Abstract
The restriction-modification (R-M) systems of many bacteria present a barrier to the stable introduction of foreign DNA. The Lyme disease spirochete Borrelia burgdorferi has two plasmid-borne putative R-M genes, bbe02 and bbq67, whose presence limits transformation by shuttle vector DNA from Escherichia coli. We show that both the bbe02 and bbq67 loci in recipient B. burgdorferi limit transformation with shuttle vector DNA from E. coli, irrespective of its dam, dcm, or hsd methylation status. However, plasmid DNA purified from B. burgdorferi transformed naïve B. burgdorferi much more efficiently than plasmid DNA from E. coli, particularly when the bbe02 and bbq67 genotypes of the B. burgdorferi DNA source matched those of the recipient. We detected adenine methylation of plasmid DNA prepared from B. burgdorferi that carried bbe02 and bbq67. These results indicate that the bbe02 and bbq67 loci of B. burgdorferi encode distinct R-M enzymes that methylate endogenous DNA and cleave foreign DNA lacking the same sequence-specific modification. Our findings have basic implications for horizontal gene transfer among B. burgdorferi strains with distinct plasmid contents. Further characterization and identification of the nucleotide sequences recognized by BBE02 and BBQ67 will facilitate efficient genetic manipulation of this pathogenic spirochete.
Borrelia burgdorferi sensu lato is a zoonotic pathogen whose natural infectious cycle alternates between a tick vector and rodent or bird reservoir hosts (1, 7, 8, 14, 32, 33, 36). Transmission of B. burgdorferi to humans occurs through the bite of an infected tick and can lead to Lyme disease, which is a major public health concern in areas of North America and Europe where B. burgdorferi is endemic (8, 53).
The genomic structure of the spirochete B. burgdorferi is unique, consisting of a linear chromosome of approximately 900 kb and more than 20 linear (lp) and circular (cp) plasmids, ranging in size from ∼5 kb to 56 kb, in the type strain B31 (9, 10, 11, 19, 42). The plasmids of B. burgdorferi are present at unit copy number relative to the chromosome (22), and some are relatively unstable during in vitro propagation (52, 57). The loss of linear plasmids lp25, lp28-1, and lp36 by strain B31 was found to correlate with the loss of infectivity in mice (20, 31, 45, 56), leading to the identification of genes carried on these plasmids that are dispensable in vitro but required in vivo during an experimental infectious cycle (21, 26, 35, 44, 47). The loss of two linear plasmids, lp25 and lp56, was shown to correlate with enhanced shuttle vector transformation, suggesting that specific lp25 and lp56 gene products present a barrier to stable introduction of foreign DNA (34). Further studies linked the transformation phenotype of B. burgdorferi strain B31 with the bbe02 and bbq67 genes on lp25 and lp56, respectively, and the putative restriction-modification (R-M) enzymes that they encode (11, 27, 29, 34). The recent demonstration by Chen and colleagues of enhanced transformation of B. burgdorferi following in vitro methylation of DNA (13) further supports the hypothesis that these B. burgdorferi plasmids encode R-M enzymes that degrade foreign DNA lacking the appropriate modification.
The barrier to foreign DNA presented by the bbe02 and bbq67 loci of B. burgdorferi implies that genomic DNA should be modified in spirochetes carrying these plasmid genes. To test this hypothesis, we compared the transformation of B. burgdorferi with shuttle vector DNA isolated from either Escherichia coli or B. burgdorferi, as outlined in Fig. 1. We also examined whether and how the presence of putative R-M genes in either the donor or recipient B. burgdorferi strain influenced transformation. Finally, we analyzed the type of modification present on DNA isolated from B. burgdorferi with different plasmid or gene contents. Our data indicate that the bbe02 and bbq67 loci of B. burgdorferi encode enzymes that both methylate endogenous DNA and restrict foreign DNA lacking these modifications. These findings have basic implications regarding horizontal gene transfer among B. burgdorferi strains with distinct plasmid contents. These results also help elucidate the molecular mechanisms underlying the relative inefficiency of genetic transformation of B. burgdorferi and suggest ways in which genetic manipulation of this pathogenic spirochete could be enhanced.
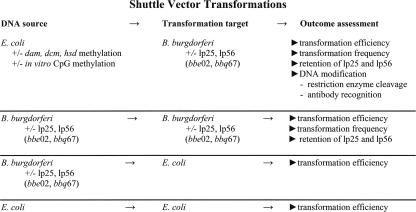
FIG. 1.
Shuttle vector transformations. Schematic representation of the various DNA sources, strains and methods used to assess the contributions of bbe02 and bbq67 to the restriction-modification (R-M) systems of B. burgdorferi.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The B. burgdorferi clones used in this study are described in Table 1. B. burgdorferi strain B31-A3 is an infectious, clonal derivative (17) of the sequenced type strain B31 (ATCC 35210) that has all the plasmids except cp9 (11, 19). Infectious clone A3-68 was derived by passage of clone A3 through a mouse, followed by single colony isolation and screening for loss of lp56. The bbe02 mutants used in this study were generated in the A3 and A3-68 backgrounds as described below. B31-A34 is a clonal derivative of serially passaged, noninfectious B31-A that lacks several plasmids, including lp25 and lp56 (26). Spirochetes were grown at 35°C in liquid Barbour-Stoenner-Kelly II (BSKII) medium (4, 28) supplemented with 6% rabbit serum (Pel Freez Biologicals, Rogers, AR). The appropriate antibiotic was added when growing bbe02 mutants or shuttle vector transformants of B. burgdorferi. Kanamycin was used at a concentration of 200 μg ml−1, streptomycin was used at 50 μg ml−1, and gentamicin was used at 40 μg ml−1. Escherichia coli Top10 cells (dam+ dcm+ hsd) (Life Technologies, Carlsbad, CA), INV110 cells (dam dcm hsd+) (Life Technologies), and strain GM161 (dam dcm+ hsd) (provided by Martin G. Marinus) were transformed with the shuttle vectors. E. coli bacteria were grown in lysogeny broth (LB) with the appropriate antibiotic (kanamycin [50 μg ml−1], spectinomycin [100 μg ml−1], or gentamicin [5 μg ml−1]).
TABLE 1.
B. burgdorferi strain B31 clones used in this study
| B. burgdorferi strain B31 clone | Plasmid content | Presence or absence of linear plasmid: |
Reference | |
|---|---|---|---|---|
| lp25 | lp56 | |||
| A3 | All plasmids present except cp9 | + | + | Elias et al. (17) |
| A3ΔBBE02 | All plasmids present except cp9 | + | + | This study |
| A3-68 | All plasmids present except cp9 and lp56 | + | − | This study |
| A3-68ΔBBE02 | All plasmids present except cp9 and lp56 | + | − | This study |
| A34 | All plasmids except cp9, lp5, lp21, lp25, lp28-1, lp28-4, cp32-6, cp32-7, lp36, and lp56 | − | − | Jewett et al. (25) |
Construction of pBBE02-Spec.
