Abstract
Variovorax paradoxus is a microorganism of special interest due to its diverse metabolic capabilities, including the biodegradation of both biogenic compounds and anthropogenic contaminants. V. paradoxus also engages in mutually beneficial interactions with both bacteria and plants. The complete genome sequence of V. paradoxus S110 is composed of 6,754,997 bp with 6,279 predicted protein-coding sequences within two circular chromosomes. Genomic analysis has revealed multiple metabolic features for autotrophic and heterotrophic lifestyles. These metabolic diversities enable independent survival, as well as a symbiotic lifestyle. Consequently, S110 appears to have evolved into a superbly adaptable microorganism that is able to survive in ever-changing environmental conditions. Based on our findings, we suggest V. paradoxus S110 as a potential candidate for agrobiotechnological applications, such as biofertilizer and biopesticide. Because it has many associations with other biota, it is also suited to serve as an additional model system for studies of microbe-plant and microbe-microbe interactions.
Variovorax paradoxus is a metabolically diverse, aerobic bacterium that engages in mutually beneficial interactions with a variety of bacteria and plants. V. paradoxus belongs to the subclass of Proteobacteria and can metabolically utilize natural compounds produced by other biota, such as acyl homoserine lactones (AHLs) (25) and alkyl/aryl-sulfonates (38). This metabolic capacity suggests that Variovorax plays an essential role in the natural cycling of biogenic chemicals. Variovorax species are also able to degrade a variety of contaminants, including pesticides and crude oil-associated S-metabolites (5, 19, 37, 41, 42, 46, 50, 51, 52), often in synergistic and mutually beneficial interactions with other bacteria. In addition, a close relative of Variovorax was found to be the central, nonphotosynthetic partner within the phototrophic consortium “Chlorochromatium aggregatum” (22). Moreover, V. paradoxus is resistant to various heavy metals, including cadmium and mercury (2).
V. paradoxus belongs to a group referred to as plant growth-promoting rhizobacteria (PGPR), which exert beneficial effects on plant growth. As a common plant symbiont found in the rhizosphere (2, 3), the metabolic diversity of V. paradoxus appears to be related to its role as a PGPR. By degrading toxic contaminants, this bacterium can prevent harm otherwise experienced by the plant and thus can promote plant growth. Strains of Variovorax can enhance the host plant's stress tolerance and disease resistance (2, 3) and aid in nutrient availability and uptake (38). The effectiveness of Variovorax as a PGPR is likely to be more potent because it also appears to be a good endophytic symbiont (34, 36, 39, 43, 44, 45, 47) and thus interacts more closely with the host plant. Conversely, endospheric habitats are known to offer microbes the advantage of a more uniform and protective niche compared to the competitive, high-stress environment of the soil (36).
The diverse metabolic capabilities of V. paradoxus make it an excellent choice for continued studies on novel biodegradation. In addition, its capacity for multiple interspecies interactions makes V. paradoxus ideal for studies of microbe-microbe and microbe-plant interactions. Despite its apparent ecological importance and potential for application, there is no published genome sequence for a strain of Variovorax. V. paradoxus S110 (Table 1) was isolated by Han from the interior of a potato plant grown in a farm of upstate New York and was identified as a degrader of AHLs, a bacterial signal molecule. In the present study, we report the complete genome sequence of V. paradoxus S110 and highlight genes that may contribute to its metabolic diversity, symbiotic lifestyles, and potential for use in biotechnological applications.
TABLE 1.
General features of V. paradoxus S110
| Feature | Genome |
|---|---|
| Size (bp) | |
| Chromosome 1 | 5,626,355 |
| Chromosome 2 | 1,128,646 |
| G+C content (%) | 67.5 |
| Total protein-coding DNA size in bp (%) | 6,177,424 (91.4) |
| Repeats (%) | 787 (0.74) |
| Gene | |
| No. of protein-coding RNAs | 6,279a |
| No. of rRNAs | 6 |
| No. of tRNAs | 61 |
| No. of genes with assigned function | 4,557 |
| No. of genes without assigned function | 1,722 |
| No. of enzymes | 1,989 |
Plus 39 ORFs in KRIBB.
MATERIALS AND METHODS
Whole-genome shotgun/pyrosequencing and sequence assembly.
Whole-genome shotgun sequencing was conducted at the Joint Genome Institute (JGI) of the U.S. Department of Energy (DOE) according to JGI protocols (http://www.jgi.doe.gov/sequencing/protocols/index.html). V. paradoxus S110 genomic DNA was isolated using the CTAB (cetyltrimethylammonium bromide) protocol as described by the JGI (http://my.jgi.doe.gov/general/index.html). The isolated genomic DNA was used to construct DNA shotgun clone libraries with insert sizes of 3, 8, and 40 kb, each of which was cloned into pUC18, pMCL200, and pCC1Fos vectors, respectively. A standard Sanger sequencing method was used to produce a high-quality draft sequence using ABI 3730 capillary sequencers. Plasmid clones were end sequenced from both sides of the library insert. In addition to the Sanger sequencing, 454 pyrosequencing was conducted to a depth of 20× coverage. The resulting sequence reads were base-called by using PHRED (9, 10). High-quality sequence reads were defined by a minimal length of 250 bp with an average value of ≥20. A total of 37,264 sequenced reads were aligned by using the PHRAP assembly tool (http://www.phrap.org) to produce primary draft assembly, which consists of contigs linked with larger scaffolds by paired-end information. The CONSED/AUTOFINISH (16, 17) software package was used for the finishing of the genome sequence. Possible mis-assemblies were corrected using Dupfinisher of bridging clones (Epicentre Biotechnologies, Madison, WI). Gaps between contigs were closed by editing in CONSED, custom primer walking, or PCR amplification (Roche Applied Sciences, Indianapolis, IN). A total of 1,251 additional reactions were necessary to close gaps and to raise the quality of the finished sequence. The completed genome sequence of V. paradoxus contained 38,521 reads, achieving an average of 7.2-fold sequence coverage per base, with an error rate of less than 1 in 100,000.
