Abstract
The genome sequence of Streptomyces ambofaciens, a species known to produce the congocidine and spiramycin antibiotics, has revealed the presence of numerous gene clusters predicted to be involved in the biosynthesis of secondary metabolites. Among them, the type II polyketide synthase-encoding alp cluster was shown to be responsible for the biosynthesis of a compound with antibacterial activity. Here, by means of a deregulation approach, we gained access to workable amounts of the antibiotics for structure elucidation. These compounds, previously designated as alpomycin, were shown to be known members of kinamycin family of antibiotics. Indeed, a mutant lacking AlpW, a member of the TetR regulator family, was shown to constitutively produce kinamycins. Comparative transcriptional analyses showed that expression of alpV, the essential regulator gene required for activation of the biosynthetic genes, is strongly maintained during the stationary growth phase in the alpW mutant, a stage at which alpV transcripts and thereby transcripts of the biosynthetic genes normally drop off. Recombinant AlpW displayed DNA binding activity toward specific motifs in the promoter region of its own gene and that of alpV and alpZ. These recognition sequences are also targets for AlpZ, the γ-butyrolactone-like receptor involved in the regulation of the alp cluster. However, unlike that of AlpZ, the AlpW DNA-binding ability seemed to be insensitive to the signaling molecules controlling antibiotic biosynthesis. Together, the results presented in this study reveal S. ambofaciens to be a new producer of kinamycins and AlpW to be a key late repressor of the cellular control of kinamycin biosynthesis.
Streptomycetes are filamentous, soil-dwelling bacteria that undergo a complex morphological differentiation correlated with a rich biochemical specialization occurring during the late stages of growth. These prokaryotes are known for their capacity to biosynthesize a vast array of important secondary metabolites used in human activities, including antibiotics, antitumor agents, immunosuppressants, antihelmenthics, and herbicides. In spite of decades of genetic studies and industrial uses, members of the genus Streptomyces have recently revealed, by means of genomic analyses, their hitherto unsuspected ability to produce further novel secondary metabolites with potentially useful activities (5, 22, 37).
In Streptomyces ambofaciens ATCC 23877, which was previously known to produce only the antibiotics congocidine (12) and spiramycin (42), the sequencing of the terminal regions of the linear chromosome (over circa 3 Mb; accession no. AM238663 and AM238664, respectively, for the left and right arms) has unveiled 14 novel secondary metabolite gene clusters (http://www.weblgm.scbiol.uhp-nancy.fr/ambofaciens/) (10). Among them, two clusters were experimentally shown to be involved in the biosynthesis of the siderophore coelichelin (3) and the pyrrole-amide congocidine (23), respectively. In our groups, the function of the duplicated type II polyketide synthase (PKS) gene cluster located in the terminal inverted repeats of the chromosome has also been unraveled. This cluster, named alp (for angucyclinone-like polyketide) is responsible for the biosynthesis of an antibacterial activity formerly called alpomycin, and a diffusible orange pigment, which is likely to be either a degradation or modification product of the antibiotic (41). The alp cluster, as previously defined (41), is composed of about 30 genes and covers approximately 34 kb of the chromosome. Based on sequence similarity, it could be divided into three regions reflecting probable sequential acquisitions by horizontal gene transfer. A first region of 12 genes (alpG to alpL2), which includes the minimal PKS genes alpA, alpB, and alpC responsible for assembling the polyketide chain and coding for a β-ketoacyl synthase (KS), a chain length factor (CLF), and an acyl carrier protein (ACP), respectively, shows strong synteny with a part of the kinamycin cluster of Streptomyces murayamaensis (accession no. AY228175). The second region contains eight genes (alpM to alpT) showing high sequence similarity to genes carried by pSLA2-L, a linear plasmid from Streptomyces rochei (33). In particular, the alpQRST locus, which encodes a second KS (alpR) and CLF (alpQ), whose role is still unknown, is syntenic with a part of the mithramycin gene cluster. Finally, a third region of six genes (alpUVWXYZ) includes alpU, alpV, alpW, and alpZ, which show similar genetic organization to genes that regulate the tylosin (type I modular PKS) biosynthetic gene cluster in Streptomyces fradiae (4, 41).
Among the alp regulatory genes, alpT, alpU, and alpV are predicted by comparison with database sequences to encode proteins from the Streptomyces antibiotic regulatory protein family (SARP), which mainly act as transcriptional activators (50). The product of the alpV gene was previously shown to be essential for the production of both the orange pigment and alpomycin by activating the transcription of the structural biosynthetic genes (1). The transcription of alpV is itself under the control of the product of alpZ, a γ-butyrolactone (GBL)-like receptor homologue from the TetR superfamily of transcription factors (for a review, see reference 43), which acts at the top of the regulatory cascade controlling the activation/repression of the alp pathway (8). Indeed, AlpZ is able to bind specific AT-rich regulatory elements (ARE) within the promoter region of alpV, thereby repressing transcription of alpV. In a manner similar to other GBL receptors (49), AlpZ is able to respond to increasing amounts of hormone-like small signaling molecules (8). Binding of the ligand to the receptor results in the dissociation of the AlpZ-ligand complex from the target DNA sequence, thereby allowing expression of the downstream gene. In the AlpZ/autoregulator system, the signaling molecule, however, most likely differs from typical GBLs as its resistance to alkaline inactivation is incompatible with the presence of a γ-butyrolactone ring (8). This autoinducer might instead belong to the 2-alkyl-4-hydroxymethylfuran-3-carboxylic acid (AHFCA) family of antibiotic biosynthesis inducers recently discovered in S. coelicolor (11; see also Discussion). In addition, AlpZ was shown to negatively control its own expression and the expression of alpW, another deduced regulatory gene whose product also shows a high overall similarity to proteins of the GBL receptor family (8). AlpW homologs act as transcriptional repressors: for example, TylQ from S. fradiae, which is involved in the regulation of tylosin production (47); BarB from Streptomyces virginiae, which regulates virginiamycin biosynthesis (32); and the pSLA2-L plasmid-encoded protein SrrB, which negatively controls lankacidin and lankamycin biosynthesis in S. rochei (2). On the basis of both their ability to interact with a small regulatory molecule partner and their pI value, the members of this class of transcriptional regulators have been divided into two groups. One group includes the true receptors, and the other group comprises pseudoreceptors (24). So far, all members of the group that have been demonstrated to bind to GBL molecules display a low pI (∼5), whereas the other group of proteins, with a higher pI (∼10), fail to respond to any tested autoregulators. Therefore, with a pI of 11.1, AlpW is likely to belong to the second group of pseudo-GBL receptors.
In this study, we report the role of AlpW in the complex regulatory cascade controlling expression of the alp gene cluster. We describe genetic and biochemical studies demonstrating that AlpW acts as a late transcriptional repressor which is able to switch off expression of the biosynthetic genes within the alp cluster during the late stages of growth. Moreover, the alpW mutant produced sufficient quantities of the metabolic products of the alp cluster to enable their identification as known members of the kinamycin family of natural products. These antibiotics, originally isolated from S. murayamaensis (21), are diazo-substituted benzo[b]fluorenes that belong to the angucyclinone family (46). They have recently attracted renewed interest because, in addition to their good activity against Gram-positive bacteria, they have potential as anticancer agents (20, 36).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains, plasmids, and cosmids used in this study are listed in Table 1. Streptomyces strains were manipulated as described previously (25, 41). Pigment and antibiotic production were assessed on or in R2 medium as described previously (1, 41). Escherichia coli strains were routinely cultivated in Luria-Bertani (LB) and SOB liquid media (44).
TABLE 1.
