Abstract
The function of the essential inner membrane protein (IMP) YidC in Escherichia coli has been studied for a limited number of model IMPs and primarily using targeted approaches. These studies suggested that YidC acts at the level of insertion, folding, and quality control of IMPs, both in the context of the Sec translocon and as a separate entity. To further our understanding of YidC's role in IMP biogenesis, we screened a random overexpression library for factors that rescued the growth of cells upon YidC depletion. We found that the overexpression of the GadX and GadY regulators of the glutamate-dependent acid resistance system complemented the growth defect of YidC-depleted cells. Evidence is presented that GadXY overexpression counteracts the deleterious effects of YidC depletion on at least two fronts. First, GadXY prepares the cells for the decrease in respiratory capacity upon the depletion of YidC. Most likely, GadXY-regulated processes reduce the drop in proton-motive force that impairs the fitness of YidC-depleted cells. Second, in GadXY-overproducing cells increased levels of the general chaperone GroEL cofractionate with the inner membranes, which may help to keep newly synthesized inner membrane proteins in an insertion-competent state when YidC levels are limiting.
Approximately 20% of the proteins encoded by the Escherichia coli genome function in the inner membrane (IM). These inner membrane proteins (IMPs) first are targeted to and then inserted into the IM, after which they fold and, if required, assemble into oligomeric complexes (51). The Sec translocon is the main protein-conducting channel used to insert IMPs (18, 52). The translocon consists of the heterotrimeric channel complex SecYEG and the accessory components YidC and SecDF YajC. It functions not only in the lateral transfer of transmembrane segments (TMs) of IMPs into the lipid bilayer but also in the vectorial translocation of protein (domains) into the periplasm.
YidC has been identified as an essential membrane factor that plays an important but poorly understood role in the biogenesis of IMPs in bacteria. YidC can act together with the SecYEG channel and also independently as a membrane insertase. Cross-linking studies have shown that YidC contacts the TMs of substrate IMPs upon their lateral exit from the Sec translocon, possibly to assist lipid partitioning (41). However, the depletion of YidC in vivo only slightly affects the levels of IMPs that follow this insertion pathway in the inner membrane, as shown for FtsQ and Lep (41). Furthermore, the membrane insertion of FtsQ in a reconstituted system does not require YidC, which also suggests that YidC is not critical for insertion per se (45). YidC can function upstream of the Sec translocon, as demonstrated for the integral inner membrane lipoprotein CyoA (subunit II of the cytochrome bo3 oxidase). Here, YidC is required for the insertion of the N-terminal region of CyoA into the membrane, followed by the translocation of the more complex C-terminal domain by the Sec translocon (4, 9, 43). While YidC can act in concert with the Sec translocon, other studies have shown that YidC also can function as an independent insertase for small proteins, such as the M13 and Pf3 phage coat proteins, the subunits a and c of the F1Fo ATPase (Foa and Foc), and subunit K of the NADH dehydrogenase complex (27, 30, 31, 46). It has been shown that both aerobic and anaerobic respiratory chain complexes are affected in YidC-depleted cells (27, 28, 46), resulting in a reduction of the proton-motive force (PMF). PMF reduction leads to a strong upregulation of the stress response protein PspA, which is part of the Psp shock response thought to help maintain the PMF upon membrane stress (5, 17, 50).
Besides YidC's role as an insertase, it is required for cotranslational folding into a stable conformation and/or assembly of multimeric IMP complexes, like MalFGK2 and MscL (23, 26, 47). The observation that YidC depletion induces the cell envelope stress response further supports a role for YidC in the folding of IMPs (35, 48). At an even later stage, a role for YidC in the quality control of IMPs has been implied from its association with the membrane protease and chaperone complex HflK/C-FtsH (42).
Although these studies have provided insight into the various roles of YidC, it remains difficult to imagine how one protein can combine the diverse functions ascribed to YidC. Pressing questions include the following: why is YidC essential, and how does YidC operate in various structural contexts, alone or connected to the Sec translocon? To gain more insight in the key processes that determine why YidC is essential, we have used an unbiased genetic screen to select factors that are able to complement the growth of cells that express YidC at sublethal levels. We found that the overexpression of gadX and gadY, regulators of the glutamic acid decarboxylase (Gad)-based acid resistance system in E. coli, rescues growth at low YidC levels and suppresses the concomitant PspA response.
MATERIALS AND METHODS
Enzymes and materials.
Restriction endonucleases and other DNA-modifying enzymes were obtained from Roche and Invitrogen. All other chemicals were supplied by Sigma. Antisera against the His tag and the hemagglutinin (HA) tag were purchased from Roche and Sigma, respectively. Antisera against YidC, PspA, Lep, and GadAB have been described previously (33, 39). Antiserum against GroEL was kindly donated by Peter Lund (University of Birmingham, Birmingham, United Kingdom).
Strains, plasmids, growth conditions, and primers.
E. coli strain Top10F′ (Invitrogen) was used for the cloning and maintenance of plasmid constructs. The YidC depletion strain FTL10 and its isogenic parental arabinose-resistant strain MC4100-A were generous gifts from Frank Sargent (University of East Anglia, Norwich, United Kingdom) (13). All strains were grown routinely in Luria Bertani (LB) medium or in M9 medium where indicated, with appropriate antibiotics. To monitor cell growth, cultures were pregrown in medium with 0.2% l-arabinose, washed in fresh medium to remove l-arabinose, and diluted as appropriate for the following growth experiments. Strains, plasmids, and primers used in this study are listed in Table 1. For the construction of plasmid pEH3.GadBC-HA, the sequence encoding GadB and GadC was PCR amplified using genomic DNA of MC4100-A and the primers GadB Forw and GadC-HA Rev. The PCR product was digested with XbaI and SacI and cloned into pEH3. The nucleotide sequence was verified by DNA sequencing.
