Abstract
The PipX factor is a regulatory protein that seems to occur only in cyanobacteria. In the filamentous, heterocyst-forming Anabaena sp. strain PCC 7120, open reading frame (ORF) asr0485, identified as the pipX gene, is expressed mainly under conditions of combined-nitrogen deprivation dependent on the global N regulator NtcA and the heterocyst-specific regulator HetR. Primer extension and 5′ rapid amplification of cDNA ends (RACE) analyses detected three transcription start points corresponding to a canonical NtcA-activated promoter (to which direct binding of NtcA was observed), an NtcA- and HetR-dependent promoter, and a consensus-type promoter, the last with putative −35 and −10 determinants. Activation of pipX took place in cells differentiating into heterocysts at intermediate to late stages of the process. Accordingly, disruption of pipX led to impaired diazotrophic growth, reduced nitrogenase activity, and impaired activation of the nitrogenase structural genes. The nitrogenase activity of the mutant was low under oxic conditions, likely resulting from inefficient protection against oxygen. In line with this, the activation of the coxB2A2C2 and coxB3A3C3 operons, encoding heterocyst-specific terminal respiratory oxidases responsible for internal oxygen removal, was deficient in the pipX mutant. Therefore, the Anabaena PipX factor shows a spatiotemporal specificity contributing to normal heterocyst function, including full activation of the nitrogenase structural genes and genes of the nitrogenase-protective features of the heterocyst.
Cyanobacteria make up a group of microorganisms characterized by a photoautotrophic mode of life relying on oxygenic photosynthesis, a process developed by ancestors of extant cyanobacteria. Nowadays, cyanobacteria are important primary producers in the oceans and thus play a crucial role in the C and N cycling of our planet (31). Many cyanobacteria, both unicellular and filamentous, are able to carry out the fixation of N2. To protect the N2 fixation machinery from O2, either coming from the external medium or produced inside the cells, some filamentous cyanobacteria, such as those of the Anabaena/Nostoc genera, produce cells called heterocysts, which are specialized in the fixation of N2 (42). In these organisms, diazotrophic growth requires the activities of two cell types, namely, vegetative cells, which fix CO2 photosynthetically, and heterocysts, which fix N2, and the intercellular exchange of the products of both processes (13). Heterocysts have specific features to minimize the concentration of oxygen in the cytoplasm that may be either structural barriers to prevent external oxygen penetration or metabolic features to avoid internal oxygen production and to remove traces of this gas. The structural barriers of the heterocyst include additional glycolipid and polysaccharide layers of the cell envelope and a narrowed septum of connection with neighboring vegetative cells. Metabolic features include a lack of activity of water-splitting photosystem II and of the Rubisco and ribulokinase enzymes involved in the photosynthetic fixation of CO2 and the expression of special terminal oxidases to cope with residual O2 (42).
Nitrogen metabolism in cyanobacteria, including the assimilation of nitrate, nitrite, and urea in undifferentiated cells, as well as heterocyst differentiation and N2 fixation, is regulated by the transcription factor NtcA, a member of the CRP class of bacterial regulators (15). NtcA together with 2-oxoglutarate (2-OG), a metabolic signal of N deficiency, binds to specific DNA sites with the sequence signature GTAN8TAC and activates or represses the expression of regulated genes (21, 35, 39, 43). At canonical (class II) activated promoters, which include an NtcA-binding site centered at about nucleotide (nt) −41 from the regulated gene transcription start and a −10 box with the consensus TAN3T sequence, NtcA together with 2-OG induces the production of transcriptionally active open promoter complexes with RNA polymerase (RNAP) (36). In the unicellular cyanobacterium Synechococcus sp. strain PCC 7942, PipX has been identified as a protein interacting with the signal transduction PII protein and with NtcA, albeit not simultaneously (8, 20). Inactivation of the pipX gene leads to diminished NtcA-dependent activation of the glnA, glnN, and nblA genes under N stress (8, 9). Thus, a role for PipX as a coactivator of NtcA has been proposed. According to the model proposed, PII controls the concentration of active PipX by sequestering it. The binding of 2-OG to PII that follows a rise in the cellular C/N ratio releases PipX, which is then available for interaction with NtcA loaded with 2-OG (8, 20, 44). The recent solution of the crystal structure of the NtcA-PipX complex has led Llácer et al. to propose a role for PipX in stabilizing the active conformation of NtcA (20). Finally, gene activation during heterocyst differentiation requires the participation of, besides NtcA, the differentiation-specific factor HetR (1, 12, 16, 27), which has been reported to bind DNA upstream of some regulated genes (17, 18).
This work represents the first study of the physiological role of the PipX factor in a filamentous, heterocyst-forming cyanobacterium. We have performed a detailed characterization of the expression of the pipX gene in Anabaena sp. strain PCC 7120 and have constructed strains with mutations in the pipX genomic region. We found that this gene is specifically activated in the cells that undergo differentiation at late stages of the process and that it is required for attaining full nitrogenase activity, for efficient protection of this enzyme from O2, and for normal diazotrophic growth.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Anabaena sp. (also known as Nostoc sp.) strain PCC 7120 was grown in BG11 (containing NaNO3 [34]), BG110 (free of combined nitrogen), or BG110 plus ammonium {BG110 containing 4 mM NH4Cl and 8 mM N-[tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid (TES)-NaOH buffer, pH 7.5} medium at 30°C under continuous illumination (75 microeinsteins m−2 s−1) in shaken (80 to 90 rpm) liquid cultures or in medium solidified with 1% Difco agar. Alternatively, cultures (referred to as bubbled cultures) were supplemented with 10 mM NaHCO3 and bubbled with a mixture of CO2 and air (1%, vol/vol). In this case, the ammonium-containing medium was supplemented with 6 mM NH4Cl and 12 mM TES-NaOH buffer (pH 7.5). The hetR mutant, Anabaena sp. strain 216 (4), the ntcA mutant, strain CSE2 (14), and the mutant strains CSV6, CSV7, CSV6-53, CSAV142, and CSAV143 generated in this work were grown in BG110 plus ammonium medium, supplemented with streptomycin (Sm) and spectinomycin (Sp) in the cases of strains CSE2, CSV6, CSAV142, and CSAV143, and with Sm, Sp, and neomycin (Nm) in the case of strain CSV6-53. Antibiotics were used at the following concentrations: for Sm, 2 to 5 μg ml−1; Sp, 2 to 5 μg ml−1; and Nm, 10 to 50 μg ml−1.
To test growth in liquid medium, cells of the corresponding strains that had been grown in BG110 plus ammonium medium (with antibiotics for the mutants) were harvested, washed with BG110 medium, and resuspended in BG110, BG11, or BG110 plus ammonium medium at 0.2 μg chlorophyll a (Chl)·ml−1. The Chl content of cultures was determined by the method of Mackinney (22). After incubation for the indicated times, 0.2-ml samples were taken and their protein contents determined (23). The growth rate constant (μ = ln2/td, where td is the doubling time) was calculated from the increase in the protein content of a culture. (The cells that contain 1 μg of Chl also contain 25 to 30 μg of protein.) To test the growth of the mutants on solid medium, cell suspensions of the different strains with similar Chl contents were prepared, drops of 10 μl were spotted on plates of BG110, BG11, or BG110 plus ammonium medium, and the plates were incubated under culture conditions.
Escherichia coli DH5α and XL1-Blue were used for plasmid constructions. They and strains HB101 and ED8654, used for conjugations with Anabaena sp., were grown in LB medium (2) supplemented, when appropriate, with antibiotics at standard concentrations.
Strain constructions.