Bacterial strains, cosmids, and plasmids used in this work
| Strain, cosmid, or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| Strains | ||
| S. ambofaciens | ||
| ATCC 23877 | Wild type | 42 |
| ΔΔalpW mutant | alpW loci on two chromosomal arms replaced by a scar | This study |
| E. coli | ||
| DH5α | General cloning strain | 19 |
| ET12567 | Nonmethylating strain used for conjugation with Streptomyces | 31 |
| BW25113 | Strain used for PCR-targeted mutagenesis | 13 |
| BL21(DE3) | Strain used for heterologous protein expression | Novagen |
| B. subtilis ATCC 6633 | Strain used as indicator in bioassays | |
| Cosmids | ||
| F6 | Cosmid from genomic library of S. ambofaciens ATCC 23877; bla neo | 29 |
| F6ΔalpW:aac(3)-IV/oriT | alpW replaced by aac(3)-IV/oriT cassette in F6; bla neo | This study |
| F6ΔalpW::scar | alpW replaced by 81-bp scar in F6; bla neo | This study |
| F6ΔalpX::scar | alpX replaced by 81-bp scar in F6; bla neo | B. Aigle, unpublished data |
| Plasmids | ||
| pIJ773 | oriT aac(3)IV | 18 |
| pIJ778 | oriT aadA | 18 |
| pIJ790 | gam bet exo cat | 18 |
| pUZ8002 | Mobilization plasmid; neo | 40 |
| BT340 | FLP recombination plasmid; flp bla cat repA101 | 13 |
| pGEM-T Easy | PCR cloning vector; bla | Promega |
| pGEM-alpWexp | bla alpW | This study |
| pGEM-alpW | bla alpW, including its own promoter region | This study |
| pGEM-alpW-loop | pGEM-alpW derivative; bla alpW with internal deletion from G217 to C288 | This study |
| pSET152 | oriT attP int aac(3)-IV | 6 |
| pSET-alpW | oriT attP int aac(3)-IV alpW, including its own promoter region | This study |
| pSET-alpW-loop | oriT attP int aac(3)-IV alpW with deletion from G217 to C288 | This study |
| pIB139 | oriT attP int aac(3)-IV ermE*p | 51 |
| pIBW | oriT attP int aac(3)-IV ermE*p alpW | This study |
| pIBW-loop | oriT attP int aac(3)-IV ermE*p alpW with deletion from G217 to C288 | This study |
| pIJ8600 | oriT attP int aac(3)-IV tipAp | 48 |
| pIJW | oriT attP int aac(3)-IV tipAp alpW | This study |
| pIJW-loop | oriT attP int aac(3)-IV tipAp alpW with deletion from G217 to C288 | This study |
| pET-12a | E. coli expression vector; bla T7 promoter | Novagen |
| pET-alpW | pET-12a derivative; bla T7 promoter alpW | This study |
| pET-alpW-loop | pET-12a derivative; bla alpW with deletion from G217 to C288 | This study |
| pSBET | neo tRNA(Arg)(AGA/AGG) | 45 |
| pTST101 | E. coli expression vector; bla malE-egfp fusion | J. Altenbuchner, personal communication |
| pTST-alpW | pTST101 derivative; bla malE-alpW fusion | This study |
bla, ampicillin resistance gene; neo, kanamycin resistance gene; aac(3)-IV, apramycin resistance gene; oriT, origin of transfer; aadA, spectinomycin and streptomycin resistance gene; gam, inhibits the host exonuclease V; bet, single-stranded DNA binding protein; exo, exonuclease promoting recombination along with bet; cat, chloramphenicol resistance gene; flp, FLP recombinase gene; repA101, thermosensitive replication origin; attP, phage ΦC31 attachment site; int, integrase gene.
Nucleic acids and protein manipulation.
Isolation, cloning, and manipulation of DNA were carried out as previously described in references 25 and 29 for Streptomyces and reference 44 for E. coli. Southern and pulsed-field gel electrophoresis analyses were performed as described in reference 30. Total RNAs were isolated by the “modified Kirby mix” method, as described in reference (25), from 2-ml samples of S. ambofaciens R2 liquid-grown cultures. Crude protein extracts or purified proteins were resolved by SDS-PAGE (12% resolving polyacrylamide gel) and were visualized by Coomassie brilliant blue staining.
PCR and RT-PCR.
Amplification of DNA fragments by PCR was performed with Taq DNA polymerase (NEB) or high-fidelity DNA polymerase Phusion (Finnzymes) according to the manufacturer's instructions. The method used for reverse transcription-PCR (RT-PCR) analysis was previously described (8). The cDNAs were obtained after reverse transcription of 4 μg of DNase I-treated total RNA with SuperScript III reverse transcriptase (Invitrogen) and high-GC-content random hexamer primers (Oligo spiking; Eurogentec). The absence of residual genomic DNA in the RNA samples was verified before addition of reverse transcriptase by 35 cycles of PCR with primers hrdB-F and hrdB-R. The sequences of primer pairs hrdB-F and hrdB-R, KSI-F and KSI-R, KSII-F and KSII-R, alpV-1 and alpV-2, alpT-1 and alpT-2, alpU-1 and alpU-2, and alpW-1 and alpW-2, which were used to amplify cDNAs for the hrdB-like gene, alpA, alpR, alpV, alpT, alpU, and alpW, respectively, are described elsewhere (8). Another set of primers, RT-alpX-1 and RT-alpX-2, was used for transcriptional analyses of the alpXW locus to assess alpW expression in the alpW deletion strain (Table 2). The negative and positive controls of PCR correspond, respectively, to addition of water and genomic DNA instead of cDNA templates.
TABLE 2.
Oligonucleotides used in this work
| Use(s) | Primer | Nucleotide sequence (5′→3′)a |
|---|---|---|
| Deletion of alpW and control of gene replacement | alpW-rep1 | AAGCACGGCAACCCACCGCAGTGACGGAAGCGAGCGATGATTCCGGGGATCCGTCGACC |
| alpW-rep2 | TCAGGGGGCACGGGGAACAGACCTTGCGGTCTTATTTCATGTAGGCTGGAGCTGCTTC | |
| CW1 | GTGGACGACGTCATCGA | |
| CW2 | AAGATGCGGCGGATACGG | |
| Complementation, overexpression, heterologous expression, and transcriptional analyses | alpW-compl-F | CGGAATTCTGGCTGATTCATGCGCGT |
| alpW-compl-R | GCTCTAGATCAGCGGGGAAAGGAGCC | |
| alpW-tip | GAAGGTACATATGGTCAGACAGGAACGTGCA | |
| MBP-AlpW1 | AAAGGATCCATGGTCAGACAGGAACGT | |
| MBP-AlpW2 | CACAAGCTTTCAGCGGGGAAAGGAGCC | |
| RT-alpX-1 | TCGCGGACCATGAACACGA | |
| RT-alpX-2 | GCATCTTCCAGCGCAACAC | |
| Deletion of the AlpW loop | Slim-Ft | CGGCGCCCGCCGGCACCGGTGATGGTCTGCAGGACGCC |
| Slim-Fs | GGTGATGGTCTGCAGGACGCC | |
| Slim-Rt | GGTGCCGGCGGGCGCCGGGACACCAGCACCCTGCAGC | |
| Slim-Rs | GGACACCAGCACCCTGCAGC | |
| Verif-loop-fwd | TGGCCGTCACCCAGGTGTGC | |
| Verif-loop-rev | TGCACTTCCACTTCGCCAGC |
For the primers alpW-rep1 and alpW-rep2, the sequences in boldface type correspond to the sequences immediately downstream and upstream of alpW that include the stop and start codons (underlined). For the primers alpWcompl-F, alpWcompl-R, alpW-tip, MBP-AlpW1, and MBP-AlpW2, the sequences in boldface type correspond, respectively, to the EcoRI, XbaI, NdeI, BamHI, and HindIII sites used for cloning.
Construction of S. ambofaciens alpW deletion strain.
The REDIRECT system (18) was used to make the in-frame deletion of the two copies of alpW in S. ambofaciens ATCC 23877, as described in previous work for deleting other genes of the alp cluster (8). The primer pair alpW-rep1 and alpW-rep2 (Table 2) was used to amplify the aac(3)-IV/oriT cassette from pIJ773 (18). Only the start and stop codons of alpW remained after deletion. Gene replacement was confirmed by Southern blotting and PCR analysis using the flanking primers CW1 (112 nucleotides [nt] upstream of the start codon of alpW) and CW2 (106 nt downstream of the stop codon of alpW). Pulsed-field gel electrophoresis analyses were carried out to rule out the presence of any chromosomal rearrangement.
Complementation of the alpW mutant.
The genes alpX and alpW are most likely to be transcribed as a single transcript since the two open reading frames are separated from each other by an intergenic region as short as 13 bp and thus would share the same promoter region. Therefore, the native promoter region located upstream of alpX and the coding sequence of alpW were directly obtained by PCR using the cosmid F6ΔalpX::scar (Table 1), in which alpX was in-frame deleted, leaving a scar of 81 bp (B. Aigle, unpublished data). Using this cosmid as the template and high-fidelity enzyme, a PCR product encompassing 98 bp upstream of the alpX start codon and the alpW coding sequence was obtained with primers alpW-compl-F and alpW-compl-R (Table 2). After terminal dATP addition, the PCR product was first ligated with pGEM-T Easy vector (Promega). The insert part of the newly generated vector pGEM-alpW was further verified by sequencing to confirm the integrity of alpW. The gel-purified EcoRI restriction fragment of pGEM-alpW that includes alpW was ligated with the pSET152 vector (6) previously digested with the same enzyme, yielding pSET-alpW. A complementation vector containing a modified version of alpW lacking an internal sequence was also prepared by cloning the EcoRI fragment from pGEM-alpW-loop (see below; Table 1) into the EcoRI site of pSET152, yielding pSET-alpW-loop. The integrative shuttle vectors pSET-alpW and pSET-alpW-loop were then introduced into the S. ambofaciens alpW deletion strain by means of intergeneric conjugal transfer. As a control, pSET152 was introduced into the wild-type and alpW deletion strains.
