Abstract
Lipopolysaccharides (LPS) are an important class of macromolecules that are components of the outer membrane of Gram-negative bacteria such as Pseudomonas aeruginosa. P. aeruginosa contains two different sugar chains, the homopolymer common antigen (A band) and the heteropolymer O antigen (B band), which impart serospecificity. The characteristics of LPS are generally assessed after isolation rather than in the context of whole bacteria. Here we used atomic force microscopy (AFM) to probe the physical properties of the LPS of P. aeruginosa strain PA103 (serogroup O11) in situ. This strain contains a mixture of long and very long polymers of O antigen, regulated by two different genes. For this analysis, we studied the wild-type strain and four mutants, ΔWzz1 (producing only very long LPS), ΔWzz2 (producing only long LPS), DΔM (with both the wzz1 and wzz2 genes deleted), and Wzy::GM (producing an LPS core oligosaccharide plus one unit of O antigen). Forces of adhesion between the LPS on these strains and the silicon nitride AFM tip were measured, and the Alexander and de Gennes model of steric repulsion between a flat surface and a polymer brush was used to calculate the LPS layer thickness (which we refer to as length), compressibility, and spacing between the individual molecules. LPS chains were longest for the wild-type strain and ΔWzz1, at 170.6 and 212.4 nm, respectively, and these values were not statistically significantly different from one another. Wzy::GM and DΔM have reduced LPS lengths, at 34.6 and 37.7 nm, respectively. Adhesion forces were not correlated with LPS length, but a relationship between adhesion force and bacterial pathogenicity was found in a mouse acute pneumonia model of infection. The adhesion forces with the AFM probe were lower for strains with LPS mutations, suggesting that the wild-type strain is optimized for maximal adhesion. Our research contributes to further understanding of the role of LPS in the adhesion and virulence of P. aeruginosa.
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that can cause several different types of infections in both community and health care settings and is especially problematic for immunocompromised and cystic fibrosis patients, as well as burn victims (23, 32). Surface structures on the bacterium are related to virulence and the ability to cause infections. They make it possible for P. aeruginosa to attach to a wide range of surfaces, as well as to interact with components of the host (20). The lipopolysaccharide (LPS) is an important virulence factor for P. aeruginosa (25). The structure of LPS consists of lipid A, which anchors the LPS to the outer membrane, the core oligosaccharide, and the highly variable O-antigen side chain, which consists of repeating saccharide units and confers serotype specificity to the bacterium. The O antigen protrudes into the environment and allows the bacterium to interact with its surroundings. There are 20 serotypes of P. aeruginosa (27), and 14 of those groups also produce the A-band saccharide known as common antigen, which is composed of d-rhamnose sugar polymer and consists of about 23 repeating units (6, 47).
LPS physical properties such as three-dimensional structure and number of repeating units contribute to bacterial adhesion (3, 37, 40). P. aeruginosa uses complicated biosynthetic mechanisms for assembly of LPS O antigen; this has been described extensively elsewhere (24). In brief, the O-antigen subunits are synthesized in the cytoplasm, attached to undecaprenol phosphate, and then flipped across the inner membrane by the Wzx translocase. Next, the O-antigen side chains are linked together by the Wzy polymerase and the Wzz proteins determine the number of subunits added. In P. aeruginosa strain PA103, the Wzz1 protein regulates the long-chain length and adds more than 30 subunits together, while the Wzz2 protein is responsible for the very long O-antigen chain length, linking more than 60 subunits together (26). The length of the side chain correlates with the length of the coiled-coil regions within the Wzz proteins, as described by Morona et al. (33). Finally, the WaaL ligase moves the completed side chain to lipid A plus the core (lipid A-core), and the whole LPS molecule is transported to the outer membrane (25). The relationships between genes encoding LPS production and the physical characteristics of P. aeruginosa LPS have been difficult to determine, in part due to the lack of a suitable noninvasive technique for measuring the long molecules on these bacteria.
Previous studies have shown that the presence of O antigen is critical for bacterial virulence. O-antigen-deficient mutants appear to be more attenuated than wild-type organisms in mouse burn, corneal, and pneumonia models of P. aeruginosa infections (11, 36, 41). They disseminate less readily from the initial site of infection, are more serum sensitive than wild-type strains (36), and also exhibit decreased adherence to epithelial cells in vitro (41). The addition of O antigen to lipid A-core is, however, not essential for the viability of the organism (25), and indeed, in P. aeruginosa, much more of the lipid A-core remains devoid of O antigen than in other Gram-negative bacteria (45). The mechanism controlling this is not known, but observations have been made that suggest that the presence of complete LPS core molecules without O antigen increases bacterial accumulation in the lung through interactions with the cystic fibrosis transmembrane conductance regulator (35).
The importance of O-antigen chain length regulator proteins for bacterial pathogenicity has also been investigated in P. aeruginosa. In this system, the Wzz1 protein plays a greater role than Wzz2 in serum resistance and also is more important in an mouse acute pneumonia model of infection (26). In the same study, mice infected with a strain having both the wzz1 and wzz2 genes insertionally inactivated survived longer than those given the wild-type strain or either of the individual wzz mutants, suggesting an additive effect of the loss of virulence.
In order to better relate P. aeruginosa LPS physical properties with virulence, we need to be able to characterize the LPS lengths on the bacterium in situ. Atomic force microscopy (AFM) is a technique widely used in microbiology to probe bacterial and fungal cell surface molecules (2, 16, 43, 46). A recent study has shown that measurement of the forces of Listeria monocytogenes adhesion to a silicon nitride AFM probe could be used to discriminate more virulent strains of this bacterium (34). The AFM functions by using a very sharp tip as a probe that can contact molecules on a microbial surface. As the tip is being brought closer to contact with the microbial cell, the tip (typically made of silicon nitride or silicon) will experience repulsive steric interactions with molecules on the surface of the microbe (8). These steric forces have been modeled by Alexander (1) and de Gennes (14) using a formulation originally developed to describe the steric repulsion between surfaces with grafted polymer layers and later adapted to AFM studies (7). By making these calculations, we can determine important physical properties of the polymers that extend from the microbial surface.
The purposes of the present study were to characterize the polymer layers of P. aeruginosa mutants that had LPSs that differed in O-antigen chain length and to determine the adhesion of each strain to a model silicon nitride probe. An advantage of using isogenic strains for this analysis, as opposed to strains of different serotypes, as we have done previously (39), is that it allows us to attribute any observed differences to the physical properties of LPS and not to any other, unrelated, differences between strains or the different O-antigen structures. We demonstrate how AFM experiments and modeling can be used to characterize bacterial LPS, since adhesion strength and LPS physical properties can be important predictors of virulence. Our findings suggest that wild-type strains have optimized expression of O-antigen chains of particular lengths and that any alteration of these lengths may decrease virulence.