To inactivate asr0485, two DNA fragments, one encompassing the 63 bp of the 5′ end of asr0485 and sequences upstream and the other encompassing the 39 bp of the 3′ end of asr0485 and 562 bp downstream, were amplified by standard PCR using as a template DNA from strain PCC 7120 and oligonucleotide pairs PX1/PX2 and PX3/PX4, respectively (all oligodeoxynucleotide primers are listed in Table 1). The two DNA fragments were cloned in pGEM-T (Promega) and, after digestion, ligated in the direct orientation separated by gene cassette C.S3, encoding Smr and Spr (6). The insert of the resulting plasmid, excised with XhoI, was transferred into plasmid pRL278, which carries an Nmr determinant and the sacB gene for positive selection (3), producing plasmid pCSV24, which was transferred by triparental mating (7) into Anabaena sp.
TABLE 1.
Oligodeoxynucleotides used in this study
| Oligonucleotide | Sequence (5′-3′) | Positions relative to the translation start of the corresponding gene |
|---|---|---|
| PX1 | GCCTCCTCGAGTATATTCTG | −654 to −637 (pipX) |
| PX2 | CATGCGGATCCGATATAG | +70 to +52 (pipX) |
| PX3 | CAGGAGTAGGATCCGCTTC | +232 to +251 (pipX) |
| PX4 | CAACTGACCCTCGAGAATATG | +576 to +556 (alr0486) |
| PX7 | GACTGAAGTTATTCGTGCTGC | +90 to +110 (alr0486) |
| PX8 | CTACCTACAAGGATGAGATCAAGG | −165 to −142 (pipX) |
| PX9 | GAAGGTGCGTTGGAAAACAC | +276 to +252 (pipX) |
| PX10 | CTACTCGATGGATCCATGAATCCAG | −10 to +8 (pipX) |
| PX11 | GCAATTAGGTCGACTGAAGGCG | +71 to +50 (alr0486) |
| PX14 | GAAAGCCAGGATTTGTTCACAAC | +76 to +98 (pipX) |
| PX17 | CTATCAATCGAGTAGGAGAATC | −1 to −22 (pipX) |
| PX18 | CAACGCTGAAAATCCCTGATGCG | −282 to −304 (pipX) |
| PX19 | CTTACAAGCTAGAAGAAGCAG | −588 to −568 (pipX) |
| PX21 | GTTTATTTGCGGTGTCCAG | −304 to −322 (pipX) |
| PX22 | GCAAATTTTACCCTTAAATG | −109 to −128 (pipX) |
| PX23 | CGCAGTGAAAGACAAATATTCG | −35 to −56 (pipX) |
| PXgfp1 | GCAATCGATTTTAATAGCTATATGAACTTAC | −614 to −584 (pipX) |
| PXgfp2 | ATCGATCGAAGGATAATTATGGCAACATGG | −415 to −392 (pipX) |
| PXgfp3 | GATATCTTGGAAGGTGCGTTGGAAAACACTC | +276 to +252 (pipX) |
| asr0485-7120-1 | GCAGATCCGATATAGCAAACC | +66 to +46 (pipX) |
| alr0486-7120-1 | ATGATTAGTTCGATCAACGAAC | +1 to +22 (alr0486) |
| alr0486-7120-4 | GGATCCGAGTAATATTGCTTAAGTACG | +687 to +661 (alr0486) |
| alr0486-7120-1F | ATGATTAGTTCGATCAACGAAC | +1 to +22 (alr0486) |
| RT-1 | CTGGTTTGAGCATCAATTCTG | +1849 to +1829 (alr0489) |
| RT-2 | CTCTAATTATCCGGTTAATGCTTAATG | +1015 to +989 (alr0486) |
| alr0487-7120-1 | CAATTTGCAGAAGGAATTCAACTAAATG | −25 to +3 (alr0487) |
| alr0488-7120-1 | GCAGATGCAGTATTCACAGACCATTC | +253 to +278 (alr0488) |
| alr0488-7120-2 | GAATGGTCTGTGAATACTGTATCTGC | +278 to +253 (alr0488) |
| alr0488-7120-3 | CGATTAAGGCTGAACGGAATC | +766 to +746 (alr0488) |
| alr0489-7120-1 | GTGGGTTTGCAGTAAGTTCAAAC | +1341 to +1318 (alr0489) |
| DB034 | ATGTCAAGGGTGACGGAAG | +1 to +19 (devB) |
| DB018 | ATTTATTAATGTCAACCACTACC | +1423 to +1400 (devB) |
| CB2-4 | CACTCTGGACTCTAATTGCTG | +20 to +40 (coxB2) |
| CB2-5 | GACGTTGTATGCAATGTCTC | +1127 to +1108 (coxB2) |
| NH1 | GTACTGCAAGGGGCGTGTGGC | −334 to −314 (nifH) |
| NH4 | CCTATTGGTAGCTTCTGCGGGC | +885 to +864 (nifH) |
| NA10 | TAGGATCCTGTTATTCCGGCATTGGGTAGG | −39 to −10 (ntcA) |
| NA11 | CAGGATATCAGTATGGGTTCACCGTCAC | +809 to +782 (ntcA) |
| CS3-1 | GGATGACCTTTGAATGACC | −518 to −537 (Sm/Sp) |
| GFP4 | CAAGAATTGGGACAACTCC | +46 to +28 (gfp-mut2) |
| CB3-6 | CGATCGCTGTGACTATTACCAG | +47 to +68 (coxB3) |
| CB3-7 | AACCAGGGTAATTAACCAAAGG | +913 to +892 (coxB3) |
To inactivate alr0486, a DNA fragment of 486 bp internal to the open reading frame (ORF), was amplified by standard PCR using, as the template, DNA from strain PCC 7120 and oligonucleotide pair PX4/PX7. This DNA fragment was cloned into HincII-digested plasmid pCSV3 (which is a derivative of the mobilizable vector pRL500 [6], in which the Apr gene has been excised with DraI and replaced by the DraI-blunt-ended gene cassette C.S3, encoding Smr and Spr, in the direct orientation), generating the suicide plasmid pCSV27, which was transferred by triparental mating (7) into Anabaena sp. strain PCC 7120.
To complement the pipX mutation of strain CSV6, a DNA fragment of 1,504 bp from positions −654 to +850 with respect to the translational start site of the gene, was amplified by PCR using primers PX1 and PX4 and strain PCC 7120 DNA. This fragment was cloned in the mobilizable Nmr-encoding vector pRL424 (6), producing plasmid pCSV53, which was transferred to strain CSV6 by conjugation. The genomic structure of the exconjugants in the pipX region was tested by PCR using the primer pairs PX8/PX11 and PX8/CS3-1.
To fuse the genes pipX and gfp, a fragment of the pipX genomic region, encompassing positions from −605 to +276 with regard to the ORF, was amplified with oligonucleotides PXgfp1 and PXgfp3 and cloned into ClaI/EcoRV-digested plasmid pCSEL22a (29), producing plasmid pCSAV171, which encodes Smr/Spr and can be mobilized into Anabaena by conjugation. Plasmid pCSAV171 bears the pipX ORF (except for the last nucleotide) fused in frame to the gfp gene. This plasmid was transferred to strains PCC 7120 and 216 by conjugation, and selection for resistance to Sm/Sp was applied. The genomic structures of the exconjugants were analyzed by PCR with the oligonucleotide pair PX1/GFP4 and by segregation with PX1/PX11 and PXgfp1/PX11. The accumulation of green fluorescent protein (GFP) was analyzed by laser confocal microscopy using a Leica HCX Plan-APO 63× 1.4 NA objective and a Leica TCS SP2 microscope (Leica, Wetzlar, Germany) as described previously (29).
DNA and RNA isolation, manipulation, and analysis.
Isolation of genomic DNA (5) and of total RNA (2) from Anabaena sp. was done as described previously. For Northern blots, 10 to 20 μg of RNA was loaded per lane and electrophoresed in denaturing 1% agarose formaldehyde gels. DNA probes were generated by PCR using strain PCC 7120 DNA and oligonucleotide primers, as follows: PX9/PX10 for pipX, PXgfp2/PX22 for the pipX leader region, alr0486-7120-1F/alr0486-7120-4 for alr0486, NA10/NA11 for ntcA, DB034/DB018 for devB, NH1/NH4 for nifH, CB2-4/CB2-5 for coxB2, and CB3-6/CB3-7 for coxB3. Hybridizations were performed as previously described (33). As a control for RNA loading and transfer efficiency, the filters were hybridized with a probe of the RNase P RNA gene (rnpB) from strain PCC 7120, which was amplified by PCR with primers Universal and Reverse and with plasmid pT7-7120 as the template (40). Probes were labeled with a DNA labeling kit (Ready to Go; Amersham Pharmacia Biotech) and [α-32P]dCTP. Radioactive areas in Northern blot hybridizations were visualized with a Cyclone storage phosphor system (Packard).
