant. In contrast, the expression of htaB (cg0470) and htaC (cg0471), coding for putative heme binding proteins which are both DtxR targets, also showed an upregulation which could be explained by the increased iron starvation response in the mutant. Keeping in mind that the mRNA level of other DtxR targets was increased in the ΔhrrA mutant strain (see Table S3 in the supplemental material), this result supported our assumption that the HrrSA system is involved in heme-dependent activation of hmuO expression and might also be involved in the heme-dependent regulation of heme uptake systems.
TABLE 3.
Comparative transcriptome analysis of C. glutamicum WT and the ΔhrrA mutanta
| Gene ID | Gene name | Annotation | ΔhrrA/wild-type mRNA ratiob |
|
|---|---|---|---|---|
| Heme | FeSO4 | |||
| Two-component systems | ||||
| cg3247 | hrrA | Two-component system, response regulator | 0.02 | 0.02 |
| cg2200 | cgtR8 | Two-component system, response regulator | 1.79 | 2.13 |
| cg2201 | cgtS8 | Two-component system, signal transduction histidine kinase | 1.99 | 2.42 |
| Heme transport/utilization | ||||
| cg0466 | htaA | Secreted heme transport-associated protein | 0.46 | 1.02* |
| cg0467 | hmuT | Hemin-binding periplasmic protein precursor | 0.57 | 1.03* |
| cg0468 | hmuU | Hemin transport system, permease protein | 0.60 | 1.04* |
| cg0469 | hmuV | Hemin transport system, ATP-binding protein | 0.86* | 1.24* |
| cg0470 | htaB | Secreted heme transport-associated protein | 9.00 | 8.71* |
| cg0471 | htaC | Secreted heme transport-associated protein | 8.06 | 5.09 |
| cg2202 | hrtB | ABC-type transport system, permease component | 10.90 | 40.79 |
| cg2204 | hrtA | ABC-type transport system, ATPase component | 10.47 | 43.93 |
| cg2445 | hmuO | Heme oxygenase | 0.06 | 0.12 |
| cg3156 | htaD | Secreted heme transport-associated protein | 0.26 | 0.63* |
| Heme-containing proteins | ||||
| cg0645 | cytP | Cytochrome P450 | 0.20 | 0.32 |
| cg2403 | qcrB | Cytochrome b, membrane protein | 0.35 | 0.47 |
| cg2404 | qcrA | Rieske iron-sulfur protein | 0.36 | 0.47 |
| cg2405 | qcrC | Cytochrome c1 | 0.36 | 0.48 |
| cg2406 | ctaE | Cytochrome aa3 oxidase, subunit 3 | 0.34 | 0.46 |
| cg2780 | ctaD | Cytochrome aa3 oxidase, subunit 1 | 0.51 | 0.62 |
| cg3141 | hmp | Flavohemoprotein | 2.74 | 1.95 |
| cg3369 | Rieske-type iron-sulfur protein | 0.57 | 0.75* | |
| Heme biosynthesis | ||||
| cg0497 | hemA | Glutamyl-tRNA reductase | 1.77* | 1.44* |
| cg0498 | hemC | Porphobilinogen deaminase | 1.62* | 1.19* |
| cg0516 | hemE | Uroporphyrinogen decarboxylase | 1.87 | 1.99 |
| cg0517 | hemY | Protoporphyrinogen oxidase | 1.90* | 2.04 |
| cg0518 | hemL | Glutamate-1-semialdehyde 2,1-aminomutase | 1.77 | 2.10 |
| cg0519 | Putative phosphoglycerate mutase | 1.59* | 1.74 | |
| cg0520 | Secreted thiol-disulfide isomerase or thioredoxin | 1.51* | 1.70 | |
| cg0522 | ccsA | Cytochrome c biogenesis protein membrane protein | 1.34* | 1.53 |
| cg0523 | Membrane protein required for cytochrome c biosynthesis | 1.37* | 1.75 | |
| cg0524 | ccsB | Cytochrome c assembly membrane protein | 1.39* | 1.90 |
| cg1734 | hemH | Ferrochelatase precursor | 2.41* | 2.20 |
This table includes all genes with a putative heme/iron-related function that are differentially expressed in the ΔhrrA mutant. A list of all genes with a ≥2-fold-altered mRNA level is provided in Table S2 in the supplemental material. Genes and operons shown to be directly regulated by HrrA have all been included in this table, irrespective of their relative expression.
