Abstract
Cells respond to external stimuli through networks of regulatory interactions. The human pathogen Mycobacterium tuberculosis responds to stress encountered during infection by arresting multiplication and implementing critical metabolic changes that lead to or sustain the nonreplicative state. Much of this differentiation program is recapitulated when M. tuberculosis cultures are subjected to gradual oxygen depletion in vitro. Here we report that hypoxic induction of critical central metabolism genes in the glyoxylate shunt (icl1) and in the methylcitrate cycle (gltA1) involves both global and local regulators. The global regulators are accessory sigma factors σB for icl1 and σE for gltA1. The local regulators are the products of two paralogous genes mapping at positions adjacent to the corresponding effector gene or operon. We call these genes lrpI and lrpG (for local regulatory protein of icl1 and gltA1). We also found that (i) each sigma factor controls the corresponding local regulator, (ii) both global and local regulators are required for effector gene induction, and (iii) the occurrence of sigma factor control of effector gene induction is independent of its control over the corresponding local regulator. Together, these data indicate that induction of icl1 and gltA1 utilizes parallel feed-forward loops with an AND input function. Both feed-forward loops are affected by σE, since this sigma factor is part of the gltA1 loop and controls sigB in the icl1 loop. Feed-forward loops may critically contribute to the cellular developmental program associated with M. tuberculosis dormancy.
Bacteria have evolved complex strategies to survive stress encountered in their habitats. Much of the complexity resides in the structure of the regulatory network that controls the stress response. Work with model microorganisms has shown that the stress response usually requires the integration of multiple signal-processing pathways by information-processing units that control downstream cascades, ultimately regulating effector gene expression (2, 13, 24). Understanding such complexity is facilitated by dissecting the regulatory network into smaller, relatively autonomous units that are amenable to experimental manipulation. These units are often identified as highly recurring network motifs (28). One such motif, termed a feed-forward loop (FFL), is defined by a transcription factor, X, that regulates a second transcription factor, Y, and by joint regulation by X and Y of a third gene/operon, Z (14). The relationships between the structure and dynamics of FFLs have been previously investigated, primarily in Escherichia coli studies (14, 28), and may have functional implications. For example, coherent type 1 FFLs, in which all regulatory interactions have a positive sign (activation), can filter noise and protect against brief input fluctuations (15). How network motifs are implicated in the virulence and adaptation of pathogenic bacteria is unknown. Here we report that parallel FFLs (each composed of a master regulator, a local regulator, and a central metabolism gene) are utilized in the stress response of two critical central metabolism genes of the human pathogen Mycobacterium tuberculosis.
M. tuberculosis is an intracellular pathogen that responds to the stress generated by the host immune response by entering a nonreplicating state associated with a chronic, asymptomatic (latent) infection. Latent infection progresses to disease when the immune response is depressed and tubercle bacilli resume growth. The response of M. tuberculosis to immunity-mediated stress includes changes in bacterial central metabolism, which have typically been revealed by transcriptional analyses of tubercle bacilli during mouse lung infection (see, for example, references 29, 30, 31, and 34). Two metabolic pathways that are involved in lipid catabolism, the glyoxylate shunt and the methylcitrate cycle, have been previously investigated in detail (19, 21, 22). Both pathways utilize the product of icl1 (aceA, or rv0467 according to the M. tuberculosis H37Rv numbering system devised by Cole et al. [3]), which exhibits isocitrate lyase activity in the glyoxylate shunt and 2-methyl-isocitrate lyase activity in the methylcitrate cycle (10). Two additional enzymes, methylcitrate dehydratase and methylcitrate synthase, which are encoded by the gltA1 operon (also named prpDC and rv1130-rv1131), are involved in the methylcitrate cycle. Upregulation of icl1 and gltA1 occurs when tubercle bacilli respond to expression of adaptive immunity in the lungs of infected mice (31, 34). Tubercle bacilli isolated from sputum of tuberculosis patients also show increased expression of icl1 relative to cultures growing exponentially in vitro (8). Thus, induction of these pathways is relevant to the bacterial state in vivo.
Regulation of icl1 and gltA1 involves accessory sigma factors (27), which direct the bacterial RNA polymerase holoenzyme to specific genes during particular growth phases or under particular stress conditions (11). Gene expression data determined by microarray hybridization show that icl1 and gltA1 are among the genes regulated by σE under conditions of detergent-mediated, cell-surface stress (17). σE also regulates sigB (19), and it is regulated by sigH under conditions of heat and oxidative stress (16, 26). Moreover, the gltA1 operon is regulated by σB when tubercle bacilli are subjected to diamide stress (7). Thus, it is likely that multiple accessory sigma factors are involved in the regulation of icl1 and gltA1. How these sigma factors interact to regulate the two effector genes (and whether additional transcriptional factors are involved) has not been established.