MATERIALS AND METHODS
DNA manipulations.
Chromosomal P. aeruginosa DNA was isolated using the Wizard Genome Prep kit (Promega, Madison, WI). Plasmid DNA was isolated using the Qiagen Miniprep kit (Qiagen, Valencia, CA). Restriction enzymes were purchased from New England Biolabs (Ipswich, MA) and used in accordance with the manufacturer's instructions. PCR was performed using the Roche Expand High Fidelity PCR system, and amplicons were purified using the Qiagen PCR or gel purification kit in accordance with the manufacturer's instructions.
Construction of mutagenesis vectors.
Creation of the deletion mutant constructs followed the protocol described by Choi and Schweizer (10). In brief, fragments encompassing regions 500 bp upstream of the start codon and downstream of the stop codon of each wzz gene were PCR amplified from serogroup O11 P. aeruginosa strain PA103 (ATCC 29260). The 1-kb FRT-flanked gentamicin (Gm) cassette from vector pPS586 was also amplified. Tails to the upstream reverse primer and the downstream forward primer for each wzz gene included a tail homologous to the FRT sites in the Gm cassette. The products were purified and used in a second round of PCR to create the deletion mutant construct via overlap extension (10). One-microliter samples of the products, i.e., the upstream region, the downstream region, and the Gm cassette, were amplified together and cloned into Topo2.1 (Invitrogen). DNA sequence analysis revealed that the FRT-flanked Gm cassette had recombined out from the construct, leaving an 85-bp scar between the designed up- and downstream regions of each wzz gene. Each deletion mutant construct was excised from Topo2.1 using the EcoRI sites and ligated into the EcoRI-cut pGPI vector, creating the pGPI-Δwzz1 and pGPI-Δwzz2 vectors. Ligations were precipitated and electroporated directly into Escherichia coli strain Sy327 (19).
Generation of deletion mutants.
Triparental matings were performed with wild-type PA103, Sy327 with either pGPI-Δwzz1 or pGPI-Δwzz2, and DH5α containing the helper plasmid pRK2013 (18). After mating, merodiploids were selected on L agar with spectinomycin at 100 μg/ml (to select against E. coli) and trimethoprim at 1,500 μg/ml (to select for the plasmid-encoded antibiotic resistance marker). After confirming the presence of both the wild type and the deletion mutant copy via PCR with the primers used to create the deletion mutant constructs, the merodiploids underwent second triparental matings with the DH5α/pRK2013 strain and DH5α containing the pDAI resolution vector. P. aeruginosa containing the resolution vector was selected for on L agar plates with spectinomycin at 100 μg/ml and tetracycline at 100 μg/ml, and colonies were screened for resolution of the chromosomal pGPI vector via PCR. Maintenance on L agar without antibiotic resulted in the loss of pDAI (19). The resulting deletion derivatives were referred to as ΔWzz1 and ΔWzz2. To construct a PA103 strain with both wzz genes deleted, the pGPI-Δwzz1 construct was mated into the ΔWzz2 strain and selection was performed as described above to generate a double deletion mutant (DΔM). All mutations were confirmed by PCR analysis and sequencing.
LPS preparation and visualization.
P. aeruginosa strains were grown overnight in Luria-Bertani (LB) broth and then diluted to an optical density at 600 nm (OD600) of 0.5, followed by pelleting of 1.5 ml. Pellets were resuspended in 200 μl of 2× sodium dodecyl sulfate (SDS) buffer (0.1 M Tris-HCl [pH 6.8], 4% β-mercaptoethanol, 4% SDS, 20% glycerol) and boiled for 5 min. After cooling, 12 μl of proteinase K (10 mg/ml) was added and samples were incubated for 3 h at 60°C. LPS samples were separated on either 8 or 12% SDS-polyacrylamide gels and transferred to nitrocellulose. Blots were analyzed with polyclonal antisera specific for P. aeruginosa serogroup O11 LPS (Accurate Chemical & Scientific, Westbury, NY), a monoclonal antibody to serogroup O11 LPS (Rougier Bio-Tech Ltd., Montreal, Quebec, Canada), or A-band LPS-specific monoclonal antibody N1F10 (purchased from J. S. Lam, University of Guelph, Guelph, Ontario, Canada). The secondary antibodies used were a goat anti-rabbit immunoglobulin G for the polyclonal antibody and a goat anti-mouse immunoglobulin M for the monoclonal antibodies. Both secondary antibodies were coupled to horseradish peroxidase (Sigma-Aldrich, St. Louis, MO), and detection was performed with Western Lightning-ECL reagent (Perkin-Elmer).
Intranasal mouse infection studies.
Bacterial strains were grown on Trypticase soy agar plates (Remel, Lenexa, KS) for 12 h at 37°C, resuspended in PBS to an OD650 of 0.5, and then diluted to obtain the desired dose in 20 μl. Six- to 8-week-old female BALB/c mice (Harlan Laboratories) were anesthetized by intraperitoneal injection with 0.2 ml of ketamine (6.7 mg/ml) and xylazine (1.3 mg/ml) in 0.9% saline, and then 10 μl of the bacterial inoculum was placed into each nostril. Mice were checked for survival three or four times a day for 1 week after infection. The University of Virginia Animal Care and Use Committee approved all of the procedures used in this work.
Bacterial cultures for AFM experiments.
Bacterial stock solutions were stored at −80°C. Bacteria were grown overnight in LB broth or on LB agar at 37°C. Overnight liquid cultures were diluted 1:100 and agitated at 37°C until mid- to late-exponential phase (corresponding to an OD600 of 0.9 to 1.2). P. aeruginosa PA103 was used as the wild-type strain. Its properties have been extensively discussed in the literature (25, 29). Wzy::GM, previously referred to as PA103 wzyPaO11::aacC1, has an LPS that contains the core plus one O-antigen unit (13).
Bacterial attachment to glass slides.
Glass slides (plain microscope slides; Corning Inc.) were cut, rinsed with ultrapure water (resistivity of 18.2 MΩ/cm and <10 ppb total organic carbon; Milli-Q plus; Millipore, Billerica, MA), and sonicated for 30 min. Slides were then immersed in 3:1 (vol/vol) HCl/NHO3 solution for 30 min, rinsed with ultrapure water, and treated with piranha solution (7:3 [vol/vol] H2SO4/H2O2) for 30 min. Glass slides were stored in ultrapure water under cover at 4°C for up to 4 weeks.