Kawabata and colleagues previously described an allelic exchange construct, pNP, used to delete 1,250 bp within the bbe02 locus on lp25, with concomitant introduction of a kanamycin resistance cassette (27). We generated a closely related plasmid, pBBE02-Spec (Spec stands for spectinomycin), in which the kanamycin cassette was replaced with a spectinomycin-streptomycin resistance cassette (flaBp::aadA) (26), as follows. The flaBp::aadA fusion was amplified (primers 1 and 2 [Table 2]) from a pCR 2.1 TOPO plasmid carrying this resistance cassette (18, 25). The flaBp::aadA PCR product and the pNP plasmid DNA (27) were digested with restriction enzymes NheI and PacI and ligated together with T4 DNA ligase (New England BioLabs, Beverly, MA). The ligation was transformed into E. coli Top10 cells (Life Technologies), and transformants were selected on LB plates containing spectinomycin (300 μg ml−1). Primers 1 and 2 (Table 2) were used to screen colonies for the flaBp::aadA cassette, and plasmid DNA was purified and sequenced from a positive clone, yielding pBBE02-Spec.
TABLE 2.
Primers used in this study
| Primer no. | Primer designationa | Sequence |
|---|---|---|
| 1 | Fla-Spec-NheI-5′ | TGT CGC TAG CTG TCG CCT CTT GTG GCT C |
| 2 | Fla-Spec-PacI-3′ | GAA TTT AAT TAA TTA TTT GCC GAC TAC CTT GG |
| 3 | BBE03-R | TTC TAG ACC AAA TAA GGC TTC CGG |
| 4 | Kan-F | TCA GAC TAA ACT GGC TGA CG |
| 5 | Kan-R | AAA CTC ACC GAG GCA GTT CC |
| 6 | Spec-F | TCC GCG CTG TAG AAG TCA CCA TTG |
| 7 | Spec-R | CCG GCA GGC GCT CCA TTG |
| 8 | Gent-F | TCT CGG CTT GAA CGA ATT GTT ACG T |
| 9 | Gent-R | GGC AGT CGC CCT AAA ACA AAG TT |
| 10 | Lp25-F | GTG CAC CTA TTG GAA AGG TC |
| 11 | Lp25-R | GGG CAT GTT GCA CAT ACG TT |
| 12 | Lp56-F | ACT ATT AAG ACG AGC AAT AAA AAG TCC A |
| 13 | Lp56-R | GAC GAA GCA AAG AAG GAT TTG GAT CAC C |
| 14 | 4278F | GCA CCC CAG GCT TTA CAC TTT ATG |
| 15 | 4278R | CGG GCC TCT TCG CTA TTA CG |
| 16 | 6188F | CCC GGG ACT TCG TGG AGG AC |
| 17 | 6188R | CCC GGA AGT TCG TGG ACA |
The orientation of the primer is indicated at the end of the primer designation as follows: F, forward; R, reverse.
Transformation of B. burgdorferi.
In order to transform the wild-type B. burgdorferi clone B31-A3 and isogenic derivatives with shuttle vectors carrying different selectable markers, we inactivated the bbe02 locus in B31-A3 and B31-A3-68 clones with either pNP or pBBE02-Spec allelic exchange constructs. In brief, 20 μg of E. coli pNP or pBBE02 plasmid DNA, 1 to 20 μg of E. coli shuttle vector DNA, or 10 μg of total plasmid DNA from B. burgdorferi shuttle vector transformants, in volumes between 5 and 10 μl, were electroporated into B. burgdorferi as previously described (17, 51). Electroporated cells were resuspended in 5 ml of BSKII medium and allowed to recover for 20 to 24 h at 35°C. Aliquots of the transformation mix were then plated in solid BSK medium, with or without the appropriate antibiotic.
Screening and characterization of bbe02 mutants.
Approximately 2 weeks after plating, colonies were screened by PCR using primers 4 and 5 (Table 2) to confirm mutants obtained with pNP or using primers 6 and 7 (Table 2) for mutants obtained with pBBE02-Spec. PCR screening conditions were as previously described (26). Colonies identified as bbe02 mutants in the B31-A3 and A3-68 background were aspirated into 10 ml of BSKII liquid medium and grown to mid-log phase. Total genomic DNA was prepared from 7 ml of culture using the Wizard genomic DNA kit (Promega, Madison, WI) per the manufacturer's instructions, and glycerol stocks were made of the remaining 3 ml. The total plasmid contents of bbe02 mutants were compared to that of the parental strain to ensure the isogenicity of the strains, using primers specific for each plasmid (17).
PCR screening of shuttle vector transformants.
Two weeks after plating, B. burgdorferi colonies were counted, and up to 40 colonies per transformation were screened by PCR (average number, 24). Primers (Table 2) specific for the antibiotic resistance cassette on the shuttle vector were used to confirm the presence of pBSV2 (kan [primers 4 and 5]), pKFSS1 (aadA [primers 6 and 7]), or pBSV2G (aacC1 [primers 8 and 9]) in transformants as previously described (26). Retention of plasmids lp25 and lp56 in shuttle vector transformants was determined by PCR using primers 10 and 11 for lp25 and primers 12 and 13 for lp56 (Table 2).
In vitro methylation of plasmid DNA.
As described previously by Chen et al. (13), between 50 and 100 μg of E. coli plasmid DNA was methylated, following the manufacturer's recommendation, with the CpG methyltransferase M.SssI (New England BioLabs or Zymo Research, Irvine, CA). Methylation was assessed by digestion of plasmid DNA with the restriction enzyme BstUI, which is blocked by CpG methylation, before and after in vitro methylation.
Isolation of plasmid DNA from B. burgdorferi.
B. burgdorferi cultures (200 ml) of shuttle vector transformants were grown to late log phase in BSKII medium. Plasmid DNA was isolated using the Hi-Speed Maxi Prep kit (Qiagen, Valencia, CA) per the manufacturer's instructions, with an additional wash of cells with HEPES-NaCl (HN) buffer (10 mM HEPES, 50 mM NaCl [pH 8.0]) prior to resuspension and lysis.
Transformation frequency and efficiency.
Transformation outcome was assessed in two ways. Transformation frequency (TF) measures what proportion of the bacterial population, after recovery from electroporation, has stably acquired the transforming DNA. This is calculated by dividing the number of transformed bacteria by the number of viable bacteria in the same volume of electroporated culture. Transformation efficiency (TE) reflects the number of transformants obtained per microgram of DNA and is calculated by dividing the number of transformants by the amount of DNA used in the electroporation. The TE for plasmid DNA purified from B. burgdorferi was calculated for the amount of relevant shuttle vector DNA, which was estimated to be approximately 1% of total B. burgdorferi plasmid DNA (6-kb shuttle vector out of 600-kb total plasmid DNA). Viable bacteria were enumerated as CFU by plating aliquots and dilutions of the electroporated culture in media containing or lacking antibiotic selection for the transforming DNA.
For both TF and TE calculations, PCR screening of colonies on plates containing antibiotics was used to distinguish true transformants (presence of transforming DNA) from spontaneous resistance mutants.
Southern blot analysis.
Restriction digestions, with restriction enzymes from New England BioLabs, were carried out per the manufacturer's instructions, using plasmid DNA isolated from B. burgdorferi shuttle vector transformants as well as plasmid DNA isolated from E. coli. Plasmid DNA was run on 0.7% agarose gels (overnight, 30 V), depurinated, denatured, and blotted onto Biotrans nylon membranes (40). DNA was cross-linked to the membrane using a UV Stratalinker 1800 (Stratagene, Santa Clara, CA) and hybridized with digoxigenin (DIG)-labeled probes (Roche, Indianapolis, IN) (Table 3) as previously described (55).
TABLE 3.