In the present work, we used a model of gradual oxygen starvation of M. tuberculosis cultures in vitro (35) to examine regulatory interactions that control expression of icl1 and gltA1 in response to bacteriostatic treatment. This in vitro model recapitulates transcriptional changes seen in tubercle bacilli during mouse lung infection (see, for example, references 29, 30, and 31). We found that induction of icl1 and gltA1 during hypoxic stress is regulated by two parallel, σE-controlled FFLs that include a global regulator (an accessory sigma factor) and a local regulator.
MATERIALS AND METHODS
Bacterial strains, reagents and media, and growth conditions.
M. tuberculosis mutants with knockouts in sigE and sigB, complemented sigE and sigB mutants, and transposon-insertion mutants in gene loci rv1129c and rv0465c were previously reported (see references 7 and 17 and http://webhost.nts.jhu.edu/target). The gene numbering of the M. tuberculosis genome is presented according to the system of Cole et al. (3). M. tuberculosis cultures were grown in Middlebrook (MB) 7H9 (liquid medium) or 7H10 (solid medium) (Difco) supplemented with 0.05% Tween 80, 0.2% glycerol, and 10% ADN (2% glucose, 5% bovine serum albumin [BSA; Sigma], 0.15 M NaCl). Aerated liquid cultures of M. tuberculosis were grown in 25-ml tubes at 37°C with magnetic-bar stirring at 450 rpm. Plates were incubated at 37°C in sealed plastic bags. Hypoxic cultures of M. tuberculosis were grown in Dubos Tween-albumin broth (Becton Dickinson). Escherichia coli XL1 Blue was grown in Luria-Bertani broth (LB; Difco), and on LB agar plates at 37°C.
Construction of rv0465c- and rv1129-complementing plasmids.
Fragments containing the coding region of rv0465c or rv1129c were amplified by PCR from M. tuberculosis genomic DNA (see Table S1 in the supplemental material for PCR primers) and cloned under the control of the constitutive groEL promoter in the replicative apramycin (Apr) resistance shuttle plasmid pMP167 (4). The resulting plasmids were named pRv0465c and pRv1129c (Table 1). E. coli transformants were selected on LB agar plates containing 40 μg/ml apramycin. Recombinant plasmids were transferred to M. tuberculosis by electroporation. Transformants were selected on Middlebrook 7H10 agar plates containing 40 μg/ml apramycin after three weeks of incubation at 37°C.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pMP167 | Mycobacterium/E. coli shuttle vector with groEL promoter; Aprr | 4 |
| pRv0465c | rv0465c gene locus cloned into pMP167 with PvuII and EcoRI | This study |
| pRv1129c | rv1129c gene locus cloned into pMP167 with PvuII and EcoRI | This study |
Gradual oxygen depletion.
M. tuberculosis cultures were grown in Dubos Tween-albumin broth at 37°C as described by Wayne and Hayes (35). In brief, mid-log cultures were diluted to an optical density at 580 nm (OD580) of 0.004 and subjected to slow stirring with a magnetic stirring bar in sealed tubes with a ratio of headspace air to medium of 0.5. Growth was monitored by turbidity measurement and CFU enumeration. At selected times, cells were harvested by centrifugation and quickly frozen in a dry-ice/alcohol bath for subsequent RNA extraction.
Enumeration of bacterial transcripts.
RNA extraction, reverse transcription, and real-time quantitative PCR (qPCR) were performed as previously described (see reference 29 and http://www.phri.org/research/res_pigennaro.asp). Briefly, bacterial cell pellets were resuspended in 1 ml of TRI reagent (Molecular Research Center, Cincinnati, OH) and disrupted by rapid mechanical lysis. Total RNA was purified in TRI reagent according to the manufacturer's instructions and stored at −80°C. Reverse transcription was performed with random hexameric primers and ThermoScript reverse transcriptase (Invitrogen, Carlsbad, CA). Enumeration of M. tuberculosis mRNAs was carried out by qPCR using gene-specific primers, molecular beacons, and AmpliTaq Gold polymerase (Applied Biosystems) in a Stratagene Mx4000 thermal cycler (Agilent Technologies). Nucleotide sequences of PCR primers and molecular beacons are listed in Table S2 in the supplemental material. The M. tuberculosis 16S rRNA copy number was used as a normalization factor to enumerate bacterial transcripts per cell, as previously described (29).
RESULTS
The hypoxic response of icl1 and gltA1.