Overexpression of alpW using a thiostrepton-inducible promoter, tipAp, or constitutive promoter, ermE*p.
Using cosmid F6 as template, the alpW coding sequence was amplified by PCR with primers alpW-tip and alpW-compl-R (Table 2), which include an NdeI site and an XbaI site, respectively. The PCR product was first cloned into pGEM-T Easy (Promega), yielding pGEM-alpWexp1. The integrity of the insert was checked by sequencing. The pGEM-alpWexp1 vector was then digested with NdeI and XbaI enzymes, and the fragment corresponding to the alpW coding sequence was gel purified and ligated into the vectors previously digested with the same enzymes, pIJ8600 (under the control of tipAp) (48) and pIB139 (under the control of ermE*p) (51), in which a typical Streptomyces ribosome binding site (RBS) sequence (AAAGGAGG) was previously inserted between the BamHI and NdeI sites of the multiple-cloning site (MCS) region (8). This yielded the plasmids pIJW and pIBW, respectively.
To obtain pIJ8600 and pIB139 derivative vectors containing the alpW coding sequence lacking the extra internal sequence (see Results), a PCR was carried out with the primer set alpW-compl-R and alpW-tip and with pGEM-alpW-loop (see below) as the template. The PCR product was then digested by NdeI and XbaI and cloned into pIJ8600 and pIB139 previously digested by the same enzymes, yielding pIJW-loop and pIBW-loop, respectively.
The conjugative and integrative vectors pIJW, pIBW, pIJW-loop, and pIBW-loop were introduced by conjugation into the wild-type and alpW deletion S. ambofaciens strains. The wild-type or ΔΔalpW strains, designated by “+pIJW1” and “+pIJW2” in Fig. 3, correspond to two independent exconjugants.
Heterologous expression and purification of recombinant AlpW and AlpW-loop proteins.
The alpW coding sequence was isolated from pIJW and pIJW-loop (Table 1) after an NdeI/BamHI digest and inserted into the expression vector pET-12a (Novagen) digested with the same enzymes, resulting in pET-alpW and pET-alpW-loop, respectively (Table 1). The alpW coding sequence contains rare translated arginine codons (AGA and AGG) in E. coli. Thus, for heterologous expression, competent E. coli BL21(DE3) cells (Novagen) were electroporated with pET-alpW along with the pSBET vector (45), which contains an arginine tRNA-encoding gene able to efficiently recognize AGA and AGG codons. An LB culture of E. coli BL21(DE3)/pSBET harboring pET-alpW or pET-alpW-loop containing ampicillin (50 μg/ml) and kanamycin (50 μg/ml) was grown at 37°C at 250 rpm, until it reached an optical density at 600 nm (OD600) of ∼0.6, at which time 0.1 mM IPTG was added for induction. After further 4 h of incubation at 37°C, cells were collected, resuspended in TE buffer (Tris-HCl 30 mM, EDTA 1 mM, pH 8) containing 200 mM NaCl, and then disrupted by sonication. After centrifugation (30 min, 16,000 rpm, 4°C), aliquots of crude protein extract containing AlpW and AlpW-loop were kept frozen at −70°C for later use in gel retardation experiments. AlpW was further purified by sequential steps using ammonium sulfate precipitation, gel filtration, and ion-exchange chromatography as described for the homologous protein AlpZ (8). SDS-PAGE confirmed the purity of AlpW, albeit in low quantity, as a predominantly single band with an apparent migration at 25 kDa.
For stabilization of AlpW, which tends to precipitate during the sequential steps of purification, AlpW was fused to the C terminus of the highly soluble maltose binding protein (MBP). The coding sequence of alpW was thus amplified by high-fidelity PCR using the MBP-AlpW1 and MBP-AlpW2 primer pair (Table 2), which include at the 5′ termini BamHI and HindIII sites, respectively, and cosmid F6 (Table 1) as the template. The PCR product after purification was cloned into pGEM-T Easy according to the recommendation of the manufacturer (Promega), and the alpW sequence was checked by sequencing. The BamHI/HindIII alpW fragment from the previous vector was cloned into the BamHI/HindIII vector part of pTST101 (J. Altenbuchner, personal communication) (Table 1), yielding pTST-alpW. For expression, E. coli DH5α was transformed with pTST-alpW and cells were grown in LB medium supplemented with ampicillin (50 μg/ml) at 37°C at 250 rpm to an OD600 of ∼0.8. Rhamnose (10 mM) was added for induction of the recombinant protein expression. After an additional 3 h of cultivation, cells were harvested by centrifugation, washed, and disrupted by sonication in a buffer containing 20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol (DTT). After centrifugation, the fusion protein MBP-AlpW present in the soluble fraction was purified to homogeneity with amylose resin. When necessary, the MBP tag was cleaved off from AlpW by using factor Xa (NEB) as recommended by the supplier.
In-frame deletion of an internal sequence within alpW.
The nucleotide sequence spanning from nt 217 to nt 288 in the alpW CDS was deleted directly on pGEM-alpW by the site-directed, ligase-independent mutagenesis (SLIM) PCR method (9). Long-tailed primers Slim-Ft and Slim-Rt and short corresponding primers Slim-Fs and Slim-Rs were designed to specifically remove the internal low-complexity sequence in alpW (Table 2). The PCR conditions used to amplify the whole plasmid pGEM-alpW were as follows: 2 min at 96°C; 10 cycles of 30 s at 95°C, 20 s at 50°C, and 4 min at 70°C followed by 15 cycles of 30 s at 95°C, 20 s at 55°C, and 4 min at 70°C; and then a final elongation step of 7 min at 70°C. The template plasmid was removed by restriction with DpnI. After transformation of E. coli DH5α with PCR products obtained by following the SLIM PCR protocol, plasmids bearing the deletion were in the first instance screened by colony PCR with flanking primers Verif-loop-fwd and Verif-loop-rev. The candidate plasmids were then confirmed by sequencing to carry the desired deletion in alpW and for the integrity of the rest of the alpW sequence, yielding pGEM-alpW-loop.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed as described elsewhere (8) with the Roche digoxigenin (DIG) gel shift kit, 2nd generation, following the manufacturer's instructions. DIG-labeled DNA probes were obtained as described in reference 8. Typically, 20 fmol of DIG-labeled probe was used in the reaction mixture. Recombinant AlpW proteins and S. ambofaciens extract containing the alpZ interactive ligand(s) in EMSA were used as described in reference 8.
Detection of metabolic products of the alp cluster by bioassay and reverse-phase HPLC.
Bioassays were carried out as previously described with Bacillus subtilis ATCC 6633 as the indicator strain, using plugs from cultures of S. ambofaciens grown on R2 agar or supernatants from cultures grown in R2 liquid (41). High-pressure liquid chromatography (HPLC) was performed as previously described (41) with a Lichrosphere RP18 column (150 by 2 mm, 5 μm; Merck).
Purification and structure elucidation of metabolic products of the alp cluster.
Semipreparative HPLC on an Agilent 1100 instrument equipped with an Agilent Zorbax Eclipse RP-C18 column (21 by 100 mm, 5 μm) monitoring absorbance at 424 nm was used to purify metabolic products of the alp cluster from a chloroform extract of the spent R2 culture supernatant of the alpW double deletion (ΔΔalpW) mutant. The elution conditions were as follows. Solvent A contained water plus 0.1% trifluoroacetic acid (TFA), and solvent B contained acetonitrile plus 0.1% TFA. The flow rate was 5 ml/min for 0 to 30 min with 60% A-40% B to 30% A-70% B, 30 to 35 min with 30% A-70% B to 100% B, 40 to 45 min with 100% B, and 45 to 60 min with 100% B to 40% A-60% B. High-resolution mass spectrometry (MS) measurements were carried out on a Bruker MaXis mass spectrometer. Nuclear magnetic resonance (NMR) experiments (1H, 13C, correlation spectroscopy [COSY], heteronuclear single-quantum coherence [HSQC], heteronuclear multiple-bond coherence [HMBC], and nuclear Overhauser effect spectroscopy [NOESY]) were performed on a Bruker Avance 500-MHz spectrometer in CDCl3 at 25°C. Residual CHCl3 was used as an internal standard for signal calibration.
RESULTS
AlpW acts as a late repressor of orange pigment and antibiotic production.