Primers used to construct probes for Southern blot analysis
| Primer pairs used to generate probes | Shuttle vector(s) detected by the PCR-generated probe |
|---|---|
| AACACTCCTTTAAATCTACAC and ATCTAATATAAACAAAACCAGT | pBSV2, pKFSS1, and pBSV2G |
| GCACCCCAGGCTTTACACTTTATG and CGGGCCTCTTCGCTATTACG | pBSV2, pKFSS1, and pBSV2G |
| CTGGCGTAATAGCGAAGAGG and CCAGGGGAAGCCGAAGTT | pKFSS1 |
| GTGCACGACGACATCATTCC and CGGCAGCGACATCCTTC | pKFSS1 |
| AGCGCCTGCCGGCCCAGTATCAG and CCCGCCCCCACGGCTGCTC | pKFSS1 |
| TCAGACTAAACTGGCTGACG and AAACTCACCGAGGCAGTTCC | pBSV2 and pKFSS1 |
Southwestern analysis of DNA modification.
Membranes containing plasmid DNA prepared as described above were also subjected to immunoblot analysis to determine the type of modification present on B. burgdorferi DNA. The membranes were blocked for 1 h in 5% nonfat dry milk in Tris-buffered saline containing 1% Tween 20 (TBS-Tween) and then incubated with one of the following primary antibodies overnight at 4°C; rabbit antisera (New England BioLabs) that recognize modified DNA containing N6-methyladenine (m6A) (1:5,000) or N4-methylcytosine (m4C) (1:10,000) or a mouse monoclonal antibody that recognizes C5-methylcytosine (m5C) (1:1,000) (Zymo Research). After the membranes were washed with TBS-Tween, secondary anti-rabbit (1:50,000) or anti-mouse (1:10,000) serum conjugated with horseradish peroxidase (Sigma-Aldrich, St. Louis, MO) was added to the membrane and incubated for 1 h. Positive signals were detected using a SuperSignal WestPico chemiluminescent-substrate kit (Thermo Scientific, Rockford, IL).
Animal infections.
Rocky Mountain Laboratories (RML) is accredited by the International Association of Assessment and Accreditation of Laboratory Animal Care. Animal protocols were prepared according to the guidelines of the National Institutes of Health and approved by the RML Animal Care and Use Committee. C3H/HeN mice (Harlan Sprague-Dawley, Indianapolis, IN), representing a uniform, inbred rodent population, were used to assess the infectivity of kanamycin- or spectinomycin-resistant bbe02 mutants derived from B31-A3 and B31-A3-68 clones (Table 1). Mouse inoculations and assessment of infection were as described previously (17, 26). Briefly, 4 mice per strain were used, and spirochetes were injected intraperitoneally (4 × 103 spirochetes) and subcutaneously (1 × 103) (17). Mice were euthanized at 3 weeks postinoculation, and infectivity was assessed by seroconversion of mice to B. burgdorferi proteins and reisolation of spirochetes from ear tissues, bladders, and rear ankle joints (20).
RESULTS
Assessing the roles of bbe02 and bbq67 in limiting B. burgdorferi shuttle vector transformations.
Previous studies from other labs have demonstrated enhanced transformation of B. burgdorferi clones lacking linear plasmids lp25 and lp56 with the shuttle vector pBSV2 (Table 4) (27, 34). Anecdotal data from our own lab suggest a difference between pBSV2 and the closely related shuttle vectors pBSV2G and pKFSS1 (Table 4), which carry different antibiotic resistance cassettes, in stable transformation of the infectious clone B31-A3, which we routinely use for genetic studies and in vivo analyses. In order to directly compare the influence of the putative R-M system encoded by lp25 and lp56 on transformations with all three shuttle vectors, we inactivated the bbe02 locus on lp25 in clones B31-A3 and B31-A3-68 (an isogenic derivative of B31-A3 that lacks lp56) with related allelic exchange constructs that confer either kanamycin resistance or spectinomycin-streptomycin resistance (see Materials and Methods). All bbe02 mutants and the B31-A3-68 strain retained infectivity in mice, consistent with previous findings (27). We have assumed that bbq67 is the only gene on lp56 that influences the transformation phenotype or R-M activity of B. burgdorferi but have not demonstrated this through gene inactivation or complementation.
TABLE 4.
Plasmids used in this study
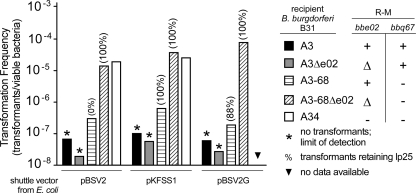
Although both bbe02 and bbq67 have been identified as R-M gene homologs, the putative endonuclease and methyltransferase activities of their products have not been defined (11, 19, 27, 34). To determine whether sequence differences between closely related shuttle vectors resulted in differential recognition by these putative R-M systems, we transformed B. burgdorferi clones containing or lacking bbe02 and bbq67 with the shuttle vectors pBSV2, pBSV2G, and pKFSS1, using plasmid DNA prepared from E. coli (dam+ dcm+ hsd). Consistent with previous findings, transformation frequencies varied among clones depending upon their bbe02 and bbq67 genotype (Fig. 2) (27, 34). No shuttle vector transformants were obtained when both loci or only bbq67 was present, whereas much higher transformation frequencies were observed when both bbe02 and bbq67 were absent in recipient B. burgdorferi. Similar transformation frequencies were obtained with all three shuttle vectors compared for the same B. burgdorferi clone (Fig. 2). However, a difference was noted between pBSV2 and the other shuttle vectors in the outcome of transformations with strain B31-A3-68 (which lacks lp56 and bbq67 but in which bbe02 was initially present); although pBSV2 transformants were recovered, they all lacked lp25, whereas most of the pBSV2G and pKFSS1 transformants in this genetic background retained lp25. These experiments were performed at least twice in all B. burgdorferi backgrounds with similar results. Figure 2 shows data from a single experiment/competence preparation for each clone transformed in parallel with all three shuttle vectors. These data indicate that the bbe02 and bbq67 loci of B. burgdorferi represent barriers to shuttle vector transformation, confirming and extending previous findings (13, 27, 34).
FIG. 2.
Transformation of B. burgdorferi with shuttle vector DNA from E. coli. Transformation frequencies of B. burgdorferi strains used in this study with shuttle vector DNAs isolated from E. coli (dam+ dcm+ hsd) were calculated by determining the proportion of viable bacteria after electroporation that contained the shuttle vector. The shuttle vectors used in each transformation are identified below the bars. Recipient B. burgdorferi strain genotypes are distinguished by the different bars identified to the right of the graph (+, intact gene; Δ, gene deletion; −, missing plasmid). An asterisk denotes the limit of detection when no transformants were obtained from a defined number of viable bacteria. The percentage of transformants retaining lp25 when initially present in the recipient clone is indicated in parentheses above the bars.