Glass slides were functionalized to facilitate bacterial attachment by using a procedure we have applied previously (30). In brief, cleaned glass slides were rinsed with methanol and immersed in 50% 3-aminopropyltrimethoxysilane in methanol for 20 min. Glass slides were then rinsed with methanol and ultrapure water. The bacterial culture (20 ml) was centrifuged at 7,000 rpm for 10 min, and pellets were washed twice with ultrapure water. To a 10-ml vial of bacterial solution we added 300 μl of 100 mM 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride and 600 μl of 40 mM N-hydroxysuccinimide and N-hydroxysulfosuccinimide. The solution was added to the aminosilane-treated glass slide and agitated at 70 rpm for 4 to 6 h to promote bacterial lawn formation.
AFM.
A Digital Instruments Dimension 3100 atomic force microscope with a Nanoscope IIIa controller (Veeco Metrology, Inc., Santa Barbara, CA) was used for all of the experiments. We used a total of five silicon nitride probes (DNP, Veeco Instruments Inc., Santa Barbara, CA) with measured spring constants of 0.12 N/m ± 10%, resonance frequencies of ∼5 kHz in liquid, and average radii of 40 nm (previously measured by electron microscopy; data not shown). Probes were immersed in 100% ethanol for 12 h and then subjected to UV treatment (365 nm) to remove organic films (4, 38). AFM experiments were performed with ultrapure water. For each force measurement, the sensitivity was determined by making a measurement on clean glass. To select bacterial cells for analysis, an image of the slide was obtained at a scanning rate of 1 Hz in the intermittent-contact mode. The tip was then positioned over the center of a bacterium and measurements were performed in the force mode. Ten force cycles were recorded for at least five different cells of each strain examined. Raw AFM data were processed in Microsoft Excel and MATLAB (MathWorks, Natick, MA) by using a procedure we previously described, which is based on a method developed by Ducker et al. (3, 15). A steric repulsion model was applied to individual force curves using nonlinear curve fitting in TableCurve 2D (Systat Software Inc., Chicago, IL) to obtain the equilibrium length of the polymer brush, its compressibility, and the spacing between the LPS molecules (see Fig. S1 in the supplemental material).
Steric model.
A model was applied to the steric repulsive forces between the bacterium and the AFM tip which arise during force measurements as the tip approaches the bacterial cell. The model we used was originally developed by Alexander and de Gennes to describe the pressure between two flat surfaces with grafted polymers. Integrating over the area to account for the spherical AFM probes, the model (22) takes the form
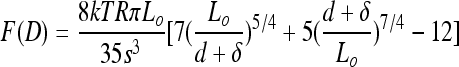 |
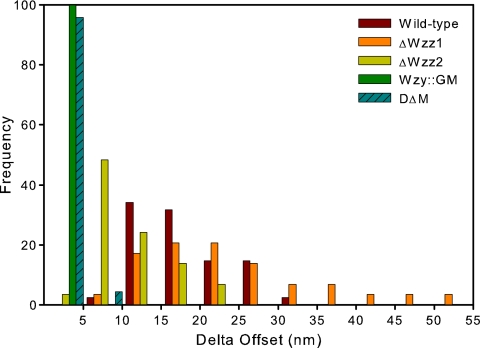
where F is the steric repulsive force measured in the AFM experiment as a function of separation distance (D), k is Boltzmann's constant, T is the absolute temperature, R is the tip radius, s is the spacing of the LPS molecules, and Lo is the equilibrium thickness of the polymer layer (LPS), which here we refer to as length. The separation distance, D, was used as the sum of the measured distance (d) and the offset distance (δ), according to the procedure of Chang et al. (9). The δ offset enables better fitting because it provides an estimate for the layer thickness at the maximal applied force in the measurement.
Zeta potential.
The zeta potential of wild-type P. aeruginosa strain PA103 and the four mutants was measured using a Zetasizer Nano ZS (Malvern Instruments, Worcestershire, United Kingdom) and a universal dip cell (ZEN1002; Malvern Instruments). Bacteria were grown to exponential phase, washed once to remove the culture medium and salts, and resuspended in ultrapure water (Milli-Q plus; Millipore, Billerica, MA). Cells were diluted to a uniform OD600, and three sets of at least 10 measurements were conducted to ensure reproducibility. Electrophoretic mobility was converted to zeta potential using the Smoluchowski equation (17).
Statistical analysis.
Survival curves for mouse infection studies were generated and statistical analysis performed using Prism 4 (GraphPad, La Jolla, CA). Steric modeling results were analyzed in SigmaPlot (Systat Software Inc., Chicago, IL) using Kruskal-Wallis one-way analysis of variance on ranks, and pairwise multiple comparisons using Dunn's method were used to determine statistically significantly different groups. Histograms were created in Microsoft Excel using manual settings for bin width and graphed in SigmaPlot.
RESULTS
Characterization of LPS mutants.
P. aeruginosa serogroup O11 strain PA103 expresses LPS O-antigen lengths that have been defined as long and very long. We have previously constructed and characterized mutants insertionally inactivated in the O-antigen chain length regulator genes wzz1 and wzz2: the wzz1 mutant had less long O antigen, while the wzz2 mutant lacked the very long O antigen (26). To verify that the effects we observed in these strains were not due to polar effects on the upstream or downstream regions of these genes, we constructed wzz1 and wzz2 deletion mutants and a double deletion mutant, DΔM. In these strains, the entire coding region of each wzz gene was deleted, leaving behind an 85-bp FRT scar, which has had no reported polar effects (10). Western blotting was performed on LPS isolated from wild-type strain PA103 and each of the mutants in order to verify the LPS phenotype. As we had observed previously (26), LPS isolated from PA103 shows a characteristic ladder pattern reflecting the addition of individual O-antigen units onto the lipid A-LPS core (Fig. 1 A). This banding pattern is not random; apart from bands in the low-molecular-weight ranges corresponding to the addition of individual subunits, there is also a preference for lengths running between 50 and 150 kDa, which we refer to as “long,” and for lengths running around 250 kDa, which we refer to as “very long” (25). Complete deletion of the wzz1 gene resulted in a mutant expressing a preference for only very long B-band O-antigen side chains, while deletion of the wzz2 gene produced mutants expressing no very long O antigen. Both single deletion mutants still had a pattern of banding in the low-molecular-weight range due to the action of the O-antigen polymerase Wzy. Simultaneous deletion of both genes resulted in a strain with a random distribution of O-antigen side chains, with proportionally more short O-antigen side chains than long O-antigen side chains. These three strains are referred to as ΔWzz1, ΔWzz2, and DΔM, respectively. As we had noted previously (26), complementation of each mutant with the corresponding wild-type gene resulted in the parental LPS phenotype, indicating that these deletions were not polar on downstream genes (unpublished observations).