Average ratio of genes showing a ≥2-fold-altered mRNA level (ΔhrrA/wild-type mRNA ratio) of C. glutamicum cells grown on CGXII minimal medium with 2.5 μM heme or 2.5 μM FeSO4 as the iron source. The mRNA ratios represent average values from three independent DNA microarray experiments (P ≤ 0.05). *, P > 0.05.
Two genes belonging neither to the DtxR regulon nor to the typical iron starvation response are hrtA and hrtB, whose mRNA level was significantly increased in the ΔhrrA mutant. In several Gram-positive species, these genes are activated in a heme-dependent manner by a two-component system located in close vicinity to hrtAB. In fact, in C. glutamicum the genes encoding the two-component system CgtSR8 are found nearby and, interestingly, both genes show a 2-fold upregulation in the ΔhrrA mutant. Thus, the altered mRNA level of hrtAB might not be a direct effect of the hrrA deletion but rather an indirect effect caused by the altered mRNA level of cgtSR8.
Besides hmuO, several other genes involved in heme metabolism were differentially expressed in the ΔhrrA mutant. Almost all genes known to be involved in heme biosynthesis showed a ca. 2-fold-increased mRNA level in the mutant, during both growth with heme and that with FeSO4. Among those are the genes encoding ferrochelatase (hemH), glutamyl-tRNA reductase (hemA), and uroporphyrinogen decarboxylase (hemE), which are the first genes of the heme biosynthesis operons hemA-hemC and hemEYL-cg0519-cg0520-ccsA-cg0523-ccsB, respectively (see also Fig. 4) (for operon prediction, see http://coryneregnet.cebitec.uni-bielefeld.de) (7). Even so, these genes are more or less equally upregulated in heme- and FeSO4-grown cells; these data reveal that HrrA is involved in the repression of the heme biosynthesis machinery in C. glutamicum.
Another group of genes related to heme metabolism which showed an altered expression in the ΔhrrA mutant were those coding for subunits I and III of cytochrome aa3 oxidase (ctaD and ctaE, respectively) and the three subunits of the cytochrome bc1 complex (qcrCAB), which presumably form an operon with ctaE (31). In the ΔhrrA mutant, the ctaE-qcrCAB and ctaD mRNA levels showed a stronger decrease in heme-grown cells (2- to 3-fold) than in cells cultivated with FeSO4 as the iron source (1.5- to 2-fold). These data suggest that expression of these genes might be positively regulated by the HrrSA system in the presence of heme in a direct or indirect manner. The cytochrome P450 and the Rieske-type iron-sulfur protein Cg3369 showed a similar expression pattern to that described for ctaE-qcrCAB and ctaD. Only the gene hmp encoding a flavohemoglobin showed an increased mRNA level in the ΔhrrA mutant. According to the results described above, HrrA might carry out a dual function in C. glutamicum by acting as an activator of genes for heme oxygenase and heme-containing proteins and as a repressor of genes involved in heme biosynthesis.
Phosphorylation of HrrA.
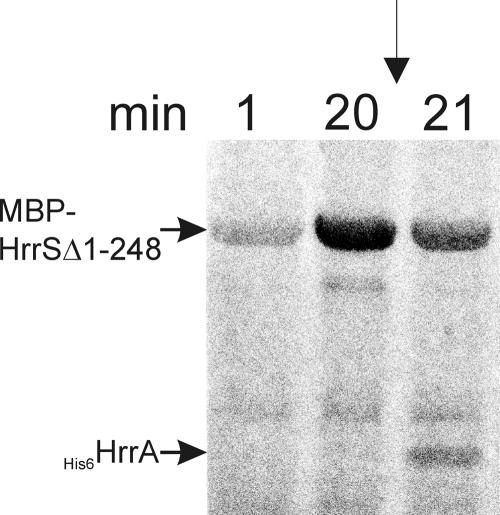
In nearly all described cases, the response regulator of a two-component system is activated by phosphorylation of a conserved aspartate residue in the receiver domain. To test the phosphorylation activities of the HrrSA system, the kinase domain of HrrS (MBP-HrrSΔ1-248) and the full-length HrrA were overexpressed in E. coli and purified via affinity chromatography. To monitor autophosphorylation of MBP-HrrSΔ1-248, the protein was incubated with [γ-33P]ATP and samples were taken at different time points and analyzed via SDS-PAGE and autoradiography. As shown in Fig. 2, MBP-HrrSΔ1-248 showed significant autophosphorylation after approximately 20 min. When HrrA was added, an efficient transfer of the phosphoryl group occurred and was detectable already after 1 min (Fig. 2). The two bands below MBP-HrrSΔ1-248 are most likely cleavage products which are often observed in the case of MBP fusion proteins.