E. coli modification of shuttle vector DNA.
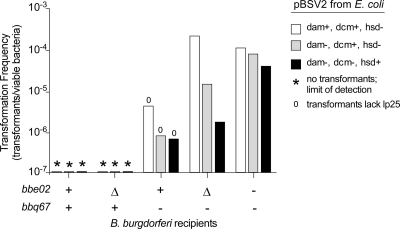
Modification of plasmid DNA by E. coli has been shown to influence subsequent transformations in other bacteria (30, 38, 39, 41). Plasmid DNA used in the initial shuttle vector comparison (Fig. 2) was prepared from E. coli containing Dam (GATC) and Dcm (CCA/TGG) methyltransferases, which modify adenine and cytosine residues, respectively, in a sequence-specific fashion. To determine whether dam, dcm, or hsd (AACAGTGC/GCACAGTT) modifications were specifically recognized by BBE02 or BBQ67, we transformed B. burgdorferi with pBSV2 shuttle vector DNA prepared from three different strains of E. coli with various methyltransferase activities. No pBSV2 transformants were obtained, regardless of the plasmid DNA methylation status, when recipient B. burgdorferi carried both bbe02 and bbq67 or either locus individually (Fig. 3). Numerous transformants of B. burgdorferi clones that lacked both bbe02 and bbq67 were obtained, and we observed no consistent differences that correlated with B. burgdorferi plasmid content and E. coli methylation (Fig. 3). Transformations were performed twice in recipients containing both bbe02 and bbq67 or only bbq67 and once in the other strains, with all three E. coli plasmid sources. We conclude that dam, dcm, and hsd modification of shuttle vector DNA by E. coli does not form the basis for recognition of foreign DNA by the bbe02 and bbq67 gene products in B. burgdorferi.
FIG. 3.
Influence of shuttle vector DNA methylation by E. coli on transformation of B. burgdorferi. B. burgdorferi clones were transformed with pBSV2 shuttle vector DNA prepared from E. coli containing or lacking Dam, Dcm, and Hsd methyltransferases. Recipient B. burgdorferi strain genotypes are indicated below the bars (+, intact gene; Δ, gene deletion; −, missing plasmid). Strain genotypes for the E. coli DNA donors are distinguished by the different bars identified to the right of the graph. Transformation frequency was calculated by determining the proportion of viable bacteria after electroporation that contained the shuttle vector. An asterisk denotes the limit of detection when no transformants were obtained from a defined number of viable bacteria. The absence of lp25 in all recovered transformants is indicated (zero), although lp25 was present in >90% of viable bacteria after electroporation. Transformations were performed twice for recipients containing both bbe02 and bbq67 or only bbq67 and once for all other recipients, in parallel with pBSV2 DNA from all three E. coli sources.
In separate experiments using in vitro-methylated DNA as described by Chen et al. (13), we noticed a potential influence of dam methylation when transforming B. burgdorferi strains carrying both bbe02 and bbq67 with the shuttle vector pBSV2. In this case, pBSV2 transformants retaining lp25 (bbe02) were obtained only when plasmid DNA was prepared from E. coli dam mutant (data not shown). This outcome was not observed with pKFSS1 or pBSV2G shuttle vectors and was not previously reported for pBSV2 by Chen et al. (13). If confirmed, this result could indicate the presence of a unique restriction site on pBSV2 whose cleavage by BBE02 is blocked by in vitro M.SssI methylation unless this modification is itself blocked by overlapping dam methylation.
Transformation efficiency of shuttle vector DNA from B. burgdorferi.
As stated previously, we wished to determine whether the putative R-M system genes of B. burgdorferi encode activities that can discriminate between cellular and foreign DNA of identical nucleotide sequence, using shuttle vector DNA prepared from both B. burgdorferi and E. coli. Several steps were taken to obtain shuttle vector transformants in B. burgdorferi genetic backgrounds that contained bbe02 and bbq67 to serve as a source of modified plasmid DNA from Borrelia. We incorporated in vitro methylation of E. coli-derived plasmid DNA with the methyltransferase M.SssI, as previously described by Chen and colleagues (13), to introduce the shuttle vectors into B. burgdorferi clones that retained linear plasmids lp25 and lp56. These B. burgdorferi transformants were used in turn as a source of Borrelia-modified shuttle vector DNA with which to transform naïve recipient B. burgdorferi. Transformation efficiencies were determined for shuttle vector DNA endogenously modified in B. burgdorferi transformants and used to transform B. burgdorferi with different bbe02 and bbq67 backgrounds, as outlined in Fig. 1.
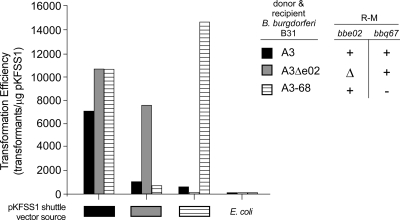
The genotypes of both the B. burgdorferi plasmid DNA source and transformed recipient influenced the observed transformation efficiency, as illustrated by the data presented in Table 5 and Fig. 4. Consistent with the hypothesis that bbq67 and bbe02 encode enzymes with both modification and restriction activities, transformation efficiency was highest with potentially fully modified plasmid DNA obtained from B. burgdorferi in which both loci were present and introduced into spirochetes in which putative restriction by BBE02 and BBQ67 was absent (Table 5 and Fig. 4; see Fig. S1 and S2 in the supplemental material). Few or no transformants were recovered when plasmid DNA was introduced from donors that lacked bbe02 or bbq67 into recipient B. burgdorferi that retained both loci. The absence of bbe02 or bbq67 in DNA donors was tolerated if the same locus was also missing in the transformed recipient. However, the presence of the bbe02 locus in recipient bacteria was particularly limiting for transformation with the shuttle vector pBSV2, even when bbe02 was present in the DNA donor (Table 5; see Fig. S1 in the supplemental material). In general, qualitatively similar results were obtained with all three shuttle vectors, but transformation efficiencies varied quantitatively with different combinations of shuttle vectors and strains (Table 5 and Fig. 4; see Fig. S1 and S2 in the supplemental material). In recipient strains that possessed lp25 and/or lp56, retention of these plasmids in transformants was 100%. Importantly, shuttle vector DNA obtained from spirochetes carrying bbq67 and bbe02 transformed recipient Borrelia much more efficiently than plasmid DNA purified from E. coli, even with in vitro M.SssI methylation, demonstrating differential recognition and restriction of foreign versus Borrelia DNA (Fig. 4; see Fig. S1 and S2 in the supplemental material).
TABLE 5.
Transformation efficiencies obtained when total plasmid DNA from a bbe02+ bbq67+ shuttle vector transformant is used for transforming various strains of B. burgdorferi within this study
| Transformation target | Transformation efficiency (no. of transformants/μg) with shuttle vector DNA from B31-A3 |
||
|---|---|---|---|
| pBSV2 | pKFSS1 | pBSV2G | |
| B31-A3 (bbe02+bbq67+) | 220 | 6,700 | 4,800 |
| B31-A3ΔBBE02 (Δbbe02 bbq67+) | 17,000 | 10,300 | 7,800 |
| B31-A3-68 (bbe02+) | 20 | 10,300 | 8,600 |
FIG. 4.
Influence of putative restriction-modification systems in B. burgdorferi on the transformation efficiency of pKFSS1 shuttle vector DNA prepared from B. burgdorferi. Three different B. burgdorferi clones, containing or lacking the bbe02 and bbq67 loci, were transformed by electroporation with B. burgdorferi plasmid DNA prepared from three different B. burdorferi clones containing the shuttle vector pKFSS1. The transformation efficiency for pKFSS1 was calculated for the fraction of total B. burgdorferi plasmid DNA that it represents. The pertinent bbe02 and bbq67 genotypes of both donor and recipient B. burgdorferi are distinguished by shading beneath the bars (plasmid source) and shading of the bars (plasmid recipient) identified to the right of the graph (+, intact gene; Δ, gene deletion; −, missing plasmid). The transformation efficiency of in vitro-methylated (M.SssI) pKFSS1 DNA from E. coli (dam+ dcm+ hsd) is included for comparison. Transformations for all donor-recipient combinations were carried out at least twice except for the following, which was carried out once: donor A3Δbbe02/recipient A3. Plasmids lp25 and lp56 were retained in all transformants when initially present in the recipient. The transformation efficiency of pKFSS1 from clone A3 into all recipients is also presented in Table 5.