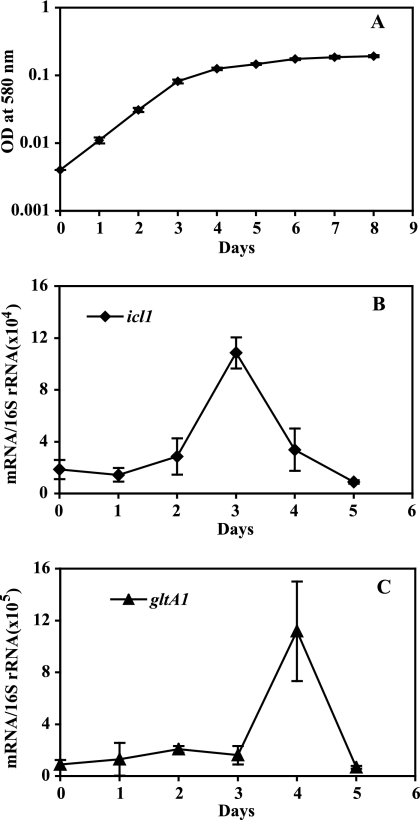
When M. tuberculosis cultures are subjected to gradual oxygen depletion, they stop growing after approximately 3 days of incubation (35) (Fig. 1A). We used this growth arrest model to investigate the expression of ic11 and gltA1. Transcript enumeration by qPCR showed that both genes were induced: icl1 peaked at day 3 and gltA1 at day 4 of oxygen depletion (5- and 10-fold induction relative to mid-log growth, respectively) (Fig. 1B and C). Thus, icl1 and gltA1 are transiently upregulated during gradual O2 depletion.
FIG. 1.
The hypoxic response of icl1 and gltA1. Cultures of M. tuberculosis were subjected to gradual O2 depletion (35). (A) Growth curve. In the Wayne model, cultures of M. tuberculosis continue to multiply for approximately 70 h and then stop growing (microaerophilia). After this time, the additional slow rise in absorbance is due to cell wall enlargement. Anaerobiosis ensues at around 200 h, with no further cell enlargement. Bacterial growth was monitored by measuring OD at 580 nm. (B and C) Enumeration of icl1 and gltA1 transcripts. Cultures subjected to gradual O2 depletion were harvested daily, RNA was extracted, and bacterial transcripts were enumerated by qPCR. Transcript copy numbers were normalized to 16S rRNA (29, 30). Three independent experiments were carried out, and means (± standard deviations [SD]) were calculated for normalized mRNA copy numbers at each time point. The results shown represent normalized copy numbers of mRNA for icl1 and gltA1 at each time point (one gene per panel, as indicated). Measurements performed using M. tuberculosis H37Rv and M. tuberculosis CDC1551 gave indistinguishable results (see Fig. S1 in the supplemental material), indicating that the hypoxic response of icl1 and gltA1 is conserved among different M. tuberculosis strains.
Sigma factor requirements for the hypoxic response of gltA1 and icl1.
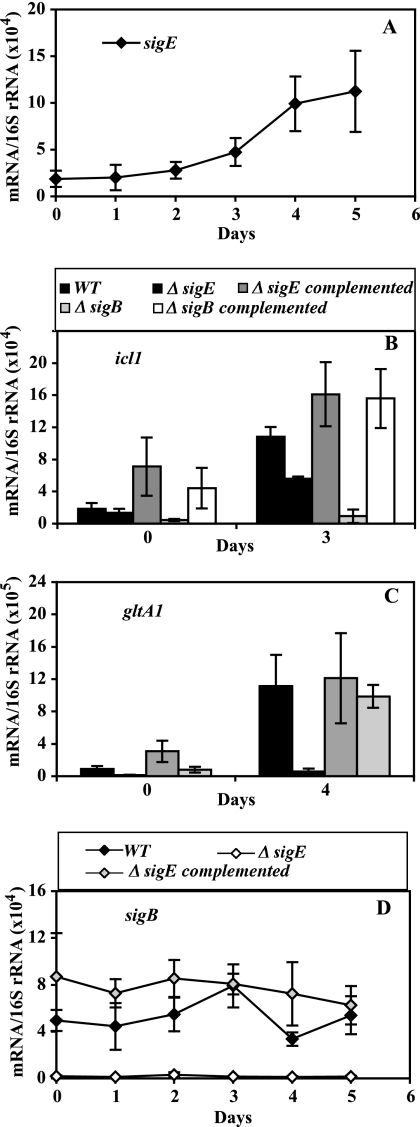
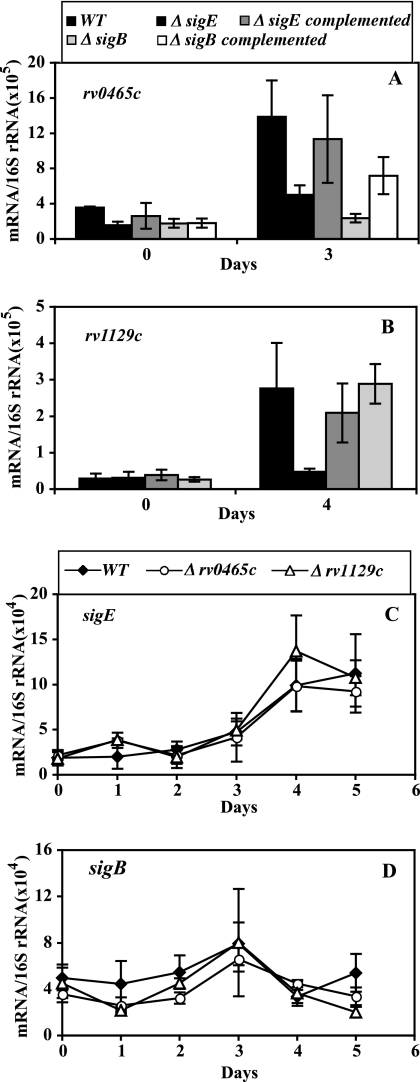
As noted above, it is likely that multiple accessory sigma factors are involved in the regulation of icl1 and gltA1. One accessory sigma factor candidate is σE, since both icl1 and gltA1 were previously described as responding to sodium dodecyl sulfate (SDS)-mediated stress in a sigE-dependent fashion (17). When sigE transcript levels were examined during hypoxia, we observed a gradual induction (4- to 5-fold increase in copy numbers at day 5 relative to mid-log growth) (Fig. 2A), which indicates a transcriptional response of sigE to hypoxia. We then tested the role of sigE in the icl1 and gltA1 hypoxic response by enumerating icl1 and gltA1 transcripts in a sigE knockout mutant. Upregulation of icl1 was reduced by the sigE mutation (48% decrease at day 3; P < 0.05 for the Student t test) (Fig. 2B), while that of gltA1 was nearly abolished (Fig. 2C).