TABLE 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Description | Reference or source |
|---|---|---|
| Strains | ||
| Top10F′ | F−mcrAΔ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL (Strr) endA1 λ− | Invitrogen |
| MC400-A | F− ΔlacU169araD139rpsL150relA1ptsFrbsflbB5301; arabinose resistant | 13 |
| FTL10 | MC4100-A ΔyidCattB::(araC+ PBADyidC+) Kanr | 13 |
| Plasmids | ||
| pBR322 | Cloning vector | 49 |
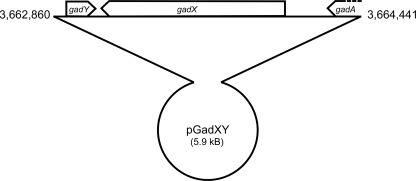
| pGadXY | pBR322 containing gadXY fragment (Fig. 1) | This study |
| pCl.CyoA-HA | cyoA ORF with C-terminal HA tag | 43 |
| pEH3.GadBC-HA | gadB ORF and gadC ORF with C-terminal HA tag | This study |
| Primers | ||
| YidC Forw | TGCTTACCCGAAAGAGCTGAA | |
| YidC Rev | AATAAACTGCGGTGAAGTTTCCA | |
| PspA Forw | AGCCACAGCTTCGGTAAACAA | |
| PspA Rev | TGCATCATCGGCTTTCAGTTC | |
| GadX Forw | CACTCATGGGCGATATTATTATTGAT | |
| GadX Rev | CGCTGCTTCTGAACGTTTTACA | |
| GadC Forw | ACGCCATTTTTGTTCATTTTAGC | |
| GadC Rev | TCTCGCGGGATGTATGTAACAG | |
| RssC Forw | GAATGCCACGGTGAATACGTT | |
| RssC Rev | AACCCACTCCCATGGTGTGA | |
| GadB forw | CCGGTCTAGAAGAAGGAGATATACATATGGATAAGAAGCAAGTAACGGATTTAAGG | |
| GadC-HA Rev | TTCCGAGCTCTTATTAGGCATAGTCTGGGACGTCATATGGATAAGATCCGTGTTTCTTGTCATTCATCAC |
Complementation of YidC depletion screen.
FTL10 cells were electroporated with a pBR322-based MC4100 ΔdnaJK tig genomic DNA library (kindly donated by Pierre Genevaux, CNRS Université Paul Sabatier, Toulouse, France) or empty vector pBR322 as a control (Fermentas). The resulting transformants were selected for growth on agar plates containing no l-arabinose or low concentrations of l-arabinose that are unable to support growth with pBR322 as indicated. From colonies that grew on the selective agar plates, plasmids were isolated and retransformed into FTL10 to confirm the rescue phenotype. The boundaries of the inserted genomic fragments in the plasmids subsequently were determined by DNA sequencing.
Quantitative real-time PCR.
Total RNA was isolated using an RNeasy minikit (Qiagen) from bacterial strains grown to mid-log phase in M9 and treated with RNase-free DNase (Qiagen) according to the manufacturer's protocol. The RNA quantity was determined by Nanodrop (ND-100 spectrophotometer), and the quality was assessed by Nanochip (Agilent Technologies). Only RNA with RNA integrity numbers (RINs) higher than 9 was used for subsequent analysis. cDNA samples were synthesized from total RNA using the SuperScript II reverse transcriptase kit (Invitrogen) with random primer oligonucleotides (Invitrogen). Quantitative real-time PCR (qPCR) was performed with each specific primer pair (Table 1) using Platinum SYBR green qPCR SuperMix-UDG (Invitrogen). The reactions were run on an ABI7500 detection system (Applied Biosystems); the fluorescence signal due to SYBR green intercalation was monitored to quantify the double-stranded DNA product formed in each PCR cycle. Melting curve analysis was performed after each qPCR to verify the specificity of each primer pair. The rrsC gene of 16S rRNA was used to normalize the cDNA input. For each time point and condition, four biological replicas were taken.
Multilevel variant statistical analysis was performed to determine the significance of each sample. The differences between each condition at different time points are listed in Table 2.
TABLE 2.
Expression levels of genes in FTL10 as analyzed by qPCR
| Cell type and gene | 30 min |
5 h |
||
|---|---|---|---|---|
| Fold change | P value | Fold change | P value | |
| Cultured without or with l-arabinose | ||||
| yidC | 5.3↓ | 0.009 | 6.8↓ | 0.005 |
| pspA | 1.2↑ | 0.632 | 22.2↑ | 0.000 |
| gadX | 1.76↓ | 0.269 | 1.6↑ | 0.362 |
| gadC | 2.66↓ | 0.055 | 1.0↓ | 0.929 |
| Overexpressing pGadXY versus control in absence of l-arabinose | ||||
| yidC | 1.8↓ | 0.238 | 1.8↓ | 0.221 |
| pspA | 1.6↓ | 0.778 | 8.4↓ | 0.001 |
| gadX | 104.9↑ | 0.000 | 42.2↑ | 0.000 |
| gadC | 9.3↑ | 0.002 | 6.7↑ | 0.004 |
Analysis of lactate in spent medium.
Bacterial strains were grown in M9, and cells were separated from the medium by centrifugation at 4°C for 5 min at 8,000 × g. The spent medium was assayed for lactate by high-performance liquid chromatography (HPLC) (LKB) with a REZEX organic acid analysis column (Phenomenex) at 45°C with 7.2 mM H2SO4 as the eluent, using an RI 1530 refractive index detector (Jasco) and AZUR chromatography software for data integration (1).
β-Galactosidase assay.
The construction of gadBC(−244/77)::lacZ, gadX(−222/+121)::lacZ, and gadY(−78/+65)::lacZ transcriptional fusions in the vector pRS415 (37) was described elsewhere (10, 38, 39). The E. coli strain FTL10, carrying the plasmids with transcriptional fusions, was grown for 5 h in LB with or without 0.2% l-arabinose. β-Galactosidase activity was determined as described by Miller (22) as 1,000 × (OD420 × 1.75 × OD550)/(OD600 × reaction time × volume), where OD420 is the optical density at 420 nm.
BN-PAGE and protein identification.
Enriched inner membrane fractions (eIM) were prepared essentially as described previously (8), with the following modifications: crude membranes were centrifuged at 4°C for 3 min at 380,000 × g to remove most of the outer membranes, after which the supernatant was used to collect eIMs by centrifugation at 4°C for 60 min at 380,000 × g. eIM samples containing equal amounts of protein (50 μg), as determined using the bicinchoninic acid protein assay kit (Pierce), were dissolved using the mild detergent n-dodecyl-β-d-maltoside as described previously (26) and subjected to blue native PAGE (BN-PAGE) using precast 3 to 12% gradient native PAGE Novex gels (Invitrogen) according to the manufacturer's protocol. After the gel was stained with Coomassie, the bands of interest were excised and prepared as described for identification by matrix-assisted laser desorption-ionization time of flight-time of flight (MALDI TOF-TOF) mass spectrometry (32).
RESULTS
Genetic screening for restored growth upon YidC depletion.