DNA methylation by BBE02 and BBQ67.
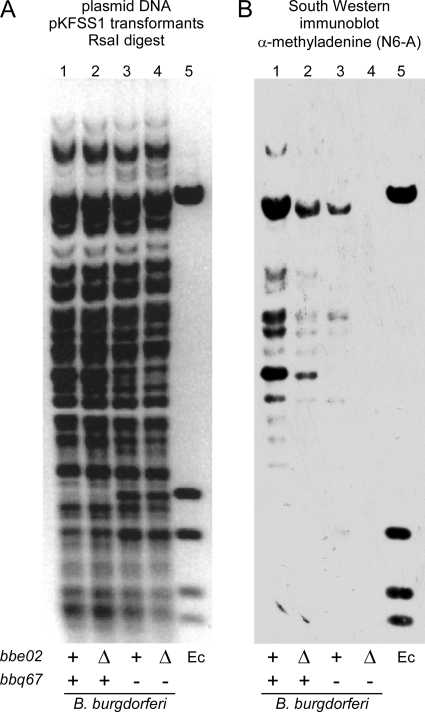
BBE02 and BBQ67 have been classified as homologs of type II R-M enzymes (48). The type II R-M system is one of four categorized R-M systems and is composed of an endonuclease-methyltransferase enzyme pair that recognizes/cleaves and modifies the same DNA sequence. There are several subdivisions within the type II systems that are based on how the enzymes recognize/cleave DNA, and fusion of the R and M genes as a single composite gene (48). Bioinformatics predictions with the software programs PROSITE and PHYRE suggest that both BBE02 and BBQ67 encode N6-adenine methyltransferases on the basis of sequence homology and conserved amino acid motifs. We wished to determine whether B. burgdorferi DNA contains methylated bases and whether BBE02 and BBQ67 influence this modification. To this end, we determined whether antisera specific for methylated adenine or methylated cytosine could recognize and bind B. burgdorferi DNA. Plasmid DNA from E. coli with defined adenine and cytosine methylation patterns was included as a control. We found that antiserum specific for N6-methyladenine recognized B. burgdorferi DNA only when it came from strains in which either bbe02 or bbq67 was present and that the signal was strongest when both genes were present in the source strain (Fig. 5). B. burgdorferi DNA was not recognized by antiserum specific for N4-methylcytosine irrespective of the strain background (data not shown). A monoclonal antibody specific for C5-methylcytosine bound B. burgdorferi DNA from all strains in a dot blot assay but had a high level of nonspecific background when used in the Southwestern format (data not shown). We conclude that B. burgdorferi DNA can contain N6-methylated adenine and that BBE02 and BBQ67 contribute to this modification, consistent with their homology to adenine methyltransferases (11, 19).
FIG. 5.
Adenine methylation of DNA by B. burgdorferi clones harboring bbe02 or bbq67. (A) RsaI digestion products of total plasmid DNA prepared from B. burgdorferi and E. coli (dam+ dcm+ hsd) carrying the shuttle vector pKFSS1, separated by gel electrophoresis, and stained with GelRed. (B) Southwestern blot analysis of plasmid DNA from B. burgdorferi and E. coli. The gel shown in panel A was transferred to a membrane and incubated with a polyclonal rabbit antiserum that recognizes methylated adenine (N6-A). The pertinent bbe02 and bbq67 genotypes of B. burgdorferi that served as sources of plasmid DNA are identified beneath the lanes (+, intact gene; Δ, gene deletion; −, missing plasmid). Lane 1, A3; lane 2, A3ΔBBE02; lane 3, A3-68; lane 4, A3-68ΔBBE02; lane 5, E. coli (Ec) (dam+ dcm+ hsd).
Identification of BBQ67 modification site.
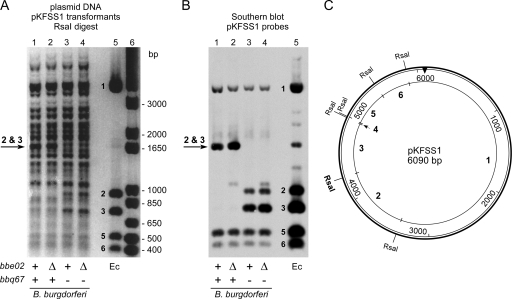
To confirm and further characterize the modification of endogenous DNA by BBE02 and BBQ67, we investigated whether B. burgdorferi DNA was resistant to cleavage by a number of potentially sensitive restriction enzymes. The only enzyme whose activity was blocked/impaired by overlapping methylation of B. burgdorferi DNA was RsaI, which recognizes and cleaves unmethylated GTAC sequences. RsaI cleavage was partially blocked when DNA was obtained from B. burgdorferi strains carrying bbq67 but was unaffected by the presence of bbe02. This is illustrated in Fig. 6 with B. burgdorferi plasmid DNA purified from pKFSS1 shuttle vector transformants in 4 different genetic backgrounds; pKFSS1 DNA from E. coli (dam+ dcm+ hsd), which is fully susceptible to RsaI digestion, is shown for comparison. One of six RsaI sites present on pKFSS1 is protected from cleavage when DNA is obtained from B. burgdorferi with bbq67, resulting in a larger ∼1.6-kb fragment instead of two smaller RsaI fragments. This RsaI site is also protected on pBSV2 and pBSV2G plasmid DNA when purified from B. burgdorferi carrying bbq67 (see Fig. S3 and S4 in the supplemental material). pBSV2G has an additional RsaI site that is protected from digestion when obtained from B. burgdorferi transformants that carry bbq67 (see Fig. S4 in the supplemental material). The presence of bbe02 in B. burgdorferi did not noticeably impair RsaI cleavage of any sites in shuttle vector, total plasmid, or genomic DNA (Fig. 6 and data not shown). We conclude that BBQ67 encodes an adenine methyltransferase that modifies a sequence including or adjacent to a subset of RsaI sites (GTAC) in these shuttle vectors.
FIG. 6.
Protection of an RsaI site on pKFSS1 by BBQ67. (A) RsaI digestion products of total plasmid DNA from B. burgdorferi and E. coli transformants carrying the shuttle vector pKFSS1, separated by gel electrophoresis, and stained with GelRed. The migration positions of size standards (in base pairs) are indicated to the right of the gel. (B) Southern blot analysis of RsaI-digested plasmid DNA from B. burgdorferi and E. coli. The gel shown in panel A was transferred to a membrane and hybridized with labeled pKFSS1 fragments. The pertinent bbe02 and bbq67 genotypes of B. burgdorferi that served as sources of plasmid DNA are identified beneath the gels (+, intact gene; Δ, gene deletion; −, missing plasmid). Lanes 1, A3; lanes 2, A3ΔBBE02; lanes 3, A3-68; lanes 4, A3-68ΔBBE02; lanes 5, E. coli (dam+ dcm+ hsd); lane 6, 1-kb Plus DNA ladder (Invitrogen). The numbering of pKFSS1 RsaI fragments from E. coli corresponds with the numbering in the schematic diagram in panel C. The arrow identifies the pKFSS1 fragment that results when cleavage of a particular RsaI site is blocked. (C) Schematic diagram of the shuttle vector pKFSS1. The RsaI sites are indicated, and fragments are numbered for comparison with the restriction profiles in panels A and B. The RsaI site indicated in bold type is protected from cleavage on plasmid DNA from B. burgdorferi carrying bbq67.