FIG. 2.
Effect of sigE and sigB inactivation on the hypoxic response of icl1 and gltA1. Wild-type and mutant cultures were subjected to gradual O2 depletion, cells were harvested, RNA was extracted, and bacterial transcripts were enumerated by qPCR, as described in the Fig. 1 legend. In all panels, means (± SD) for each time point were calculated from data from three independent experiments. For the duration of the treatment, CFU counts of wild-type and mutant strains were indistinguishable (see Fig. S2 in the supplemental material). Each panel presents data for one gene. (A) The hypoxic response of sigE. The results shown represent normalized copy numbers of the sigE transcript in wild-type cultures during 5 days of a hypoxic time course. (B and C) The hypoxic response of effector genes. The results shown represent normalized copy numbers of icl1 (B) and gltA1 (C) at mid-log growth (day 0) and at the day of peak gene activation (day 3 for icl1 and day 4 for gltA1) in wild-type, sigE and sigB mutant, and complemented mutant strains. (D) The hypoxic response of sigB. The results shown represent normalized copy numbers of the sigB transcript in wild-type, sigE mutant, and complemented sigE mutant cultures during 5 days of a hypoxic time course.
The partial effect of sigE inactivation on icl1 induction suggested that additional sigma factors might be involved in the network regulating icl1. A potential candidate was sigB, since sigB contains a sigE-dependent promoter (17, 32) and because sigB levels are decreased in a sigE mutant during logarithmic growth, SDS-mediated stress (17), and hypoxia (Fig. 2D). We found that a sigB knockout mutation nearly abolished induction of icl1 (but not of gltA1) during hypoxia (Fig. 2B and C). Thus, induction of gltA1 requires a functional sigE, whereas induction of icl1 requires sigB function. Given the effect of sigE on sigB expression, the weaker effect on icl1 induction of sigE loss of function relative to that of sigB suggests that sigE might control the icl1 response via sigB.
Two local regulatory factors and their requirement for the hypoxic response of gltA1 and icl1.
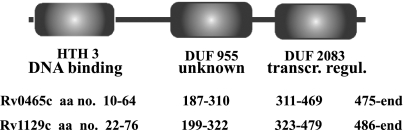
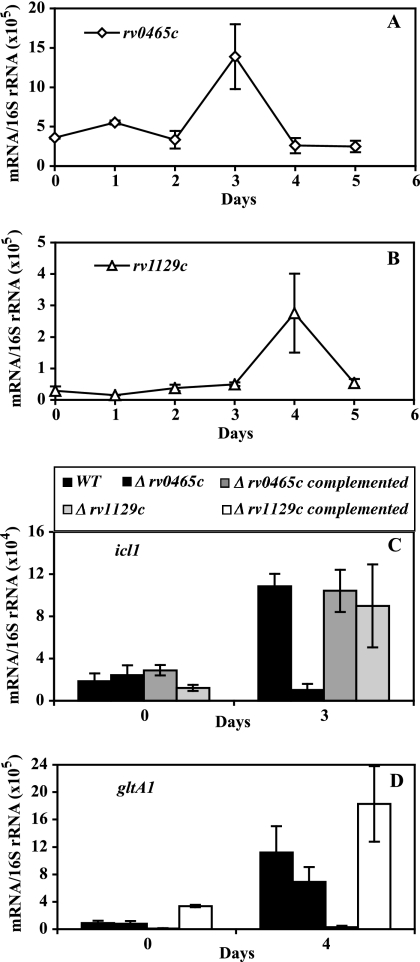
Among the gene loci regulated by σE in SDS-stressed cultures are rv0465c and rv1129c (17). These two gene loci are annotated as transcription factors (http://genolist.pasteur.fr/TubercuList/) and map at positions adjacent to (but on the opposite strand from) icl1 (rv0467) and gltA1 (the gltA1 operon includes rv1130 and rv1131, which encode two enzymes of the methylcitrate cycle [22]), respectively. The deduced amino acid sequences share 47% identity and a three-domain architecture that includes DNA-binding motifs (Fig. 3). These observations raised the possibility that rv0465c and rv1129c are paralogous gene loci involved in the regulation of the adjacent effector genes. When we measured the relative levels of abundance of rv0465c and rv1129c transcripts during hypoxia, we found that each gene was upregulated at the same time as the corresponding adjacent effector gene (Fig. 4A and B; compare with Fig. 1B and C). When we used qPCR to assess the hypoxic response of icl1 and gltA1 in transposon-insertion mutants of rv0465c and rv1129c, we found that the absence of rv0465c and rv1129c abolished induction of icl1 and gltA1, respectively (Fig. 4C and D). Thus, rv0465c and rv1129c are required for the hypoxic response of the adjacent effector gene. Moreover, their activity is specific to the target, since deletion of either regulator had little if any effect on the expression of the distant target gene.