To gain insight in the function(s) of YidC, we performed a genetic screen to identify factors that suppress the lethality observed upon the depletion of YidC in E. coli strain FTL10 in which yidC is under the control of an l-arabinose promoter (13). Chromosomal DNA of E. coli was partially digested with the restriction enzyme Sau3AI, ligated in the vector pBR322, and introduced into FTL10. FTL10 transformed with empty pBR322 was unable to grow on plates that contained no or low concentrations of l-arabinose (0.001 to 0.01% range). FTL10 cells transformed with the genetic library also were plated on plates with no or low concentrations of l-arabinose. Plasmids were extracted from clones that grew on these plates. After confirming the phenotype upon the retransformation of the plasmids into fresh FTL10 competent cells, the 5′ and 3′ ends of the inserts were sequenced.
Only one clone was obtained that was able to grow in the absence of l-arabinose. Not surprisingly, this clone contained the yidC gene, validating our screening conditions (data not shown). In contrast, screening in the presence of a low concentration of l-arabinose (0.001%) resulted in numerous colonies. The sequence analysis of 26 clones revealed that 10 of them contained overlapping inserts containing at least the gadX and gadY genes. The plasmid of a clone that contained only the gadXY genes (pGadXY) was further analyzed (Fig. 1).
FIG. 1.
Schematic representation of the pGadXY construct containing gadY, gadX, and partial gadA ORFs.
The presence of pGadXY partially complements growth and represses the PspA response.
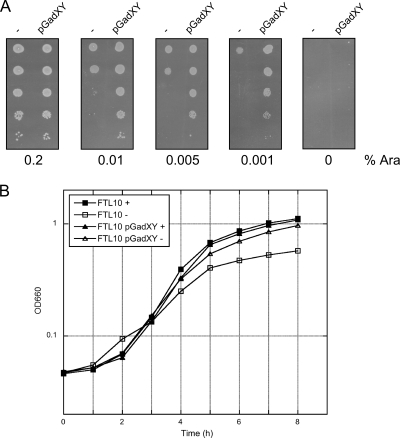
We first studied in more detail the effect of pGadXY on growth when YidC was depleted. FTL10 cells harboring pGadXY or pBR322 were grown to mid-log phase in the presence of l-arabinose (to sustain YidC expression) and then spotted in serial dilutions on solid LB medium containing different concentrations of l-arabinose (Fig. 2A). The pGadXY plasmid was not able to support growth on plates without l-arabinose, indicating that the complete absence of YidC was not complemented by the plasmid. However, cells harboring pGadXY efficiently supported growth on LB containing as low as 0.001% l-arabinose (the concentration used in the screening), whereas cells harboring pBR322 already were strongly affected in growth at 0.01% l-arabinose. In addition, we analyzed the effect of YidC depletion in liquid culture by diluting cells grown to mid-log phase in M9 medium containing no or 0.2% l-arabinose. The FTL10 cells with pGadXY were less affected in growth upon YidC depletion than control cells (Fig. 2B). The difference in growth was less pronounced when cells were cultured in LB medium (data not shown). Apparently the presence of pGadXY allows the cells to overcome the deleterious effect of YidC depletion, likely as a result of the higher levels of gadX and gadY.
FIG. 2.
GadXY overexpression partially complements growth of YidC depleted FTL10 cells on solid and in liquid media. (A) Serial dilution of FTL10 cells containing pGadXY or the control vector pBR322 (−). Cells were grown to mid-log phase in liquid LB medium. Tenfold serial dilutions of the cultures were prepared and spotted on LB plates supplemented with increasing amounts of l-arabinose (Ara). (B) Growth curve of FTL10 and FTL10 containing pGadXY. Cells were grown in liquid M9 medium in the presence (+) or absence (−) of 0.2% l-arabinose. Absorbance was measured every hour at the OD660 for a period of 8 h.
The Gad system plays an important role in glutamate-dependent acid resistance and includes two glutamate decarboxylase isoforms, GadA and GadB, which consume intracellular protons by mediating the decarboxylation of glutamate to γ-aminobutyric acid (GABA), and the inner membrane-based antiporter GadC, which exchanges the intracellular GABA for extracellular glutamate (6, 53). The expression of these effector proteins is upregulated in response to low pH and stationary-phase stress conditions via a complex interplay of various regulatory proteins and RNAs, including those encoded by gadX and gadY. The gadX gene encodes an AraC-like transcription factor that plays a critical role in the activation of Gad effector genes (36, 39). The gadY gene, adjacent to gadX (Fig. 1), is transcribed in the opposite direction and encodes a small noncoding RNA (ncRNA) that interacts with the 3′-untranslated region (UTR) of gadX mRNA to stabilize it (25). To examine the effect of gadX or gadY individually, most of the coding region of gadX or gadY was deleted in pGadXY, and the resulting constructs were tested for their ability to restore the growth of FTL10 upon the depletion of YidC. The presence of the plasmids containing the individual genes showed only partial effects, but intact gadX and gadY both were required for the optimal restoration of growth (results not shown). A plausible explanation for the partial rescuing effect of gadY on its own is that gadY exerts a stabilizing effect on the endogenous gadX mRNA (25). The overexpression of gadX, cloned under the control of an inducible promoter, led to a severe inhibition of cell growth in both wild-type (WT) and YidC-depleted cells. This toxic effect probably resulted from the excessive expression of the GadX activator protein, which has been observed previously (39). For these reasons, we decided to continue with pGadXY as isolated in the screen.
A possible explanation for the rescue phenotype is that GadXY overproduction stabilizes YidC, and therefore less l-arabinose is needed to maintain sufficient YidC levels in the cells, which would lead to residual YidC levels in the absence of l-arabinose. To study the effects of GadXY overproduction on YidC expression, we analyzed the levels of YidC in FTL10 (plus or minus pGadXY) cells grown for 4 h at various concentrations of l-arabinose by Western blotting. As shown in Fig. 3A, YidC was depleted to comparable low levels in both control and pGadXY-complemented FTL10 cells grown with low l-arabinose concentrations. Furthermore, qPCR analysis showed that after 30 min of the growth of FTL10 in the absence of l-arabinose, the mRNA level of YidC was 5-fold lower, and after 5 h 7-fold lower, compared to that of cells grown in the presence of l-arabinose (Table 2). Furthermore, there was no significant difference in yidC mRNA levels in YidC-depleted cells with or without pGadXY (Table 2). We therefore conclude that GadXY overexpression does not lead to altered levels of YidC in FTL10.
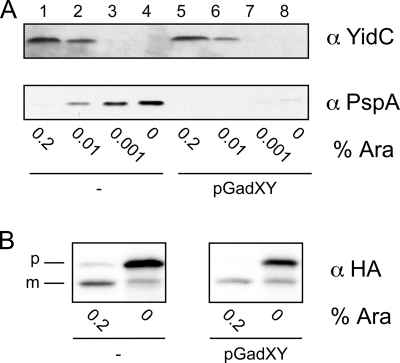
FIG. 3.