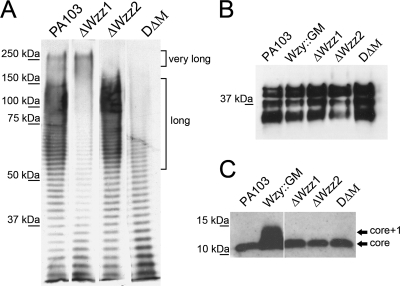
FIG. 1.
Western blot assay of LPSs isolated from P. aeruginosa PA103 and mutants. Isolated LPS was separated by either 8% (A) or 12% (B and C) SDS-PAGE and immunoblotted using (A) polyclonal P. aeruginosa serotype O11 antibody on purified LPS, (B) monoclonal antibody to the A-band common antigen, and (C) monoclonal antibody to serotype O11. Wild-type PA103 produces both long and very long B-band O-antigen side chains, while ΔWzz1 and ΔWzz2 express only very long and long LPS chains, respectively. Wzy::GM has only one O-antigen subunit attached to the lipid A-core (13).
There was no obvious defect in A-band production in any of the mutants compared to the wild-type strain (Fig. 1B). These results are consistent with prior studies on the role of wzz proteins in the LPS production pathway (13).
Steric modeling and zeta potential results.
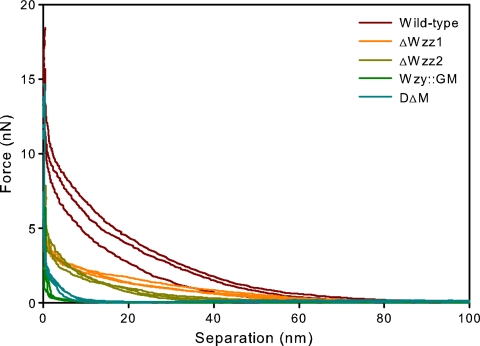
Approach curves obtained from AFM experiments exhibited increased repulsion force with increasing LPS length across the wild type and the four mutants studied (Fig. 2). This effect illustrates the contribution of the LPS brush in interactions between the sample and the AFM tip—bacterial cells having only a very short LPS layer on their outer membrane behave much more like a stiff surface, while cells with a longer LPS layer show a smoother increase in force with decreasing distance.
FIG. 2.
Representative approach curves obtained from the PA103 wild-type strain and the four LPS mutants. Repulsive forces between the sample and the AFM tip increase with increasing LPS length.
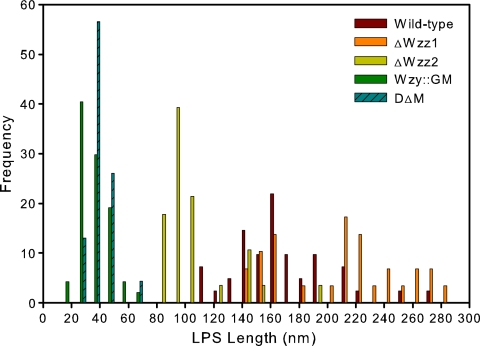
After the AFM experiments were performed, the LPS lengths were calculated from the modified Alexander and de Gennes (AdG) steric model. As a control for these experiments, we used Wzy::GM (13), a PA103 mutant expressing an LPS core plus one subunit of the O antigen (Fig. 1C). As anticipated, this strain showed the shortest LPS length, with an average of 34.6 nm (Fig. 3). The LPS length in DΔM with no O-antigen chain length preference was calculated to be 37.7 nm. The average LPS lengths for ΔWzz2 and ΔWzz1 were 108 and 212 nm, respectively. For the wild-type strain, we detected primarily very long chains (>150 nm), which ranged up to 280 nm (Fig. 3), but there were also some long chains observed, as well (90 to 140 nm). Statistical analysis of the LPS length results showed significantly different comparisons (P < 0.05) for all treatments, except for the wild-type strain versus the ΔWzz1 mutant and Wzy::GM versus DΔM.
FIG. 3.
Histogram showing the length distribution of the LPSs of PA103 and the four mutants. DΔM and Wzy::GM show the shortest lengths resulting from interactions with A-band LPS molecules. Following is ΔWzz2 with the long O-antigen side chain preserved. The wild type and ΔWzz1 (lacking long O-antigen side chains but still possessing the very long ones) express LPSs of the greatest length.
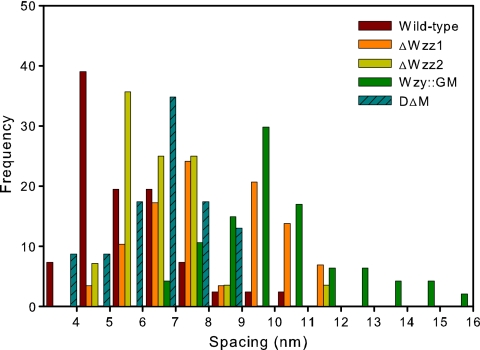
The smallest spacing was observed for the wild-type strain, and the largest was observed for the Wzy::GM mutant (Fig. 4). The ΔWzz1 and ΔWzz2 mutants had similar spacing values, and both were larger than the ones we observed for PA103 (Table 1).
FIG. 4.
Histogram of the spacing values obtained from the AdG model.
TABLE 1.
Summary of LPS physical properties based on the application of steric modeling to AFM force data and zeta potential measurements
| Strain | Force (nN) | LPS length (nm) | Spacing (nm) | δ offset (nm) | Zeta potential (mV) |
|---|---|---|---|---|---|
| Wild type | 1.18 ± 0.07a | 171 ± 5 | 5.7 ± 0.3 | 18.5 ± 0.9 | −10.2 ± 0.5 |
| ΔWzz1 | 0.60 ± 0.03 | 212 ± 8 | 8.4 ± 0.4 | 25 ± 2 | −9.2 ± 0.3 |
| ΔWzz2 | 0.63 ± 0.04 | 108 ± 5 | 6.6 ± 0.3 | 11.9 ± 0.9 | −3.6 ± 0.5 |
| Wzy::GM | 0.72 ± 0.03 | 34.6 ± 1.4 | 10.3 ± 0.3 | 0.70 ± 0.09 | −45.0 ± 0.5 |
| DΔM | 1.10 ± 0.05 | 37.7 ± 1.6 | 6.4 ± 0.3 | 2.0 ± 0.3 | −12.0 ± 0.3 |
Each value shown represents the mean ± the standard error of the mean.