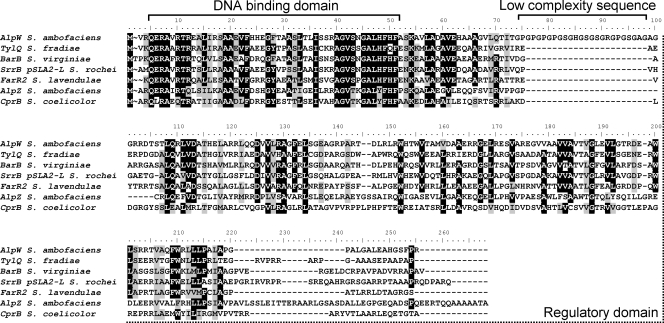
Within the subcluster of regulatory genes, the product of alpW was assigned by comparison with databases as a member of the TetR transcriptional regulator family (41). AlpW is homologous to pathway-specific repressors involved in the regulation of antibiotic biosynthetic gene clusters in Streptomyces such as TylQ (45% identity, 53% similarity) from S. fradiae (4), BarB (43% identity, 54% similarity) from S. virginiae (26), FarR2 (41% identity, 54% similarity) from S. lavendulae (28), or CprB (29% identity, 47% similarity) from S. coelicolor (38) (Fig. 1). In comparison with these homologues, AlpW intriguingly bears an additional sequence of low complexity, localized between the N-terminal helix-turn-helix DNA binding domain and the regulatory domain of the protein. This spacer was deleted to assess its possible function (see below).
FIG. 1.
Amino acid sequence alignment of AlpW and characterized homologues from the protein databases. The DNA-binding and regulatory domains were assigned by comparison with the sequence of CprB, a homologue of known structure (34). The low-complexity region of AlpW is indicated.
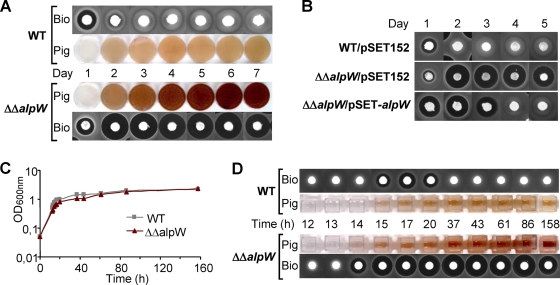
To determine the role of alpW in the regulation of antibiotic and orange pigment biosynthesis, the two copies of alpW were removed by in-frame deletion. The genomic DNA isolated from three independent mutants was analyzed by PCR and Southern analysis (data not shown). This confirmed the presence of the mutation in both terminal inverted repeats (TIRs). Pulsed-field gel electrophoresis was used to rule out the formation of large genomic rearrangements (data not shown). The resulting strains lacking alpW and designated “ΔΔalpW” strains, showed growth and morphological characteristics identical to those of the parent strain on SFM, HT, R2YE, and R2 agar plates (data not shown) and in R2 liquid medium (Fig. 2 C), indicating that AlpW is not involved in the growth and morphological differentiation of the bacteria. As the alp cluster genes were previously shown to be associated with the production of an orange pigment and an antibacterial activity, their appearance during growth on solid R2 medium and in liquid R2 medium, a suitable medium for detection of both compounds, was thoroughly explored (Fig. 2). Plugs from agar plates and supernatant from liquid-grown cultures of the mutant and parent strains were assessed during a time course for their ability to inhibit the growth of Bacillus subtilis. On surface-grown cultures, both the wild-type and the mutant strains started to produce the antibiotic and the light orange pigment after 1 day of incubation (Fig. 2A). The colorless antibiotic is believed to be rather rapidly converted by either degradation or modification into the orange pigment, which shows no bioactivity against B. subtilis (41). In the parent strain, the antibacterial activity disappeared after the second day of growth, revealing that the production of the antibiotic ceases after approximately 24 h. In contrast, the ΔΔalpW strains continued to produce the antibiotic activity during a >7-day period (Fig. 2A). In addition, the mutant produced a dark red-brown pigment which likely results from the accumulation of the orange pigment observed in wild-type cultures (the intensity of which remained unchanged once antibacterial activity had disappeared).
FIG. 2.
Effect of alpW deletion on production of diffusible pigment and antibiotic activity. (A) Pigment (Pig) and antibiotic (Bio) syntheses were assessed on R2 plates over 7 days in the wild-type (WT) and alpW double deletion (ΔΔalpW) strains. The photos were taken from below the plate. Inhibition of B. subtilis growth was visualized by the dark halo surrounding the agar plug. (B) Antibiotic production assays in the complemented ΔΔalpW strain carrying the pSET152 derivative pSET-alpW in comparison with the WT/pSET152 and ΔΔalpW/pSET152 control strains (C) Growth curves of the WT and ΔΔalpW strains in R2 liquid cultures. (D) Bioactivity and pigment production associated with the liquid fermentation in panel C.
To ensure that the absence of alpW in the ΔΔalpW strain was responsible for the production of the dark red-brown pigment and continuous production of the antibiotic, complementation experiments were carried out by reintroducing a copy of alpW under the control of its own promoter into the chromosomal ΦC31 attachment site by using the plasmid pSET-alpW (see Materials and Methods). It is noteworthy that alpX (a deduced carboxyl transferase) and alpW likely constitute an operon, given that the deduced translational initiation codon of alpW is only 13 nucleotides downstream of the stop codon of alpX. Therefore, the alpW coding sequence and the associated promoter region of alpXW were amplified from a cosmid in which alpX was in-frame deleted (see Materials and Methods). As controls, both the parent and ΔΔalpW strains were transformed with the empty pSET152 vector. The ΔΔalpW strains bearing the vector alone showed enhanced production of the antibiotic and the pigment (Fig. 2B) (similar to that observed for the ΔΔalpW strains) compared to the wild-type strain harboring pSET152. The complemented ΔΔalpW/pSET-alpW strain showed slightly higher levels of pigment production than the wild-type strain but lower than that of the ΔΔalpW/pSET152 strain (data not shown). Similarly, antibiotic production in the ΔΔalpW/pSET-alpW strain ceased after 3 days instead of at least 6 days in the ΔΔalpW/pSET152 strain (Fig. 2B). The partial restoration to the wild-type phenotype resulting from the reintroduction of a single ectopic copy of alpW supports the involvement of alpW in the repression of pigment and antibiotic production.
The unrelenting production of the antibiotic and pigment in the ΔΔalpW mutant strains was also observed during growth in R2 liquid cultures (Fig. 2C and D). The production of both compounds in the S. ambofaciens wild-type and ΔΔalpW strains occurred in the late transition phase, but whereas it was limited to a short period in the wild type, production was increased to at least 6 days in the mutant strain (Fig. 2D). The presence of the compound associated with the antibiotic activity during this late production period was verified by HPLC analyses (data not shown).
The product of alpW thus seems to act as a late repressor of antibiotic and pigment biosynthesis, shutting off expression of the alp gene cluster in the late stages of growth.
Overexpression of alpW abrogates pigment and antibiotic production.
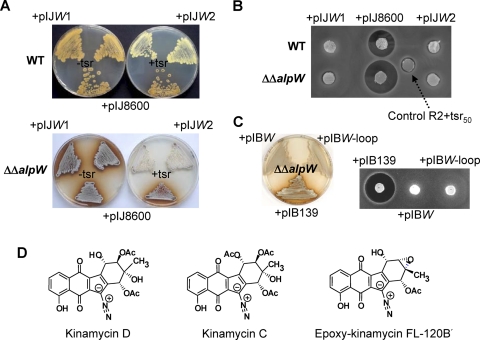
To further confirm its role as a repressor, AlpW was overexpressed in both S. ambofaciens wild-type and ΔΔalpW strains by inserting the alpW coding sequence under the control of either the thiostrepton-inducible tipAp promoter or the strong, constitutive ermE*p promoter, into the conjugative and integrative pIJ8600 (48) and pIB139 (51) vectors, yielding pIJW and pIBW, respectively. As a control, the empty pIJ8600 and pIB139 vectors were introduced in the S. ambofaciens wild-type and ΔΔalpW strains. In the absence of thiostrepton, both the wild-type and ΔΔalpW strains carrying pIJW produced the pigment (Fig. 3 A) and antibiotic (data not shown) in a manner similar to that of the control strains on agar plates. However, induction of alpW expression by addition of thiostrepton to the plate resulted in the abrogation of antibiotic and pigment production in the wild-type strain bearing pIJW (Fig. 3A and B). This inhibitory effect was even more striking when the overexpression construct was introduced into the antibiotic- and pigment-overproducing ΔΔalpW strain. The production of both compounds was completely blocked when the exconjugants were grown in the presence of thiostrepton. In comparison, the ΔΔalpW strain, which carries the empty pIJ8600 vector, still produced both compounds even in the presence of thiostrepton (Fig. 3A and B). Similarly, the wild-type (not shown) and ΔΔalpW strains carrying pIBW failed to produce the antibiotic and the associated pigment, unlike the control strains (Fig. 3C). Together, these results imply that AlpW plays a crucial role in the repression of antibiotic and pigment production.