GadXY overexpression represses the PspA response in YidC-depleted cells without restoring YidC insertase function. (A) YidC and PspA protein levels in FTL10 cells containing pGadXY or control vector pBR322 (−). Cells were grown for 4 h in LB medium with various concentrations of l-arabinose. Cell lysates were analyzed by SDS-PAGE and Western blotting using antisera against YidC and PspA. (B) Steady-state analysis of CyoA-HA processing in FTL10 cells harboring pGadXY or control vector pBR322 (−). CyoA-HA processing was analyzed by SDS-PAGE and Western blotting using an antiserum against HA. The precursor and mature forms are indicated as p and m, respectively.
The depletion of YidC has been shown to induce a massive expression of PspA, a stress protein that is believed to respond to the dissipation of the PMF (5). This in turn is caused by the impaired membrane insertion of CyoA and Foc, subunits that are essential for the functional assembly of the cytochrome bo3 oxidase and F1Fo ATPase complex, respectively (46). To investigate if GadXY overexpression is involved in suppressing this strong PspA response, lysates from FTL10 cells (with or without pGadXY) were analyzed for PspA content by Western blotting (Fig. 3A). Strikingly, PspA showed little upregulation in FTL10 cells harboring pGadXY (lanes 5 to 8) despite the low levels of YidC, in contrast to what is observed in the control cells (lanes 1 to 4). Moreover, qPCR analysis revealed a strong upregulation of pspA mRNA (22-fold up after 5 h) in FTL10 cells (Table 2), while this upregulation was reduced in cells complemented with pGadXY (8-fold down after 5 h) (Table 2). Taken together, these data demonstrate that GadXY overexpression strongly represses the PspA response upon YidC depletion. Apparently, the PspA response is not a compulsory consequence of YidC depletion.
The presence of GadXY does not restore insertion of CyoA and Foc.
As shown above, GadXY overexpression does not prevent the depletion of YidC in FTL10 cells grown without l-arabinose, but it does significantly repress the PspA response. One possible explanation is that GadXY overexpression leads to the improved insertion or assembly of YidC-dependent subunits of the respiratory chain complexes, thereby relieving the loss of PMF and subsequent PspA response. In concept, GadXY could do so by improving the insertase function of the remaining YidC under depletion conditions or by stimulating YidC-independent alternative insertion mechanisms. To examine the influence of pGadXY presence on the insertase function of YidC, we monitored the processing of CyoA, which depends solely on YidC for its initial insertion in the inner membrane (4, 9, 43). The membrane insertion of the lipoprotein CyoA can conveniently be monitored by analyzing the cleavage of its signal peptide by the lipoprotein-specific signal peptidase II. A plasmid expressing HA-tagged CyoA (44) was transformed into FTL10 containing either pGadXY or pBR322. Cells were grown in the absence or presence of l-arabinose, and CyoA-HA processing was followed by Western blotting using anti-HA antibodies. Consistently with previous data, the processing of CyoA-HA was impaired upon YidC depletion (Fig. 3B). The processing of CyoA-HA remained hampered in the presence of pGadXY under these steady-state conditions, suggesting that the insertase function of YidC is not improved by GadXY or that alternative mechanisms cannot complement this function. In addition, the absorption spectra of reduced versus oxidized eIM fractions that were prepared from WT, YidC-depleted, and pGadXY-containing cells demonstrated that the loss of cytochrome bo3 oxidase (46) was not rescued by the presence of pGadXY (results not shown).
Only a few natural substrates are known that use YidC as an independent insertase. If CyoA and Foc were the only substrates of YidC that are critical for maintaining the PMF and the viability of the cell under the experimental conditions, the overexpression of both proteins might restore the PMF and bypass the requirement of YidC for growth. However, the overexpression of either CyoA or Foc in FTL10 cells depleted of YidC did not result in the recovery of growth (data not shown). Apparently, YidC's lethality was not bypassed by CyoA and Foc overexpression and spontaneous insertion in the absence of YidC.
Overexpression of GadXY but not GadBC complements the growth defect and PspA response in YidC-depleted cells.
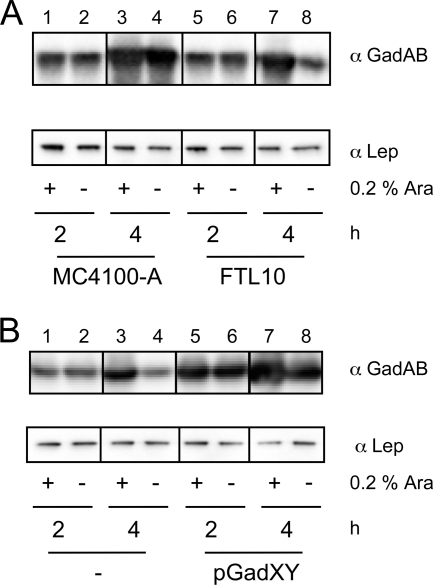
As discussed above, neither improved YidC stability nor improved YidC insertase activity are likely explanations for the positive effect of GadXY on YidC-depleted cells. We therefore considered the explanation that the overproduction of GadXY mitigated indirect effects of YidC depletion that might be related to acidic stress. To examine this, we first studied the effects of YidC depletion on gadY, gadX, and gadBC transcription by the analysis of reporter gene activity using lacZ transcriptional fusions to the promoters of gadY, gadX, and gadBC. The β-galactosidase activities of all fusions were decreased by ∼50% in YidC-depleted cells compared to those of control cells (Table 3). We did not observe significant differences in gadX and gadC mRNA levels in the qPCR analysis (Table 2). However, Western blot analysis revealed that GadAB levels were increased in FTL10, in the presence of l-arabinose, and in its parental strain MC4100-A (Fig. 4A, lanes 3, 4, and 7) (6). In contrast, FTL10 cells grown for 4 h in the absence of l-arabinose to deplete YidC showed decreased GadAB levels, in line with data from β-galactosidase assays (Fig. 4A, lane 8, and Table 3).
TABLE 3.
β-Galactosidase activity (in Miller units) as directed by gad gene promoters
| Promotera | FTL10 + Ara | FTL10 − Ara |
|---|---|---|
| PgadY (−78/+65) | 1,379 | 767 (56%) |
| PgadX (−222/+121) | 1,899 | 1,150 (60%) |
| PgadBC (−244/+77) | 1,767 | 910 (52%) |
Numbering refers to the transcriptional start site of each gene and indicates the length of the DNA regions used to generate the lacZ fusions.