The δ offset parameter is related to the compressibility of the polymer layer, relative to the cantilever stiffness, and also dependent on the AFM tip radius. We found that δ was the smallest for Wzy::GM and DΔM, which were not statistically significantly different. The δ offset was the largest for ΔWzz1, the mutant with the longest O antigen, at 25 nm. The wild-type and ΔWzz2 strains, with δ offset parameters of 18.5 and 11.9 nm, respectively, fell in the middle range of values (Fig. 5). The δ offset parameters of the wild type compared to those of ΔWzz2 and those of the wild type compared to those of ΔWzz1 were not statistically significantly different from one another (P > 0.05).
FIG. 5.
LPS layer compressibility (δ offset) histogram. DΔM and Wzy::GM are the most compressible, followed, respectively, by ΔWzz2, wild-type PA103, and ΔWzz1.
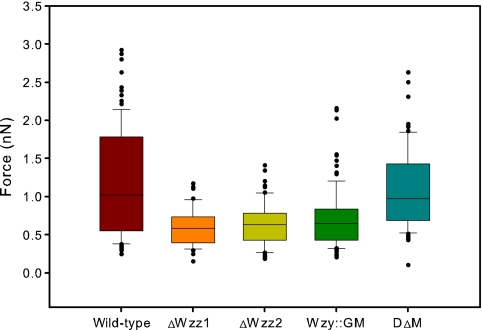
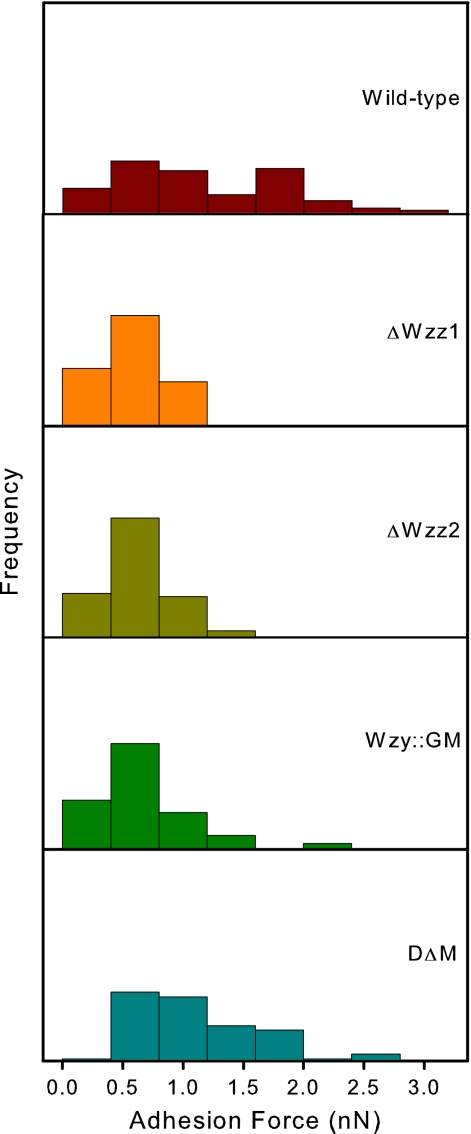
Adhesion forces measured by AFM were highest for the wild type and DΔM (Fig. 6). Although the medians for this pair were similar and the statistical tests showed that they were not significantly different from one another, the distributions show some differences (Fig. 7). We observed a broader distribution of adhesion forces for the wild-type strain than for the double mutant. The mutants ΔWzz1, ΔWzz2, and Wzy::GM all showed lower similarly distributed adhesion forces (Fig. 6 and 7). The average adhesion force for Wzy::GM was slightly higher than for the ΔWzz1 and ΔWzz2 strains (Table 1). However, the adhesion forces for these three mutants were not statistically significantly different (P > 0.05).
FIG. 6.
Box plot of adhesion data obtained from AFM experiments. Wild-type strain PA103 shows the highest average force, while all LPS mutations lower the force of adhesion to the Si3N4 tip. Of the four mutants, DΔM shows the highest absolute force.
FIG. 7.
Histogram of force data obtained from AFM experiments. PA103 shows the broadest distribution range, while the force values obtained from ΔWzz1, ΔWzz2, and Wzy::GM show similar Gaussian distribution patterns. The frequency along the vertical axis ranges from 0 to 100% for each of the treatments.
The zeta potential was highest for the ΔWzz2 mutant (−3.6 mV) and decreased in the order ΔWzz1, wild-type PA103, DΔM, and Wzy::GM (Table 1). The Wzy::GM mutant was significantly different from the other strains studied, with a zeta potential of −45.03 mV. All pairwise comparisons showed statistically significant differences (P < 0.05), except for ΔWzz1 and the wild type.
Virulence studies.
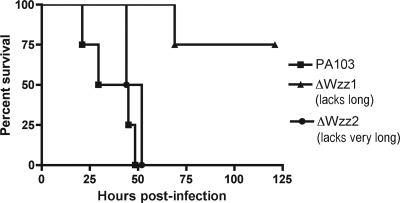
Previous work has demonstrated a correlation between adhesion forces and increased virulence in L. monocytogenes (19). To determine if the measured adhesion forces could serve as a predictor of virulence for the P. aeruginosa Wzz mutants, we tested each of the deletion mutants in an acute pneumonia model of infection using BALB/c mice. When receiving a dose of 7 × 105 CFU, mice infected with wild-type PA103 all succumbed to the infection by 50 h (Fig. 8). Mice infected with ΔWzz2 also succumbed to the infection in around 50 h. Mice infected with the ΔWzz1 mutant survived the longest, with only one mouse out of four succumbing to the infection by the end of the experiment. A second experiment, using a lower dose averaging 2 × 105 CFU, was also performed which included the double deletion mutant (see Fig. S2 in the supplemental material). At this lower dose, all four of the PA103-infected mice died by 50 h. One mouse out of the four infected with the ΔWzz2 mutant survived, while two mice out of the four survived when infected with the ΔWzz1 mutant. At this dose, the double deletion mutant was completely avirulent, with no mice succumbing to the infection. In both cases, the survival curve of mice infected with ΔWzz1 was significantly different (P < 0.05) from that of mice infected with PA103, while the survival curve of mice infected with ΔWzz2 was not. In the second experiment, the survival curve of mice infected with the double deletion mutant was also significantly different from that of mice infected with wild-type strain PA103.