FIG. 2.
Autophosphorylation of the kinase domain MBP-HrrSΔ1-248 and phosphoryl group transfer to the response regulator HrrA. Purified MBP-HrrSΔ1-248 (12 μM final concentration) was incubated with [γ-33P]ATP for 20 min (A). Then the purified response regulators HrrA was added (12 μM final concentration), resulting in a 2-fold dilution of MBP-HrrSΔ1-248 and the ATP. The samples were incubated at room temperature for a further 60 min. At the indicated time points, samples were taken, mixed with SDS loading buffer, and stored on ice (see Materials and Methods). Finally, these samples were separated by SDS-PAGE and the dried gels were analyzed with a phosphorimager.
Identification of direct target genes of HrrA.
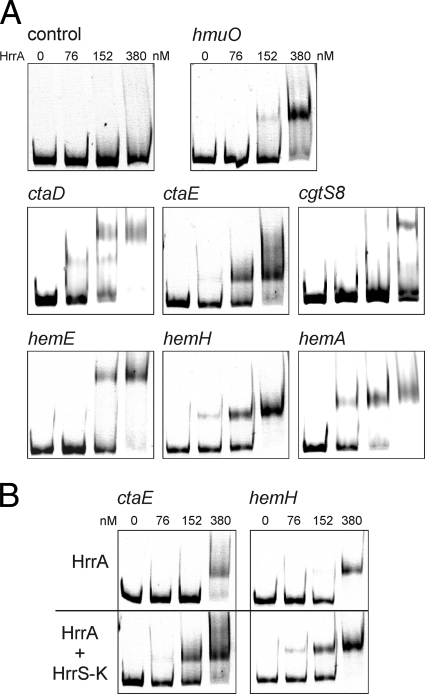
In order to test whether some of the genes showing an altered mRNA level in the ΔhrrA mutant are direct target genes of this response regulator, binding of purified HrrA protein to the promoter regions of selected genes with a putative heme-related function was tested in gel shift assays. As shown in Fig. 3 A, HrrA was able to bind to the promoter regions of hmuO, hemA, hemE, hemH, ctaD, and ctaE. Depending on the DNA fragment tested, a complete shift was observed at a 20- to 40-fold molar excess (300 to 600 nM) of HrrA. No binding was observed when HrrA was incubated with the promoter regions of other genes which showed an altered mRNA level in the ΔhrrA mutant, including htaA, htaB, and htaD (putative heme binding proteins), cg0640 (ferredoxin), and cytP (cytochrome P450), or with the promoter regions of genes (sdhC, narK, and pck) that served as a negative control. A weak but significant binding was also observed with the promoter region of cgtS8. However, further experiments have to verify whether HrrA directly regulates expression of the cgtSR8 two-component system (Fig. 3). All gel shifts shown in Fig. 3A were carried out with HrrA protein that had been phosphorylated by preincubation with ATP and the purified HrrS kinase domain (MBP-HrrSΔ1-248), as described in Materials and Methods. We decided to do so since we observed an approximately 2-fold-increased binding affinity of the phosphorylated HrrA protein in comparison to that of unphosphorylated HrrA for the DNA fragments tested (Fig. 3B). Figure 3B shows two examples: ctaE, which is presumably activated by HrrA; and hemH, which is presumably repressed by HrrA. In both cases, the difference is not striking but significant and was also observed with the other HrrA target promoters. The comparatively weak effect might be due to an incomplete phosphorylation of HrrA upon incubation with MBP-HrrSΔ1-248. Nevertheless, in vivo small differences in the binding affinity of a transcriptional regulator may already significantly alter gene expression.