FIG. 3.
Effect of overexpression of alpW in the wild-type (WT) and ΔΔalpW strains. (A) Two clones of the WT and ΔΔalpW strains carrying the overexpression construct pIJW (designated pIJW1 and pIJW2) are shown (photos are taken from below for the WT strain and from above for the ΔΔalpW strain) after 8 days of growth at 30°C. −tsr, no addition of thiostrepton to the medium; +tsr, 12.5 μg/ml of thiostrepton included as an inducer. (B) The antibiotic production by the WT and ΔΔalpW strains was tested against B. subtilis from R2 culture plates supplemented with 50 ng/ml of thiostrepton. The control plug was taken from a noninoculated R2 plate supplemented with 50 ng/ml of thiostrepton. (C) ΔΔalpW strains carrying the overexpression plasmids pIB139 (control), pIBW, and pIBW-loop were assessed for pigment production (left) and antibiotic production (right). The picture (left) was taken from above after 5 days of incubation at 30°C. Agar plugs (right) were taken from plates inoculated with the ΔΔalpW strain containing pIB139, pIBW, or pIBW-loop after 5 days of incubation at 30°C. (D) Structures of the known kinamycins identified as metabolic products of the alp cluster.
Identification of the metabolic products of the alp cluster as kinamycins.
The antibiotic products of the alp cluster accumulate for only a few hours after entry into the transition phase in wild-type S. ambofaciens. This hindered attempts to isolate sufficient quantities of the antibiotics for structure elucidation. Thus, we investigated isolation of the antibiotics from the deregulated ΔΔalpW mutant strain, which produces them continuously. This allowed us to purify sufficient material for structural elucidation. High-resolution MS (HRMS) analyses of the purified antibiotics gave their molecular formulae as C22H18N2O9 (calculated for C22H18N2O9Na+, 477.0905; found, 477.0895), C24H20N2O10 (calculated for C24H20N2O10Na+, 519.1010; found, 519.1003), and C20H14N2O7 (calculated for C20H14N2O7Na+, 417.0593; found, 417.0587) (see Fig. S1 to S3 in the supplemental material). These correspond to the molecular formulae of kinamycins D and C, and FL-120B′, respectively (Fig. 3D). One-dimensional (1D) and 2D NMR spectroscopic analyses of the two major compounds confirmed that they are indeed kinamycin D and FL-120B′, as suggested by the HRMS data (see Fig. S4 and S5 and Tables S1 and S2 in the supplemental material).
AlpW negatively regulates the expression of the essential pathway-specific activator gene alpV.
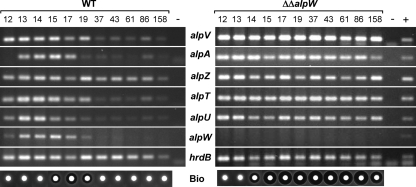
To validate the negative regulatory role of AlpW, transcriptional studies were conducted in parallel with the alpW mutant and wild-type strains (Fig. 4). Transcripts from the β-ketoacyl synthase gene, alpA, and the five regulatory genes alpT, alpU, alpV, alpZ, and alpW were detected by RT-PCR experiments using total RNA isolated from mycelial samples harvested at different time points during growth in R2 liquid medium (Fig. 2C). The transcript of the major and essential sigma factor gene hrdB, which is expressed at an almost constant level during growth, was used as an internal control (Fig. 4). In the parent strain, the expression of the alpA biosynthetic gene was induced during the late transition phase and persisted until entry into the stationary phase, as previously observed (1, 8, 41). In the alpW mutant, alpA transcripts were also detected from the transition phase, but in contrast to the wild type, the transcripts were detected 1 h earlier and were strongly maintained through the stationary phase. This observation concurs with the continuous production of antibiotic activity in the alpW mutant. This maintenance of antibiotic production is most likely caused by derepression of the pathway-specific activators of the alp cluster, especially alpV, but also alpT and alpU, because expression of these genes persisted in the ΔΔalpW strain. In contrast, the transcription of the alpZ gene in the absence of AlpW was not significantly affected. As expected, the alpW transcript was not detected in the alpW disruption strain since the whole coding sequence of alpW (except for the start and stop codons) was removed during the construction of this mutant. Nevertheless, the expression of alpW could be indirectly addressed by monitoring the expression of alpX, which is located directly upstream of alpW in the same operon. Whereas the transcription of alpX and alpW (Fig. 4) decreased after entry into the stationary phase in the wild type, the transcription of alpX was maintained at a constant high level during the late stages of growth in the ΔΔalpW mutant (data not shown), suggesting that AlpW has a negative regulatory effect on its own expression. Together these results indicate that AlpW represses transcription of alpW, as well as the pathway-specific activator genes.
FIG. 4.
Comparative transcriptional studies of selected alp genes in the wild-type and ΔΔalpW strains by reverse transcriptase PCR. Transcripts of hrdB (major and essential sigma factor gene; positive control), alpV (essential SARP activator gene), alpA (β-ketoacyl synthase gene involved in both orange pigment and kinamycin biosynthesis), alpZ (repressor gene), alpT (SARP gene), alpU (SARP gene), and alpW from the WT and alpW mutant strains were analyzed by RT-PCR with 25 cycles of PCR on cDNA generated from RNA isolated at various times during growth (represented by the curves shown in Fig. 2C). Negative (−) and positive (+) controls for the PCR are indicated. The bioactivity (Bio) against B. subtilis observed at each time point is shown below each lane.
AlpW shares DNA binding motifs with AlpZ.
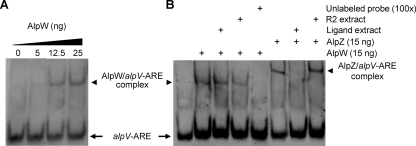
In our previous study, the GBL-like receptor protein AlpZ was shown to repress transcription of the other regulatory genes in the alp cluster through binding to specific AT-rich DNA sequences located within their promoter regions (8). These DNA motifs were identified by comparison with a consensus ARE sequence that is typically recognized by GBL receptors (14, 27). They were localized upstream of alpZ, (AREZ), alpV (AREV), and the alpXW operon (AREXW) and in the divergent promoter region of alpT and alpU (AREU) (see Fig. S6 in the supplemental material). In order to explore the ability of AlpW to bind to these specific sequences and determine whether it could interfere with the AlpZ-dependent regulation by binding at the same sites, various DIG-labeled probes (40 bp to 392 bp) bearing the ARE sequences and used in previous work to determine AlpZ DNA targets (8) were tested along with purified recombinant AlpW in gel mobility shift assays. Thus, AlpW was expressed and purified as a recombinant protein in E. coli cells. Most of the recombinant protein expressed from the pET12a vector appeared to be unstable and precipitated in solution. To circumvent this insolubility issue, alpW was appended to the highly soluble maltose binding protein (MBP) sequence of malE by means of pTST101 (J. Altenbuchner, personal communication) and purified to homogeneity by affinity chromatography on amylose resin. Prior to gel shift analyses, the MBP was cleaved from AlpW by the endoprotease factor Xa (although the DNA binding abilities of AlpW were not impaired by the MBP tag when assayed [data not shown]). No DNA binding activity was observed with DNA fragments containing the AREU motif (see Fig. S7 in the supplemental material). Also, no DNA-protein complex was observed in gel shift analyses with purified recombinant AlpZ (8), suggesting that the divergent promoter region of alpU and alpT is not under the direct control of either the AlpZ or AlpW transcriptional repressors. However, when DNA fragments encompassing the AREV, AREZ, and AREXW motifs were used in the EMSA, retardation of the migration of these probes was observed in the presence of modest amounts of recombinant AlpW (12.5 ng), as typified by the data for the 40-bp alpV-ARE probe shown in Fig. 5 A (see Fig. S7 in the supplemental material for EMSA with alpZ- and alpXW-ARE probes). Moreover, competition assays with an excess of the corresponding unlabeled probes (i.e., addition of the unlabeled probe after incubation of the labeled probe with AlpW) showed no shift in mobility, demonstrating that AlpW binds specifically to these ARE motifs (Fig. 5B, lane 5; and see Fig. S7 in the supplemental material).
FIG. 5.
DNA binding activity of AlpW examined by EMSA. (A) Twenty femtomoles of the 40-bp DIG-labeled oligonucleotide probe alpV-ARE encompassing the ARE sequence located in the alpV promoter region was incubated with increasing amounts of recombinant AlpW. (B) Effect of addition of unlabeled probe and concentrated culture supernatant extract containing the AlpZ-binding ligand (“Ligand extract”) on DNA-bound AlpW. AlpZ was used as a positive control to demonstrate the disrupting activity of the extract containing the AlpZ-binding ligand. This extract was obtained from late-transition-phase culture of the WT strain as described in reference 8. “R2 extract” corresponds to solvent extract of noninoculated R2 medium. Competition with excess of unlabeled probe (100×; 2 pmol) confirmed specificity of AlpW DNA binding activity.