FIG. 8.
Survival of mice infected with wild-type PA103 and the ΔWzz mutants. The doses used for infection were as follows: PA103, 6.8 × 105 CFU; ΔWzz1, 7.5 × 105 CFU; ΔWzz2, 6.7 × 105 CFU. Four mice were included per group. Curves were plotted using the Kaplan-Meier method, and curve comparisons were done using the log-rank test. The PA103 and ΔWzz1 survival curves were significantly different from each other (P < 0.05), while the PA103 and ΔWzz2 curves were not significantly different.
DISCUSSION
AFM has become an important tool for characterizing the physical properties of bacterial surface molecules, including LPS, proteins, extracellular polymeric substances, and flagella (5, 31, 42). In some cases, the force of adhesion between a bacterium and an AFM tip has been shown to correlate with LPS length, such as we observed on several strains of E. coli interacting with a silicon nitride AFM probe (39). For P. aeruginosa in the present study, we did not see a direct correlation with LPS length and forces of adhesion to silicon nitride (see Fig. S3A in the supplemental material). However, there are important differences between the E. coli strains used previously and the P. aeruginosa strains used in the present study. In general, P. aeruginosa LPS cores have been found to be uncapped in about 80% of the cases (12), while for E. coli, only ∼10% of LPS molecules are uncapped (21). E. coli was also only compared across strains, such as by comparing E. coli O157:H7 (which has long LPS) to E. coli ML35 (with no O antigen and thus very short LPS). For different E. coli strains, we generally observed that the ones with longer LPS exhibited higher forces of adhesion to silicon nitride. For the P. aeruginosa mutants studied here, comparison of isogenic strains revealed more subtle differences which can be attributed specifically to the presence and/or presentation of LPS on the bacterial cell surface and not to differences in LPS composition.
Western blotting suggests that A-band LPS is longer than most of the O antigen in the DΔM strain (see Fig. S4 in the supplemental material). The A band would therefore mask the LPS of the DΔM strain, as well as the LPS core plus one O-antigen unit of the Wzy::GM mutant. Our AFM experiments and steric modeling results confirm that the top LPS layer on the Wzy::GM and DΔM mutants is indeed composed of the same type of molecules. Both mutants showed very similar LPS length and compressibility parameters which were also not significantly different. The decreased spacing in DΔM is consistent with the expected presence of underlying B-band LPS molecules. Thus, we conclude that the longest LPS in the Wzy::GM and DΔM strains is likely composed of A-band molecules.
The wild-type strain exhibited the broadest distributions of both LPS length and adhesion forces. Any of the introduced mutations narrowed the adhesion force distribution (Fig. 7). The LPS length distribution only became very narrow when nearly all of the LPS was removed, as was the case for the Wzy::GM mutant, or when the O-antigen chain length preference was lost, as in the case of DΔM (Fig. 3). This can be attributed to the presence of A-band LPS, which covers the B-band molecules on the surface of these two mutants. Overall, the wild type was the most adhesive to silicon nitride and adhesion decreased with any mutations introduced. This suggests that the wild-type strain is already optimized to have the maximum adhesion in nature.
Although LPS length and adhesion force were not well correlated with one another, some of the other physical properties showed interdependence. LPS length showed a positive correlation with delta offset (R2 = 0.65) in that δ increased as Lo increased (see Fig. S3C in the supplemental material). This is the result of the AFM tip making a stiff contact further away from the cell membrane with an increase in the thickness of the LPS layer. LPS length and spacing showed a weak correlation for the O-antigen-expressing strains (see Fig. S3B in the supplemental material). The reason for this interdependence is not clear.
The relative values of the parameters of the AdG model were consistent with previous biological findings and expectations. The strongest agreement was observed in the Lo parameter of the fitted equation. The median LPS lengths of the wild type and ΔWzz1 showed no statistically significant differences, as the AFM tip interacts only with the surface of the LPS layer, which in both cases is built up of the very long B-band LPS. Still, ΔWzz1 showed slightly longer LPS molecules, because removing the long O-antigen side chain would result in an excess amount of saccharide O-antigen subunits. The double mutant and Wzy::GM were also not significantly different from each other, as in this case, the A-band chains mask the underlying B-band molecules and thus form the topmost LPS layer, which we measured to have a length of about 36 nm.
The δ offset parameter of the AdG equation is an experimental variable and is dependent on both the cantilever stiffness and the tip radius. Assuming that these specifications are similar for the different AFM probes that we used, the δ offset can provide insight into the LPS layer's physical properties. The median δ offset values for the wild type, ΔWzz1, and ΔWzz2 showed no significant difference from each other, indicating that the O-antigen part of the LPS molecule has uniform compressibility, which is independent of the synthesis mechanism. The median values for DΔM and Wzy::GM were also not statistically significantly different, suggesting that the A-band LPS molecules have the same compressibility throughout the different mutants as well.
Bacteria, like most other microbes and cells, are negatively charged. The magnitude of the charge, however, is dependent upon the bacterial surface structures and is related to the cell's ability to attach to various substrates (44). P. aeruginosa LPS varies among the different serogroups, and its chemical structure affects the bacterial surface charge. P. aeruginosa PA103 LPS core consists mainly of neutral sugars but has several negatively charged sites such as 3-deoxy-d-manno-octulosonic acid residues and phosphate groups. The A band, composed of d-rhamnose trisaccharide repeating units, and the B-band LPS, consisting of trisaccharide units of one glucose and two N-acetylfucosamine residues, are electroneutral (28). Our zeta potential measurements are consistent with the expected charge distribution on the P. aeruginosa surface. The effect of the negatively charged LPS core molecules is greatest for the Wzy::GM mutant due to its lack of O antigen (zeta potential of −45.0 ± 0.5 mV). These charges are then masked by the addition of O-antigen repeating units for the wild type and the other three mutants, as indicated by the increase in zeta potential. Studies aimed at exploring the effect of LPS mutation on attachment to various biotic and abiotic surfaces could further elucidate the role of LPS structure in bacterial adhesion.