FIG. 3.
Identification of direct target genes of HrrA in C. glutamicum. (A) DNA fragments (500 bp) covering the promoter region of hmuO, ctaE, cgtS8, hemE, hemH, and hemA were incubated without or with a 5, 10, and 25 molar excess of (partially) phosphorylated HrrA protein (0 to 380 nM), as described in Materials and Methods. After incubation, samples were separated on a 10% nondenaturating polyacrylamide gel and stained with Sybr green I. DNA fragments covering the promoter region of cytP or pck served as a negative control. (B) DNA fragments containing the promoter region of a gene postulated to be activated by HrrA (ctaE) and one repressed by HrrA (hemH) were incubated with equal amounts of unphosphorylated HrrA (upper gels) or HrrA that had been phosphorylated by preincubation with MBP-HrrSΔ1-248 (HrrS-K) and ATP. These examples clearly demonstrate increased binding affinity of the phosphorylated response regulator to its target promoters.
DISCUSSION
Most organisms have evolved complex regulatory networks ensuring an adequate supply of iron, which is essential for central metabolic processes, such as the tricarboxylic acid (TCA) cycle, respiration, and DNA biosynthesis. Especially in pathogenic bacteria, several heme-responsive regulatory systems have been described consisting of classic transcriptional regulators (30, 33, 53), two-component systems (5, 20, 34, 42, 43), and extracytoplasmic sigma factors (22, 24, 48). In this work, we investigated the role of the HrrSA two-component system in the regulation of iron homeostasis in the Gram-positive soil bacterium C. glutamicum. Recent studies on the homologous system HrrSA in C. diphtheriae provided detailed insights into the heme-dependent regulation of hmuO and hemA, which were both postulated to be direct targets of the response regulator HrrA (4, 5, 39). We used DNA microarray analysis of a ΔhrrA deletion mutant and DNA-protein interaction studies with purified HrrA protein to demonstrate a direct regulation of at least 20 genes encoding proteins involved in heme metabolism in C. glutamicum. Based on our results, it can be deduced that the response regulator HrrA directly activates expression of the hmuO gene coding for heme oxygenase, an enzyme catalyzing the degradation of the tetrapyrrole ring to release iron. Furthermore, HrrA acts as a heme-dependent activator of ctaD and the putative operon ctaE-qcrBAC. These genes encode subunits I (CtaD) and III (CtaE) of cytochrome aa3 oxidase and the three subunits of the cytochrome bc1 complex (QcrCAB). Previous studies revealed that these two complexes form a supercomplex in C. glutamicum (32). Each of the proteins CtaD, QcrB, and QcrC contains two heme moieties (heme a and a3 in the case of CtaD, two heme b moieties in the case of QcrB, and two heme c moieties in the case of QcrC). Thus, HrrSA not only is involved in the release of heme iron by the activation of heme oxygenase but also activates expression of heme-containing protein complexes of the respiratory chain when heme is available. A regulation of heme-containing proteins has not yet been described for the C. diphtheriae HrrSA system; however, repression of the gene hemA encoding glutamyl-tRNA reductase, the first enzyme of heme biosynthesis, was recently reported (5). Besides the hemA-hemC operon, our results provide evidence that HrrA, in fact, is involved in the repression of almost the whole set of genes involved in heme biosynthesis in C. glutamicum (except hemB and hemD). Based on these data, it can be assumed that HrrSA downregulates energy-demanding heme biosynthesis in C. glutamicum when heme can be acquired by the uptake of extracellular heme sources.