FIG. 3.
Architecture of the proteins encoded by rv0465c and rv1129c. The deduced amino acid sequences of Rv0465c and Rv1129c were interrogated against an integrated database of protein domains and functional sites with the InterProScan package (http://www.ebi.ac.uk/Tools/InterProScan/). The analysis revealed that the two proteins belong to a protein family sharing a three-domain architecture: HTH_3 is a helix-turn-helix, DNA-binding motif; DUF955 (IPR010359) is a domain of unknown function; and DUF2083 (IPR018653) is annotated as a predicted transcriptional regulator found in the XRE family of prokaryotic transcriptional regulatory proteins (http://dbtbs.hgc.jp/ver1/tfactable.html). Rv0465c and Rv1129c are members of a ubiquitous protein family and are widely distributed in Mycobacteriaceae (see Fig. S3 in the supplemental material).
FIG. 4.
Characterization of adjacent regulatory factors. O2-depleted culture growth, RNA extraction, and mRNA enumeration by qPCR were conducted as described in the Fig. 1 legend. For the duration of the treatment, CFU counts of wild-type and mutant strains were indistinguishable (see Fig. S2 in the supplemental material). Each panel presents data for one gene. (A and B) The hypoxic response of rv0465c and rv1129c. The results shown represent normalized copy numbers of mRNA for rv0465c and rv1129c at each time point. (C and D) The hypoxic response of effector genes in wild-type and rv0465c and rv1129c transposon-insertion mutant strains. The results shown represent normalized copy numbers of icl1 and gltA1 at mid-log growth (day 0) and the day of peak gene activation (day 3 for icl1 and day 4 for gltA1) in wild-type and rv0465c and rv1129c mutant strains and the corresponding complemented mutant strains. Means (± SD) for each time point were calculated from data from three independent experiments.
Interactions between sigma factors and rv0465c and rv1129c in the hypoxic response of icl1 and gltA1.
We tested the requirement for sigE and sigB in the induction of rv0465c and rv1129c during hypoxic stress. Induction of rv0465c was abolished by a sigB mutation and was decreased almost 50% by a sigE mutation (Fig. 5A), which are results comparable with those obtained for icl1 induction with these two mutant strains (Fig. 2B). Similarly, induction of rv1129c was abolished by a sigE mutation and unaffected by a sigB mutation (Fig. 5B), as seen with gltA1 (Fig. 2C). Thus, induction of the local regulators rv0465c and rv1129c in hypoxic cultures required functional sigB and sigE, respectively, which is similar to the results seen with induction of the corresponding effector genes. To fully examine the regulatory relationships between local regulators and sigma factors, we also tested the effect of mutations in rv0465c and rv1129c on the hypoxic response of sigE and sigB. We found no effect caused by the local regulators on these sigma factors (Fig. 5C and D), indicating that the local regulators exert no feedback regulation on the master regulators (i.e., the sigma factors).
FIG. 5.
Regulatory interactions between sigE, sigB, rv0465c, and rv1129c. O2-depleted cultures, daily harvesting, and mRNA enumeration by qPCR were conducted as described in the Fig. 1 legend. Means (± SD) for each time point were calculated from data from three independent experiments. Each panel presents data for one gene. (A and B) Effect of sigE and sigB inactivation on the hypoxic response of rv0465c and rv1129c. Transcripts were enumerated and data are presented as described in the Fig. 2B and C legends. (C and D) Effect of rv0465c and rv1129c inactivation on the hypoxic response of sigE and sigB. Transcripts were enumerated and data are presented as described in the Fig. 2A to D legends.