Genome analysis and gene annotation.
Genome analyses were conducted by the Computational Biology at the Oak Ridge National Laboratory (ORNL) in the United States and also by the Korea Research Institute of Bioscience and Biotechnology (KRIBB) in the Republic of Korea. For the purpose of initial automatic annotation, gene prediction was performed with the gene modeling program Prodigal (http://prodigal.ornl.gov) for all contigs/replicons, using default settings that permit overlapping genes and using ATG, GTG, and TTG as potential start codons. The identified genes were searched against the nonredundant (NR) protein database at the GenBank by using BLAST analysis. Based on the similarity searches, predicted functions were assigned to individual genes with a cutoff E-value of 1 E−05. Pfam and TIGRFam were used to investigate protein domains with the default cutoff options. PRIAM was used to detect enzyme-coding genes with a cutoff value of 1.0 E−20. Cluster of Orthologous Group (COG) was also searched with the default cutoff value. tRNAs and rRNAs were identified by using tRNAscan-SE and RNAmmer1.2, respectively, with the prokaryotic default setting. RepeatMasker and CrossMatch were used in order to examine repeat sequences. For more precise detection of potentially useful open reading frames (ORFs), KRIBB used YACOP (47) to combine the multiple gene prediction results from Criticia, Glimmer (28), and ZCURVE (20). Finally, the automatically annotated genes were manually inspected and corrected by BLASTX analysis with a cutoff range of E-value < 1.0 E−10 against the NCBI, PDB, and UniProtKB databases.
Gene network/pathway analysis.
Predicted and annotated gene sequences were analyzed for similarity with the enzyme database of KEGG followed by assignment of each gene into KEGG pathway chart. These analyses were conducted independently by JGI and KRIBB for the purpose of complementary integration of both data. Based on individual analysis results of the KEGG pathways, integrated biochemical pathway maps were constructed, which demonstrated characteristic physiological features in assimilation and degradation metabolisms of S110. The existence of a certain pathway was determined and integrated when component genes within the corresponding pathway had been completely identified. We allowed exceptions, in special cases, to combine pathways with the integrated map only when a significant portion of genes could be identified in the corresponding pathway.
Data submission.
The complete genomic nucleotide sequence of V. paradoxus S110 was submitted to the GenBank under accession numbers NC012791 and NC012792. The data for the genome analysis are also available at the ORNL website (http://genome.ornl.gov/microbial/vpar_s110).
RESULTS
General genome features.
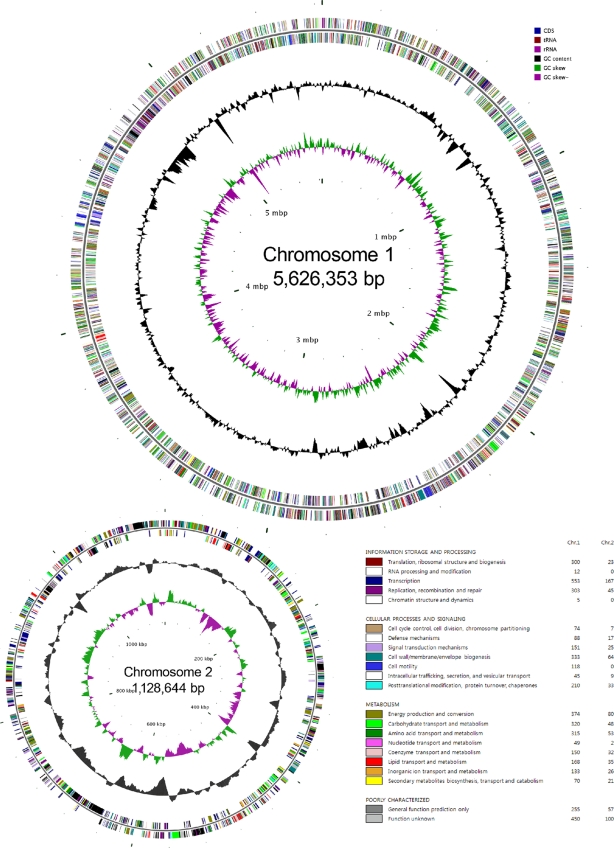
The genome of V. paradoxus S110 has a total of 6,754,997 bp and is revealed to separate into two chromosomes (chromosomes 1 and 2) (Fig. 1) and an average G+C content of 67.4%. Bioinformatic analyses were conducted independently by both the JGI in the United States and the KRIBB in South Korea for the purpose of integrating both data sets (see Materials and Methods for details). According to the analyses, the genome contains 6,279 predicted ORFs. Coding regions cover 91.4% of the whole-genome sequence. Biological roles were assigned to 4,557 genes of the predicted coding sequences based on similarity searches. The remaining coding sequences were classified as proteins with hypothetical or unknown function (see Table S1 in the supplemental material). More detailed information can be found on the genome website at Computational Biology at the ORNL (http://genome.ornl.gov/microbial/vpar_s110/18feb09/). The results of gene prediction by JGI differed from those of KRIBB in the number of identified ORFs. The number of genes predicted by KRIBB was slightly higher, presumably due to the use of the YACOP gene prediction algorithm, which combined three different gene modeling programs: Criticia, Glimmer, and ZCURVE.
