Abstract
Bdellovibrio bacteriovorus HD100 is an obligate predatory bacterium that attacks and invades Gram-negative bacteria. The predator requires living bacteria to survive as growth and replication take place inside the bacterial prey. It is possible to isolate mutants that grow and replicate outside prey bacteria. Such mutants are designated host or prey independent, and their nutritional requirements vary. Some mutants are saprophytic and require prey extracts for extracellular growth, whereas other mutants grow axenically, which denotes the formation of colonies on complete medium in the absence of any prey components. The initial events leading to prey-independent growth are still under debate, and several genes may be involved. We selected new mutants by three different methods: spontaneous mutation, transposon mutagenesis, and targeted gene knockout. By all approaches we isolated mutants of the hit (host interaction) locus. As the relevance of this locus for the development of prey independence has been questioned, we performed whole-genome sequencing of five prey-independent mutants. Three mutants were saprophytic, and two mutants could grow axenically. Whole-genome analysis revealed that the mutation of a small open reading frame of the hit locus is sufficient for the conversion from predatory to saprophytic growth. Complementation experiments were performed by introduction of a plasmid carrying the wild-type hit gene into saprophytic mutants, and predatory growth could be restored. Whole-genome sequencing of two axenic mutants demonstrated that in addition to the hit mutation the colony formation on complete medium was shown to be influenced by the mutations of two genes involved in RNA processing. Complementation experiments with a wild-type gene encoding an RNA helicase, RhlB, abolished the ability to form colonies on complete medium, indicating that stability of RNA influences axenic growth.
Bdellovibrio bacteriovorus HD100 is an obligate predatory bacterium which requires live Gram-negative bacteria as prey for growth and replication (26, 36). The predatory life cycle is characterized by two phases: an intracellular growth and replication phase within the invaded bacteria and an extracellular phase in which free-living predators hunt putative prey. In the latter phase, termed attack phase, the predators attach nonspecifically to surfaces. Attachment also occurs to abiotic surfaces or biological substrates; however, the attachment to a potential prey bacterium causes an as-yet-unknown signal which enables the attacker to invade and begin the intracellular growth phase. In this phase the invader grows out into a filament which terminates its elongation depending on prey factors. It then divides into progeny cells which in turn hunt for new prey.
The interaction of predator and prey is crucial for the predatory life cycle, and a number of experiments were performed to identify potential prey factors involved in this process. When concentrated bacterial cell extracts of prey bacteria were added to media, some wild-type bdellovibrios could be tricked into a growth and replication phase (13, 16). However, none of these experiments established conclusive evidence of which growth factors or signal molecules were responsible. It was suggested that at least two kinds of prey signals are necessary for the completion of the life cycle. The first signal triggers the attack-phase bacteria to switch to the growth phase, and the second signal initiates the DNA synthesis (15).
From the beginning of bdellovibrio research, spontaneous mutants could be isolated from every predatory strain that could grow and replicate in the absence of prey bacteria. These mutants were termed host independent (HI) and arose with frequencies of 10−6 and 10−7 (6, 32). Such mutants—now mostly designated prey independent (PI)—are often pleiomorphic and consist of vibrio-shaped and spiral-shaped cells which can be morphologically extended in length. However, prey-independent mutants are difficult to interpret, as their nutritional requirements vary (38). Prey-independent mutants are mostly initially facultative predacious retaining the ability to invade living bacteria. However, the plaques formed by prey-independent strains are smaller and more turbid than those of the wild type. Some mutants can be cultivated only on prey cell extracts or heat-killed cells and do not form single isolated colonies when plated on complete medium. However, on complete medium a small number of colonies can arise which are able to grow in the absence of bacterial prey or prey extract.
It was suggested to distinguish between type I and type II host-independent (prey-independent) mutants (38). Type I mutants form isolated colonies only on nutrient agar plates supplemented with bacterial cell extracts, while type II mutants can grow on standard complete medium. When prey-independent mutants are selected, the majority of the mutants are type I mutants and roughly 1% of all PI mutants belong to type II. The isolation frequency (10−6 to 10−7) for type I mutants suggests that they arise through a single mutation event. Type II colonies are isolated with a frequency of 10−8 to 10−9, which seems to be too high a frequency for the occurrence of two independent mutation events (38).