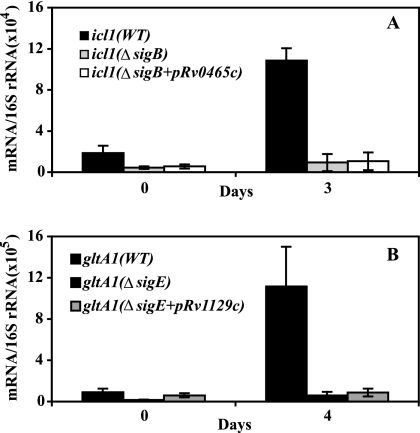
We next asked whether the observed dependence of effector gene induction on the sigma factor (sigB for icl1 and sigE for gltA1) reflected a requirement for rv0465c and rv1129c induction. To address this question, we examined the sigma factor requirement for induction of the effector gene in the presence or absence of a constitutively expressed copy of the corresponding local regulator. We introduced a plasmid expressing rv0465c (or rv1129c) controlled by the strong constitutive groEL promoter in a sigB (or sigE) knockout mutant and measured the transcript abundance of icl1 (or gltA1). Ectopic expression of the local regulator failed to bypass the relevant sigma factor requirement for icl1 or gltA1 induction in hypoxic cultures (Fig. 6A and B). That the plasmids used in these experiments expressed functional Rv0465c and Rv1129c proteins was demonstrated by the ability of the same plasmids to complement the corresponding mutations (Fig. 4C and D). Thus, the requirement for a sigma factor to induce icl1 and gltA1 is independent of the regulation of the local regulator by the same sigma factor.
FIG. 6.
Effect of constitutive expression of local regulators on icl1 and gltA1 hypoxic response in sigma factor mutants. Derivatives of plasmid pMP167 expressing rv0465c or rv1129c under the control of the constitutive groEL promoter (indicated as pRv0465c and pRv1129c) were introduced in sigB or sigE knockout mutant strains. Cultures were harvested at daily intervals, and mRNA enumeration by qPCR was conducted as described in the Fig. 1 legend. Each panel presents data for one gene (panel A, icl1; panel B, gltA1). The results shown represent the mid-log time point and the time points corresponding to peak gene induction (day 3 for icl1 and day 4 for gltA1).
DISCUSSION
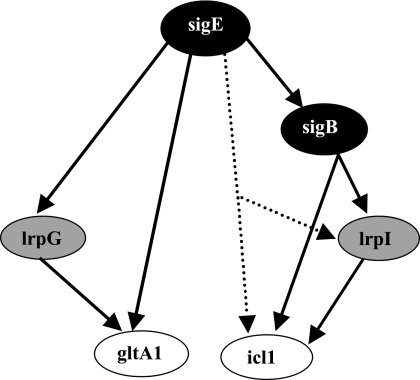
In the present report, we provide genetic evidence that the response to stress of two central metabolism genes of M. tuberculosis, icl1 and gltA1, is controlled by a global regulator and by a regulator located near the effector gene/operon. The global regulators are sigma factors, σB for icl1 and σE for gltA1. The local regulators are the products of rv0465c and rv1129c. To underscore their role in regulating adjacently located effector genes, we name these genes lrpI (local regulatory protein of icl1, or rv0465c) and lrpG (local regulatory protein of gltA1, or rv1129c). The data also show that (i) induction of either effector requires both global and local regulators and (ii) the global regulator controls both local regulator and effector. These regulatory interactions are not explained by a simple cascade in which the global regulator controlled the local regulator that in turn induced the effector operon, because high-level, constitutive expression of the local regulator failed to bypass the requirement for the global regulator. In contrast, the data demonstrate the existence of nested feed-forward loops (5, 28) in which an accessory sigma factor regulates a second, local regulator and both factors jointly regulate the target gene/operon (icl1 or gltA1) (Fig. 7). Both FFLs appear to be of coherent type 1 (all regulatory interactions have a positive sign) and exhibit AND logic (both global and local regulators are needed to induce expression of the effector gene) (14).
FIG. 7.
Feed-forward loops controlling icl1 and gltA1 hypoxic induction. The figure shows the regulatory interactions revealed in the present work. The sigE-sigB interaction is taken from the literature (17, 32). The colors of the ovals containing the gene names represent gene classes in the network structure as follows: black, global regulators; gray, local regulators; white, effector genes. The dotted line indicates a potential direct interaction between sigE and lrpI/icl1, a possibility that cannot be excluded by our genetic data.
Three-node, coherent type 1 FFLs do not fully account for the observed behavior of icl1 and gltA1 in response to hypoxia. A coherent type 1 FFL is expected per se to filter noise and reject transient input stimuli (14), whereas it should maintain the response for persistent input stimuli. However, even though hypoxia persisted in our experimental setting and sigE transcript levels gradually increased (at least through day 5; Fig. 2A), the hypoxic induction of gltA1 and icl1 was transient (Fig. 1B and C), with both transcripts returning to near-basal levels by day 5. Thus, the downturn of the icl1 and gltA1 hypoxic response strongly implies the existence of yet-unrevealed downregulatory interactions. One candidate for downregulation is the two-component system mprAB (36), since an mprAB mutant exhibits elevated levels of gltA1 (but not of icl1) at day 5 of hypoxia, the time when gltA1 is downregulated in wild-type cells (data not shown). This result is consistent with the increased abundance of gltA1 (but not icl1) transcripts seen in an mprA mutant relative to wild-type cells during SDS stress (23). Another possibility is that, as expression levels of lrpI and lrpG increase, the corresponding gene products become repressors, thus turning the FFLs from coherent to incoherent (many transcription factors function both as activators and repressors, for example, the bacteriophage lambda repressor [25]). The idea of dual regulation by the lrp genes is supported by the recent observation that the product of rv0465c (lrpI) represses icl1 during growth of M. tuberculosis on glucose (20). Thus, studying gene expression in the context of network structure should help unravel additional regulatory interactions and reconstruct increasingly complex networks.