FIG. 4.
Upregulation of GadAB expression upon GadXY overexpression. Cells were grown in LB medium, and samples were taken after 2 and 4 h and analyzed by SDS-PAGE and immunoblotting using antisera against GadAB and Lep. (A) FTL10 and its parental strain MC4100-A; (B) FTL10 containing pGadXY or control vector pBR322.
We further analyzed the effect of GadXY overproduction on GadAB protein levels (Fig. 4B). As expected, the levels of GadAB were higher in cells with pGadXY after 2 h of growth than the basal levels present in FTL10 cells harboring the control plasmid pBR322 (compare lanes 5 and 6 to 1 and 2). After 4 h of growth, the levels of GadAB still were higher in cells harboring pGadXY than in cells with the control plasmid (compare lanes 7 and 8 to 3 and 4), even in the absence of YidC (lane 8). Additional analysis of gadC and gadX mRNA levels by qPCR showed their increase in FTL10 with pGadXY (Table 2). Taken together, these data indicate that in YidC-depleted cells the Gad response is not switched on, but it can be reestablished by the overexpression of GadXY. The reason for the reduced GadB levels remains unexplained, but it is of interest that GadC levels were observed to be 20-fold lower in YidC-depleted inverted membrane vesicles (IMVs), hinting at a direct involvement of YidC in GadC biogenesis (J.-W. de Gier, personal communication).
Assuming that the depletion of YidC might lead to the acidification of the cells, the inability to mount a Gad response and the low levels of GadC could be an explanation for the growth defect upon YidC depletion. Following this scenario, the overexpression of GadBC would alleviate this effect. Therefore, we introduced a pEH3 plasmid containing the gadBC operon under the control of the lac promoter in FTL10. However, the overexpression of GadBC did not influence the growth of the cells or the induction of the PspA response when YidC was depleted (data not shown). Hence, other as-yet unidentified factors of the poorly characterized GadXY regulon likely are important causes of the observed effects. This also would explain why the gadBC operon was not among the plasmids selected in our genetic screen.
To rule out the hypothesis that YidC-depleted cells were experiencing acid stress, we measured the internal pH of WT, YidC-depleted, and pGadXY cells using methods previously described (34). The internal pH of wild-type cells grown in the presence of 0.2% l-arabinose was slightly acidic (pH 6.7 ± 0.11), while cells depleted for YidC for 4 h had an internal pH of 8.1 ± 0.57. In the presence of pGadXY and YidC depletion, the internal pH was 7.4 ± 0.14. The internal pH values measured here would not have a suppressive effect on the growth of E. coli and are within the range of values reported for cells growing at neutral to mildly acidic pH. During YidC depletion, we noted no significant change in the external pH of the growth medium (i.e., pH ranged from 6.4 to 7.2), supporting the proposal that YidC-depleted cells were not experiencing either external or internal pH stress.
YidC-depleted cells shift to fermentation.
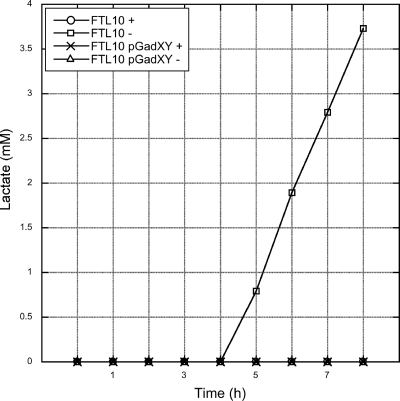
When a low YidC level in cells is accompanied by a decrease in cytochrome bo3 oxidase and the F1Fo ATPase, the capacity of the cell to reduce O2 would be rapidly impaired. This could affect the ratio of ubiquinone and ubiquinol (2), resulting in a switch to fermentation as a last resort to generate ATP and to oxidize NADH. In aerobic batch conditions with glucose as the sole carbon source, E. coli cells that lack respiratory capacity secrete lactate into the medium (1). To study this phenomenon, we determined the levels of lactate using HPLC in spent M9 medium, supplemented with 0.2% glucose, from YidC-depleted and control FTL10 cells. We found that lactate was present in medium from cells that were grown for prolonged times under YidC depletion conditions (Fig. 5). Medium from control cells did not contain lactate. Strikingly, FTL10 cells harboring pGadXY did not secrete lactate independently of the presence of YidC, indicating that the presence of pGadXY prevented the shift to fermentation, possibly by increasing the nonrespiratory NADH-oxidizing capacity.
FIG. 5.
GadXY overexpression results in the cessation of lactate secretion in spent medium. Cells were grown in M9 medium in the presence (+) or absence (−) of l-arabinose. Samples were taken every hour. Cells were removed by centrifugation from the medium before the lactate levels in the medium were determined by HPLC.
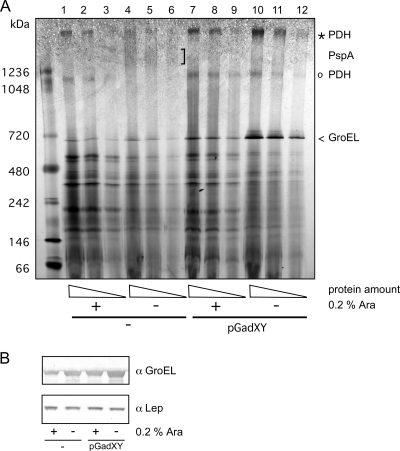
The protein profiles of eIMs isolated from FTL10 cells grown with l-arabinose were compared to those of cells grown without l-arabinose using BN-PAGE. The gel showed clearly reduced amounts of a high-molecular-weight protein complex (Fig. 6A, indicated by an open circle) in YidC-depleted cells. To identify the proteins in this complex, it was excised from the gel and subjected to peptide mass fingerprinting. AceE, AceF, and LpdA were unambiguously identified in the complex. They form the large (4.5-MDa) multienzyme pyruvate dehydrogenase complex (PDH) that catalyzes the oxidative decarboxylation of pyruvate with the concomitant formation of acetyl-coenzyme A (CoA). Importantly, the complex is known to be downregulated upon the accumulation of NADH, again indicating that the expression of GadXY results in higher nonrespiratory NADH-oxidizing capacity (7). Another high-molecular-weight complex (Fig. 6A, indicated by an asterisk) also was identified as PDH by peptide mass fingerprinting and appeared to be reduced in eIM isolated from YidC-depleted cells, but to a lesser degree than the smaller PDH (sub)complex. In contrast, upon the overexpression of GadXY, the PDH complexes remained detectable upon YidC depletion. Taken together, these data suggest that as a result of YidC depletion, cells were unable to reoxidize the NADH pool and, therefore, showed a switch to fermentation, as evidenced by the secretion of lactate. However, the cells that overexpressed GadXY continued to form PDH complexes, indicative of NADH oxidation, and, therefore, did not show the concomitant lactate fermentation.