DISCUSSION
In this study we have investigated the roles of putative R-M genes in the type strain of the Lyme disease spirochete, B. burgdorferi B31. Although steady progress has been made since B. burgdorferi was first transformed in 1994, genetic manipulation of B. burgdorferi remains inefficient, particularly when working with infectious clones that retain most of the plasmids (50). Bioinformatics analyses, coupled with transformation of strain B31 clones with various plasmid backgrounds, previously pinpointed the bbe02 and bbq67 plasmid loci, on lp25 and lp56, respectively, as likely components of the R-M system of B. burgdorferi (11, 19, 29, 34). Kawabata and colleagues highlighted the roles of these gene products in limiting stable introduction of exogenous DNA by inactivation of bbe02 in a B. burgdorferi clone that lacked linear plasmid lp56, which dramatically increased the transformation efficiency of shuttle vector DNA from E. coli (27). They and we have presumed that bbq67 is the only gene on lp56 that influences transformation phenotype, but a formal demonstration of this is needed. A subsequent study demonstrated that CpG methylation of foreign plasmid DNA enhanced transformation of B. burgdorferi carrying lp56, consistent with an R-M system encoded by bbq67 (13). High-throughput genetic transformation techniques, like transposon mutagenesis, have been effective only in B31 strains that lack lp25 and lp56 (6, 54).
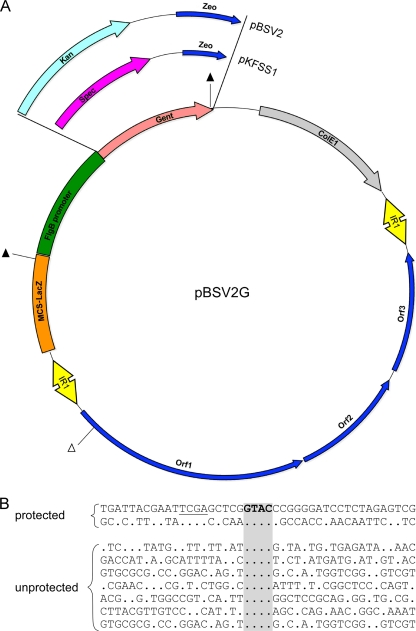
We extended the observation that bbe02 and bbq67 limit stable introduction of foreign DNA into B. burgdorferi, using three closely related shuttle vectors that differ primarily in the selectable markers they carry (Fig. 7 A). Transformation with all three plasmids prepared from E. coli was decreased by the presence of bbe02 and bbq67 (or the lp25 and lp56 plasmids that carry them) in targeted recipient bacteria, although the pBSV2 shuttle vector appeared to be more sensitive than pKFSS1 and pBSV2G to the barrier presented by bbe02 (Fig. 2). The bbh09 gene on lp28-3 is a homolog of bbe02 (19, 34), yet we have not detected any influence of lp28-3 on shuttle vector transformations in unrelated studies (data not shown). This may indicate that these shuttle vectors do not possess any sites recognized/cleaved by BBH09 or that the enzyme is not active or made in these clones. Transformation studies in other bacteria have shown that methylated plasmid DNA from E. coli can be restricted unless the recipient bacterium contains a similar methyltransferase activity (12, 37, 38, 39, 41). Hughes and Johnson (23) detected Dam methylation of DNA by the relapsing fever spirochete Borrelia hermsii but did not detect Dam methylation by most B. burgdorferi strains, including B31. We did not observe a major influence on transformation of B. burgdorferi by modification of shuttle vector DNA by Dam, Dcm, or Hsd methyltransferases in E. coli (Fig. 3). However, transformation of B. burgdorferi carrying both bbe02 and bbq67 with the shuttle vector pBSV2 was successful only when plasmid DNA was obtained from E. coli lacking dam and subsequently modified in vitro with the CpG methyltransferase M.SssI.
FIG. 7.
Differences between shuttle vectors and sequences surrounding cleaved versus protected RsaI sites. (A) Schematic diagram depicting the differences between the three shuttle vectors used in this study. ▴, protected RsaI restriction sites on pBSV2G; Δ, unprotected RsaI restriction site on pBSV2G; IR1, inverted repeat 1; ColE1, E. coli origin of replication; MCS-LacZ, alpha-peptide and multiple-cloning site (MCS) of pUC18; Orf1, Orf2, and Orf3, plasmid replication genes from cp9 of B. burgdorferi; Zeo, zeocin resistance marker; Kan, kanamycin resistance gene; Spec, spectinomycin-streptomycin resistance gene (aadA); Gent, gentamicin resistance gene (aacC1). (B) Alignment of nucleotide sequences surrounding the protected and unprotected RsaI sites (unique and shared) on shuttle vectors pBSV2G, pKFSS1, and pBSV2. Nucleotides that are identical to those in the top line are indicated by periods. The top line shows the protected RsaI site in the MCS shared by all three shuttle vectors. The RsaI restriction site, GTAC, is shown in bold type and shaded. The underlined sequence, TCGA, indicates the conservation of this sequence upstream of the RsaI cleavage site in both protected sequences.
The data presented in this study demonstrate for the first time that the bbe02 and bbq67 gene products both modify endogenous DNA and restrict foreign DNA lacking similar modification in a sequence-specific fashion, as predicted for R-M systems. This conclusion was supported by experiments in which increased transformation efficiencies were obtained when B. burgdorferi was both the source and recipient of transforming shuttle vector DNA (Fig. 4). The transformation barrier presented by bbe02 and bbq67 in recipient bacteria was overcome if shuttle vector DNA came from B. burgdorferi donors that also carried these loci. The inferred modification of shuttle vector DNA by bbe02 and bbq67 gene products did not limit transformation into recipient B. burgdorferi, consistent with recognition and cleavage of distinct and unmethylated DNA sequences by these putative R-M systems. The highest transformation efficiencies were obtained with B. burgdorferi recipients that lack bbe02 or bbq67 (Fig. 4 and Table 5), suggesting incomplete modification and only partial protection of restriction sites on endogenous shuttle vector DNA by bacteria carrying bbe02 and bbq67. Significantly, transformation of B. burgdorferi containing bbe02 and bbq67 was dramatically enhanced with plasmid DNA obtained from B. burgdorferi relative to shuttle vector DNA from E. coli, even with in vitro CpG modification (Fig. 4). Together these results suggest that the bbe02 and bbq67 loci of B. burgdorferi encode R-M enzymes that recognize and cleave distinct unmodified sequences.
NCBI and PROSITE analyses identified bbe02 and bbq67 as homologs of adenine methyltransferase. We have provided the first direct evidence supporting this designation using an antiserum that recognizes DNA containing N6-methyladenine. Antibody binding was observed with DNA from B. burgdorferi containing bbe02 or bbq67 when used in a Southwestern blot format (Fig. 5). Dot blot assays with the same antiserum, in which genomic DNA from these strains was focused in a single spot, indicated the presence of weak adenine methylation in the strain lacking both bbe02 and bbq67 (lp56) (data not shown). This result suggests that B. burgdorferi may contain additional adenine methyltransferases but with more limited activities than BBE02 and BBQ67.