Relating LPS physical properties and adhesion force to bacterial pathogenicity can provide a convenient in vitro way to discriminate between virulent and avirulent strains. The silicon nitride AFM probe is not truly representative of the complex pathogen-host interactions, but it has been successfully used as a model surface. Previous work has found correlations between LPS length and adhesion force (39) and between adhesion force and virulence (34) in different strains of the same species using a silicon nitride cantilever. In this work, we compared isogenic LPS mutants and did not find a correlation between LPS length and virulence, but we noted a relationship between adhesion force and virulence. Wzz deletion mutants lacking the preferences for wild-type lengths of the O-antigen side chain were tested in a mouse acute pneumonia model of infection. At 7 × 105 CFU, mice infected with the ΔWzz2 mutant did not show any significant difference from mice infected with wild-type PA103, while mice infected with the ΔWzz1 mutant exhibited much longer survival times. This shows that LPS length cannot be reliably used as a predictor of virulence, because the biological differences among the mutants overrule the effect of variations in LPS layer physical conformation. At a lower dose averaging 2 × 105 CFU, the ΔWzz2 mutant exhibited a slight attenuation compared to the wild type, although statistical tests could not determine a significant difference due to the obstacles in testing a larger number of mice. These results suggest that adhesion force can be used as a predictor of virulence at low doses, which in fact more accurately represent natural infection settings. The wild-type PA103 proved to be the most virulent and also showed the highest adhesion forces, while ΔWzz1 and ΔWzz2 showed lower force profiles and, respectively, attenuated and slightly attenuated pathogenicity. We speculate that the difference between the wild-type and ΔWzz2 strains will be even more pronounced at lower infection doses. At 2 × 105 CFU, the double deletion mutant proved to be completely avirulent, suggesting an additive effect of the removal of both long and very long O-antigen side chains. Other studies have demonstrated that LPS rough mutants exhibit much higher 50% lethal doses than the respective wild-type organisms (11). No comparison of adhesion force magnitude can be made between the O-antigen-expressing strains (wild-type PA103 and the ΔWzz1 and ΔWzz2 mutants) and the mutants in which A-band LPSs make up the topmost part of the LPS layer (Wzy::GM and DΔM), as the interaction forces in the two groups arise from contact with chemically different molecules. We can, however, predict, based on the adhesion force profiles of the two mutants, that Wzy::GM will be more attenuated than DΔM, although it will be difficult to determine if subtle differences exist, given the avirulence of the DΔM mutant.
The deletion mutants created in this study exhibit a more pronounced phenotype than the insertional mutants created previously (26) with respect to the decreased presentation of O-antigen chains of particular lengths, which facilitated AFM experiments and modeling and might have also led to a further reduction in virulence. However, when the deletion mutants generated here were tested in the acute pneumonia model at the same dose as that previously used (26), very similar survival curves were obtained. This indicates that the insertional mutants already altered the presentation of O-antigen chain length enough that virulence was decreased. Further attenuation was not detected, even when there was a complete lack of O antigen of these particular lengths.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (R01 AI068112 to J.B.G.). E.N.K. was partly supported by the National Institutes of Health through University of Virginia Infectious Diseases training grant AI07406.
We thank Joshua Strauss, Paola Pinzón-Arango, and Yuanyuan Tao for their support.
Footnotes
Published ahead of print on 10 December 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alexander, S. 1977. Adsorption of chain molecules with a polar head a scaling description. J. Phys. II (Paris) 38:983-987. [Google Scholar]
- 2.Andre, G., et al. 2010. Imaging the nanoscale organization of peptidoglycan in living Lactococcus lactis cells. Nat. Commun. 1(3):1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atabek, A., and T. A. Camesano. 2007. An atomic force microscopy study of the effect of lipopolysaccharides and extrapolymeric substances on the adhesion of Pseudomonas aeruginosa. J. Bacteriol. 189:8503-8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolon, D. A., and C. O. Kunz. 1972. Ultraviolet depolymerization of photoresist polymers. Polym. Eng. Sci. 12:109-111. [Google Scholar]
- 5.Boonaert, C. J. P., P. G. Rouxhet, and Y. F. Dufrêne. 2000. Surface properties of microbial cells probed at the nanometre scale with atomic force microscopy. Surf. Interface Anal. 30:32-35. [Google Scholar]
- 6.Burrows, L. L., H. L. Rocchetta, and J. S. Lam. 2000. Assembly pathways for biosynthesis of A-band and B-band lipopolysaccharide in Pseudomonas aeruginosa, p. 127-143. In R. R. Doyle (ed.), Glycomicrobiology. Kluwer Academic/Plenum Publishers, New York, NY.
- 7.Butt, H.-J., M. Kappl, H. Mueller, and R. Raiteri. 1999. Steric forces measured with the atomic force microscope at various temperatures. Langmuir 15:2559-2565. [Google Scholar]
- 8.Camesano, T. A., and B. E. Logan. 2000. Probing bacterial electrosteric interactions using atomic force microscopy. Environ. Sci. Technol. 34:3354-3362. [Google Scholar]
- 9.Chang, D. P., N. I. Abu-Lail, F. Guilak, G. D. Jay, and S. Zauscher. 2008. Conformational mechanics, adsorption, and normal force interactions of lubricin and hyaluronic acid on model surfaces. Langmuir 24:1183-1193. [DOI] [PubMed] [Google Scholar]
- 10.Choi, K. H., and H. P. Schweizer. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cryz, S. J., Jr., T. L. Pitt, E. Furer, and R. Germanier. 1984. Role of lipopolysaccharide in virulence of Pseudomonas aeruginosa. Infect. Immun. 44:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darveau, R. P., and R. E. W. Hancock. 1983. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J. Bacteriol. 155:831-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean, C. R., et al. 1999. Characterization of the serogroup O11 O-antigen locus of Pseudomonas aeruginosa PA103. J. Bacteriol. 181:4275-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Gennes, P. G. 1987. Polymers at an interface: a simplified view. Adv. Colloid Interface Sci. 27:189-209. [Google Scholar]
- 15.Ducker, W. A., T. J. Senden, and R. M. Pashley. 1992. Measurement of forces in liquids using a force microscope. Langmuir 8:1831-1836. [Google Scholar]
- 16.Dufrêne, Y. F. 2010. Atomic force microscopy of fungal cell walls: an update. Yeast 27:465-471. [DOI] [PubMed] [Google Scholar]
- 17.Elimelech, M., X. Jia, J. Gregory, and R. Williams. 1998. Particle deposition and aggregation measurement, modelling, and simulation: measurement, modeling and simulation. Butterworth-Heinemann, Oxford, England.
- 18.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flannagan, R. S., T. Linn, and M. A. Valvano. 2008. A system for the construction of targeted unmarked gene deletions in the genus Burkholderia. Environ. Microbiol. 10:1652-1660. [DOI] [PubMed] [Google Scholar]
- 20.Fletcher, M. 1996. Bacterial attachment in aquatic environments: a diversity of surfaces and adhesion strategies, p. 1-24. In M. Fletcher (ed.), Bacterial adhesion: molecular and ecological diversity. Wiley-Liss, Inc., New York, NY.