FIG. 1.
Genomes of V. paradoxus S110 chromosomes 1 and 2. The outer circle represents coding region with colors of COG categories, rRNA, and tRNA. The black circle represents GC contents, the green color shows the GC skew on the leading strand, and the pink color circle represents GC skew on the lagging strand. This figure was drawn with CGview. GC skew was calculated as (G − C)/(G + C), where G is the number of G's and C is the number of C's. The COG colors are defined (or categories) and the numbers of related genes are given at the bottom right of the figure.
The origin of replication (ori C-type), which was analyzed with the web tool Ori-Finder (14), was identified in chromosome 1 only, which is located between 796699…797176 and 5626103…5626136. No transposons or insertion sequence elements were detected in either replicon.
A simple comparison of gene ratios in different KEGG categories to the relative sizes of the two replicons suggests that core cellular functions are primarily found on chromosome 1 (see Table S1 in the supplemental material). Genes for chromosome partition (D), recombination and repair (KEGG group L), motility (N), nucleotide metabolism (F), intracellular trafficking (U), and translation (J) are particularly overrepresented in chromosome 1. This is compatible with the notion of chromosome 2 arising as an accessory factor. The only KEGG functional groups present on chromosome 2 in a higher proportion were transcription factors (K), secondary metabolite production (Q), energy conversion (C), and lipid metabolism (I).
Carbon metabolism.
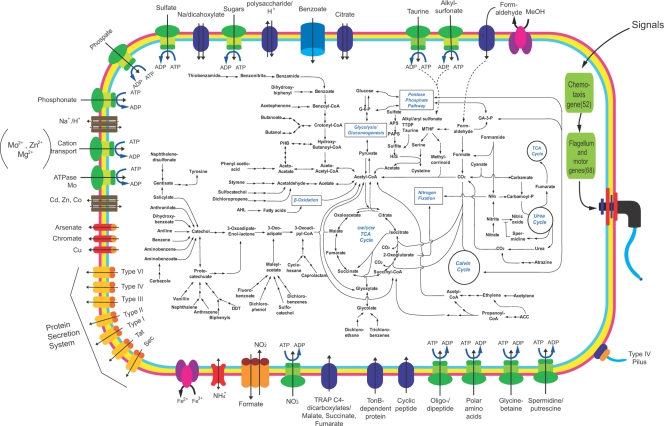
The metabolic pathways of strain S110 were predicted through comparison to known bacterial genes. Figure 2 illustrates the predicted metabolic pathways and shows associations between assimilatory and degradative pathways (see Fig. S1 in the supplemental material).
FIG. 2.
Schematic summary of metabolic strategies in V. paradoxus S110. The depicted pathways were predicted based on the genomic data of V. paradoxus S110 analyzed by the JGI and KRIBB. Details are available in Fig. S1 in the supplemental material. The figure demonstrates integration of a range of anabolic and carbolic metabolisms, along with membrane-bound transport systems. These include the assimilation of carbon, nitrogen, and sulfur compounds and the degradation and subsequent utilization of xenobiotic contaminants in correlation with the central pathways that are recognized to operate in this microorganism. Individual pathways are denoted by single-headed arrows, while reversible pathways are denoted by double-headed arrows. Abbreviations: cw, clockwise (for oxidative TCA cycle); ccw, counter-clockwise (for reductive TCA cycle); AHL, acyl homoserine lactone; GA3P, glyceraldehyde-3-phosphate; G6P, glucose-6-phosphate; MTHF, 5,10-methylenetetrahydrofolate; PHB, polyhydroxy butyrate; APS, adenylyl sulfate; PAPS, 3-phosphoadenylyl sulfate; TTDP, thioether 3,3-thiodipropionic acid.
The genome of strain S110 contains a complete central carbon metabolism including glycolysis/gluconeogenesis, a tricarboxylic acid (TCA) cycle, and a pentose phosphate pathway (PPP). The pentose phosphate pathway apparently contains an auxiliary route which converts d-glucose into 6-phospho-d-gluconate, which is one of the PPP intermediates (see Fig. S1A in the supplemental material). There is also evidence for the glyoxylate pathway, since putative genes encoding the two key enzymes, isocitratelyase and malate synthase, are present (see Table S2 in the supplemental material).
S110 carries one complete and three incomplete sets of genes for CO2 fixation. It possesses genes consistent with CO2 fixation by the Calvin-Benson cycle (Fig. 2 and see Fig. S1A and Vapar_Table S2 in the supplemental material). One set of RuBisco-encoding genes (Vapar_3031 and Vapar_3032, which have >84% identity with Methylibium petroleiphilum PM1), one more RuBisco-encoding gene (Vapar_1945), and a complete set of Calvin cycle-associated genes were found, suggesting that S110 can grow autotrophically (see Fig. S1A and Table S2 in the supplemental material). However, any candidate genes encoding light-harvesting pigments were not identified.
Three genes encoding aerobic carbon monoxide dehydrogenase (CODH) are present in S110 (Vapar_1594, Vapar_1598, and Vapar_2379), all of which are very similar to that of the aerobe Polaromonas sp. strain JS666 (up to 83% similarity). These aerobic CODHs catalyze CO oxidation (1), which may contribute to the unique symbiotic lifestyle of S110 by relieving the toxicity of CO in surrounding biota. Strangely, we were unable to identify any genes encoding a hydrogenase in the S110 genome, even though V. paradoxus is known to use H2 as an electron donor (27).