Interestingly, transcriptome analysis and in vitro gel shift experiments suggested direct repression of the two-component system CgtSR8 by HrrA. In a number of distinct Gram-positive bacteria, a microsynteny is observed at this genomic locus (43): two genes encoding a prototypical two-component system are located in close vicinity to hrtAB encoding an ABC transporter that was supposed to be required for coping with high, toxic hemin concentrations (42-44). The two-component systems encoded by these genes, HssRS in Staphylococcus aureus and Bacillus anthracis and ChrSA in C. diphtheriae, were recently shown to be involved in the activation of hrtAB expression in the presence of heme (6, 42, 44). Sequence analysis revealed a high sequence identity of CgtSR8 to the ChrSA and HrrSA systems of C. diphtheriae: thus, a similar function of CgtSR8 in control of hrtAB transcription (cg2204 and cg2202) and potentially other heme-related targets can be envisaged. Since the potential HrrA binding site seems to be located in the intergenic region of cgtS8-hrtA, a direct regulation of hrtAB by HrrA cannot be excluded based on our data. However, the significance of HrrA binding to this region needs to be confirmed by additional studies as our gel shift experiments revealed a comparably low binding affinity of HrrA to the corresponding fragment. Further experiments will focus on the investigation how C. glutamicum HrrSA and CgtSR8 are involved in transcription control of their own promoters as well as hrtAB in response to heme availability.
As expected for a heme-dependent two-component system, our transcriptome studies with a ΔhrrA deletion mutant revealed a stronger effect on the mRNA level of hmuO, ctaD, and ctaE-qcrCAB in heme-grown cells than in cells cultivated on FeSO4, when the system is supposed to be less active (Table 3). This was not the case for the mRNA ratio observed for heme biosynthesis genes or cgtSR8, which are also supposed to be directly regulated by HrrSA, but show rather equally altered mRNA levels in both experiments. In C. diphtheriae, hmuO and hemA expression is regulated by intensive cross talk of the two-component systems HrrSA and ChrSA (5). In C. glutamicum, a putative cross talk with other systems, such as CgtSR8, might also play an important role in heme-dependent regulation of some target genes and might partially compensate for hrrSA deletion.
Although the C. diphtheriae and C. glutamicum HrrSA proteins show high similarity with regard to sequence identity and function, a striking difference between the systems is that in C. glutamicum hrrA expression is repressed by the global iron regulator DtxR, which also downregulates expression of the hmu operon, htaB, htaC, and htaD (49) (Fig. 4). In contrast, neither hrrSA nor chrSA in C. diphtheriae was reported to be controlled by DtxR and inspection of the corresponding promoter regions failed to reveal the presence of a DtxR binding motif.
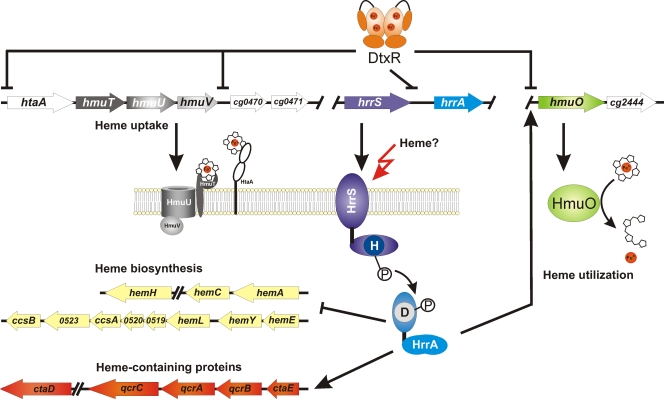
FIG. 4.
Model for the regulation of heme homeostasis by the two-component system HrrSA in C. glutamicum. Under conditions of sufficient iron supply, the global iron regulator DtxR represses expression of the response regulator hrrA, heme uptake systems (htaA and hmuTUV), and heme oxygenase (hmuO), an enzyme involved in the release of heme iron. Under iron limitation, expression of hrrA increases. In this study, we could show that the two-component system HrrSA of C. glutamicum is involved in the heme-dependent activation of hmuO and genes encoding heme-containing protein complexes of the respiratory chain. Furthermore, HrrA directly represses transcription of genes coding for heme biosynthesis enzymes (for operon prediction, see http://coryneregnet.cebitec.uni-bielefeld.de). The signal sensed by the sensor kinase HrrS is not yet known, but heme itself or a heme-related metabolite is likely. Not included in this model is the postulated repression of the CgtSR8 two-component system by HrrSA, which is currently under investigation and might disclose a further level of complexity in this regulatory network.