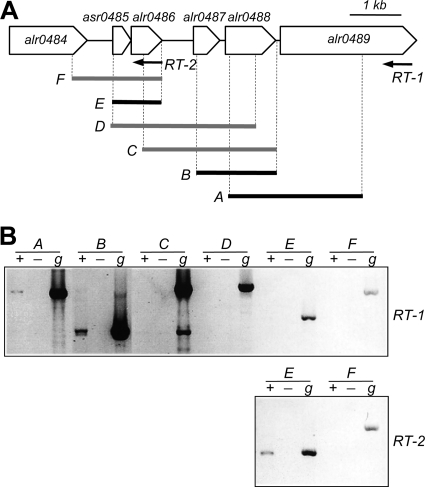
For reverse transcriptase PCR (RT-PCR) experiments, 10 μg of strain PCC 7120 total RNA was mixed with 40 pmol of oligonucleotide RT-1 or RT-2 in the presence of 10 mM Tris-HCl (pH 8.0), 150 mM KCl, and 1 mM EDTA and heated for 10 min at 85°C and then at 50°C for 1 h for annealing. The extension reactions were carried out at 47°C for 1 h in a final volume of 32 μl containing the whole annealing reaction mixture, 0.25 mM each deoxynucleoside triphosphate, 100 U of reverse transcriptase (Superscript II; Invitrogen), and the buffer recommended by the transcriptase provider. To control for the presence of contaminating DNA, samples containing 10 μg of RNA were processed as described above but with reverse transcriptase omitted (−RT control). PCR was carried out with 2 to 5 μl of a retrotranscription mixture (or −RT control mixture) as the template and the following oligonucleotide pairs as the primers: alr0488-7120-1/alr0489-7120-1 (for the segment named A in Fig. 1), alr0487-7120-1/alr0488-7120-3 (segment B), alr0486-7120-1/alr0488-7120-3 (segment C), PX14/alr0488-7120-2 (segment D), PX14/RT-2 (segment E), and PX1/RT-2 (segment F). Samples containing the same oligonucleotides and strain PCC 7120 DNA as the template were run in parallel and used as controls. PCR was performed by standard procedures, and the PCR products were resolved by electrophoresis in 0.7% agarose gels.
FIG. 1.
RT-PCR analysis of the expression of the alr0484-alr0489 gene cluster. (A) The gene cluster and Anabaena ORF names (19) are depicted. (B) Retrotranscription was carried out with oligonucleotide primer RT-1 or RT-2 (initial positions indicated by the start of the corresponding arrow) and with RNA isolated from bubbled ammonium-grown cultures incubated for 6 h in the absence of combined nitrogen. The positions of the primers used for amplification correspond to the ends of the segments indicated in panel A, which are depicted in black, indicating amplification by RT-PCR, or in gray, indicating no amplification. + or −, RNA samples subjected or not to retrotranscription; g, total genomic DNA used as the template for amplification with the corresponding primers.
Primer extension analyses were performed as previously described (26) with the oligonucleotide primers asr0485-7120-1, PX17, PX18, and PX21. 5′ rapid amplification of cDNA ends (RACE) was performed as described previously (25) in samples treated or not treated with tobacco acid pyrophosphatase (TAP), using for retrotranscription the gene-specific primer PX17 and for PCR the RNA adaptor primer and one of the gene-specific primers PX17, PX22, and PX23. DNase I footprinting assays were performed as described in reference 36 with purified NtcA. Radioactive areas of the gels were visualized and quantified with a Cyclone storage phosphor system (Packard).
Glycolipid analysis and nitrogenase activity.
For glycolipid analysis, filaments of Anabaena sp. grown in BG11 medium (in the presence of Sm and Sp for the CSV6 mutant) were harvested, washed with nitrogen-free medium, and incubated in bubbled BG110 cultures for 24 h. Filaments were then harvested and washed with buffer 1 containing 50 mM imidazole and 0.5 mM EDTA (pH 8.0), and lipids were extracted with chloroform-methanol (2:1, vol/vol), concentrated under N2, and chromatographed on thin layers of silica gel (28). Heterocyst glycolipids were identified as described previously (41).
Nitrogenase activity was determined by the acetylene reduction technique in filaments incubated for 24 h in BG110 medium as described previously (24). For assays under micro-oxic conditions, 10 μM DCMU [(3-3,4-dichlorophenyl)-1,1-dimethylurea] was added to the cell suspension and the flask containing the cells was sealed with a rubber stopper, bubbled with argon for 3 min, and further incubated under culture conditions for 1 h before the assay was started by addition of acetylene.
RESULTS
Identification and regulated expression of the Anabaena pipX gene.
A similarity search using as a query the sequence of the PipX protein from Synechococcus sp. strain PCC 7942 against the whole genomic sequence of Anabaena sp. strain PCC 7120 (19) identified ORF asr0485. This ORF encodes a product of 92 amino acids exhibiting 63% identity to the 89-amino-acid Synechococcus PipX protein, but it did not show similarity to any noncyanobacterial gene product in the databases. In the genome of strain PCC 7120, asr0485 is flanked by ORFs alr0484 and alr0486, both encoding hypothetical proteins. The latter overlaps asr0485 by 4 nucleotides (Fig. 1A).
We analyzed the expression of the Anabaena gene cluster depicted in Fig. 1A by RT-PCR using 2 distinct primers for retrotranscription and different primer pairs for PCR to determine the transcriptional units in this genomic region. The RNA was extracted from ammonium-grown filaments incubated for 6 (Fig. 1B) or 12 (not shown) hours in the absence of combined nitrogen (i.e., in the absence of any nitrogen source other than atmospheric nitrogen). The pattern of amplification products obtained indicates the existence of transcripts overlapping pipX and alr0486, on one hand, and alr0487, alr0488, and alr0489 on the other hand. Evidence for transcripts overlapping alr0484 and pipX or alr0486 and alr0487 was not obtained.