The gene network revealed by our genetic work requires biochemical characterization to assess protein-DNA interactions at the regulatory sequences of the genes in the network. Published data agree with our genetic results, since a σE recognition sequence is found upstream of the gltA1 operon (17), the product of rv0465c binds to DNA upstream of icl1 (20), and we have mapped a sigA or sigB promoter (it is not possible to discriminate between these two consensus sequences [27]) upstream of icl1 (R. Manganelli and M. L. Gennaro, unpublished data). In particular, while our data clearly show that the effect of sigB on lrpI and icl1 expression is more pronounced than that of sigE, we cannot exclude the possibility of a direct interaction between sigE and lrpI or icl1 (as sketched in Fig. 7).
The dynamics and the regulation of the icl1 and gltA1 stress response revealed by our work fit well with the physiology of M. tuberculosis dormancy. The involvement of sigE as a common regulatory node may reflect a requirement for coordinated expression of icl1 and gltA1, since the products of these two genes participate in the methylcitrate cycle (22), a metabolic pathway for the assimilation of propionyl coenzyme A (CoA) generated during lipid catabolism. Moreover, icl1 and gltA1 are critical for the metabolic remodeling associated with stress-induced growth arrest of M. tuberculosis (31); thus, it seems appropriate that their upregulation should be controlled by network structures, such as coherent FFLs, which reject transient input signals (15). Additional regulatory interactions may have evolved to turn off expression of icl1 and gltA1, even when the input stimulus persists, to help implement the overall metabolic shutdown associated with dormancy. Indeed, the transient nature of icl1 and gltA1 induction should be physiologically relevant, since it is also seen during mouse lung infection (31). Moreover, peak induction of icl1 precedes that of gltA1 in the murine model as well (31), thus providing another similarity between the in vivo and in vitro situations. A more general consideration is that a role for FFLs in M. tuberculosis dormancy is consistent with the notion that these network structures are involved in cellular differentiation processes in other microorganisms, such as Bacillus subtilis sporulation (6).
The general stress response in bacteria, which affords cross-protection against multiple stresses, typically entails global regulators (reviewed in reference 13). We propose σE as a central regulator of the M. tuberculosis stress response. The sigE gene is induced during growth in human macrophages and in response to various stress conditions in vitro, such as detergent- and vancomycin-mediated cell surface stress, pH stress, heat shock, oxidative stress (reviewed in reference 27), and hypoxia (this work). Various signal-processing and stress-responsive factors, such as the mprAB two-component system (36) in M. tuberculosis and M. smegmatis (12, 33) and the accessory sigma factor σH (16, 26), regulate sigE transcription during heat and oxidative stress. We postulate that σE acts as an information-processing unit that connects upstream signaling systems to downstream networks that regulate effector functions. A critical role for σE in the response of M. tuberculosis to stress is consistent with the observations that sigE knockout mutants were severely attenuated for growth in one mouse model (18) and for virulence in another (1) and that they also grew poorly in macrophages and in dendritic cells (9, 18).
The genetic findings reported above open the way to future research to characterize protein-DNA interactions involved in the FFLs that regulate icl1 and gltA1 and to identify additional regulatory interactions implicated in the observed gene expression patterns. We intend also to examine whether the FFL motif applies to icl1 and gltA1 regulation under different sets of stress conditions and whether it is utilized to regulate the stress response of other metabolic genes of M. tuberculosis. Moreover, we plan to address the nature of the physiological signal(s) activating and subsequently repressing the response of these FFLs, which we expect to show to be related to stress rather than associated with changes of carbon source (31). Reconstructing networks regulating the stress response should help to identify nodes that are critical to the M. tuberculosis survival program and to target them with drugs and vaccines.
Supplementary Material
Acknowledgments
We thank Patricia Fontan and Issar Smith for providing sigma factor knockout mutants and complemented mutants and, in particular, for sharing a sigB mutant prior to publication; the TARGET initiative at the Johns Hopkins University for rv0465c and rv1129c transposon-insertion mutants; Oleg Igoshin and Eduardo Sontag for discussions on FFL structure and dynamics; and Karl Drlica, David Dubnau, and Issar Smith for critical reading of the manuscript.
N.B. is the recipient of a grant from the U.S. Civilian Research and Development Foundation. The work was supported by NIAID grants to L.S. and M.L.G. and by a European Union grant (TB PAN-NET FP7-223681) to G.B. and M.L.G.