FIG. 6.
Effect of GadXY overproduction on the levels of pyruvate dehydrogenase complex and GroEL in the IM. (A) BN-PAGE analysis of eIMs isolated from FTL10 cells grown with or without l-arabinose in the presence or absence of pGadXY. Comparable protein amounts were loaded on gels. The indicated bands were identified by MALDI-TOF-TOF mass spectrometry. (B) Cell lysates were analyzed by SDS-PAGE followed by Western blotting using antisera against GroEL and Lep.
GadXY overexpression leads to a decreased level of PspA complexes and increased levels of GroEL complexes.
Two other observations were made from the BN-PAGE analysis. First, in a high-molecular-mass band of >1.2 MDa, which was detected in the YidC-depleted eIMs, PspA was identified as the main constituent of that complex by peptide mass fingerprinting. Consistently, purified PspA has been shown to oligomerize in ∼1-MDa complexes (12). The diffuse, high-molecular-weight PspA complex was absent from the eIM fraction obtained from YidC-depleted cells overexpressing GadXY, in accordance with data from the Western blotting of whole-cell lysates (Fig. 3A). Second, a protein complex around 0.7 MDa was strongly increased in intensity in eIMs from the YidC-depleted cells upon GadXY overexpression. It was identified as the general heat shock chaperone GroEL by peptide mass fingerprinting. GroEL is a homotetradecamer of 57-kDa subunits that are arranged as two heptameric rings stacked back to back (20). Western blot analysis showed that YidC-depleted cells had slightly upregulated GroEL content, while GadXY-overexpressing cells had strongly increased levels of GroEL compared to the unaltered level of the YidC-independent IMP leader peptidase (Lep) (Fig. 6B). Although GroEL is not known to be part of the GadX regulon, it has been reported that GadY overexpression results in the upregulation of GroEL (25). However, the sole overexpression of GroEL and GroES from an inducible expression plasmid appeared insufficient to restore growth upon YidC depletion (results not shown).
DISCUSSION
YidC is an essential E. coli IMP that plays an important but poorly defined role in the biogenesis of IMPs (18, 52). So far, targeted approaches have revealed a plethora of YidC functions that seem almost too varied to be combined in one protein. Here, we have screened a random genetic library to identify factors that improve the survival of YidC-depleted cells and may reveal why YidC is an essential protein. The gadX and gadY genes were selected multiple times in the screen and were found to partly compensate for the growth defect and the PspA response that accompanies YidC depletion (46). The genes encode regulatory proteins of the major acid resistance system in E. coli (25, 36, 38), hinting at an unexpected link between YidC and the acid stress response.
The gadX gene encodes a transcriptional regulator belonging to the AraC family, while gadY, encoding a small ncRNA, stabilizes the gadX transcript (25, 36). GadX regulates transcription of the glutamate decarboxylase isozymes GadA and GadB, which convert glutamate to GABA while consuming a proton from the cytoplasm (53). The antiporter GadC expels the decarboxylated products in exchange for glutamate. The resulting net export of protons is thought to increase the internal pH to sustain membrane integrity, normal physiological processes, and protein stability. The control of this acid resistance system is reported to involve at least 15 regulatory factors whose functions respond to the environmental pH, growth phase, medium composition, and oxygen levels. The regulators include the AraC-like transcription factors GadX, GadW, and YdeO; the LuxR-like transcriptional regulator GadE; the two-component regulatory system EvgAS; the alternative sigma factor RpoS; TorRS;Crp; H-NS; Dps; TrmE; and three small ncRNAs, GadY, DsrA, and GcvB (11, 15, 16, 19, 21, 29, 40). Why so many regulators and control circuits are needed is not clear, but it probably reflects a need to connect the induction of this system to many facets of cell physiology. The complexity is underscored by the fact that the regulon of GadX contains many genes that have no direct link with acid resistance (15). In addition, GadX enhances multidrug resistance by activating the mdtEF efflux genes (24). Notably, the Gad response is not exclusively a consequence of acidic stress but also occurs upon salt stress and in the stationary growth phase (6).
Considering this complexity, it is intriguing that gadXY was repeatedly selected in our screen. Other regulators of the acid stress response, such as gadE or gadW, were not selected, although our screen may not have been exhaustive. Furthermore, members of the GadX regulon, like GadBC, also were not selected, nor were other genes encoding amino acid decarboxylases and acid stress regulators (data not shown). Apparently, only the overexpression of GadXY leads to a response of effector proteins that together have a beneficial effect on YidC-depleted cells. Consistently, while we demonstrated the upregulation of the main resistance effectors in response to acid stress, GadBC alone could not compensate for the growth defect upon YidC depletion.
We ruled out that the rescuing effect of the pGadXY plasmid was due to a direct influence on YidC expression or stability. Furthermore, GadXY overexpression did not improve the processing of the YidC-dependent IMP CyoA, nor were the levels of the cytochrome bo3 oxidase restored, suggesting that the insertase function of YidC was not restored as a result of GadXY overexpression. Although we cannot exclude that the Sec-associated YidC functions are improved by GadXY, these functions are less important to sustain cell growth (41, 45). Therefore, it is more likely that GadXY overexpression alleviates negative secondary effects of the absence of the essential YidC protein.
Previous studies have shown that YidC-depleted cells suffer from a reduced PMF, which triggers a strong PspA response. This is probably a consequence of the fact that YidC is strictly required for the membrane insertion of CyoA and Foc (4, 9, 43, 46), which are critical subunits of the cytochrome bo3 oxidase and F1Fo ATPase, respectively. The absence of these proteins may result in the accumulation of NADH. Indicative for the accumulation of NADH is the strongly regulated multienzyme PDH complex that connects glycolysis with the tricarboxylic acid cycle, which was much less abundant in YidC-depleted cells. This limits the capacity of the cell to convert pyruvate to acetyl-CoA, and therefore the only remaining alternative to generate ATP, although less efficient, is a shift to lactic acid fermentation. Indeed, we detected the secretion of lactate in YidC-depleted cells, indicating that such a shift had occurred. In contrast, cells that overproduced GadXY from the pGadXY plasmid retained higher levels of the PDH complex and did not secrete lactate. However, it remains unclear how the switch to fermentation is prevented in the GadXY-overexpressing cells.