Further support for adenine methylation by the bbq67 gene product was suggested by protection of certain RsaI sites on plasmid DNA in strains containing bbq67 (Fig. 6; see Fig. S3 and S4 in the supplemental material). RsaI cleavage is blocked by adenine methylation of a GTAC restriction site or by overlapping cytosine methylation of CGTAC or GTACG sequences (43). The latter sequences, however, are not among the protected RsaI sites present on B. burgdorferi shuttle vectors (Fig. 7B). We also have not observed any protection of RsaI sites on shuttle vector DNA following in vitro CpG or GpC methylation (data not shown). Rocha et al. (49) suggested that GTAC could be a potential restriction site for endonucleases in Borrelia based on the palindromic avoidance of this sequence in the B. burgdorferi genome. The cleavage by RsaI of unprotected GTAC sites on B. burgdorferi DNA indicates that GTAC cannot comprise the entire sequence recognized and modified by the bbq67 gene product. A shared feature of the sites protected from RsaI cleavage is a TCGA sequence five bases upstream of the GTAC RsaI site, which suggests that it may contribute to the recognition sequence (Fig. 7B). We found no evidence that any of the other potential restriction sites mentioned by Rocha and colleagues (49) could be blocked by BBQ67 or BBE02 modification (data not shown). Modification of GTAC sequences has been reported in another spirochete, Leptospira (46).
A search of the NCBI database indicated that all B. burgdorferi strains in the database have a homolog of bbe02, whereas only a subset of these strains have a homolog of bbq67. REBASE, the database for genes that encode R-M enzymes, identified homologs of a cytosine methyltransferase (GCGC) in available B. burgdorferi sequences, including strains 64b, 118, and 156a. B. burgdorferi strain N40 also carries this homolog (S. Casjens, personal communication). No homologs of this methyltransferase are present in B. burgdorferi B31. NCBI databases indicate that those strains possessing a bbq67 homolog do not possess a homolog of the GCGC methyltransferase and vice versa. HhaI, which cleaves GCGC but is blocked by methylation of this sequence, was efficient in digesting DNA from strain B31 but not from strain N40 (data not shown), indicating the occurrence of two different R-M systems in these B. burgdorferi strains.
Modification of endogenous DNA permits bacteria to distinguish self from nonself and block introduction of foreign DNA (5). A question arises regarding the selective advantage of different R-M genes carried by individuals within the same species. Jeltsch has proposed that diversification of R-M systems within a species leads to different biotypes, which then leads to adaptation to different ecological niches (24). This protection from foreign DNA, albeit from within the same species, would avoid dilution of advantageous traits for a particular niche that arose after diversifying from a common ancestor. The array of mammalian and avian hosts that B. burgdorferi sensu lato infects could represent distinct biological niches, but currently there are no data linking particular plasmids (and the R-M systems they carry) with the adaptation of a strain to a specific host. The presence of multiple plasmid-borne R-M systems in the segmented genome of B. burgdorferi, the heterogeneity in plasmid content among B. burgdorferi strains, and the horizontal transfer of plasmid sequences between spirochetes, all need to be taken into account when considering the role of R-M systems in B. burgdorferi.
Transformation studies in other bacteria have clearly shown that endogenously modified plasmid DNA can more efficiently transform strains possessing the same R-M system (2, 3, 15, 37, 39). The data provided by this study, as well as previous work by other investigators, indicate that a primary limitation to transformation of B. burgdorferi with shuttle vector DNA from E. coli is the barrier presented by the endogenous plasmid-borne R-M systems of B. burgdorferi. This transformation barrier can be overcome by inactivation of the R-M loci or loss of the plasmids that carry these loci in recipient B. burgdorferi, which can complicate or impede subsequent studies. Obtaining plasmid DNA from an E. coli host that also expresses the R-M genes of the recipient bacterium is an approach that has been used to introduce foreign DNA into other bacteria (58). Our results clearly show that for such an approach to work in wild-type B. burgdorferi strain B31 clone, both the bbe02 and bbq67 loci need to be taken into account. Identification of the sequences recognized by BBE02 and BBQ67 in modifying and cleaving DNA would enhance genetic manipulation of infectious B. burgdorferi clones in the B31 strain background. Future work to address the R-M properties of BBE02 and BBQ67 include overexpressing and purifying recombinant forms of these proteins. Coupled with genome sequences, similar studies with the putative R-M genes in other B. burgdorferi strains are needed to advance genetic studies in potentially different biotypes of this pathogenic spirochete and to explore the biological significance of gene transfer between strains.
Supplementary Material
Acknowledgments
We thank Hiroki Kawabata and Steven Norris for the gift of the pNP plasmid used for inactivation of bbe02, Qiang Chen and John Leong for communicating the in vitro CpG methylation protocol prior to publication, Martin Marinus for providing E. coli strain GM161, New England BioLabs for providing antisera that recognize methylated DNA, Joe Hinnebusch, Paul Beare, Robert Blumenthal, and members of the Rosa lab for helpful comments, and Gary Hettrick and Anita Mora for graphic design.
This research was supported by the Intramural Research Program of the NIH, NIAID.
Footnotes
Published ahead of print on 30 December 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anderson, J. F., and L. A. Magnarelli. 1984. Avian and mammalian hosts for spirochete-infected ticks and insects in a Lyme disease focus in Connecticut. Yale J. Biol. Med. 57:627-641. [PMC free article] [PubMed] [Google Scholar]
- 2.Ando, T., et al. 2000. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol. Microbiol. 37:1052-1065. [DOI] [PubMed] [Google Scholar]
- 3.Aras, R. A., A. J. Small, T. Ando, and M. J. Blaser. 2002. Helicobacter pylori interstrain restriction-modification diversity prevents genome subversion by chromosomal DNA from competing strains. Nucleic Acids Res. 30:5391-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 5.Blumenthal, R. M., and X. Cheng. 2002. Restriction-modification systems, p. 177-226. In U. N. Streips and R. E. Yasbin (ed.), Modern microbial genetics, 2nd ed. Wiley, New York, NY.