Especially for organisms with a pathogenic lifestyle, heme and heme-containing proteins such as hemoglobin represent important iron sources during host colonization. In this work, it was demonstrated for the first time that the nonpathogenic soil bacterium C. glutamicum is able to use hemin and hemoglobin (data not shown) as the sole iron source, thereby reaching the same growth rate and final OD600 as cells cultivated on an equivalent concentration of FeSO4. Among the genes upregulated in heme-grown C. glutamicum cells was the hmu operon encoding a putative ABC hemin transporter, HmuTUV (17). The system shows striking similarity to the HmuTUV transporter of C. diphtheriae, which was shown to be important for the utilization of heme or hemoglobin as the iron source (15, 26). Recently, further hemin binding proteins (HtaA and HtaB) were identified that are involved in heme uptake of C. diphtheriae (1). Our DNA microarray analysis showed an upregulation of four putative heme-transport associated protein genes (hta-like), namely htaA (cg0466), htaB (cg0470), htaC (cg0471), and htaD (cg3156). Common features of Hta-like proteins are an N-terminal signal peptide, one or two HtaA domains (PF04213; two in the case of Cg0466), and a putative transmembrane helix at their C terminus. These typical structural features were found in C. glutamicum HtaB and HtaC as well as C. diphtheriae HtaA and HtaB or the heme binding proteins Shr and Shp of Streptococcus pyogenes (3, 28). In the case of C. glutamicum HtaA, a C-terminal transmembrane helix is predicted; none is present in HtaD. Based on the studies with C. diphtheriae, we can assume that these Hta proteins act as membrane-anchored or -associated hemin receptors which directly scavenge hemin from the environment and transfer it to the HmuT lipoprotein. HmuT then passes hemin to the permease HmuU, which together with the ATPase HmuV, catalyzes hemin uptake. Surprisingly, a C. glutamicum mutant missing the whole hmu operon and the genes htaB and htaC was still able to grow on hemin as the iron source (Fig. 1). However, for many organisms, including C. diphtheriae, S. pyogenes, and Yersinia enterolytica, it has been reported that mutations in the hemin transport systems did not result in a complete loss of the ability to use heme as the iron source (1, 3, 46). This indicates the existence of alternative heme uptake possibilities that can compensate for the absence of the Hmu ABC transporter to some extent. Besides heme uptake, heme oxygenase is required to make use of heme iron, and our analysis of a ΔhmuO mutant confirmed that this enzyme is important for utilization of exogeneous heme, but probably also for recycling of heme synthesized by C. glutamicum itself.
So far, several systems have been described which control expression of genes involved in heme utilization dependent on heme availability. Based on our current model, the response regulator HrrA of C. glutamicum functions as an activator of genes involved in heme utilization (heme oxygenase) and of genes encoding heme-containing components of the respiratory chain. On the other hand, HrrA represses transcription of heme biosynthesis genes (Fig. 4). Altogether, the investigation of the HrrA regulon adds to our understanding of the hierarchical regulatory network controlling iron homeostasis in C. glutamicum and highlights the importance of heme-dependent gene regulation in nonpathogenic bacteria.
Supplementary Material
Acknowledgments
We thank Tino Polen for great support of DNA microarray handling and analysis.