An explanation of the beneficial effect of GadXY on YidC-depleted cells might lie in a compensation of intracellular acidification as a consequence of fermentation, which produces weak acids such as acetate and lactate. Under normal circumstances, the F1Fo ATPase would counter intracellular acidification by pumping out protons at the expense of ATP. However, in YidC-depleted cells the F1Fo ATPase is compromised too. Intracellular pH measurements indicated that YidC-depleted cells were not suffering from internal acid stress. In contrast, the intracellular pH was more alkaline than that of the WT and returned to neutral values in the presence pGadXY. Consistently, the sole overproduction of GadBC, the effectors that are upregulated by GadXY to bring about a net export of protons, could not compensate for the growth reduction upon the depletion of YidC, supporting the hypothesis that the inhibition of growth was not related to cytoplasmic acidification.
It also has been proposed that the exchange of GABA for glutamate contributes to a functional PMF (14, 34), explaining the reduced PspA response in the YidC-depleted but GadXY-overexpressing cells. However, as mentioned, the upregulation of the glutamate-dependent acid response system is unlikely to be the only reason for the improved fitness of YidC-depleted cells. Other genes in the GadX regulon probably are involved as well. Interestingly, genes encoding chaperones and proteases (ycgG, yehA, yhcA, and lon) also are induced by GadX (15). In addition, the heat shock chaperone GroEL copurified with membranes and was upregulated upon GadXY overexpression (Fig. 6). GroEL is known to act on a wide range of unfolded or partially folded proteins, and with the assistance of GroES, it helps them to reach their fully folded and active state (20). GroEL levels are increased upon YidC depletion, and it has been shown that part of GroEL is redistributed to the membrane under these conditions (48). When YidC levels are low, GroEL might be recruited to the IM through interaction with IMPs that are targeted but not yet inserted or by binding to unfolded domains of improperly assembled IMPs that accumulate near the membrane. GroEL may keep such unfolded proteins in an insertion-competent state while they anticipate their insertion into the membrane by the limited amount of YidC available. In addition, GroEL has been implicated in the targeting of proteins to the Sec translocon based on its affinity for SecA (3). The further upregulation of GroEL expression by GadY ncRNA (either directly or indirectly via GadX) (25) might increase the fitness of YidC-depleted cells by preventing IMPs from aggregating near the membrane. This eventually could allow them to insert into the membrane, albeit at a lower efficiency. Overall, we have revealed here, for the first time, that the lethality of YidC depletion may involve the acid stress response, the response to a drop in PMF, and protein quality control. The intricate relationship between YidC and these processes will be the subject of further study.
Acknowledgments
We thank W. Bitter, J. W. de Gier, and K. Krab for helpful discussions and the critical reading of the manuscript, C. Zonneveld and M. de Boer for help with the statistical analysis, G. Koningstein for assistance with the mass spectrometry, and M. Wagner for assistance with the cytochrome absorption measurements. Pierre Genevaux is thanked for discussions and acknowledged for the generous supply of the genomic library.
Footnotes
Published ahead of print on 7 January 2011.
REFERENCES
- 1.Bekker, M., S. de Vries, A. Ter Beek, K. J. Hellingwerf, and M. J. de Mattos. 2009. Respiration of Escherichia coli can be fully uncoupled via the nonelectrogenic terminal cytochrome bd-II oxidase. J. Bacteriol. 191:5510-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bekker, M., et al. 2007. Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology 153:1974-1980. [DOI] [PubMed] [Google Scholar]
- 3.Bochkareva, E. S., M. E. Solovieva, and A. S. Girshovich. 1998. Targeting of GroEL to SecA on the cytoplasmic membrane of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 95:478-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Celebi, N., L. Yi, S. J. Facey, A. Kuhn, and R. E. Dalbey. 2006. Membrane biogenesis of subunit II of cytochrome bo oxidase: contrasting requirements for insertion of N-terminal and C-terminal domains. J. Mol. Biol. 357:1428-1436. [DOI] [PubMed] [Google Scholar]
- 5.Darwin, A. J. 2005. The phage-shock-protein response. Mol. Microbiol. 57:621-628. [DOI] [PubMed] [Google Scholar]
- 6.De Biase, D., A. Tramonti, F. Bossa, and P. Visca. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32:1198-1211. [DOI] [PubMed] [Google Scholar]
- 7.de Kok, A., A. F. Hengeveld, A. Martin, and A. H. Westphal. 1998. The pyruvate dehydrogenase multi-enzyme complex from Gram-negative bacteria. Biochim. Biophys. Acta 1385:353-366. [DOI] [PubMed] [Google Scholar]
- 8.De Vrije, T., J. Tommassen, and B. De Kruijff. 1987. Optimal posttranslational translocation of the precursor of PhoE protein across Escherichia coli membrane vesicles requires both ATP and the protonmotive force. Biochim. Biophys. Acta 900:63-72. [DOI] [PubMed] [Google Scholar]
- 9.du Plessis, D. J., N. Nouwen, and A. J. Driessen. 2006. Subunit a of cytochrome o oxidase requires both YidC and SecYEG for membrane insertion. J. Biol. Chem. 281:12248-12252. [DOI] [PubMed] [Google Scholar]
- 10.Giangrossi, M., S. Zattoni, A. Tramonti, D. De Biase, and M. Falconi. 2005. Antagonistic role of H-NS and GadX in the regulation of the glutamate decarboxylase-dependent acid resistance system in Escherichia coli. J. Biol. Chem. 280:21498-21505. [DOI] [PubMed] [Google Scholar]
- 11.Gong, S., Z. Ma, and J. W. Foster. 2004. The Era-like GTPase TrmE conditionally activates gadE and glutamate-dependent acid resistance in Escherichia coli. Mol. Microbiol. 54:948-961. [DOI] [PubMed] [Google Scholar]
- 12.Hankamer, B. D., S. L. Elderkin, M. Buck, and J. Nield. 2004. Organization of the AAA(+) adaptor protein PspA is an oligomeric ring. J. Biol. Chem. 279:8862-8866. [DOI] [PubMed] [Google Scholar]
- 13.Hatzixanthis, K., T. Palmer, and F. Sargent. 2003. A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol. Microbiol. 49:1377-1390. [DOI] [PubMed] [Google Scholar]
- 14.Higuchi, T., H. Hayashi, and K. Abe. 1997. Exchange of glutamate and gamma-aminobutyrate in a Lactobacillus strain. J. Bacteriol. 179:3362-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hommais, F., et al. 2004. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology 150:61-72. [DOI] [PubMed] [Google Scholar]