The growth on complete medium without the addition of bacterial cell extract is termed axenic growth; thus, type II mutants are designated axenic mutants. Type I mutants which require cellular prey extracts for growth are designated saprophytic. In the first genetic studies with B. bacteriovorus a conjugation procedure was developed for mutational analysis and a genomic region was identified that seemed to be involved in the development of host independence (6, 7). In this pioneering work, it was shown that a short open reading frame (ORF) within a small genetic region, the host-interaction locus (hit), was mutated in different prey-independent strains, leading to major changes in the resulting gene product. The hit locus is probably part of a genetic cluster responsible for type IV pilus formation and adherence that might play an important role in the attachment and invasion process (26, 31). All prey-independent mutants analyzed in the original study (7) belonged to the type II mutants and had mutations in the hit ORF. Reintroduction of the wild-type hit locus into the mutants was performed, resulting in enhanced plaque formation on living bacteria. The ability to grow axenically was only slightly affected (7).
The involvement of the hit locus in the development of prey-independent growth was questioned later, as prey-independent mutants with an unaltered hit locus were reported (4, 23). Although some recent studies were published on the role of genes involved in the interaction of predator and prey, e.g., motility genes, chemotaxis genes, and pilus genes (12, 20, 21, 39), the function of the hit locus has remained obscure. However, in a recent work a transcriptional analysis of the expressed genes during the predatory cycle was published and compared to the transcription of genes of prey-independent mutants. The mutants used in that study were carefully chosen and contained mutations in the hit locus (22). A recent study also showed that 89% of prey-independent mutants of B. bacteriovorus 109J have an altered hit locus (40).
The processes leading to prey independency remain one of the main questions of Bdellovibrio research. As the relevance of the hit locus for the development of the saprophytic lifestyle has remained unclear, we decided to generate new prey-independent mutants and analyze these by performing whole-genome sequencing. Additionally, we performed genetic experiments such as transposon mutagenesis, targeted knockout of genes, and complementation experiments to obtain further information of the genetic changes which take place during the development from predatory to saprophytic and axenic growth.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani (LB) medium (liquid or agar) overnight at 37°C. The experiments were carried out with B. bacteriovorus HD100 and the derivative strain B. bacteriovorus HD100-S1, which is a spontaneous streptomycin-resistant and prey-dependent strain. The streptomycin resistance is mediated by a mutation of the gene Bd2981 (rpsL) altering one amino acid in the ribosomal protein S12 (34). The two predatory strains were cultivated either in liquid cultures with E. coli prey or on double-layer agar plates. In the latter case, prey bacteria were resuspended within the top layer (30). Stationary-phase prey bacteria were harvested by centrifugation and dissolved in the same volume of Bdellovibrio buffer (3 mM ammonium acetate, 3 mM CaCl2, 3 mM MgCl2, pH 7.5). For liquid cultures this suspension was directly inoculated with B. bacteriovorus and shaken at 28°C until complete lysis of the prey (1 to 3 days).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| B. bacteriovorus | ||
| HD100 | Wild type | DSM50701 |
| M1 | Saprophytic HD100 mutant | This study |
| M1.1 | Axenic derivative of M1 | This study |
| M2 | Saprophytic HD100 mutant | This study |
| M2.1 | Axenic derivative of M2 | This study |
| M3 | Saprophytic HD100 mutant | This study |
| HD100-S1 | Streptomycin-resistant derivative of HD100 | BfRa,b |
| M4/Strep | Saprophytic HD100-S1 mutant | This study |
| M5/Strep | Saprophytic HD100-S1 mutant | This study |
| M6/Strep | Saprophytic HD100-S1 mutant | This study |
| M7/Strep | Saprophytic HD100-S1 mutant | This study |
| M8/Strep | Saprophytic HD100-S1 mutant | This study |
| M9/Strep | Saprophytic HD100-S1 mutant | This study |
| M10/Strep | Saprophytic HD100-S1 mutant | This study |
| M11/Strep | Saprophytic HD100-S1 mutant | This study |
| M11.1 | Axenic derivative of M11/Strep | This study |
| M11.2 | Axenic derivative of M11/Strep | This study |
| KO-1 | Cmr, hit knockout, saprophytic HD100 mutant | This study |
| E. coli | ||
| K-12 | DSM423 | |
| JC3272 | K-12, streptomycin resistant | 22a |
| DH5α pBR329 | Apr Tcr Cmr | BfRa |
| K-12 Gene Hogs | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG fhuA::IS2 | Invitrogen, Darmstadt, Germany |
| CB3537 | O:48,H21 | BfRa |
| S17-1λ | recA thiO hsdR−M− integrated plasmid RP4-2-Kn::Tn7/TC::M | DSM9526 |
| Other | ||
| Yersinia enterocolitica | DSM13030 | |
| Yersinia pseudotuberculosis | ATCC 29833 | |
| Salmonella enterica serovar Braenderup | BfRa | |
| Vibrio cholerae | ATCC 14733 | |
| Proteus mirabilis | DSM50903 | |
| Pseudomonas aeruginosa | ATCC 27853 | |
| Citrobacter freundii CB1614 | BfRa | |
| Serratia marcescens | BfRa | |
| Plasmids | ||
| pSUP404.2 | p15A replicon, RSF1010 replicon, Kmr Cmr, oriT | 25a |
| pNR16 | hit knockout construct in pK18mob/sacB | This study |
| pNR22 | Insertion of HD100_Bd0108 (hit) into pSUP404.2 via the restriction sites NheI and HindIII | This study |
| pNR29 | Insertion of Bd3461 (HD100) into pSUP404.2 via the restriction sites NheI and HindIII | This study |
| pK18mobsacB | lacZ, ColE1 origin of replication, oriT sacB Kmr | 29 |
| pLITMUS38 | M13 origin, pUC19 origin, lacZ Apr | New England Biolabs, Ipswich, MA |
| pBR329 | pMB1 replicon; Apr Tcr Cmr | 7a |
BfR strain collection.