The AlpW DNA binding activity is not regulated by a signaling molecule.
The first characterized GBL-like receptor homologue in the alp cluster, AlpZ, was established to respond to a specific ligand detectable in the supernatant of early-transition-phase S. ambofaciens cultures shortly before the onset of antibiotic biosynthesis (8). In this process, the ligand was able to dissociate DNA-bound AlpZ from its cognate DNA sequences. However, the high pI value of 11.1 for AlpW suggests that it falls into the class of pseudo-GBL receptors that, unlike authentic autoregulator receptors whose pI is low (∼5), have never been shown to respond to small signaling molecules (24). To address whether AlpW is nevertheless able to respond to the signaling molecule that controls AlpZ, extracts of culture supernatants containing the AlpZ-binding ligand were tested in gel shift assays together with purified recombinant AlpW. Unlike in the control experiment in which purified recombinant AlpZ was used in place of AlpW (Fig. 5B, lane 7), addition of the ligand extract failed to dissociate AlpW from the alp-ARE probes (the data for alpV-ARE are shown in Fig. 5B, lane 3; those for alpZ-ARE and alpXW-ARE are shown in Fig. S7 in the supplemental material). Thus, AlpW appears to be insensitive to the ligand found in S. ambofaciens culture extracts that controls AlpZ, indicating that the activity of AlpW is unlikely to be regulated by an autoregulator molecule, at least under the conditions tested.
The low-complexity additional sequence in AlpW plays no essential role.
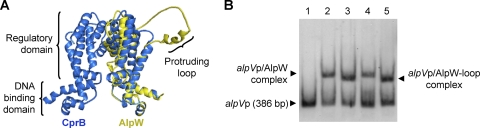
Compared to other homologues in the databases, AlpW harbors an additional low-complexity sequence consisting of GP/GA/GS/GR repeats (Fig. 1). This extra sequence is hypothesized to be located between the DNA binding domain and the regulatory domain in an externally protruding loop, from a model of the 3D structure of AlpW based on the X-ray crystal structure of the AlpW homolog CprB (34) (Fig. 6 A).
FIG. 6.
Analysis of the additional low-complexity sequence in AlpW. (A) Superposition of the 3D structure representation of the native AlpW (yellow; monomer) and the homologous pseudoautoregulator receptor CprB (blue; dimer), the structure of which was determined by X-ray crystallography (34). (B) Comparative DNA binding analysis of AlpW and AlpW-loop by EMSA. The DIG-labeled probe alpVp (386 bp) bearing the AlpW recognition sequence AREV was incubated in the absence (lane 1) or presence of soluble extracts containing recombinant AlpW (lanes 2 and 4) or AlpW-loop (lanes 3 and 5). Concentrated extracts containing the AlpZ-binding ligand were added to the mixture (lanes 4 and 5).
In order to determine if this extra sequence plays a significant role in the regulatory mechanism of the protein, the corresponding nucleotide sequence was first removed in the alpW-containing plasmid pGEM-alpW by the SLIM PCR method (9). The mutated copy of alpW under the control of its own promoter (pSET-alpW-loop), the strong constitutive ermE*p promoter (pIBW-loop), and the thiostrepton-inducible tipAp promoter (pIJW-loop) were introduced into the wild-type or ΔΔalpW strains, and the phenotype of the strains carrying the engineered alpW allele was compared with that of the strains harboring the equivalent plasmids bearing the native alpW sequence. No significant difference was observed between the strains carrying the native and modified versions of AlpW in terms of antibiotic or pigment production (see Fig. 3C for comparison of pIBW and pIBW-loop in the ΔΔalpW strain). The altered AlpW lacking the repetitive sequence was still able to complement the alpW mutant and to block the production of the pigment and the antibiotics in both the parent and alpW deletion strains. The results of these in vivo assays indicate that the additional low-complexity sequence does not impair the function of AlpW. Moreover, AlpW without the repetitive sequence was overexpressed in E. coli in a manner similar to that of the parent protein (using pET12a; see Fig. S8 in the supplemental material), and total soluble extracts were directly assayed in gel shift experiments (Fig. 6B). As expected from the results of the in vivo complementation experiments, the modified regulator was still able to bind to its DNA recognition sequences and was still unaffected by the ligand that binds to AlpZ (Fig. 6B). Consequently, under the conditions tested, this additional sequence does not appear to play any essential role in the regulatory function of AlpW.
DISCUSSION
In spite of 5 decades of study and industrial utilization, S. ambofaciens was known to produce only two antibiotics: the macrolide spiramycin and the pyrrole-amide congocidine. Sequence analysis of the alp cluster (recently identified in S. ambofaciens by a genome-mining approach) suggests that it may direct biosynthesis of a third antibiotic belonging to the angucyclinone class. Comparative phenotypic analysis of the wild-type strain and a mutant strain in which the alp genes encoding a minimal type II PKS have been deleted eventually led to the discovery of a novel antibacterial activity and a diffusible orange pigment that are metabolic products of the alp cluster (41). However, during the fermentation of wild-type S. ambofaciens strain ATCC 23877, the production of the antibacterial activity was limited to a short period during the growth cycle. This narrow window of antibiotic activity could be explained by the cessation of its biosynthesis after entry into the stationary phase of growth in liquid medium or after the onset of morphological differentiation in surface-grown cultures, along with its degradation or modification to an “inactive” pigmented form. Consequently, the purification of sufficient material for structural elucidation has been challenging. The deregulation of the biosynthetic pathway by altering regulatory genes, and especially in this case the deletion of alpW, led to a strain which persisted in antibiotic production after initial onset of its biosynthesis, allowing sufficient material to be purified for structure elucidation. Three bioactive compounds were purified from ethyl acetate extracts of culture supernatants by semipreparative HPLC and were shown by HRMS and 1D and 2D NMR spectroscopy to be known members of the kinamycin family of antitumor antibiotics (16). Thus, S. ambofaciens is the fifth actinobacterium reported to produce kinamycins, along with S. murayamaensis, a Saccharothrix sp., an unidentified actinomycete, and Streptomyces chattanoogensis subsp. taitungensis (16). A partial gene cluster believed to direct the biosynthesis of kinamycins in S. murayamaensis has been cloned and sequenced (accession no. AY228175). Heterologous expression of this gene cluster led to the production of known intermediates in kinamycin biosynthesis (dehydrorabelomycin, kinobscurinone, and stealthin C) and the shunt metabolites kinafluorenone and seongomycin (17). Therefore, we report for the first time, the complete sequence of the kinamycin biosynthetic gene cluster (the alp cluster of S. ambofaciens). It should be noted that the previously defined alp cluster (41) appears not to contain all of the genes expected to be required for kinamycin biosynthesis. Several genes flanking the originally defined alp cluster are potential candidates for the “missing” genes and are currently under investigation. Access to the complete kinamycin biosynthetic gene cluster will provide the opportunity to generate new kinamycin derivatives by genetic manipulation, which might have superior antitumor activity.
The results obtained in the present work also extend our knowledge of the complex regulatory mechanisms controlling kinamycin biosynthesis in S. ambofaciens. During the vegetative growth phase of the bacteria, the alp biosynthetic pathway is repressed by the GBL-like receptor AlpZ. This transcriptional repressor, which acts early in development, exerts its action by binding to a specific recognition site in the promoter region of target genes. The activation of the cluster is then triggered by a signaling molecule, which is detectable in the supernatant of late-exponential growth cultures. This signaling molecule is resistant to alkaline hydrolysis, unlike typical GBLs, suggesting that it does not contain a γ-butyrolactone (8). It is tempting to speculate that this molecule could belong to the recently described 2-alkyl-4-hydroxymethylfuran-3-carboxylic acid (AHFCA) family of signaling molecules that induce production of methylenomycin antibiotics in S. coelicolor. AHFCAs contain a common furan core, which is resistant to alkali treatment, in place of the common butyrolactone core of GBLs (11). In the autoregulatory system encoded within the methylenomycin biosynthetic gene cluster on the SCP1 plasmid of S. coelicolor, two genes, mmyR and mmfR, adjacent to the AHFCA biosynthetic genes, encode homologues of GBL receptors. AHFCAs appear to induce their own biosynthesis through an autoregulatory loop and the biosynthesis of methylenomycins by abolishing the DNA binding activity of the heterodimeric MmyR/MmfR repressor complex (39). Thus, the two types of inducer, GBLs and AHFCAs, seem to function in a similar manner by interacting with specific receptors. In the S. ambofaciens autoregulator system, the inducer is able to displace AlpZ from its DNA targets, hence relieving repression of the downstream genes, including alpV, alpW, and alpZ itself. The derepression of alpV, one of the essential pathway-specific activator-encoding genes, in turn allows induction of expression of the biosynthetic genes, eventually leading to the assembly of kinamycins. Nevertheless, at the same time, the transcription of alpW is also derepressed. The product of alpW, which has been shown here to act as a transcriptional repressor, can bind to the ARE sequence in the promoter region of alpV, thus preventing the initiation of its transcription. The transcription of the two other SARP-encoding genes, alpT and alpU, is likely to be indirectly downregulated by AlpW, because no binding of AlpW to the divergent promoter region between these genes was observed. AlpW has also been shown here to bind in vitro to the ARE boxes present in the promoter region of alpZ and alpW. However, in comparative transcriptional analyses, the absence of AlpW does not seem to affect the transcription of alpZ, suggesting that AlpW does not primarily target the alpZ promoter region. The absence of an in vivo role for AlpW in modulating the expression of alpZ needs, however, to be confirmed.