Footnotes
Published ahead of print on 30 December 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ando, M., T. Yoshimatsu, C. Ko, P. J. Converse, and W. R. Bishai. 2003. Deletion of Mycobacterium tuberculosis sigma factor E results in delayed time to death with bacterial persistence in the lungs of aerosol-infected mice. Infect. Immun. 71:7170-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balázsi, G., and Z. N. Oltvai. 2005. Sensing your surroundings: how transcription-regulatory networks of the cell discern environmental signals. Sci. STKE 2005:pe20. [DOI] [PubMed] [Google Scholar]
- 3.Cole, S. T., et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 4.Consaul, S. A., et al. 2005. An unusual mutation results in the replacement of diaminopimelate with lanthionine in the peptidoglycan of a mutant strain of Mycobacterium smegmatis. J. Bacteriol. 187:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobrin, R., Q. K. Beg, A. L. Barabasi, and Z. N. Oltvai. 2004. Aggregation of topological motifs in the Escherichia coli transcriptional regulatory network. BMC Bioinformatics 5:10. doi: 10.1186/1471-2105-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eichenberger, P., et al. 21 September 2004, posting date. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:1664-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontán, P. A., et al. 2009. The Mycobacterium tuberculosis sigma factor sigmaB is required for full response to cell envelope stress and hypoxia in vitro, but it is dispensable for in vivo growth. J. Bacteriol. 191:5628-5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garton, N. J., et al. 2008. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 5:634-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giacomini, E., et al. 2006. Infection of human dendritic cells with a Mycobacterium tuberculosis sigE mutant stimulates production of high levels of interleukin-10 but low levels of CXCL10: impact on the T-cell response. Infect. Immun. 74:3296-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gould, T. A., H. van de Langemheen, E. J. Munoz-Elias, J. D. McKinney, and J. C. Sacchettini. 2006. Dual role of isocitrate lyase 1 in the glyoxylate and methylcitrate cycles in Mycobacterium tuberculosis. Mol. Microbiol. 61:940-947. [DOI] [PubMed] [Google Scholar]
- 11.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57:441-466. [DOI] [PubMed] [Google Scholar]
- 12.He, H., R. Hovey, J. Kane, V. Singh, and T. C. Zahrt. 2006. MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J. Bacteriol. 188:2134-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mangan, S., and U. Alon. 2003. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. U. S. A. 100:11980-11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangan, S., A. Zaslaver, and U. Alon. 2003. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol. 334:197-204. [DOI] [PubMed] [Google Scholar]
- 16.Manganelli, R., et al. 2002. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:365-374. [DOI] [PubMed] [Google Scholar]
- 17.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor sigma E: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 18.Manganelli, R., et al. 2004. The extracytoplasmic function sigma factor sigma(E) is essential for Mycobacterium tuberculosis virulence in mice. Infect. Immun. 72:3038-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKinney, J. D., et al. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 20.Micklinghoff, J. C., et al. 2009. Role of the transcriptional regulator RamB (Rv0465c) in the control of the glyoxylate cycle in Mycobacterium tuberculosis. J. Bacteriol. 191:7260-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muñoz-Elías, E. J., and J. D. McKinney. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11:638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muñoz-Elías, E. J., A. M. Upton, J. Cherian, and J. D. McKinney. 2006. Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol. Microbiol. 60:1109-1122. [DOI] [PubMed] [Google Scholar]
- 23.Pang, X., et al. 2007. Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology 153(Pt. 4):1229-1242. [DOI] [PubMed] [Google Scholar]
- 24.Price, C. W. 2000. Protective function and regulation of the general stress response in Bacillus subtilis and related Gram-positive bacteria, p. 179-197. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, DC.
- 25.Ptashne, M., et al. 1980. How the lambda repressor and cro work. Cell 19:1-11. [DOI] [PubMed] [Google Scholar]
- 26.Raman, S., et al. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigue, S., R. Provvedi, P. E. Jacques, L. Gaudreau, and R. Manganelli. 2006. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 30:926-941. [DOI] [PubMed] [Google Scholar]
- 28.Shen-Orr, S. S., R. Milo, S. Mangan, and U. Alon. 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31:64-68. [DOI] [PubMed] [Google Scholar]
- 29.Shi, L., Y. J. Jung, S. Tyagi, M. L. Gennaro, and R. J. North. 2003. Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc. Natl. Acad. Sci. U. S. A. 100:241-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi, L., et al. 2005. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc. Natl. Acad. Sci. U. S. A. 102:15629-15634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi, L., et al. 2010. Carbon flux rerouting during Mycobacterium tuberculosis growth arrest. Mol. Microbiol. 78:1199-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song, T., S. E. Song, S. Raman, M. Anaya, and R. N. Husson. 2008. Critical role of a single position in the −35 element for promoter recognition by Mycobacterium tuberculosis SigE and SigH. J. Bacteriol. 190:2227-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sureka, K., et al. 2007. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol. Microbiol. 65:261-276. [DOI] [PubMed] [Google Scholar]
- 34.Timm, J., et al. 2003. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc. Natl. Acad. Sci. U. S. A. 100:14321-14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zahrt, T. C., and V. Deretic. 2001. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc. Natl. Acad. Sci. U. S. A. 98:12706-12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.