- 6.Botkin, D. J., et al. 2006. Identification of potential virulence determinants by Himar1 transposition of infectious Borrelia burgdorferi B31. Infect. Immun. 74:6690-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgdorfer, W. 1984. Discovery of the Lyme disease spirochete and its relationship to tick vectors. Yale J. Biol. Med. 57:71-76. [PMC free article] [PubMed] [Google Scholar]
- 8.Burgdorfer, W., et al. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317-1319. [DOI] [PubMed] [Google Scholar]
- 9.Casjens, S., M. Delange, H. L. Ley III, P. Rosa, and W. M. Huang. 1995. Linear chromosomes of Lyme disease agent spirochetes: genetic diversity and conservation of gene order. J. Bacteriol. 177:2769-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casjens, S., and W. M. Huang. 1993. Linear chromosomal physical and genetic map of Borrelia burgdorferi, the Lyme disease agent. Mol. Microbiol. 8:967-980. [DOI] [PubMed] [Google Scholar]
- 11.Casjens, S., et al. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 12.Chen, C. K., C. M. Boucle, and H. P. Blaschek. 1996. Factors involved in the transformation of previously non-transformable Clostridium perfringens type B. FEMS Microbiol. Lett. 140:185-191. [DOI] [PubMed] [Google Scholar]
- 13.Chen, Q., et al. 2008. In vitro CpG methylation increases the transformation efficiency of Borrelia burgdorferi strains harboring the endogenous linear plasmid lp56. J. Bacteriol. 190:7885-7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donahue, J. G., J. Piesman, and A. Spielman. 1987. Reservoir competence of white-footed mice for Lyme disease spirochetes. Am. J. Trop. Med. Hyg. 36:92-96. [DOI] [PubMed] [Google Scholar]
- 15.Donahue, J. P., D. A. Israel, R. M. Peek, M. J. Blaser, and G. G. Miller. 2000. Overcoming the restriction barrier to plasmid transformation of Helicobacter pylori. Mol. Microbiol. 37:1066-1074. [DOI] [PubMed] [Google Scholar]
- 16.Elias, A. F., et al. 2003. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J. Mol. Microbiol. Biotechnol. 6:29-40. [DOI] [PubMed] [Google Scholar]
- 17.Elias, A. F., et al. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank, K. L., S. F. Bundle, M. E. Kresge, C. H. Eggers, and D. S. Samuels. 2003. aadA confers streptomycin resistance in Borrelia burgdorferi. J. Bacteriol. 185:6723-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser, C. M., et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 20.Grimm, D., et al. 2004. Experimental assessment of the roles of linear plasmids lp25 and lp28-1 of Borrelia burgdorferi throughout the infectious cycle. Infect. Immun. 72:5938-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimm, D., et al. 2005. Defining plasmids required by Borrelia burgdorferi for colonization of tick vector Ixodes scapularis (Acari: Ixodidae). J. Med. Entomol. 42:676-684. [DOI] [PubMed] [Google Scholar]
- 22.Hinnebusch, J., and A. G. Barbour. 1992. Linear- and circular-plasmid copy numbers in Borrelia burgdorferi. J. Bacteriol. 174:5251-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes, C. A., and R. C. Johnson. 1990. Methylated DNA in Borrelia species. J. Bacteriol. 172:6602-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeltsch, A. 2001. The cytosine N4-methyltransferase M.PvuII also modifies adenine residues. Biol. Chem. 382:707-710. [DOI] [PubMed] [Google Scholar]
- 25.Jewett, M. W., et al. 2007. Genetic basis for retention of a critical virulence plasmid of Borrelia burgdorferi. Mol. Microbiol. 66:975-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jewett, M. W., et al. 2007. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol. Microbiol. 64:1358-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawabata, H., S. J. Norris, and H. Watanabe. 2004. BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect. Immun. 72:7147-7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly, R. 1971. Cultivation of Borrelia hermsii. Science 173:443-444. [DOI] [PubMed] [Google Scholar]
- 29.Kong, H., et al. 2000. Functional analysis of putative restriction-modification system genes in the Helicobacter pylori J99 genome. Nucleic Acids Res. 28:3216-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwak, J., H. Jiang, and K. E. Kendrick. 2002. Transformation using in vivo and in vitro methylation in Streptomyces griseus. FEMS Microbiol. Lett. 209:243-248. [DOI] [PubMed] [Google Scholar]
- 31.Labandeira-Rey, M., and J. T. Skare. 2001. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect. Immun. 69:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane, R. S., D. M. Berger, L. E. Casher, and W. Burgdorfer. 1994. Experimental infection of Columbian black-tailed deer with the Lyme disease spirochete. J. Wildl. Dis. 30:20-28. [DOI] [PubMed] [Google Scholar]
- 33.Lane, R. S., J. Piesman, and W. Burgdorfer. 1991. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu. Rev. Entomol. 36:587-609. [DOI] [PubMed] [Google Scholar]
- 34.Lawrenz, M. B., H. Kawabata, J. E. Purser, and S. J. Norris. 2002. Decreased electroporation efficiency in Borrelia burgdorferi containing linear plasmids lp25 and lp56: impact on transformation of infectious B. burgdorferi. Infect. Immun. 70:4798-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawrenz, M. B., R. M. Wooten, and S. J. Norris. 2004. Effects of vlsE complementation on the infectivity of Borrelia burgdorferi lacking the linear plasmid lp28-1. Infect. Immun. 72:6577-6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine, J. F., M. L. Wilson, and A. Spielman. 1985. Mice as reservoirs of the Lyme disease spirochete. Am. J. Trop. Med. Hyg. 34:355-360. [DOI] [PubMed] [Google Scholar]
- 37.Lyutzkanova, D., M. Stoilova-Disheva, and V. Peltekova. 2001. The restriction-modification system in Streptomyces flavopersicus. Folia Microbiol. (Praha) 46:119-122. [DOI] [PubMed] [Google Scholar]
- 38.Macaluso, A., and A. M. Mettus. 1991. Efficient transformation of Bacillus thuringiensis requires nonmethylated plasmid DNA. J. Bacteriol. 173:1353-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacNeil, D. J. 1988. Characterization of a unique methyl-specific restriction system in Streptomyces avermitilis. J. Bacteriol. 170:5607-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Marrero, R., and S. L. Welkos. 1995. The transformation frequency of plasmids into Bacillus anthracis is affected by adenine methylation. Gene 152:75-78. [DOI] [PubMed] [Google Scholar]
- 42.Miller, J. C., et al. 2000. A second allele of eppA in Borrelia burgdorferi strain B31 is located on the previously undetected circular plasmid cp9-2. J. Bacteriol. 182:6254-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson, M., E. Raschke, and M. McClelland. 1993. Effect of site-specific methylation on restriction endonucleases and DNA modification methyltransferases. Nucleic Acid Res. 21:3139-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purser, J. E., et al. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol. 48:753-764. [DOI] [PubMed] [Google Scholar]
- 45.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. U. S. A. 97:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ralph, D., Q. Que, J. L. Van Etten, and M. McClelland. 1993. Leptospira genomes are modified at 5′-GTAC. J. Bacteriol. 175:3913-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Revel, A. T., et al. 2005. bptA (bbe16) is essential for the persistence of the Lyme disease spirochete, Borrelia burgdorferi, in its natural tick vector. Proc. Natl. Acad. Sci. U. S. A. 102:6972-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts, R. J., et al. 2003. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 31:1805-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rocha, E. P., A. Danchin, and A. Viari. 2001. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res. 11:946-958. [DOI] [PubMed] [Google Scholar]
- 50.Rosa, P. A., F. Cabello, and D. S. Samuels. 2010. Genetic manipulation of Borrelia burgdorferi, p. 189-219. In D. S. Samuels and J. D. Radolf (ed.) Borrelia: molecular biology, host interaction and pathogenesis. Caister Academic Press, Norfolk, England.
- 51.Samuels, D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwan, T. G., W. Burgdorfer, and C. F. Garon. 1988. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect. Immun. 56:1831-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steere, A. C. 2001. Lyme disease. N. Engl. J. Med. 345:115-125. [DOI] [PubMed] [Google Scholar]
- 54.Stewart, P. E., J. Hoff, E. Fischer, J. G. Krum, and P. A. Rosa. 2004. Genome-wide transposon mutagenesis of Borrelia burgdorferi for identification of phenotypic mutants. Appl. Environ. Microbiol. 70:5973-5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart, P. E., R. Thalken, J. L. Bono, and P. Rosa. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39:714-721. [DOI] [PubMed] [Google Scholar]
- 56.Strother, K. O., and A. de Silva. 2005. Role of Borrelia burgdorferi linear plasmid 25 in infection of Ixodes scapularis ticks. J. Bacteriol. 187:5776-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, Y., C. Kodner, L. Coleman, and R. C. Johnson. 1996. Correlation of plasmids with infectivity of Borrelia burgdorferi sensu stricto type strain B31. Infect. Immun. 64:3870-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasui, K., et al. 2009. Improvement of bacterial transformation efficiency using plasmid artificial modification. Nucleic Acids Res. 37:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.