Spontaneous mutant isolated by Dominik Schwudke (personal communication).
Saprophytic strains derived from B. bacteriovorus HD100 are able to grow on heat-killed prey bacteria. For this purpose, stationary-phase prey bacteria were harvested by centrifugation, dissolved in the same volume of Bdellovibrio buffer, and autoclaved for 20 min at 121°C. The axenic B. bacteriovorus mutants M1.1, M2.1, and M11.1 and M11.2 are derivatives of the saprophytic M1, M2, and M11/Strep, respectively. Axenic strains are able to grow in standard bacteriological medium (PYE consisting of 10.0 g liter−1 peptone, 3.0 g liter−1 yeast extract, pH 6.8; liquid cultures or agar; 12 g liter−1). All bdellovibrio strains were stored at −80°C in either prey suspension or PYE supplemented with 30% glycerol.
Isolation of saprophytic and axenic B. bacteriovorus mutant strains.
For the isolation of saprophytic mutants, the wild-type strain B. bacteriovorus HD100 and its streptomycin-resistant derivative HD100-S1 were used. Saprophytic mutants were generated either according to the protocol described by Seidler and Starr (32) or in compliance with the protocol from Shilo and Bruff (35). While Seidler and Starr used a streptomycin-resistant strain for the isolation of saprophytic mutants, Shilo and Bruff described the separation of the small bdellovibrios from their larger hosts by filtration through a Millipore filter (0.45-μm pore size). All of the saprophytic mutants were grown on heat-killed E. coli K-12 cells with (in the case of HD100-S1 derivatives, 25 μg ml−1) or without streptomycin (derivatives of HD100). After 3 to 4 days of incubation at 28°C, small white colonies were visible. Each colony was surrounded by a zone of lysis in the lawn of dead prey cells. These initial mutants were termed B. bacteriovorus M1, M2, and M3, when derived from HD100, and M4/Strep to M11/Strep, when derived from HD100-S1 (Table 1).
Axenic mutants were derived by repeated subcultivation of the mutant strains M1, M2, and M11/Strep, which were spread on agar plates containing standard bacteriological medium (PYE). Mutants M1.1, M2.1, M11.1, and M11.2 (Table 1) grew by forming isolated colonies on PYE.
Plasmid and chromosomal DNA preparation.
Plasmids used in this work are listed in Table 1. Plasmid preparations of B. bacteriovorus and E. coli were carried out using the Gene JetTM plasmid miniprep kit (Fermentas, St. Leon-Rot, Germany). Chromosomal DNA of B. bacteriovorus was prepared from purified predator cells by a modified cetyltrimethylammonium bromide (CTAB) extraction procedure (3, 27).
Amplification of DNA by PCR.
For PCR, 0.5 μl Taq polymerase (5 U μl−1), 5 μl deoxynucleoside triphosphates (dNTPs) (2.5 mM each), 3 μl magnesium chloride (50 mM), 5 μl reaction buffer, 5 μl forward and reverse primer (5 μM each), and 1 μl template DNA (0.1 to 1.0 ng) in a 50-μl reaction volume were used. All chemicals were obtained from Bioline (Luckenwalde, Germany). The PCRs were performed with an initial denaturation step of 95°C for 5 min and 35 cycles of denaturation at 95°C for 30 s, annealing for 30 s, and elongation at 72°C for various times. Amplified PCR fragments were purified for either cloning or sequencing using the MSBSpin PCRapace kit from Invitek (Berlin, Germany). Oligonucleotide primers were designed using the Accelrys (DS) Gene program, version 2.5 (Accelrys, Inc., San Diego, CA).