Unlike that of AlpZ, the binding activity of AlpW seems not to be regulated by a ligand. This observation is in agreement with the fact that AlpW belongs to the subset of pseudoautoregulator receptors on the basis of its high pI value (24). Therefore, in spite of the presence of the AlpZ-binding ligand, the accumulation of AlpW in the cell results in the repression of the alp cluster, a process that the early repressor AlpZ is obviously not able to perform in isolation. Ultimately, the repression of the whole biosynthetic pathway is nevertheless hypothesized to occur through the reintervention of AlpZ, given that the level of transcription of alpW in the wild-type strain decreases dramatically after entry into the stationary phase. Unless AlpW is particularly stable, it seems likely that AlpZ can replace AlpW at the ARE sites, because high levels of alpZ transcripts are still detectable during the late stages of growth. In this process, the disappearance of the ligand during the late stages of growth (8) as a result of its dilution and/or the concomitant downregulation of the gene(s) involved in its biosynthesis could help AlpZ to regain control of the alp gene cluster. This downregulation could be another key element in the cessation of the kinamycin biosynthesis and could involve AlpW. Indeed, in the ΔΔalpW mutant strain, although the transcription of alpZ does not seem to be impaired, AlpZ appears unable to turn off the expression of the biosynthetic genes. Therefore, we speculate that another role of AlpW would be to repress the expression of the gene(s) involved in the production of the AlpZ-binding ligand. In the absence of AlpW, the ligand would be continuously synthesized during the late stage of growth, thus preventing AlpZ from binding to its DNA targets and therefore acting as a transcriptional repressor. Alternatively or concomitantly, a transcription factor such as AlpV (or AlpT or AlpU) could act positively on the expression of the structural gene(s) responsible for the production of the ligand. A strong expression of alpV is maintained throughout the stationary phase of growth in the ΔΔalpW strain, and this could result in the extended period of kinamycin production observed in this mutant. Further analyses are now under way to clarify the complex regulatory cascade of kinamycin production.
With the exception of AlpW, there is only one other example of a pseudo-GBL receptor acting during the late stage of antibiotic production by switching off this production. It has recently been shown in S. coelicolor that ScbR2, a pseudo-GBL receptor encoded within the cpk gene cluster (a type I PKS gene cluster), may act in a negative-feedback mechanism to suppress expression of the cpk cluster and therefore to limit the biosynthesis of its products, a yellow pigment and an antibacterial compound (15). Other characterized homologues of AlpW—for instance, BarB and TylQ—do not play a similar role. BarB participates in an early step of repression of virginiamycin biosynthesis by retarding virginiamycin production by a few hours (32). In the regulation of tylosin biosynthesis, the expression of tylQ, the product of which precociously represses expression of the activator gene tylR, is inactivated by TylP, the “true” GBL receptor, to allow activation of the biosynthetic pathway (7, 47).
Compared to its homologues, AlpW contains an additional amino acid sequence of low complexity. In spite of the presence of this extra sequence, which is hypothesized to form a prominent loop, the global organization of AlpW appears to be conserved. Our model of the 3D structure of the protein indicates that neither the DNA binding helix-turn-helix domain nor the dimerization and regulatory domains of the protein seem to be displaced, because the loop is oriented away from these domains (Fig. 6A). Consistent with this, the comparative analyses of the native protein and the variant lacking the entire loop sequence did not reveal any significant role for this sequence in vitro or in vivo. Remarkably, an AlpW homologue found in another strain of S. ambofaciens (DSM 40697) also possesses a very similar low-complexity spacer which differs only by the presence of eight more amino acid residues (see Fig. S9 in the supplemental material). Regarding the evolution of the “GBL receptor” family, it has been proposed that the acquisition of the receptor function followed evolution of the DNA-binding activity of these repressors (the DNA binding domains belong to the TetR family of transcriptional repressors) (35). Therefore, the presence of the spacer in AlpW located between the DNA-binding and regulatory domains could be an evolutionary relic left over from a domain-shuffling event. Such an additional sequence is found in one other member of the GBL autoregulator receptor family, namely, AvaR3 (SAV3703) from Streptomyces avermitilis, the role of which is yet to be elucidated (see Fig. S9 in the supplemental material). However, it should be noted that most of the additional sequence in AvaR3 is not of low complexity in comparison with the corresponding sequence in AlpW.
The remarkable variability in terms of regulation of secondary metabolite biosynthetic pathways in Streptomycetes is typified in this study: the regulation of kinamycin biosynthesis in S. murayamaensis is thought to differ drastically from that of S. ambofaciens. Indeed, the kinR gene within the partial sequence of the kinamycin biosynthetic gene cluster of S. murayamaensis encodes a putative TetR/AcrR family transcriptional regulator. A homologue of this gene is absent from the alp cluster. Likewise, orf8, another putative TetR regulatory gene, does not have any homologues in the alp cluster. Conversely, none of the subclustered regulatory genes alpT, alpU, alpV, alpW, and alpZ is found within the kin cluster. We therefore conclude that, in two Streptomyces species that produce similar secondary metabolites, the control of their biosynthesis can differ remarkably. In S. ambofaciens (and more generally in Streptomyces species), the terminal regions of the linear chromosome contain nonessential genes often involved in the secondary metabolism of the bacterium. These regions, in comparison to the core region of the chromosome, are highly variable between species of Streptomyces and are thus believed to be “hot spots” for chromosomal rearrangements and integration of exogenous DNA to allow acquisition and rapid evolution of new genes and gene clusters. In this context, the alp cluster, which appears to be structured in distinct modules, might represent an example of a biosynthetic pathway that has been built through the assembly of groups of genes acquired by horizontal gene transfer.
Supplementary Material
Acknowledgments
R.B., L.S., M.V.M., and C.C. were supported by ActinoGEN, an integrated project funded by the European Commission under the 6th framework program (FP6-5224). The Bruker MaXis mass spectrometer used in this research was obtained, through Birmingham Science City: Innovative Uses for Advanced Materials in the Modern World (West Midlands Centre for Advanced Materials Project 2), with support from Advantage West Midlands (AWM) and in part funded by the European Regional Development Fund (ERDF).
We thank Jean-Michel Girardet (Université de Lorraine, France) for help with HPLC analyses and Joe Altenbuchner (University of Stuttgart, Germany) and Peter Leadlay (University of Cambridge, United Kingdom) for the kind gifts of pTST101 and pIB139, respectively.