Although V. paradoxus has demonstrated the ability to enhance methanotrophic activity in a consortium (12), genome analysis confirms that S110 does not contain particulate or soluble methane monooxygenase (MMO). Four alcohol dehydrogenases (Vapar_700, Vapar_3040, Vapar_5018, and Vapar_6242), which predicted to be homologs of methanol dehydrogenase genes, are also unlikely to be involved in methanol oxidation.
V. paradoxus is catabolically diverse, utilizing a wide range of naturally occurring compounds as sole growth substrates (1, 4, 6, 8, 12, 13, 15, 21, 24, 29-31, 34, 40, 49). Variovorax species have also been observed to degrade a variety of contaminants (5, 19, 37, 41, 42, 46, 50-52), including the herbicide atrazine (41). The S110 genome contains an array of genes supporting similar metabolic diversity, although many of the pathways were incompletely identified. Aspects of putative xenobiotic metabolism are summarized in Fig. 2 (see also Fig. S1B in the supplemental material for details).
Interestingly, V. paradoxus engages in synergistic and mutually beneficial interactions with other bacteria during the biodegradation of certain compounds. For instance, strains of Variovorax are able to degrade capsaicin (13) and do so most effectively when operating in defined consortia containing organisms such as Pseudomonas putida. Other compounds that are degraded by this species through synergistic interactions with other bacterial species include linuron (5), simazine (37), atrazine (41), 2,4-dinitrotoluene (42), and sulfolane (19). However, we were unable to identify the genes involved in these unique synergistic activities through genome analysis.
Sulfur metabolism.
V. paradoxus is known to have the ability to metabolize sulfur compounds such as alkyl/aryl-sulfonates (key components of the sulfur present in agricultural soils) (38) and thioether 3,3-thiodipropionic acid (a widely used antioxidant and stabilizer in food) (4). As expected, the S110 genome contains an alkane monooxygenase ssuD gene (Vapar_2976 and Vapar_3871) for alkylsulfonates and an oxidoreductase asfA gene for aryl-sulfonates (38) (Vapar_1895; see Table S3 in the supplemental material). V. paradoxus is known to metabolize thioether 3,3-thiodipropionic acid and a strain of Variovorax is the only known microbe capable of utilizing both the carbon and the sulfur of this compound (4). The key enzyme involved in this metabolic pathway is 3-mercaptopropionate dioxygenase, a cysteine dioxygenase homologue. This gene was indeed present in S110 (Vapar_3875). The second enzyme involved in the pathway, a family III acyl-coenzyme A transferase, was also present (Vapar_5877, Vapar_5651, Vapar_4713, Vapar_6156, Vapar_4307, and Vapar_4973). The S110 genome has four genes encoding taurine (2-aminoethanesulfonate, a naturally occurring amino acid analog) dioxygenases (Vapar_5979, Vapar_3747, Vapar_0978, and Vapar_3825) that oxidize and generate sulfite and aminoacetaldehyde. There are also eight genes encoding taurine transporter-related proteins (see Table S3 in the supplemental material).
Sulfite is produced during the metabolism of these sulfur compounds, which can be either assimilated by plants and other associated microbes or reduced to hydrogen sulfide by sulfite reductase (Vapar_1174 and Vapar_2067). Hydrogen sulfide is in turn used as an electron donor by organisms such as green sulfur phototrophs. This type of closed-loop sulfur cycle is illustrated by the well-known photosynthetic consortium C. aggregatum, which contains a Variovorax strain as a heterotrophic partner (22).
Metal resistance.
Resistance to the toxicity of heavy metals is an important trait for microbes present at contaminated sites as symbiotic partners with plants. V. paradoxus is known to be tolerant to a number of heavy metals such as arsenate, chromate, Hg, Cu, Cd, Zn, Co, and Ni (2). Bioinformatic analysis confirmed that S110 contains multiple metal resistance elements (Fig. 2 and see Table S4 in the supplemental material), supporting its potential use as a PGPR agent to relieve host plant stress at contaminated sites.
Tolerance to arsenate in S110 is probably mediated via arsenic extrusion by ArsRBC (Vapar_1962, arsR; Vapar_1960 and Vapar_1961, arsCB). ArsC is an arsenic reductase responsible for the transformation of As(V) into As(III), and ArsR is a repressor that responds to As(III) and Sb(III) (35). Arsenite may be extruded by a carrier protein, Acr3 (Vapar_1963), energized by the membrane potential (35). Chromate detoxification appears to be mediated by four chromosomal copies of chrA in chromosome 1 (Vapar_5759, Vapar_0316, Vapar_2215, and Vapar_2214), belonging to the CHR family of transporter genes.
Mercury detoxification is likely to be mediated by mercuric reductase (Vapar_5924) in S110, although the other typical mer operon genes, including regulatory and transport genes, are missing. Copper may be removed from S110 by Cu export systems or metal-transporting P1-type ATPases (Vapar_0030 and Vapar_0031). One other gene (Vapar_5687) may also be involved in the detoxification to Cu.
The S110 genome encodes a chemiosmotic antiporter efflux system similar to CzcCBA of R. metallidurans, conferring resistance to Cd, Zn, and Co (29). This system is encoded by two copies of czcA (Vapar_3555 and Vapar_6299), one copy of czcB (Vapar_3556), and two copies of czcC (Vapar_3557). Additional proteins potentially involved in metal transport include 15 genes for nickel and 2 for cobalt (see Table S4 in the supplemental material). Interestingly, these metal resistance regions were not associated with transposases, indicating that S110 acquired these genes at an early time point in its evolution.