- 21.Goldman, R. C., and L. Leive. 1980. Heterogeneity of antigenic-side-chain length in lipopolysaccharide from Escherichia coli O111 and Salmonella typhimurium LT2. Eur. J. Biochem. 107:145-153. [DOI] [PubMed] [Google Scholar]
- 22.Israelachvili, J. N. 1992. Intermolecular and surface forces, 2nd edition. Academic Press, New York, NY.
- 23.Kerr, K. G., and A. M. Snelling. 2009. Pseudomonas aeruginosa: a formidable and ever-present adversary. J. Hosp. Infect. 73:338-344. [DOI] [PubMed] [Google Scholar]
- 24.King, J. D., D. Kocincova, E. L. Westman, and J. S. Lam. 2009. Lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 15:261-312. [DOI] [PubMed] [Google Scholar]
- 25.Kintz, E., and J. B. Goldberg. 2008. Regulation of lipopolysaccharide O antigen expression in Pseudomonas aeruginosa. Future Microbiol. 3:191-203. [DOI] [PubMed] [Google Scholar]
- 26.Kintz, E., J. M. Scarff, A. DiGiandomenico, and J. B. Goldberg. 2008. Lipopolysaccharide O-antigen chain length regulation in Pseudomonas aeruginosa serogroup O11 strain PA103. J. Bacteriol. 190:2709-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knirel, Y. A., O. V. Bystrova, N. A. Kocharova, U. Zahringer, and G. B. Pier. 2006. Conserved and variable structural features in the lipopolysaccharide of Pseudomonas aeruginosa. J. Endotoxin Res. 12:324-336. [DOI] [PubMed] [Google Scholar]
- 28.Langley, S., and T. J. Beveridge. 1999. Effect of O-side-chain-lipopolysaccharide chemistry on metal binding. Appl. Environ. Microbiol. 65:489-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, P. V. 1973. Exotoxins of Pseudomonas aeruginosa. I. Factors that influence the production of exotoxin A. J. Infect. Dis. 128:506-513. [DOI] [PubMed] [Google Scholar]
- 30.Liu, Y., and T. A. Camesano. 2008. Immobilizing bacteria for atomic force microscopy imaging or force measurements in liquids, p. 163-188. In T. A. Camesano and C. M. Mello (ed.), Microbial surfaces. American Chemical Society symposium series, vol. 984. American Chemical Society, Chicago, IL. [Google Scholar]
- 31.Lower, B. H., R. Yongsunthon, F. P. Vellano, and S. K. Lower. 2005. Simultaneous force and fluorescence measurements of a protein that forms a bond between a living bacterium and a solid surface. J. Bacteriol. 187:2127-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051-1060. [DOI] [PubMed] [Google Scholar]
- 33.Morona, R., L. Van Den Bosch, and C. Daniels. 2000. Evaluation of Wzz/MPA1/MPA2 proteins based on the presence of coiled-coil regions. Microbiology 146(Pt. 1):1-4. [DOI] [PubMed] [Google Scholar]
- 34.Park, B. J., T. Haines, and N. I. Abu-Lail. 2009. A correlation between the virulence and the adhesion of Listeria monocytogenes to silicon nitride: an atomic force microscopy study. Colloids Surf. B Biointerfaces 73:237-243. [DOI] [PubMed] [Google Scholar]
- 35.Pier, G. B., et al. 1996. Role of mutant CFTR in hypersusceptibility of cystic fibrosis patients to lung infections. Science 271:64-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Priebe, G. P., et al. 2004. The galU Gene of Pseudomonas aeruginosa is required for corneal infection and efficient systemic spread following pneumonia but not for infection confined to the lung. Infect. Immun. 72:4224-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shephard, J. J., D. M. Savory, P. J. Bremer, and A. J. McQuillan. 2010. Salt modulates bacterial hydrophobicity and charge properties influencing adhesion of Pseudomonas aeruginosa (PA01) in aqueous suspensions. Langmuir 26:8659-8665. [DOI] [PubMed] [Google Scholar]
- 38.Sowell, R. R., R. E. Cuthrell, D. M. Mattox, and R. D. Bland. 1974. Surface cleaning by ultraviolet radiation. J. Vac. Sci. Technol. 11:474-475. [Google Scholar]
- 39.Strauss, J., N. A. Burnham, and T. A. Camesano. 2009. Atomic force microscopy study of the role of LPS O-antigen on adhesion of E. coli. J. Mol. Recognit. 22:347-355. [DOI] [PubMed] [Google Scholar]
- 40.Strauss, J., A. Kadilak, C. Cronin, C. M. Mello, and T. A. Camesano. 2010. Binding, inactivation, and adhesion forces between antimicrobial peptide cecropin P1 and pathogenic E. coli. Colloids Surf. B Biointerfaces 75:156-164. [DOI] [PubMed] [Google Scholar]
- 41.Tang, H. B., et al. 1996. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect. Immun. 64:37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Touhami, A., M. H. Jericho, J. M. Boyd, and T. J. Beveridge. 2006. Nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa pili by using atomic force microscopy. J. Bacteriol. 188:370-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Mei, H. C., J. de Vries, and H. J. Busscher. 2010. Weibull analyses of bacterial interaction forces measured using AFM. Colloids Surf. B Biointerfaces 78:372-375. [DOI] [PubMed] [Google Scholar]
- 44.van Loosdrecht, M. C. M., J. Lyklema, W. Norde, G. Schraa, and A. J. B. Zehnder. 1987. Electrophoretic mobility and hydrophobicity as a measure to predict the initial steps of bacterial adhesion. Appl. Environ. Microbiol. 53:1898-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilkinson, S. G., and L. Galbraith. 1975. Studies of Lipopolysaccharides from Pseudomonas aeruginosa. Eur. J. Biochem. 52:331-343. [DOI] [PubMed] [Google Scholar]
- 46.Wright, C. J., M. K. Shah, L. C. Powell, and I. Armstrong. 2010. Application of AFM from microbial cell to biofilm. Scanning 32:134-149. [DOI] [PubMed] [Google Scholar]
- 47.Yokota, S., et al. 1987. Characterization of a polysaccharide component of lipopolysaccharide from Pseudomonas aeruginosa IID 1008 (ATCC 27584) as d-rhamnan. Eur. J. Biochem. 167:203-209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.