Footnotes
Published ahead of print on 30 December 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aigle, B., X. Pang, B. Decaris, and P. Leblond. 2005. Involvement of AlpV, a new member of the Streptomyces antibiotic regulatory protein family, in regulation of the duplicated type II polyketide synthase alp gene cluster in Streptomyces ambofaciens. J. Bacteriol. 187:2491-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa, K., S. Mochizuki, K. Yamada, T. Noma, and H. Kinashi. 2007. Gamma-butyrolactone autoregulator-receptor system involved in lankacidin and lankamycin production and morphological differentiation in Streptomyces rochei. Microbiology 153:1817-1827. [DOI] [PubMed] [Google Scholar]
- 3.Barona-Gomez, F., et al. 2006. Multiple biosynthetic and uptake systems mediate siderophore-dependent iron acquisition in Streptomyces coelicolor A3(2) and Streptomyces ambofaciens ATCC 23877. Microbiology 152:3355-3366. [DOI] [PubMed] [Google Scholar]
- 4.Bate, N., A. R. Butler, A. R. Gandecha, and E. Cundliffe. 1999. Multiple regulatory genes in the tylosin biosynthetic cluster of Streptomyces fradiae. Chem. Biol. 6:617-624. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, S. D., et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 6.Bierman, M., et al. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 7.Bignell, D. R., N. Bate, and E. Cundliffe. 2007. Regulation of tylosin production: role of a TylP-interactive ligand. Mol. Microbiol. 63:838-847. [DOI] [PubMed] [Google Scholar]
- 8.Bunet, R., et al. 2008. Regulation of the synthesis of the angucyclinone antibiotic alpomycin in Streptomyces ambofaciens by the autoregulator receptor AlpZ and its specific ligand. J. Bacteriol. 190:3293-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu, J., P. E. March, R. Lee, and D. Tillett. 2004. Site-directed, ligase-independent mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 32:e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choulet, F., et al. 2006. Evolution of the terminal regions of the Streptomyces linear chromosome. Mol. Biol. Evol. 23:2361-2369. [DOI] [PubMed] [Google Scholar]
- 11.Corre, C., L. Song, S. O'Rourke, K. F. Chater, and G. L. Challis. 2008. 2-Alkyl-4-hydroxymethylfuran-3-carboxylic acids, antibiotic production inducers discovered by Streptomyces coelicolor genome mining. Proc. Natl. Acad. Sci. U. S. A. 105:17510-17515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosar, C., L. Ninet, S. Pinnert-Sindico, and J. Preud'Homme. 1952. Trypanocide action of an antibiotic produced by a Streptomyces. C. R. Hebd. Seances Acad. Sci. 234:1498-1499. [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folcher, M., et al. 2001. Pleiotropic functions of a Streptomyces pristinaespiralis autoregulator receptor in development, antibiotic biosynthesis, and expression of a superoxide dismutase. J. Biol. Chem. 276:44297-44306. [DOI] [PubMed] [Google Scholar]
- 15.Gottelt, M., S. Kol, J. P. Gomez-Escribano, M. Bibb, and E. Takano. 2010. Deletion of a regulatory gene within the cpk gene cluster reveals novel antibacterial activity in Streptomyces coelicolor A3(2). Microbiology 156:2343-2353. [DOI] [PubMed] [Google Scholar]
- 16.Gould, S. J. 1997. Biosynthesis of the kinamycins. Chem. Rev. 97:2499-2510. [DOI] [PubMed] [Google Scholar]
- 17.Gould, S. J., S. T. Hong, and J. R. Carney. 1998. Cloning and heterologous expression of genes from the kinamycin biosynthetic pathway of Streptomyces murayamaensis. J. Antibiot. (Tokyo) 51:50-57. [DOI] [PubMed] [Google Scholar]
- 18.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 20.Hasinoff, B. B., et al. 2006. Kinamycins A and C, bacterial metabolites that contain an unusual diazo group, as potential new anticancer agents: antiproliferative and cell cycle effects. Anticancer Drugs 17:825-837. [DOI] [PubMed] [Google Scholar]
- 21.Hata, T., S. Omura, Y. Iwai, A. Nakagawa, and M. Otani. 1971. A new antibiotic, kinamycin: fermentation, isolation, purification and properties. J. Antibiot. (Tokyo) 24:353-359. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda, H., et al. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 23.Juguet, M., et al. 2009. An iterative nonribosomal peptide synthetase assembles the pyrrole-amide antibiotic congocidine in Streptomyces ambofaciens. Chem. Biol. 16:421-431. [DOI] [PubMed] [Google Scholar]
- 24.Kawachi, R., et al. 2000. Identification of an AfsA homologue (BarX) from Streptomyces virginiae as a pleiotropic regulator controlling autoregulator biosynthesis, virginiamycin biosynthesis and virginiamycin M1 resistance. Mol. Microbiol. 36:302-313. [DOI] [PubMed] [Google Scholar]
- 25.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Fundation, Norwich, United Kingdom.
- 26.Kinoshita, H., et al. 1997. Butyrolactone autoregulator receptor protein (BarA) as a transcriptional regulator in Streptomyces virginiae. J. Bacteriol. 179:6986-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinoshita, H., T. Tsuji, H. Ipposhi, T. Nihira, and Y. Yamada. 1999. Characterization of binding sequences for butyrolactone autoregulator receptors in streptomycetes. J. Bacteriol. 181:5075-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitani, S., et al. 2008. Identification of genes involved in the butyrolactone autoregulator cascade that modulates secondary metabolism in Streptomyces lavendulae FRI-5. Gene 425:9-16. [DOI] [PubMed] [Google Scholar]
- 29.Leblond, P., et al. 1996. The unstable region of Streptomyces ambofaciens includes 210 kb terminal inverted repeats flanking the extremities of the linear chromosomal DNA. Mol. Microbiol. 19:261-271. [DOI] [PubMed] [Google Scholar]
- 30.Leblond, P., F. X. Francou, J. M. Simonet, and B. Decaris. 1990. Pulsed-field gel electrophoresis analysis of the genome of Streptomyces ambofaciens strains. FEMS Microbiol. Lett. 60:79-88. [DOI] [PubMed] [Google Scholar]
- 31.MacNeil, D. J., et al. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 32.Matsuno, K., Y. Yamada, C. K. Lee, and T. Nihira. 2004. Identification by gene deletion analysis of barB as a negative regulator controlling an early process of virginiamycin biosynthesis in Streptomyces virginiae. Arch. Microbiol. 181:52-59. [DOI] [PubMed] [Google Scholar]
- 33.Mochizuki, S., et al. 2003. The large linear plasmid pSLA2-L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol. Microbiol. 48:1501-1510. [DOI] [PubMed] [Google Scholar]
- 34.Natsume, R., Y. Ohnishi, T. Senda, and S. Horinouchi. 2004. Crystal structure of a gamma-butyrolactone autoregulator receptor protein in Streptomyces coelicolor A3(2). J. Mol. Biol. 336:409-419. [DOI] [PubMed] [Google Scholar]
- 35.Nishida, H., Y. Ohnishi, T. Beppu, and S. Horinouchi. 2007. Evolution of gamma-butyrolactone synthases and receptors in Streptomyces. Environ. Microbiol. 9:1986-1994. [DOI] [PubMed] [Google Scholar]
- 36.O'Hara, K. A., G. I. Dmitrienko, and B. B. Hasinoff. 2010. Kinamycin F downregulates cyclin D3 in human leukemia K562 cells. Chem. Biol. Interact. 184:396-402. [DOI] [PubMed] [Google Scholar]
- 37.Ohnishi, Y., et al. 2008. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190:4050-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onaka, H., T. Nakagawa, and S. Horinouchi. 1998. Involvement of two A-factor receptor homologues in Streptomyces coelicolor A3(2) in the regulation of secondary metabolism and morphogenesis. Mol. Microbiol. 28:743-753. [DOI] [PubMed] [Google Scholar]
- 39.O'Rourke, S., et al. 2009. Extracellular signalling, translational control, two repressors and an activator all contribute to the regulation of methylenomycin production in Streptomyces coelicolor. Mol. Microbiol. 71:763-778. [DOI] [PubMed] [Google Scholar]
- 40.Paget, M. S., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pang, X., et al. 2004. Functional angucycline-like antibiotic gene cluster in the terminal inverted repeats of the Streptomyces ambofaciens linear chromosome. Antimicrob. Agents Chemother. 48:575-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinnert-Sindico, S. 1954. Une nouvelle espece de Streptomyces productrice d'antibiotiques: Streptomyces ambofaciens n. sp. caractères culturaux. Ann. Inst. Pasteur (Paris) 87:702-707. [PubMed] [Google Scholar]
- 43.Ramos, J. L., et al. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69:326-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Schenk, P. M., S. Baumann, R. Mattes, and H. H. Steinbiss. 1995. Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. Biotechniques 19:196-198, 200. [PubMed] [Google Scholar]
- 46.Seaton, P. J., and S. J. Gould. 1989. New products related to kinamycin from Streptomyces murayamaensis. II. Structures of pre-kinamycin, keto-anhydrokinamycin, and kinamycins E and F. J. Antibiot. (Tokyo) 42:189-197. [DOI] [PubMed] [Google Scholar]
- 47.Stratigopoulos, G., and E. Cundliffe. 2002. Expression analysis of the tylosin-biosynthetic gene cluster: pivotal regulatory role of the tylQ product. Chem. Biol. 9:71-78. [DOI] [PubMed] [Google Scholar]
- 48.Sun, J., G. H. Kelemen, J. M. Fernandez-Abalos, and M. J. Bibb. 1999. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145:2221-2227. [DOI] [PubMed] [Google Scholar]
- 49.Takano, E. 2006. Gamma-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 9:287-294. [DOI] [PubMed] [Google Scholar]
- 50.Wietzorrek, A., and M. Bibb. 1997. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 25:1181-1184. [DOI] [PubMed] [Google Scholar]
- 51.Wilkinson, C. J., et al. 2002. Increasing the efficiency of heterologous promoters in actinomycetes. J. Mol. Microbiol. Biotechnol. 4:417-426. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.