Symbiotic lifestyle.
V. paradoxus has been found in both the rhizosphere and endosphere. Analysis of the S110 genome has revealed unique properties that presumably give Variovorax the ability to adapt in such habitats.
Iron acquisition is essential for competition in the rhizosphere, as well as in endophytic growth, and its active transport is of particular importance (30). As with other Gram-negative bacteria, S110 acquires iron via ferric-siderophore complexes whose uptake is controlled by TonB-dependent proteins. Twenty-four genes encoding proteins related to this type of iron transport are widespread over the genome. Sixteen genes that putatively function in siderophore biosynthesis have been identified in the S110 genome, and an additional 12 ORFs are potentially involved in iron transport and homeostasis (see Table S5 in the supplemental material).
Fitness in the plant-associated lifestyle may also be related to metabolic capacity. The genome of strain S110 carries a broad spectrum of genes for carbon source utilization, including a large number of transporters (Fig. 2). For example, the genome contains at least 143 members of the ATP-binding cassette (ABC) transporter superfamily.
Surface characteristics of bacteria are important for a plant-associated lifestyle. Migration over a surface is typically mediated by a combination of pili, flagella, and a wetting agent to reduce surface tension (49). V. paradoxus produces a polar flagellum, and strain S110 contains numerous genes for type IV pili, which are known to be involved in endophytic colonization of plants (6). The S110 genome contains 32 genes encoding proteins that appears to be a complete set required for type IV pilus (tfp) assembly and regulation, such as pilins, proteins involved in pilus translocation and assembly, signal peptidase, and NTP-binding protein required for pilus retraction (see Table S5 in the supplemental material). In addition, two groups of nonribosomal peptide synthases with strong homology to the serrawettin W1 synthetase SwrW from Serratia marcescens are present in V. paradoxus S110 (Vapar_3742, Vapar_3743, Vapar_3744, Vapar_3746, Vapar_3184, and Vapar_3185). Either or both of these groups of genes may be involved in the synthesis of a lipopeptide wetting agent. Five genes encoding filamentous hemagglutinin, believed to be involved in plant attachment (18), are also present (see Table S5 in the supplemental material). Chemotaxis is of prime importance in adhesion to the plant surface (8). S110 has a complex chemosensory protein system composed of 45 genes (see Table S5 in the supplemental material), including a chemotaxis cluster (i.e., cheYZABW and cheR in P. aeruginosa PAO1) (40). The chemotactic capability of Variovorax also plays a key role in the motility of a well-known photosynthetic consortium C. aggregatum (22), which will be further discussed below.
V. paradoxus can use 1-aminocyclopropane-1-carboxylate (ACC) as a carbon and nitrogen source. ACC is the immediate precursor of the plant hormone ethylene, which plays an important role in plant growth and development (3). S110 contains a gene encoding ACC deaminase (Vapar_5099) that cleaves ACC to ketobutyrate and ammonia, which are carbon and nitrogen sources for the bacterium. In addition, the S110 genome contains a gene encoding spermidine synthase (Vapar_5266) and its transporter gene (Vapar_4696). Spermidine is a polyamine phytohormone that affects plant growth and development during both mitosis and meiosis. We could not identify genes involved in indole-3-acetic acid (IAA) synthesis, but a few putative genes involved in IAA transport system were found (see Table S5 in the supplemental material). Moreover, the principal detoxification mechanism of cyanide in higher organisms, the enzyme rhodanese, is found in the genome of strain S110 (see Table S5 in the supplemental material). Cyanide is produced by hydrolysis of plant cyanogenic glycosides and is a potent cytotoxin that works via the inhibition of cytochrome oxidases in the mitochondrial electron transport chain.
One mechanism of plant defense against pathogens is the production of reactive oxygen species (ROS); superoxide, hydroperoxyl radical, hydrogen peroxide, and hydroxyl radical species (21). The S110 genome encodes many proteins to protect the cell from the ROS: five superoxide dismutases, five catalases, nine peroxidases, and eight hydroperoxide reductases (see Table S5 in the supplemental material). In addition, S110 possesses 21 putative glutathione S-transferase (GST) or GST domain/family proteins compared to 12 in the endophyte Klebsiella pneumoniae 342 and 7 in E. coli K-12 (see Table S5 in the supplemental material).
Traits that contribute to a phytopathogenic lifestyle are not well represented in the S110 genome. The genome appears to lack genes involved in the production of toxins and common hydrolytic enzymes that macerate plant cell wall polymers, such as pectinases and cellulases. One exception is a gene that encodes β-glycosidase (Vapar_3136), which was reported to play a role in the endophytic colonization in Azoarcus (24). The S110 genome also contains seven genes encoding feruloylesterases, which hydrolyze feruloyl polysaccharides (see Table S5 in the supplemental material). Feruloyl polysaccharides are critical in directing plant cell wall cross-linking and in limiting plant degradation by microbes. This paucity of macerating enzymes is likely to serve for plant host compatibility, while the few enzymes that are present contribute to establishment of an endophytic association (24). Even though the genome possesses type III and type IV secretion systems (Fig. 2) that are often considered common among pathogens, these intensive transport systems may rather suggest its benign interactions with the plants.