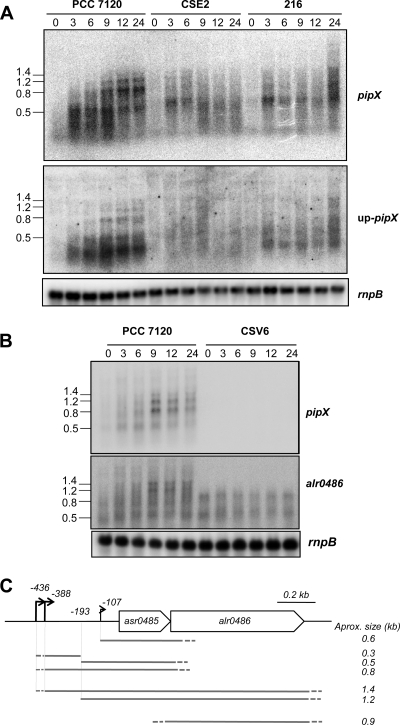
The pattern of expression, as well as the nitrogen regulation, of pipX was studied by Northern blot analysis using RNA isolated from filaments of strain PCC 7120, an ntcA mutant (strain CSE2 [14]), and a hetR mutant (strain 216 [4]) grown with ammonium and incubated in the absence of combined nitrogen (Fig. 2). In the wild-type strain, the pipX probe detected RNA species of ca. 0.5, 0.8, 1.2, and, weakly, 1.4 kb (the pipX ORF is 279 nt long), whose abundances were very low in the ammonium-grown cultures and increased upon combined-nitrogen deprivation. Whereas the 0.5-kb transcript seems to accumulate early after ammonium withdrawal, the accumulations of the larger (0.8-, 1.2-, and 1.4-kb) transcripts were higher after 9 to 12 h of combined-nitrogen deprivation and were lower in both the ntcA and hetR mutant strains (Fig. 2A). When a probe covering the whole alr0486 ORF was used, we detected RNA species of ca. 1.4 and 1.2 kb, whose abundances increased upon combined-nitrogen deprivation, and an RNA species of ca. 0.9 kb, which was faintly detected (Fig. 2B). Since pipX and alr0486 are, respectively, 279 and 668 nt long, the regulated 1.4- and 1.2-kb transcripts hybridizing with both probes may include the messages of both pipX and, at least in part, alr0486, whereas the regulated transcripts of ca. 0.5 and 0.8 kb hybridizing to pipX and the transcript of ca. 0.9 kb hybridizing to alr0486 would represent independent transcripts of those ORFs, respectively. Given that the transcripts of 1.4 and 1.2 kb are longer than the two ORFs together, we checked whether they covered additional sequences upstream of pipX. When a DNA fragment covering positions from −415 to −109 bp with respect to the pipX ORF (see below) was used as a probe (probe up-pipX) in Northern assays (Fig. 2A), the four RNA species previously detected with the probe of the pipX ORF were seen. This observation indicates that these transcripts include leader sequences upstream of pipX. Additionally, a smaller RNA species, faintly detected with the probe from within the pipX ORF, was clearly seen with the upstream probe. This RNA species could correspond to an abortive transcript, to a degradation product of RNAs initiated upstream of pipX, or to a small RNA from the pipX leader region.
FIG. 2.
Northern analysis of the expression of asr0485 (pipX)-alr0486. (A and B) RNAs isolated from bubbled cultures of Anabaena sp. strain PCC 7120, strain CSE2 (ntcA), strain 216 (hetR), and strain CSV6 (pipX) grown with ammonium (lanes 0) and incubated in the absence of combined nitrogen for the times indicated (in hours) were electrophoresed and hybridized with the probes indicated at the right, which were generated by PCR (see Materials and Methods). Deduced sizes (in kilobases) of some apparent transcripts are indicated at the left. Hybridization with an rnpB gene probe was used as a loading and transfer control. (C) Schematic transcription profile of the asr0485-alr0486 gene cluster. Transcription start sites are indicated by arrows, whereas the −193 position is proposed to correspond to a processed 5′ RNA end. Transcripts are indicated by gray horizontal lines, in which a dashed end indicates ambiguity in the determination of its precise location.
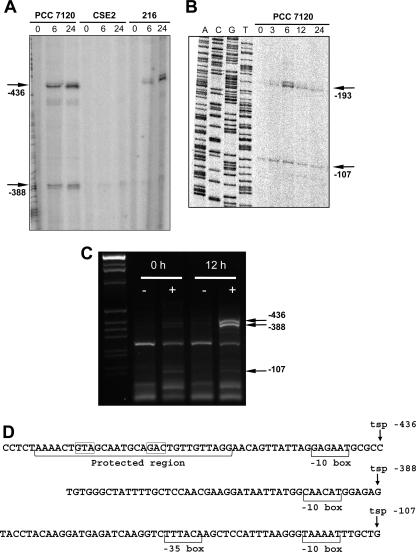
In order to analyze the molecular basis of the regulation of the expression of pipX, the location of putative transcription start points (TSPs) was sought. Primer extension analysis was performed with RNA extracted from cultures of strains PCC 7120, CSE2, and 216 grown on ammonium and incubated in the absence of combined nitrogen. In the wild type, several 5′ transcript ends were detected using primers upstream or within the 5′ region of pipX. A 5′ end corresponding to the C located 436 nucleotides upstream of the ORF was detected with primers PX18 (Fig. 3A) and PX21 (not shown). Its signal increased upon combined-nitrogen deprivation, and it was detected in the hetR strain but not in the ntcA mutant.
FIG. 3.
Primer extension and 5′ RACE analyses of the pipX gene promoter region. (A) Primer extension analysis was carried out with primer PX18 and with RNAs isolated from bubbled cultures of Anabaena sp. strain PCC 7120, strain CSE2 (ntcA), and strain 216 (hetR) grown with ammonium (lanes 0) and incubated in the absence of combined nitrogen for the times indicated (in hours). The positions of the −436 and −388 RNA 5′ ends are indicated. (B) Primer extension analysis carried out with primer asr0485-7120-1 and RNA isolated from bubbled cultures of Anabaena sp. strain PCC 7120 grown with ammonium (lanes 0) and incubated in the absence of combined nitrogen for the times indicated (in hours). The positions of the −193 and −107 5′ RNA ends are indicated. A sequence ladder of the same DNA region is shown at the left. (C) 5′ RACE analysis was carried out with RNA isolated from bubbled cultures of Anabaena sp. strain PCC 7120 grown with ammonium (0 h) and incubated in the absence of combined nitrogen (12 h) treated (+) or not (−) with TAP, as indicated, with PX17 as the gene-specific primer for the PCR step. The putative 5′ positions of some bands are indicated. Size standards (DNA molecular weight marker X; Roche) are shown in the left lane. (D) DNA sequences upstream from three identified pipX TSPs (see the text). The NtcA-protected region upstream of TSP −436 and putative −10 and −35 boxes are indicated. Conserved triplets of the NtcA binding site are boxed.
With the same oligonucleotides, a second 5′ transcript end was identified, corresponding to a G at position −388. The transcripts with this end increased in abundance upon combined-nitrogen deprivation and were not detected in the ntcA mutant or the hetR mutant (Fig. 3A). With primers PX17 (not shown) and asr0485-7120-1 (Fig. 3B), two more 5′ ends were detected; one of them corresponded to G at position −193, whose abundance increased upon combined-nitrogen deprivation (Fig. 3B) and decreased in both the ntcA and the hetR mutant (not shown), and the other corresponded to position −107 and was detected under all tested conditions both in the wild type (Fig. 3B) and the mutants (not shown). To ascertain whether all these 5′ RNA ends represent true TSPs or degradation products of transcripts originating upstream, 5′ RACE analyses were performed with RNA samples isolated from ammonium-grown cultures incubated or not for 12 h in the absence of combined nitrogen and treated or not with TAP (Materials and Methods). PCR products of the sizes expected for the 5′ ends at the −436 and −388 positions were detected with the gene-specific primers PX17 (Fig. 3C), PX22, and PX23 (not shown) and for that at −107 with primers PX17 (Fig. 3C) and PX23 (not shown), but only in the TAP-treated samples, implying that these ends represent true TSPs. Fully consistent with the above-described primer extension results, the intensities of the bands corresponding to the −436 and −388 TSPs, but not the −107 TSP, were much higher in the samples of cells incubated in the absence of combined nitrogen than in the cells coming directly from the ammonium cultures. On the other hand, a PCR product of the size expected for the 5′ end at −193 was detected with primers PX23 and PX22 (not shown). This band was always faint and appeared with similar intensities in the TAP-treated and untreated samples, suggesting that the RNA species with this end represent processed products. Sequencing of the corresponding PCR products showed ends at −434, −387, and −107, in good agreement with the transcript ends determined by primer extension.