Footnotes
Published ahead of print on 7 January 2011.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allen, C. E., and M. P. Schmitt. 2009. HtaA is an iron-regulated hemin binding protein involved in the utilization of heme iron in Corynebacterium diphtheriae. J. Bacteriol. 191:2638-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anzaldi, L. L., and E. P. Skaar. 2010. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect. Immun. 78:4977-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates, C. S., G. E. Montanez, C. R. Woods, R. M. Vincent, and Z. Eichenbaum. 2003. Identification and characterization of a Streptococcus pyogenes operon involved in binding of hemoproteins and acquisition of iron. Infect. Immun. 71:1042-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibb, L. A., N. D. King, C. A. Kunkle, and M. P. Schmitt. 2005. Analysis of a heme-dependent signal transduction system in Corynebacterium diphtheriae: deletion of the chrAS genes results in heme sensitivity and diminished heme-dependent activation of the hmuO promoter. Infect. Immun. 73:7406-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibb, L. A., C. A. Kunkle, and M. P. Schmitt. 2007. The ChrA-ChrS and HrrA-HrrS signal transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae. Infect. Immun. 75:2421-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibb, L. A., and M. P. Schmitt. 2010. The ABC transporter HrtAB confers resistance to hemin toxicity and is regulated in a hemin-dependent manner by the ChrAS two-component system in Corynebacterium diphtheriae. J. Bacteriol. 192:4606-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bott, M., and A. Niebisch. 2003. The respiratory chain of Corynebacterium glutamicum. J. Biotechnol. 104:129-153. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, J., M. N. Oza, and J. R. Murphy. 1990. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. U. S. A. 87:5968-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brazma, A., et al. 2001. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29:365-371. [DOI] [PubMed] [Google Scholar]
- 10.Brocker, M., and M. Bott. 2006. Evidence for activator and repressor functions of the response regulator MtrA from Corynebacterium glutamicum. FEMS Microbiol. Lett. 264:205-212. [DOI] [PubMed] [Google Scholar]
- 11.Brocker, M., S. Schaffer, C. Mack, and M. Bott. 2009. Citrate utilization by Corynebacterium glutamicum is controlled by the CitAB two-component system through positive regulation of the citrate transport genes citH and tctCBA. J. Bacteriol. 191:3869-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brune, I., et al. 2006. The DtxR protein acting as dual transcriptional regulator directs a global regulatory network involved in iron metabolism of Corynebacterium glutamicum. BMC Genomics 7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burkovski, A. 2008. Corynebacteria: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 14.D'Aquino, J. A., J. Tetenbaum-Novatt, A. White, F. Berkovitch, and D. Ringe. 2005. Mechanism of metal ion activation of the diphtheria toxin repressor DtxR. Proc. Natl. Acad. Sci. U. S. A. 102:18408-18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drazek, E. S., C. A. Hammack, and M. P. Schmitt. 2000. Corynebacterium diphtheriae genes required for acquisition of iron from haemin and haemoglobin are homologous to ABC haemin transporters. Mol. Microbiol. 36:68-84. [DOI] [PubMed] [Google Scholar]
- 16.Eggeling, L., and M. Bott. 2005. Handbook of Corynebacterium glutamicum. Academic Press, Inc., Boca Raton, FL.
- 17.Frunzke, J., and M. Bott. 2008. Regulation of iron homeostasis in Corynebacterium glutamicum, p. 241-266. In A. Burkovski (ed.), Corynebacteria: genomics and molecular biology. Horizon Scientific Press, Norwich, United Kingdom.
- 18.Frunzke, J., V. Engels, S. Hasenbein, C. Gätgens, and M. Bott. 2008. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol. Microbiol. 67:305-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 20.Ito, Y., et al. 2009. Heme-dependent autophosphorylation of a heme sensor kinase, ChrS, from Corynebacterium diphtheriae reconstituted in proteoliposomes. FEBS Lett. 583:2244-2248. [DOI] [PubMed] [Google Scholar]
- 21.Keilhauer, C., L. Eggeling, and H. Sahm. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175:5595-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King, N. D., A. E. Kirby, and T. D. Connell. 2005. Transcriptional control of the rhuIR-bhuRSTUV heme acquisition locus in Bordetella avium. Infect. Immun. 73:1613-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinoshita, S., S. Udaka, and M. Shimono. 1957. Studies on the amino acid fermentation. I. Production of L-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 3:193-205. [PubMed] [Google Scholar]