To survive, compete, and persist in a competitive environment or to colonize a particular host, many bacteria have adopted a cooperative group behavior known as quorum sensing (QS). The best-studied QS signals are AHLs in Gram-negative bacteria. QS systems are involved in the regulation of a wide range of bacterial functions, including pathogenesis (11). The S110 genome possesses genes encoding AHL synthase (Vapar_5808) and its cognate transcription regulator (five genes, including Vapar_5809), even though its biological function is currently unknown. V. paradoxus is also known to have the ability to degrade various AHLs (11, 25), which are likely to be mediated by a putative AHL acylase (Vapar_3883) similar to one identified in Ralstonia sp. strain XJ12B (26). This putative acylase gene was first identified in this genome analysis. The ability of quorum quenching (QQ) and potential QS might indicate that Variovorax, like Agrobacterium tumefaciens, has a QS signal turnover system (52).
DISCUSSION
The V. paradoxus S110 has become the first genome sequenced from the genus of Variovorax. This genome has revealed diverse metabolic capabilities and a symbiotic lifestyle, which suggest possible future application of this microbe as a model for bacterial symbiosis and PGPR. The present study supports and extends various laboratory observations of this species and genus reported previously (2, 3, 4, 22, 25, 38).
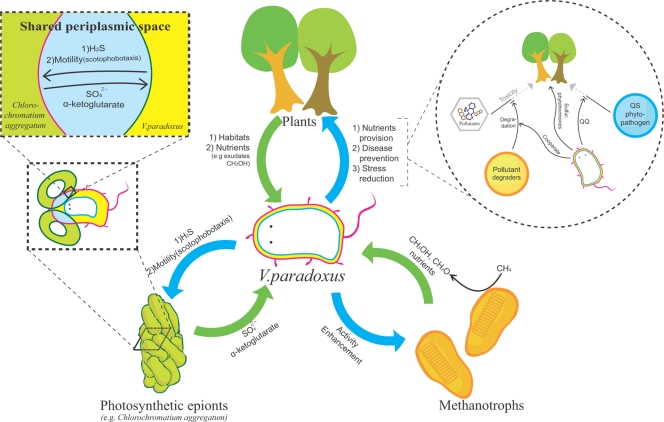
The genomic analysis has revealed that V. paradoxus S110 is uniquely equipped with a combination of features seen in autotrophic and heterotrophic lifestyles. These traits provide S110 and Variovorax in general with superb adaptability to ever-changing environmental conditions, allowing the microorganism to survive and prosper and also to survive independently in a symbiotic relationship (Fig. 3). However, we were unable to identify the genes involved in the synergistic interactions with other bacteria during biodegradations, which had been reported from physiological experiments.
FIG. 3.
Schematic representation illustrating the symbiotic interactions of V. paradoxus S110 with plants and other bacteria. The epibiont cells and central bacterial cell are known to share the periplasmic space through which signals and nutrients are directly exchanged. A wide variety of pollutants are known to be more efficiently degraded when V. paradoxus is included as an active component of a pollutant-degrading microbial consortium. Abbreviations: QS, quorum sensing; QQ, quorum quenching.
C. aggregatum, an intriguing phototrophic consortium with a relative of Variovorax as the core partner (ca. 95% similarity), is well adapted to low-sulfide conditions (32). One reason why this unique consortium is successful is that a limited resource is maximized through sulfur cycling between phototrophic epibionts and core bacterium (i.e., V. paradoxus) (Fig. 3). In addition, the epibionts are thought to benefit from the motility provided by the core bacterium, enabling the consortium to display chemotaxis toward sulfide (32). The central Variovorax-like bacterium may also benefit from carbon sources (e.g., α-ketoglutarate) secreted by the sulfur bacterium (32). The presence of these predicted functions is consistent with the metabolic interactions within the consortium.
As confirmed from previous reports and supported by the genome analysis in the present study, Variovorax strains such as S110 living within or on the plant enhances the host's stress tolerance and disease resistance and aids in nutrient availability and uptake (Fig. 3). By degrading toxic contaminants, this bacterium can effectively defend plants from adverse effects of chemicals and thereby promote plant growth, linking the metabolic diversity of Variovorax with its role as a PGPR.
The genome analysis also supports desulfurization of sulfonates as a critical factor in the plant growth-promoting effect of V. paradoxus. In fact, the genus Variovorax appears to be a key player in the mineralization of carbon-bound sulfur, allowing the cycling of soil sulfate between organic and inorganic forms (38). These bacteria may contribute to plant growth in soils with low sulfur availability. Desulfonating bacteria such as Variovorax in the soil and the rhizo-/endosphere can promote plant growth in sulfate-limited soils in much the same way that nitrogen-fixing bacteria make nitrogen available for plants (38).
The analysis of the V. paradoxus S110 genome suggests that it is a crucial species in maintaining robust microbial communities by converting diverse substrates into cell biomass, enhancing the activity of microbial metabolic consortia, and promoting the growth of plants. However, the basic physiology of Variovorax in culture and in the environment has only begun to be probed. We anticipate that the comprehensive genome analysis conducted in the present study will serve to elucidate the genomic organization and function and allow diverse array of utilization of this bacterial species in the fields of phytoremediation and crop production, for example, as a biofertilizer and/or a biopesticide. In addition, these results highlight the utility of S110 as a bacterial paradigm for continued studies of microbe-plant and microbe-microbe interactions.
Supplementary Material
Acknowledgments
We thank all of the people involved in this project. We especially thank an anonymous reviewer for helpful comments.