Primers Bd9 (5′-CTAGCTAGCAGAAGGTGATTATATGAAAA-3′) and Bd32 (5′-AGGAAGCTTTACTGTCTTCCAGTCCCGGCTTTC-3′) for construction of the hit gene of pNR22 generated a PCR product consisting of a 318-bp fragment of Bd0108 (coding region and Shine-Dalgarno sequence) plus additional restriction sites for NheI and HindIII (underlined). Primers Bd3461fwdNhe (5′-CCTGCTAGCTTGAAATTCTCAGAATTGAA-3′) and Bd3461revHind (5′-TGGAAGCTTATTAAGAGAACAGACGTTTG-3′) for construction of the rhlB gene of pNR29 generated a PCR product consisting of a 1,650-bp fragment of Bd3461 (coding region and Shine-Dalgarno sequence) plus additional restriction sites for NheI and HindIII (underlined). The primers used for analyses of mutations are shown in Table S2 in the supplemental material. Oligonucleotides were supplied by Metabion (Martinsried, Germany).
Mating experiments.
Matings were performed with the donor strain E. coli S17-1λ harboring the chromosomally integrated RP4 transfer region and with the B. bacteriovorus HD100 wild-type strain as well as the mutant strains M11, M1, M11.1, and M11.2 as recipients as described previously (27). To isolate single bacterial cells, several dilutions of the fully grown transconjugant cultures were spread on double-layer or PYE agar plates and single plaques/colonies were picked.
DNA sequencing and computer analysis.
The sequencing of all plasmids and PCR products was carried out by Qiagen sequencing services (Hilden, Germany). The obtained sequences were analyzed using the Lasergene program SeqMan (DNASTAR, Inc., Madison, WI). Sequence translations were performed using the program Accelrys (DS) Gene (Accelrys, Inc., San Diego, CA).
Whole-genome sequencing was performed with the Roche 454 genome sequencer FLX system. Libraries for 454 sequencing were prepared from cDNA with the GS FLX Titanium general library preparation kit. The obtained 454 libraries were immobilized on beads and clonally amplified using the GS FLX Titanium LV emPCR kit (Lib-L). The libraries were then sequenced using the GS FLX Titanium sequencing kit XLR70 and the GS FLX Titanium PicoTiterPlate kit. All kits used were purchased from Roche and used according to the manufacturer's protocol.
For the alignment of the reads and the analysis of the data, the Consed-finishing package was used. Data for each mutant library was individually aligned against wild-type B. bacteriovorus HD100 using cross_match. Results were inspected and manually curated using the Consed-finishing package (14).
Transposon mutagenesis.
The insertion of the plasmid-encoded Tn5 transposon with a chloramphenicol resistance gene was performed by matings between the E. coli donor strain S17-1λ carrying the pMiniCm plasmid (10) and the B. bacteriovorus strain HD100 as the recipient. The π protein-dependent R6K origin of replication is not recognized by B. bacteriovorus replication enzymes; therefore, selection of chloramphenicol-resistant bdellovibrios is indicative of the insertion of the transposon into the Bdellovibrio genome. To isolate saprophytic growing mutant strains, the transconjugants were spread on double-layer agar plates with 12.5 μg ml−1 chloramphenicol containing heat-killed E. coli within the top layer.
To identify the insertion site within the bdellovibrio genome, the chromosomal DNA of 21 saprophytic growing B. bacteriovorus HD100 was isolated, digested with HindIII, and ligated into the vector pLITMUS38 (NEB, Ipswich, MA). After electroporation using the E. coli Gene Hogs strain (Invitrogen, Darmstadt, Germany), a screening for chloramphenicol-resistant transformants was carried out and plasmid DNA was prepared from these transformants. Sequencing was performed from the 5′ and 3′ ends of the Cm resistance gene using primers Bd23 (5′-CCTAAAATGTTTTTACGA-3′) and Bd24 (5′-CCGTCTGTGATGGCTTCCAT-3′) directed into the flanking DNA regions, which allowed the identification of the cassette insertion site.
Construction of a Bd0108 knockout mutant.
The plasmid pK18mobsacB was used for the construction of a suicide vector (29). Conjugation mediated by the broad-host-range plasmid RP4 (9) is suitable for the transfer from E. coli S17-1λ to B. bacteriovorus HD100. The ColE1 origin of replication of this plasmid is not functional in B. bacteriovorus, and, therefore, selection of chloramphenicol-resistant bdellovibrios is indicative for the insertion of the knockout construct into the Bdellovibrio genome. Furthermore, pK18mobsacB carries the sacB gene (33), which confers sucrose sensitivity and can be used to detect double-crossover events (37).