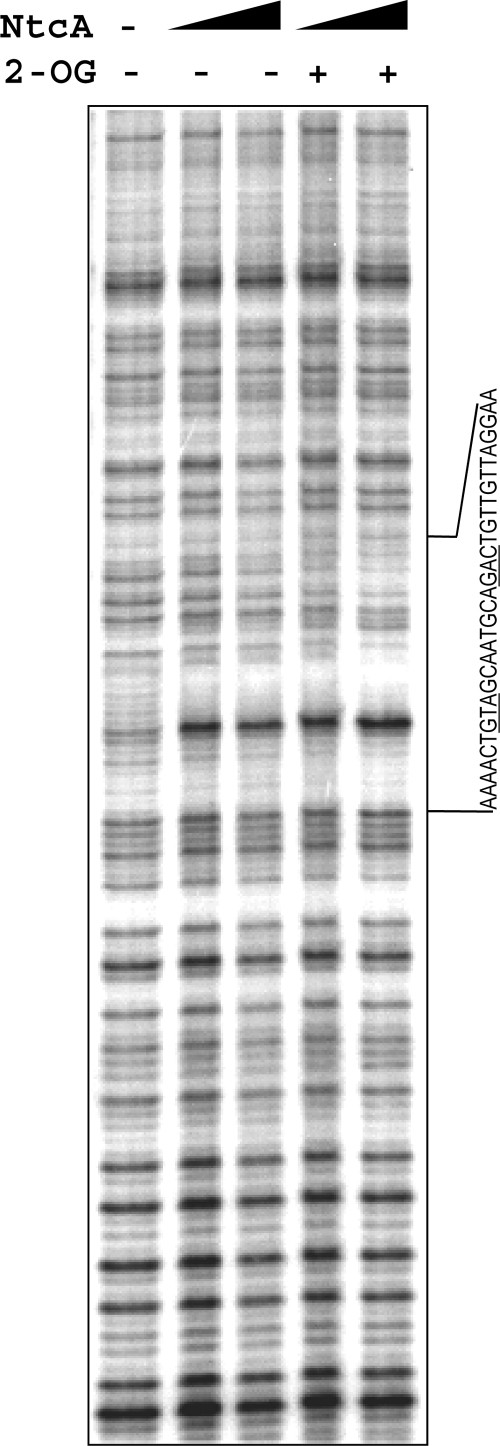
Direct binding of purified NtcA to a DNA fragment (from bp −588 to −282 with regard to the ORF) including sequences just upstream of the −436 TSP was assayed by DNase I footprinting. Figure 4 shows changes in the digestion pattern produced by the presence of NtcA with or without 2-OG. These changes included a general protection of the sequence between 23 and 52 nucleotides upstream of the −436 TSP (a region that includes a putative NtcA-binding site with the sequence GTAGCAATGCAGAC) and hypersensitivity of the thymine located 45 nucleotides upstream of that TSP. Indeed, NtcA-induced hypersensitivity at T of the GTA triplet of the NtcA recognition site has been previously noted (e.g., reference 30 and see below).
FIG. 4.
DNase I footprinting of the promoter region of the pipX gene. DNase I protection assays were carried out with purified NtcA (90 or 180 nM) and a DNA fragment of the pipX promoter region amplified by PCR using oligonucleotides PX18 (unlabeled) and PX19 (32P labeled), in the presence or absence of 0.6 mM 2-OG. The sequence of the NtcA-protected region, including the GTA/GAC triplets (underlined), is indicated.
Expression of the pipX gene in different cell types.
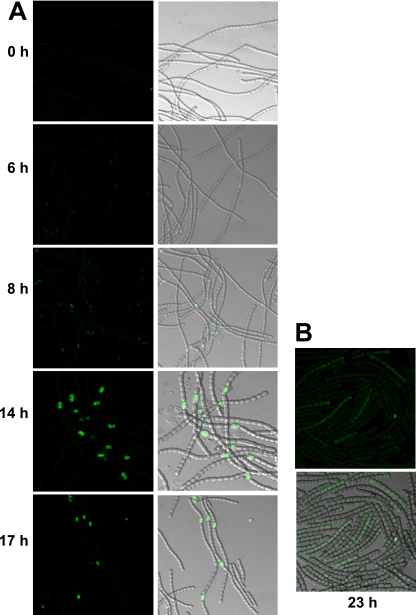
Transcription of pipX from NtcA- and HetR-regulated promoters suggested differential expression of this gene in different cell types. Expression of pipX along the filament was studied using GFP as a reporter. The gfp-mut2 gene was inserted, maintaining the translational frame, just before the stop codon of pipX in the chromosomes of strains PCC 7120 and 216, the latter of which does not show heterocyst differentiation (see Materials and Methods). The selected clones (strains CSAV142 and CSAV143, respectively) had incorporated the construct by single crossover, thus keeping an intact copy of the PpipX-pipX region. Fluorescence from GFP was analyzed in ammonium-grown filaments subjected to different periods of combined-nitrogen deprivation (Fig. 5). In the two strains, very low fluorescence was detected in the ammonium-grown filaments (0 h). In strain CSAV142, fluorescence was observed to increase from ca. 6 to 14 h after a combined-nitrogen step down, mainly confined to specific, semiregularly spaced cells identified as proheterocysts (Fig. 5A). No comparable increase in fluorescence was observed in strain CSAV143. In this strain, some fluorescence increase was observed at later times in some filaments but without apparent spatial specificity (Fig. 5B).
FIG. 5.
Spatiotemporal expression of pipX. GFP fluorescence and merged differential inference contrast (DIC)/GFP fluorescence images of filaments of strains CSAV142 (pipX-gfp in strain PCC 7120) (A) and CSAV143 (pipX-gfp in strain 216) (B), grown with ammonium and incubated in the absence of combined nitrogen for the indicated times.
Inactivation of pipX.
In order to study the function of pipX in Anabaena, the construction of strains carrying inactivated versions of this ORF was undertaken. Plasmid pCSV24 was constructed (Materials and Methods) to consist of two noncontiguous fragments of the pipX-alr0486 genomic region of strain PCC 7120 joined by gene cassette C.S3, encoding resistance to Sm and Sp, cloned in the conjugative vector pRL278 (3). This vector carries an Nmr determinant and the sacB gene, conferring sensitivity to sucrose (Suc), for positive selection. Clones exhibiting Smr/Spr, resistance to Suc, and sensitivity to Nm are expected to have incorporated the transferred construct by double crossover with the recipient's chromosome, replacing the wild-type pipX gene with a mutant version in which 177 bp from inside the reading frame was replaced with gene cassette C.S3. PCR and restriction analyses showed a number of clones exhibiting such a genomic structure. One of those clones, which according to Southern analysis (not shown) exhibited no wild-type chromosome, was selected and named strain CSV6. Consistent with Southern analysis, no transcript of pipX could be detected by Northern analysis in strain CSV6 (Fig. 2B). It should be pointed out, however, that a faint band that may correspond to the wild-type allele was observed when the genomic structure of strain CSV6 was analyzed by prolonged (35-cycle) PCR, even after repeated rounds of filament fragmentation by sonication and growth in the presence of ammonium, Sm, and Sp. Thus, we cannot ascertain that strain CSV6 is completely devoid of wild-type asr0485 alleles, but in any case, expression of this gene was negligible in the mutant.
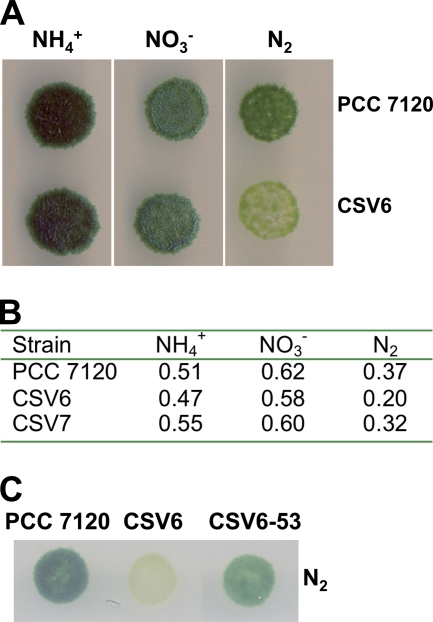
The phenotype of strain CSV6 was analyzed with regard to growth with different nitrogen sources, heterocyst differentiation, and expression of nitrogenase activity. In the presence of combined nitrogen, either nitrate or ammonium, the growth of strain CSV6 was similar to that of strain PCC 7120 both in liquid and solid media. However, in the absence of combined nitrogen, strain CSV6 showed a yellowish color indicative of N deficiency, and its growth rate in liquid medium was about half that of the wild-type strain (Fig. 6). When observed under the optical microscope, diazotrophic cultures of strain CSV6 showed many short filaments, frequently with one terminal heterocyst, and detached cells, which are rare in the wild type. Some longer filaments were also evident, but in them the vegetative cell intervals between heterocysts were longer than in the wild-type filaments. Under the electron microscope, mature heterocysts of strain CSV6 appeared similar to wild-type heterocysts (not shown). Consistently, heterocyst-specific glycolipids could be detected by thin-layer chromatography in strain CSV6 after 24 h of incubation in the absence of combined nitrogen (not shown).