Sequencing conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. J.-I.H. was supported, in part, by a startup fund provided by KAIST. J.C.S. was supported by the Defense Threat Reduction Agency and the U.S. Army Research Office grant W911NF-07-0077.
J.-I.H. and H.-K.C. wrote the manuscript; S.-W.L. did the main part of bioinformatics work; J.K. and P.M.O. performed data analysis and participated in writing the manuscript; S.L.L. and J.O. were involved in strain isolation and characterization; J.R.L., C.-G.H., S.Y.L., T.K., and J.C.S. contributed to the genome analysis; G.O., L.G., and C.H. conducted sequencing and draft annotation.
Footnotes
Published ahead of print on 23 December 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allison, N., J. Turner, and R. Wait. 1995. Degradation of homovanillate by a strain of Variovorax paradoxus via ring hydroxylation. FEMS Microbiol. Lett. 134:213-219. [DOI] [PubMed] [Google Scholar]
- 2.Belimov, A. A., et al. 2005. Cadmium-tolerant plant growth-promoting bacteria associated with the roots of Indian mustard (Brassica juncea L. Czern.). Soil Biol. Biochem. 37:241-250. [Google Scholar]
- 3.Belimov, A. A., et al. 2009. Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase yield of plants grown in drying soil via both local and systemic hormone signaling. New Phytol. 181:413-423. [DOI] [PubMed] [Google Scholar]
- 4.Bruland, N., J. H. Wübbeler, and A. Steinbüchel. 2009. 3-Mercaptopropionate dioxygenase, a cysteine dioxygenase homologue, catalyzes the initial step of 3-mercaptopropionate catabolism in the 3,3-thiodipropionic acid-degrading bacterium Variovorax paradoxus. J. Biol. Chem. 284:660-672. [DOI] [PubMed] [Google Scholar]
- 5.Dejonghe, W., et al. 2003. Synergistic degradation of linuron by a bacterial consortium and isolation of a single linuron-degrading Variovorax strain. Appl. Environ. Microbiol. 69:1532-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dörr, J., T. Hurek, and B. Reinhold-Hurek. 1998. Type IV pili are involved in plant-microbe and fungus-microbe interactions. Mol. Microbiol. 30:7-17. [DOI] [PubMed] [Google Scholar]
- 7.Reference deleted.
- 8.Elmerich, C., and W. E. Newton. 2007. Associative and endophytic nitrogen-fixing bacteria and cyanobacterial associations. Springer, Dordrecht, Netherlands.
- 9.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 10.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 11.Faure, D., and Y. Dessaux. 2007. Quorum sensing as a target for developing biocontrol strategies toward the plant pathogen Pectobacterium. Eur. J. Plant Pathol. 119:353-365. [Google Scholar]
- 12.Flagan, S., W. K. Ching, and J. R. Leadbetter. 2003. Arthrobacter strain VAI-A utilizes acyl-homoserine lactone inactivation products and stimulates quorum signal biodegradation by Variovorax paradoxus. Appl. Environ. Microbiol. 69:909-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flagan, S. F., and J. R. Leadbetter. 2006. Utilization of capsaicin and vanillylamine as growth substrates by Capsicum (hot pepper)-associated bacteria. Environ. Microbiol. 8:560-565. [DOI] [PubMed] [Google Scholar]
- 14.Gao, F., and C.-T. Zhang. 2008. Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinform. 9:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodhue, C. T., and E. E. Snell. 1966. The bacterial degradation of pantothenic acid. I. Overall nature of the reaction. Biochemistry 5:393-398. [DOI] [PubMed] [Google Scholar]
- 16.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, D., C. Desmarais, and P. Green. 2001. Automated finishing with Autofinish. Genome Res. 11:614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottig, N., B. S. Garavaglia, C. G. Garofalo, E. G. Orellano, and J. Ottado. 2009. A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS One 4:e4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greene, E. A., P. H. Beatty, and P. M. Fedorak. 2000. Sulfolane degradation by mixed cultures and a bacterial isolate identified as a Variovorax sp. Arch. Microbiol. 174:111-119. [DOI] [PubMed] [Google Scholar]
- 20.Guo, F., H. Ou, and C. Zhang. 2003. ZCURVE: a new system for recognizing protein-coding genes in bacterial and archaeal genomes. Nucleic Acids Res. 31:1780-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammond-Kosack, K. E., and J. D. Jones. 1996. Resistance gene-dependent plant defense responses. Plant Cell 8:1773-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanzler, B. E., K. R. Pfannes, K. Vogl, and J. Overmann. 2005. Molecular characterization of the nonphotosynthetic partner bacterium in the consortium “Chlorochromatium aggregatum.” Appl. Environ. Microbiol. 71:7434-7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reference deleted.
- 24.Krause, A., et al. 2006. Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat. Biotechnol. 24:1385-1391. [DOI] [PubMed] [Google Scholar]
- 25.Leadbetter, J. R., and E. P. Greenberg. 2000. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 182:6921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, Y. H., et al. 2003. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 47:849-860. [DOI] [PubMed] [Google Scholar]
- 27.Maimaiti, J. Y., et al. 2007. Isolation and characterization of hydrogen-oxidizing bacteria induced following exposure of soil to hydrogen gas and their impact on plant growth. Environ. Microbiol. 9:435-444. [DOI] [PubMed] [Google Scholar]
- 28.McHardy, A. C., A. Goesmann, A. Puhler, and F. Meyer. 2004. Development of joint application strategies for two microbial gene finders. Bioinformatics 20:1622-1631. [DOI] [PubMed] [Google Scholar]
- 29.Mergeay, M., et al. 2003. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: toward a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 27:385-410. [DOI] [PubMed] [Google Scholar]
- 30.Neilands, J. B. 1995. Siderophores: structure and function of microbial iron-transport compounds. J. Biol. Chem. 270:26723-26726. [DOI] [PubMed] [Google Scholar]
- 31.Nishino, S. F., and J. C. Spain. 2006.Biodegradation of 3-nitrotyrosine by Burkholderia sp. strain JS165 and Variovorax paradoxus JS171. Appl. Environ. Microbiol. 72:1040-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Overmann, J., and K. Schubert. 2002. Phototrophic consortia: model systems for symbiotic interrelations between prokaryotes. Arch. Microbiol. 177:201-208. [DOI] [PubMed] [Google Scholar]
- 33.Reference deleted.