- 24.Kirby, A. E., N. D. King, and T. D. Connell. 2004. RhuR, an extracytoplasmic function sigma factor activator, is essential for heme-dependent expression of the outer membrane heme and hemoprotein receptor of Bordetella avium. Infect. Immun. 72:896-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kocan, M., et al. 2006. Two-component systems of Corynebacterium glutamicum: deletion analysis and involvement of the PhoS-PhoR system in the phosphate starvation response. J. Bacteriol. 188:724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunkle, C. A., and M. P. Schmitt. 2003. Analysis of the Corynebacterium diphtheriae DtxR regulon: identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J. Bacteriol. 185:6826-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laub, M. T., and M. Goulian. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41:121-145. [DOI] [PubMed] [Google Scholar]
- 28.Lei, B., et al. 2002. Identification and characterization of a novel heme-associated cell surface protein made by Streptococcus pyogenes. Infect. Immun. 70:4494-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Möker, N., et al. 2004. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 54:420-438. [DOI] [PubMed] [Google Scholar]
- 30.Mourino, S., C. R. Osorio, M. L. Lemos, and J. H. Crosa. 2006. Transcriptional organization and regulation of the Vibrio anguillarum heme uptake gene cluster. Gene 374:68-76. [DOI] [PubMed] [Google Scholar]
- 31.Niebisch, A., and M. Bott. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 175:282-294. [DOI] [PubMed] [Google Scholar]
- 32.Niebisch, A., and M. Bott. 2003. Purification of a cytochrome bc-aa3 supercomplex with quinol oxidase activity from Corynebacterium glutamicum. Identification of a fourth subunit of cytochrome aa3 oxidase and mutational analysis of diheme cytochrome c1. J. Biol. Chem. 278:4339-4346. [DOI] [PubMed] [Google Scholar]
- 33.Ochsner, U. A., Z. Johnson, and M. L. Vasil. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146:185-198. [DOI] [PubMed] [Google Scholar]
- 34.Ortiz de Orue Lucana, D., and M. R. Groves. 2009. The three-component signalling system HbpS-SenS-SenR as an example of a redox sensing pathway in bacteria. Amino Acids 37:479-486. [DOI] [PubMed] [Google Scholar]
- 35.Polen, T., and V. F. Wendisch. 2004. Genomewide expression analysis in amino acid-producing bacteria using DNA microarrays. Appl. Biochem. Biotechnol. 118:215-232. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., P. MacCallum, and D. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Schaaf, S., and M. Bott. 2007. Target genes and DNA-binding sites of the response regulator PhoR from Corynebacterium glutamicum. J. Bacteriol. 189:5002-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schäfer, A., et al. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt, M. P. 1999. Identification of a two-component signal transduction system from Corynebacterium diphtheriae that activates gene expression in response to the presence of heme and hemoglobin. J. Bacteriol. 181:5330-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt, M. P. 1997. Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infect. Immun. 65:4634-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweitzer, J. E., M. Stolz, R. Diesveld, H. Etterich, and L. Eggeling. 2009. The serine hydroxymethyltransferase gene glyA in Corynebacterium glutamicum is controlled by GlyR. J. Biotechnol. 139:214-221. [DOI] [PubMed] [Google Scholar]
- 42.Stauff, D. L., and E. P. Skaar. 2009. Bacillus anthracis HssRS signaling to HrtAB regulates heme resistance during infection. Mol. Microbiol. 72:763-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stauff, D. L., and E. P. Skaar. 2009. The heme sensor system of Staphylococcus aureus. Contrib. Microbiol. 16:120-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stauff, D. L., V. J. Torres, and E. P. Skaar. 2007. Signaling and DNA-binding activities of the Staphylococcus aureus HssR-HssS two-component system required for heme sensing. J. Biol. Chem. 282:26111-26121. [DOI] [PubMed] [Google Scholar]
- 45.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 46.Stojiljkovic, I., and K. Hantke. 1994. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13:719-732. [DOI] [PubMed] [Google Scholar]
- 47.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 48.Vanderpool, C. K., and S. K. Armstrong. 2004. Integration of environmental signals controls expression of Bordetella heme utilization genes. J. Bacteriol. 186:938-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wennerhold, J., and M. Bott. 2006. The DtxR regulon of Corynebacterium glutamicum. J. Bacteriol. 188:2907-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wennerhold, J., A. Krug, and M. Bott. 2005. The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J. Biol. Chem. 280:40500-40508. [DOI] [PubMed] [Google Scholar]
- 51.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 52.Wilks, A., and M. P. Schmitt. 1998. Expression and characterization of a heme oxygenase (HmuO) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. J. Biol. Chem. 273:837-841. [DOI] [PubMed] [Google Scholar]
- 53.Yang, J., et al. 2006. Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol. Microbiol. 60:427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.