FIG. 6.
Growth of strains CSV6, CSV7, and CSV6-53 with different N sources. (A, C) Cell suspensions of Anabaena sp. strain PCC 7120, strain CSV6, and strain CSV6-53, as indicated, grown with ammonium (with antibiotics in the case of the mutants) and washed with combined-nitrogen-free medium, were used to inoculate (12 ng Chl per spot [A]; 100 ng Chl per spot [C]) plates of BG11 (NO3−) or of BG110 medium supplemented (NH4+) or not (N2) with ammonium; the plates were incubated for 14 days (A) or 10 days (C) under culture conditions (see Materials and Methods for details). (B) Cell suspensions of Anabaena sp. strain PCC 7120, strain CSV6, and strain CSV7, as indicated, grown with ammonium (and Sm and Sp in the cases of the mutants) and washed with combined-nitrogen-free medium, were used to inoculate liquid cultures in Erlenmeyer flasks containing 25 ml medium with the indicated N source, which were incubated under culture conditions. Aliquots were withdrawn from the cultures at different times to determine their protein content. The values are specific growth rate constants (per day) and are averages from two (CSV6, CSV7) or four (PCC 7120) independent experiments with similar results.
Nitrogenase activity was measured under oxic and micro-oxic conditions in strain CSV6 in comparison to that of the wild type. Both under micro-oxic and oxic conditions, activity levels were lower in strain CSV6 than in the wild type (e.g., 15 and 27 nmol ethylene·μg Chl−1·h−1, respectively, in a representative experiment under micro-oxic conditions), but the difference was considerably larger under oxic conditions (2 and 19 nmol ethylene·μg Chl−1·h−1 for strains CSV6 and PCC 7120, respectively). The low nitrogenase activity of strain CSV6 under oxic conditions can explain its impaired diazotrophic growth.
Strain CSV6 was complemented with a wild-type copy of the pipX gene cloned in the Kmr-encoding mobilizable plasmid pRL424 (6). Kmr exconjugant CSV6-53 had integrated into the chromosome the whole transferred plasmid by a single recombination event, keeping the inactivated version of pipX present in strain CSV6 (not shown). Although CSV6-53's capacity for diazotrophic growth was still somewhat lower than that of the wild type, it was considerably higher than that of its parental strain, CSV6 (the growth rate constant for strain CSV6-53 was 86% of that of the wild type in a representative experiment in which the growth rate constant of CSV6 was 57% of that of the wild type; see also Fig. 6C). Thus, the defect in strain CSV6 can be ascribed to the mutation introduced in the pipX gene.
On the other hand, to check for possible polar effects of the mutation introduced into strain CSV6, expression of the downstream ORF alr0486 was studied in this strain in comparison to that of the wild type. Figure 2B shows that in strain CSV6, the RNA bands of ca. 1.4 and 1.2 kb, previously interpreted as originating upstream of pipX in the wild type, were not produced. Instead, the band of ca. 0.9 kb was stronger in strain CSV6. Thus, in the pipX mutant, ORF alr0486 is expressed, albeit with a pattern different from that taking place in the wild type.
To evaluate the possibility that the phenotype of strain CSV6 results from the observed alteration in the expression pattern of ORF alr0486, a knockout mutant of this gene was generated. We constructed plasmid pCSV27 (see Materials and Methods), consisting of an internal fragment of alr0486 of strain PCC 7120 cloned into the pCSV3 plasmid, which includes gene cassette C.S3, encoding resistance to Sm and Sp. Plasmid pCSV27 was transferred to strain PCC 7120 by conjugation with selection for resistance to Sm and Sp in the presence of ammonium. Smr/Spr clones were expected to have incorporated the transferred construct into alr0486 by single recombination, with the recipient's chromosome splitting the alr0846 sequence in halves at both sides of the integration point. Southern analysis (not shown) showed that all of a number of selected clones exhibited such a genomic structure, keeping no wild-type chromosome. One of them was named strain CSV7. The growth rate of strain CSV7 was similar to that of the wild type with nitrate or ammonium and considerably higher than that of strain CSV6 under diazotrophic conditions (Fig. 6B). The nitrogenase activity of strain CSV7 was similar to that of strain PCC 7120 under micro-oxic conditions (29 and 27 nmol ethylene·μg Chl−1·h−1, respectively, in a representative experiment) and considerably higher than that of strain CSV6 (2, 14, and 19 nmol ethylene·μg Chl−1·h−1 for strains CSV6, CSV7, and PCC 7120, respectively) under oxic conditions. Thus, the phenotype of strain CSV6 could not be explained by a polar effect resulting in impaired expression of alr0486.
Role of PipX in the expression of genes involved in heterocyst differentiation.
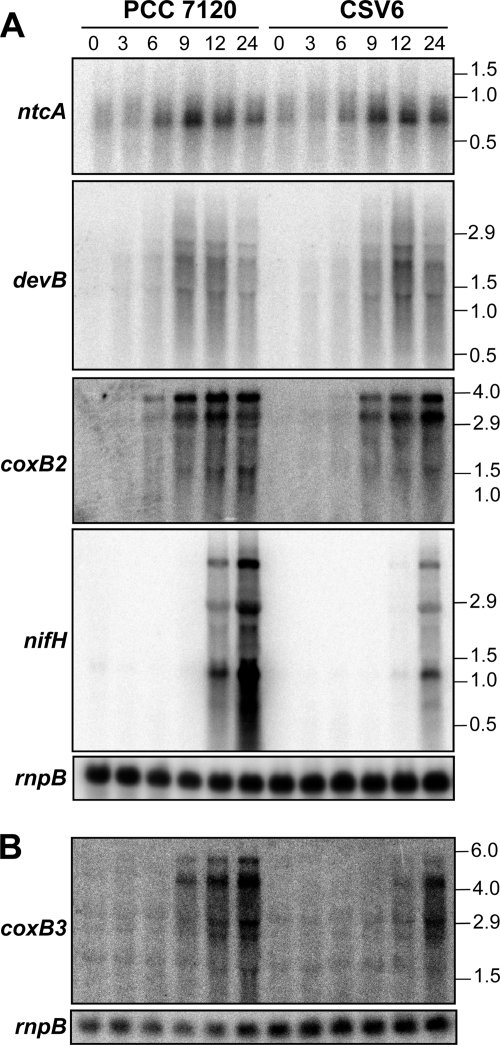
Because pipX was preferentially expressed in heterocysts and strain CSV6 showed impaired diazotrophic growth, we checked the expression of a number of genes involved in heterocyst maturation and function. As shown in Fig. 7A, no significant differences in ntcA expression were observed between the wild type and the CSV6 strain. The devBCA operon, which encodes an ABC-type exporter involved in deposition of the heterocyst envelope (10, 11), also showed similar expression profiles in the PCC 7120 strain and in the CSV6 mutant (Fig. 7A). In contrast, the expression of the nitrogenase structural genes nifHDK was retarded in the mutant and reached lower levels than in the wild type (20% of the levels in the wild type after 24 h of combined-nitrogen deprivation in the representative experiment whose results are shown in Fig. 7A). This is consistent with the lower nitrogenase activity of the mutant. On the other hand, expression of nifHDK in strain CSV7 was similar to that in the wild type (not shown), confirming that the low activity observed in strain CSV6 is due to the inactivation of the pipX gene and not to a polar effect on the downstream gene. Finally, the expression of the coxB2A2C2 and coxB3A3C3 operons, which encode heterocyst-specific terminal respiratory oxidases that help to maintain a micro-oxic ambience in the cytoplasm by active oxygen consumption (37), was impaired, albeit not to the same extent. Activation of coxB2A2C2 was significantly retarded in the mutant compared to in the wild-type strain, although at 24 h of combined-nitrogen deprivation, they both reached similar levels (Fig. 7A). Activation of coxB3A3C3 was severely retarded, resulting in 50 and 80% of the wild-type levels at 12 and 24 h, respectively, of combined-nitrogen deficiency (Fig. 7B).