- 34.Reiter, B., and A. Sessitsch. 2006. Bacterial endophytes of the wild flower Crocus albiflorus analyzed by characterization of isolates and by a cultivation-independent approach. Can. J. Microbiol. 52:140-149. [DOI] [PubMed] [Google Scholar]
- 35.Rosen, B. P. 2002. Transport and detoxification systems for transition metals, heavy metals and metalloids in eukaryotic and prokaryotic microbes. Comp. Biochem. Physiol. 133:689-693. [DOI] [PubMed] [Google Scholar]
- 36.Ryan, R. P., et al. 2008. Bacterial endophytes: recent developments and applications. FEMS Microbiol. Lett. 278:1-9. [DOI] [PubMed] [Google Scholar]
- 37.Santiago-Mora, R., F. Martin-Laurent, R. de Prado, and A. R. Franco. 2005. Degradation of simazine by microorganisms isolated from soils of Spanish olive fields. Pest Manag. Sci. 61:917-921. [DOI] [PubMed] [Google Scholar]
- 38.Schmalenberger, A., et al. 2008. The role of Variovorax and other Comamonadaceae in sulfur transformations by microbial wheat rhizosphere communities exposed to different sulfur fertilization regimes. Environ. Microbiol. 10:1486-1500. [DOI] [PubMed] [Google Scholar]
- 39.Sessitsch, A., B. Reiter, and G. Berg. 2004. Endophytic bacterial communities of field-grown potato plants and their plant growth-promoting and antagonistic abilities. Can. J. Microbiol. 50:239-249. [DOI] [PubMed] [Google Scholar]
- 40.Shitashiro, M., et al. 2005. Identification of chemosensory proteins for trichloroethylene in Pseudomonas aeruginosa. J. Biosci. Bioeng. 99:396-402. [DOI] [PubMed] [Google Scholar]
- 41.Smith, D., S. Alvey, and D. E. Crowley. 2005. Cooperative catabolic pathways within an atrazine-degrading enrichment culture isolated from soil. FEMS Microbiol. Ecol. 53:265-273. [DOI] [PubMed] [Google Scholar]
- 42.Snellinx, Z., S. Taghavi, J. Vangronsveld, and D. van der Lelie. 2003. Microbial consortia that degrade 2,4-DNT by interspecies metabolism: isolation and characterization. Biodegradation 14:19-29. [DOI] [PubMed] [Google Scholar]
- 43.Sturz, A. V., et al. 2005. Variation in antibiosis ability, against potato pathogens, of bacterial communities recovered from the endo- and exoroots of potato crops produced under conventional versus minimum tillage systems. Can. J. Microbiol. 51:643-654. [DOI] [PubMed] [Google Scholar]
- 44.Sturz, A. V., et al. 2001. Weeds as a source of plant growth promoting rhizobacteria in agricultural soils. Can. J. Microbiol. 47:1013-1024. [DOI] [PubMed] [Google Scholar]
- 45.Surette, M. A., A. V. Sturz, R. R. Lada, and J. Nowak. 2003. Bacterial endophytes in processing carrots (Daucuscarota L. var. sativus): their localization, population density, biodiversity and their effects on plant growth. Plant Soil 253:381-390. [Google Scholar]
- 46.Suyama, T., H. Hosoya, and Y. Tokiwa. 1998. Bacterial isolates degrading aliphatic polycarbonates. FEMS Microbiol. Lett. 161:255-261. [DOI] [PubMed] [Google Scholar]
- 47.Tech, M., and R. Merkl. 2003. YACOP: enhanced gene prediction obtained by a combination of existing methods. In Silico Biol. 3:441-451. [PubMed] [Google Scholar]
- 48.Reference deleted.
- 49.Verstraeten, N., et al. 2008. Living on a surface: swarming and biofilm formation. Trends Microbiol. 16:496-506. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Y. P., and J. D. Gu. 2006. Degradability of dimethyl terephthalate by Variovorax paradoxus T4 and Sphingomonas yanoikuyae DOS01 isolated from deep-ocean sediments. Ecotoxicology 15:549-557. [DOI] [PubMed] [Google Scholar]
- 51.Willumsen, P. A., J. E. Johansen, U. Karlson, and B. M. Hansen. 2005. Isolation and taxonomic affiliation of N-heterocyclic aromatic hydrocarbon-transforming bacteria. Appl. Microbiol. Biotechnol. 67:420-428. [DOI] [PubMed] [Google Scholar]
- 52.Young, R. F., S. M. Cheng, and P. M. Fedorak. 2006. Aerobic biodegradation of 2,2′-dithiodibenzoic acid produced from dibenzothiophene metabolites. Appl. Environ. Microbiol. 72:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.