- 16.Jin, Y., R. M. Watt, A. Danchin, and J. D. Huang. 2009. Small noncoding RNA GcvB is a novel regulator of acid resistance in Escherichia coli. BMC Genomics 10:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleerebezem, M., W. Crielaard, and J. Tommassen. 1996. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J. 15:162-171. [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn, A. 2009. From the Sec complex to the membrane insertase YidC. Biol. Chem. 390:701-706. [DOI] [PubMed] [Google Scholar]
- 19.Lease, R. A., and S. A. Woodson. 2004. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 344:1211-1223. [DOI] [PubMed] [Google Scholar]
- 20.Lund, P. A. 2009. Multiple chaperonins in bacteria-why so many? FEMS Microbiol. Rev. 33:785-800. [DOI] [PubMed] [Google Scholar]
- 21.Masuda, N., and G. M. Church. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48:699-712. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 23.Nagamori, S., I. N. Smirnova, and H. R. Kaback. 2004. Role of YidC in folding of polytopic membrane proteins. J. Cell Biol. 165:53-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishino, K., Y. Senda, and A. Yamaguchi. 2008. The AraC-family regulator GadX enhances multidrug resistance in Escherichia coli by activating expression of mdtEF multidrug efflux genes. J. Infect. Chemother. 14:23-29. [DOI] [PubMed] [Google Scholar]
- 25.Opdyke, J. A., J. G. Kang, and G. Storz. 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186:6698-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pop, O. I., et al. 2009. YidC is required for the assembly of the MscL homopentameric pore. FEBS J. 276:4891-4899. [DOI] [PubMed] [Google Scholar]
- 27.Price, C. E., and A. J. Driessen. 2010. Conserved negative charges in the transmembrane segments of subunit K of the NADH:ubiquinone oxidoreductase determine its dependence on YidC for membrane insertion. J. Biol. Chem. 285:3575-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price, C. E., and A. J. Driessen. 2008. YidC is involved in the biogenesis of anaerobic respiratory complexes in the inner membrane of Escherichia coli. J. Biol. Chem. 283:26921-26927. [DOI] [PubMed] [Google Scholar]
- 29.Richard, H., and J. W. Foster. 2004. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J. Bacteriol. 186:6032-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samuelson, J. C., et al. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637-641. [DOI] [PubMed] [Google Scholar]
- 31.Samuelson, J. C., et al. 2001. Function of YidC for the insertion of M13 procoat protein in Escherichia coli: translocation of mutants that show differences in their membrane potential dependence and Sec requirement. J. Biol. Chem. 276:34847-34852. [DOI] [PubMed] [Google Scholar]
- 32.Sauri, A., et al. 2009. The Bam (Omp85) complex is involved in secretion of the autotransporter haemoglobin protease. Microbiology 155:3982-3991. [DOI] [PubMed] [Google Scholar]
- 33.Scotti, P. A., et al. 2000. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19:542-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shepherd, M., G. Sanguinetti, G. M. Cook, and R. K. Poole. 2010. Compensations for diminished terminal oxidase activity in Escherichia coli: cytochrome bd-II-mediated respiration and glutamate metabolism. J. Biol. Chem. 285:18464-18472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimohata, N., S. Nagamori, Y. Akiyama, H. R. Kaback, and K. Ito. 2007. SecY alterations that impair membrane protein folding and generate a membrane stress. J. Cell Biol. 176:307-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin, S., et al. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 37.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 38.Tramonti, A., M. De Canio, and D. De Biase. 2008. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol. Microbiol. 70:965-982. [DOI] [PubMed] [Google Scholar]
- 39.Tramonti, A., P. Visca, M. De Canio, M. Falconi, and D. De Biase. 2002. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tucker, D. L., et al. 2003. Genes of the GadX-GadW regulon in Escherichia coli. J. Bacteriol. 185:3190-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urbanus, M. L., et al. 2001. Sec-dependent membrane protein insertion: sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2:524-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Bloois, E., et al. 2008. Detection of cross-links between FtsH, YidC, HflK/C suggests a linked role for these proteins in quality control upon insertion of bacterial inner membrane proteins. FEBS Lett. 582:1419-1424. [DOI] [PubMed] [Google Scholar]
- 43.van Bloois, E., G. J. Haan, J. W. de Gier, B. Oudega, and J. Luirink. 2006. Distinct requirements for translocation of the N-tail and C-tail of the Escherichia coli inner membrane protein CyoA. J. Biol. Chem. 281:10002-10009. [DOI] [PubMed] [Google Scholar]
- 44.van Bloois, E., G. Koningstein, H. Bauerschmitt, J. M. Herrmann, and J. Luirink. 2007. Saccharomyces cerevisiae Cox18 complements the essential Sec-independent function of Escherichia coli YidC. FEBS J. 274:5704-5713. [DOI] [PubMed] [Google Scholar]
- 45.van der Laan, M., N. Nouwen, and A. J. Driessen. 2004. SecYEG proteoliposomes catalyze the Deltaphi-dependent membrane insertion of FtsQ. J. Biol. Chem. 279:1659-1664. [DOI] [PubMed] [Google Scholar]
- 46.van der Laan, M., et al. 2003. A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc. Natl. Acad. Sci. U. S. A. 100:5801-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner, S., et al. 2008. Biogenesis of MalF and the MalFGK(2) maltose transport complex in Escherichia coli requires YidC. J. Biol. Chem. 283:17881-17890. [DOI] [PubMed] [Google Scholar]
- 48.Wang, P., A. Kuhn, and R. E. Dalbey. 2010. Global change of gene expression and cell physiology in YidC-depleted Escherichia coli. J. Bacteriol. 192:2193-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watson, N. 1988. A new revision of the sequence of plasmid pBR322. Gene 70:399-403. [DOI] [PubMed] [Google Scholar]
- 50.Weiner, L., and P. Model. 1994. Role of an Escherichia coli stress-response operon in stationary-phase survival. Proc. Natl. Acad. Sci. U. S. A. 91:2191-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie, K., and R. E. Dalbey. 2008. Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat. Rev. Microbiol. 6:234-244. [DOI] [PubMed] [Google Scholar]
- 52.Yuan, J., J. C. Zweers, J. M. van Dijl, and R. E. Dalbey. 2010. Protein transport across and into cell membranes in bacteria and archaea. Cell Mol. Life Sci. 67:179-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao, B., and W. A. Houry. 2010. Acid stress response in enteropathogenic gammaproteobacteria: an aptitude for survival. Biochem. Cell Biol. 88:301-314. [DOI] [PubMed] [Google Scholar]