FIG. 7.
Expression of several genes involved in heterocyst differentiation in strain CSV6 compared to strain PCC 7120. Northern assays were carried out with RNA isolated from Anabaena sp. strain PCC 7120 or strain CSV6 (asr0485::C.S3) grown with ammonium (lanes 0) and incubated in the absence of combined nitrogen for the times indicated (in hours) and with probes for the indicated genes generated by PCR (see Materials and Methods). Size markers (kb) are indicated at the right. Hybridization with an rnpB gene probe was used as a loading and transfer control. (A and B) Hybridization with the same RNA preparations on different filters.
DISCUSSION
In the genome of Anabaena sp. strain PCC 7120, asr0485 encodes a homolog of the PipX factor of the unicellular cyanobacterium Synechococcus sp. strain PCC 7942. PipX homologs are widespread in cyanobacteria but do not exist outside this phylum. Physiological differences between Anabaena and Synechococcus are relevant, especially those concerning nitrogen metabolism and adaptation to different nitrogen regimes. For instance, while Synechococcus may be subjected to prolonged periods of nitrogen stress due to deficiency in combined-nitrogen sources, for Anabaena, nitrogen starvation is instead transient, lasting for the time that heterocysts take to differentiate. Therefore, important differences are also expected in the operation of the elements involved in signal transduction of nitrogen stimuli. Our results indicate that in Anabaena sp. strain PCC 7120, (i) pipX expression is induced by combined-nitrogen deficiency and occurs preferentially in differentiating heterocysts, (ii) transcription of pipX takes place from a complex promoter region containing promoters controlled by NtcA and HetR, and (iii) inactivation of the pipX gene provokes slow diazotrophic growth and low nitrogenase activity due to impaired expression of at least the nitrogenase structural genes and of the gene clusters coxB2A2C2 and coxB3A3C3, encoding terminal respiratory oxidases involved in nitrogenase protection against oxygen in the heterocyst.
Information on the transcription profile of pipX was not available for unicellular or heterocyst-forming cyanobacteria. Here we have shown that in Anabaena sp. strain PCC 7120, the expression of pipX is activated upon combined-nitrogen deprivation influenced by the global N regulator NtcA and the heterocyst differentiation regulator HetR (Fig. 2). Using the GFP as a reporter of pipX expression, activation is observed to take place mainly in specific, semiregularly spaced cells in the wild-type strain but not in the hetR mutant (Fig. 5). Taken together, these results indicate that expression of pipX takes place mainly in an NtcA- and HetR-dependent manner at intermediate to late stages of the differentiation process (under our laboratory conditions, heterocyst differentiation was completed within 24 h of incubation in the absence of combined nitrogen). Like a number of other Anabaena genes expressed during heterocyst differentiation (12), pipX appears to be expressed from a complex promoter region encompassing several in-tandem promoters. A TSP localized at position −107 of the pipX ORF is constitutively expressed and is preceded by recognizable −10 and −35 determinants (Fig. 3D), thus representing a consensus-type promoter. In addition, two N-regulated TSPs for the pipX gene could be identified. The use of the −436 TSP requires NtcA, and the DNA sequence upstream of it conforms to a canonical class II NtcA-activated promoter (Fig. 3D) (15, 21). Indeed, purified NtcA has been shown to bind to and to protect from DNase I digestion sequences upstream from this TSP. Like other class II NtcA-activated promoters of Anabaena sp. strain PCC 7120 (16), this pipX promoter does not show a requirement for HetR. The −388 TSP is NtcA and HetR dependent and, as such, is expected to contribute to the increase of pipX expression observed after ca. 9 h of N deprivation (Fig. 2), which is localized mainly to proheterocysts (Fig. 5). As in the case of other Anabaena promoters directing localized gene expression in proheterocysts, no promoter determinants could be recognized here other than an imperfect −10 box (Fig. 3D). On the other hand, no consensus −10 box or NtcA-binding sequence could be recognized upstream from position −193. Thus, consistently with 5′ RACE results, the detected regulated RNA species with its 5′ end at this position appears to result from the processing of transcripts initiated at the regulated upstream promoters.
A schematic of the transcription profile of the pipX-alr0486 cluster of Anabaena sp. strain PCC 7120, which is consistent with the RT-PCR, Northern blot, primer extension, and 5′ RACE analyses included in this work, is presented in Fig. 2C. The pipX gene, which is very weakly expressed in the presence of ammonium, is activated upon N deprivation according to a time-specific cascade of promoter utilization that generates transcripts of different lengths. Transcripts differ in their 5′ ends, depending on the promoter from which they originate or the point where they are processed, but may also differ in their 3′ ends, depending on premature termination.
Strain CSV6 bearing an inactivated pipX gene is impaired specifically in diazotrophic growth. The phenotype of this strain results from disruption of the pipX gene and not from a polar effect on the downstream gene, since the phenotype of CSV6 is not reproduced by an insertion in alr0486, and besides, the wild-type phenotype can be rescued by complementation of CSV6 with the pipX gene. Filaments of strain CSV6 grown diazotrophically show low levels of nitrogenase activity under anoxic conditions, which is consistent with the diminished expression of the nifHDK genes with respect to their expression in the wild type. The deficiency in nitrogenase activity levels in the CSV6 mutant is more severe under oxic than under anoxic conditions, additionally indicating inefficient oxygen protection of the nitrogenase. However, we have not found evidence for alteration of heterocyst structural barriers to oxygen, including the cell envelope (which shows a normal appearance in electron micrographs [not shown]), the glycolipid content of the heterocyst cell wall (which seems to be normal when analyzed by thin-layer chromatography [not shown]), and the expression of the devBCA operon, encoding a glycolipid exporter (which does not show significant differences in CSV6 with respect to PCC 7120). In contrast, the expression of the coxB2A2C2 and the coxB3A3C3 operons, encoding heterocyst-specific terminal respiratory oxidase complexes (37), which are involved in the removal of internal oxygen by reduction, is impaired (albeit to a different extent). Both cox operons are specifically expressed in the heterocyst, and although none of them is essential, a double mutant is incapable of diazotrophic growth and does not exhibit nitrogenase activity under oxic conditions (37, 38). Concomitant impairment of both operons in the CSV6 mutant would likely compromise the oxygen protection of the nitrogenase, leading to a partial inactivation of the enzyme under oxic conditions. It is worth noting that cox3, whose expression is more severely affected in CSV6, has probably a more important role than cox2 in the oxygen protection of the nitrogenase in the heterocysts (38).
In Synechococcus sp. strain PCC 7942, PipX has been proposed to be an accessory protein for NtcA-mediated transcription activation (8, 20). Indeed, several genes or activities subjected to NtcA regulation show a reduced activation in a pipX mutant (8, 9). However, in the presence of 2-OG, NtcA is able to bind DNA and activate transcription in vitro in the absence of PipX both at Synechococcus (35) and Anabaena (36) promoters. We show here that in vivo expression of two ntcA-dependent genes, devB and ntcA, is not affected in the CSV6 mutant but that the expression of three operons, nifHDK, cox2, and cox3, is impaired. This observation suggests that, at least in Anabaena sp. PCC 7120, some ntcA-dependent genes require PipX for full activation but that other ntcA-dependent genes seem to be independent of this factor.
As we have shown here, the Anabaena pipX gene is expressed mainly in the (pro)heterocysts. Because in these cells the expression of the glnB gene (encoding the PII regulator) is turned down (32), an effect of PipX sequestration by PII is minimized. This leaves a mostly active PipX protein in differentiating cells, consistent with the phenotype of the CSV6 mutant, which is defective in heterocyst function. Thus, in filamentous heterocyst-forming cyanobacteria, the function of the PipX factor appears adapted to the distinct physiology of these organisms, reinforcing the expression of late heterocyst-specific genes to allow full levels of nitrogen fixation under oxic environments.
Acknowledgments
We thank Iris Maldener for electron microscopy and Enrique Flores for fruitful discussions.
This work was supported by grants BFU2007-60457, BFU2007-66589, and BFU2010-17980, cofinanced by FEDER, from the Ministerio de Ciencia e Innovación (Spain).
Footnotes
Published ahead of print on 30 December 2010.
REFERENCES
- 1.Aldea, M. R., K. Kumar, and J. W. Golden. 2008. Heterocyst development and pattern formation, p. 75-90. In S. C. Winans and B. L. Bassler (ed.), Chemical communication among bacteria. ASM Press, Washington, DC.
- 2.Ausubel, F. M., et al. 2010. Current protocols in molecular biology. Greene Publishing & Wiley-Interscience, New York, NY.
- 3.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 4.Buikema, W. J., and R. Haselkorn. 1991. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 5:321-330. [DOI] [PubMed] [Google Scholar]
- 5.Cai, Y., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elhai, J., and C. P. Wolk. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68:119-138. [DOI] [PubMed] [Google Scholar]
- 7.Elhai, J., and C. P. Wolk. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167:747-754. [DOI] [PubMed] [Google Scholar]
- 8.Espinosa, J., K. Forchhammer, S. Burillo, and A. Contreras. 2006. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 61:457-469. [DOI] [PubMed] [Google Scholar]
- 9.Espinosa, J., K. Forchhammer, and A. Contreras. 2007. Role of the Synechococcus PCC 7942 nitrogen regulator protein PipX in NtcA-controlled processes. Microbiology 153:711-718. [DOI] [PubMed] [Google Scholar]
- 10.Fiedler, G., M. Arnold, S. Hannus, and I. Maldener. 1998. The DevBCA exporter is essential for envelope formation in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 27:1193-1202. [DOI] [PubMed] [Google Scholar]
- 11.Fiedler, G., A. M. Muro-Pastor, E. Flores, and I. Maldener. 2001. NtcA-dependent expression of the devBCA operon, encoding a heterocyst-specific ATP-binding cassette transporter in Anabaena spp. J. Bacteriol. 183:3795-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores, E., and A. Herrero. 2010. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol. 8:39-50. [DOI] [PubMed] [Google Scholar]
- 13.Flores, E., A. Herrero, C. P. Wolk, and I. Maldener. 2006. Is the periplasm continuous in filamentous multicellular cyanobacteria? Trends Microbiol. 14:439-443. [DOI] [PubMed] [Google Scholar]
- 14.Frías, J. E., E. Flores, and A. Herrero. 1994. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 14:823-832. [DOI] [PubMed] [Google Scholar]
- 15.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183:411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrero, A., A. M. Muro-Pastor, A. Valladares, and E. Flores. 2004. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol. Rev. 28:469-487. [DOI] [PubMed] [Google Scholar]
- 17.Higa, K. C., and S. M. Callahan. 2010. Ectopic expression of hetP can partially bypass the need for hetR in heterocyst differentiation by Anabaena sp. PCC 7120. Mol. Microbiol. 77:562-574. [DOI] [PubMed] [Google Scholar]
- 18.Huang, X., Y. Dong, and J. Zhao. 2004. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc. Natl. Acad. Sci. U. S. A. 101:4848-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko, T., et al. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213. [DOI] [PubMed] [Google Scholar]
- 20.Llácer, J. L., et al. 2010. Structural basis for the regulation of NtcA-dependent transcription by proteins PipX and PII. Proc. Natl. Acad. Sci. U. S. A. 107:15397-15402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luque, I., E. Flores, and A. Herrero. 1994. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 13:2862-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackinney, G. 1941. Absorption of light by chlorophyll solutions. J. Biol. Chem. 140:315-322. [Google Scholar]
- 23.Markwell, M. A. K., S. M. Hass, L. L. Bieber, and N. E. Tolbert. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206-210. [DOI] [PubMed] [Google Scholar]
- 24.Montesinos, M. L., A. Herrero, and E. Flores. 1995. Amino acid transport systems required for diazotrophic growth in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 177:3150-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muro-Pastor, A. M., E. Flores, and A. Herrero. 2009. NtcA-regulated heterocyst differentiation genes hetC and devB from Anabaena sp. strain PCC 7120 exhibit a similar tandem promoter arrangement. J. Bacteriol. 191:5765-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muro-Pastor, A. M., A. Valladares, E. Flores, and A. Herrero. 1999. The hetC gene is a direct target of the NtcA transcriptional regulator in cyanobacterial heterocyst development. J. Bacteriol. 181:6664-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muro-Pastor, A. M., A. Valladares, E. Flores, and A. Herrero. 2002. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol. Microbiol. 44:1377-1385. [DOI] [PubMed] [Google Scholar]
- 28.Nichols, B. W., and B. J. B. Wood. 1968. New glycolipid specific to nitrogen-fixing blue-green algae. Nature 217:767-768. [Google Scholar]
- 29.Olmedo-Verd, E., A. M. Muro-Pastor, E. Flores, and A. Herrero. 2006. Localized induction of the ntcA regulatory gene in developing heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 188:6694-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olmedo-Verd, E., A. Valladares, E. Flores, A. Herrero, and A. M. Muro-Pastor. 2008. Role of two NtcA-binding sites in the complex ntcA gene promoter of the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 190:7584-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Partensky, F., W. R. Hess, and D. Vaulot. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paz-Yepes, J., E. Flores, and A. Herrero. 2009. Expression and mutational analysis of the glnB genomic region in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 191:2353-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paz-Yepes, J., A. Herrero, and E. Flores. 2007. The NtcA-regulated amtB gene is necessary for full methylammonium uptake activity in the cyanobacterium Synechococcus elongatus. J. Bacteriol. 189:7791-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 35.Tanigawa, R., et al. 2002. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc. Natl. Acad. Sci. U. S. A. 99:4251-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valladares, A., E. Flores, and A. Herrero. 2008. Transcription activation by NtcA and 2-oxoglutarate of three genes involved in heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 190:6126-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valladares, A., A. Herrero, D. Pils, G. Schmetterer, and E. Flores. 2003. Cytochrome c oxidase genes required for nitrogenase activity and dyazotrophic growth in Anabaena sp. PCC 7120. Mol. Microbiol. 47:1239-1249. [DOI] [PubMed] [Google Scholar]
- 38.Valladares, A., I. Maldener, A. M. Muro-Pastor, E. Flores, and A. Herrero. 2007. Heterocyst development and diazotrophic metabolism in terminal respiratory oxidase mutants of the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 189:4425-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vázquez-Bermúdez, M. F., A. Herrero, and E. Flores. 2002. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 512:71-74. [DOI] [PubMed] [Google Scholar]
- 40.Vioque, A. 1997. The RNase P RNA from cyanobacteria: short tandemly repeated repetitive (STRR) sequences are present within the RNase P RNA gene in heterocyst-forming cyanobacteria. Nucleic Acids Res. 25:3471-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winkenbach, F., C. P. Wolk, and M. Jost. 1972. Lipids of membranes and of the cell envelope in heterocysts of a blue-green alga. Planta 107:69-80. [DOI] [PubMed] [Google Scholar]
- 42.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 43.Zhao, M.-X., et al. 2010. Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate. Proc. Natl. Acad. Sci. U. S. A. 107:12487-12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao, M.-X., et al. 2010. Crystal structure of the cyanobacterial signal transduction protein PII in complex with PipX. J. Mol. Biol. 402:552-559. [DOI] [PubMed] [Google Scholar]