To knock out the hit gene we used the Bd0108 gene disrupted by a Cmr cassette (for a description, see “Transposon mutagenesis” above) as a template and amplified and inserted this construct via the restriction sites BamHI and HindIII into pK18mobsacB (Bd29, 5′-GCGGATCCTATGGGAAGCCATCTGTAGTTA-3′, and Bd30, 5′-GCGAAGCTTATAAAACTCTCCTTT TTTCTTAC-3′) The primers amplify 696 bp of the wild-type DNA containing the hit sequence centrally (306 bp; accession no. NC_005363), while in the saprophytic mutant the PCR product is enlarged by the Cmr cassette and has a size of 1,695 bp. Sequence analyses of the insertion site showed that the Cmr cassette was integrated into the coding region of the hit gene flanked by six duplicated nucleotides corresponding to positions 193 to 198 of the coding sequence. Matings were performed using the donor strain E. coli S17-1λ and the recipient B. bacteriovorus HD100, and transconjugants were screened for the insertion of the knockout construct via PCR.
Transconjugants were twice cultured with live E. coli DH5α[pBR329] with 12.5 μg ml−1 chloramphenicol in liquid cultures. Thereafter, transconjugants were transferred onto double-layer agar plates containing 5% sucrose in the bottom layer plus an additional 5% sucrose and live E. coli DH5α[pBR329] within the top layer. After 5 days of incubation, turbid plaques became visible. Several plaques were picked and cultivated in liquid media again. After 2 to 3 days of cultivation, these were checked by PCR and found to be merodiploid for the hit region (two PCR products with sizes of 696 bp and 1,695 bp). One culture containing a very strong PCR signal for the insertion of the knockout construct (larger PCR product) was chosen and plated again onto double-layer agar plates. In this case, small colonies could be observed within the zone of prey lysis. Some of these colonies were picked and examined under light microscope, showing morphological aberrances similar to those of saprophytic strains.
The resulting colony-forming bacteria were plated on autoclaved prey bacteria. PCR analyses indicated that the PCR product accounting for the knockout construct was dominant. To eliminate the last merodiploid bacteria, the cultivation on double-layer agar plates containing dead E. coli K-12 and 5% sucrose was repeated. PCR amplification revealed that only the knockout construct was present and the small PCR product which is generated by the wild-type hit gene was absent (see Fig. 4). This PCR product was not detectable after repeated cultivation.
Southern blot and hybridization.
Chromosomal DNA from HD100 and M11/Strep or plasmid DNA (pNR22) were digested with restriction enzymes (EcoRV and HindIII) and analyzed by agarose gel electrophoresis. The DNA fragments were transferred to Hybond N+ membranes (Amersham Pharmacia, Freiburg, Germany) by capillary blotting with 0.4 M NaOH. Hybridization probes for the hit gene were generated by PCR using the primers Bd9 and Bd32 (see above) and fluorescein-labeled dNTPs (PCR fluorescein labeling mix; Roche, Mannheim, Germany). Hybridization was performed as previously described (28) Detection of hybridization signals was obtained by chemiluminescence using antifluorescein conjugate (Perkin Elmer Life Sciences Inc., Boston, MA) and DNA Thunder chemiluminescence reagent (Perkin Elmer Life Sciences Inc., Boston, MA).
RESULTS
Isolation and phenotypic characterization of prey-independent B. bacteriovorus mutant strains.
To study the molecular events that enable B. bacteriovorus to grow on heat-killed bacteria, new mutants of B. bacteriovorus HD100 were isolated. The selection was performed with the wild-type strain and the derivative HD100-S1, a spontaneous streptomycin-resistant mutant. Strain HD100-S1 was used because an efficient and widely used protocol for the isolation of prey-independent mutants is based on the application of streptomycin to eliminate prey bacteria (32). Finally, 11 mutants were obtained for further analyses. Three are derived from the HD100 wild type (M1 to M3), and the remaining eight originate from the streptomycin-resistant Bdellovibrio strain HD100-S1 (M4/Strep to M11/Strep; Table 1).
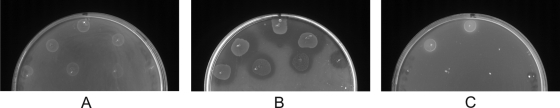
All 11 mutants were facultatively predatory and able to grow on heat-killed prey cells by forming visible colonies on double-layer agar plates, which were surrounded by a zone of lysis in the lawn of dead prey cells (Fig. 1). The colonies and zone of lysis increased slowly in size for 3 to 4 days. By using different kinds of heat-killed prey bacteria (Serratia marcescens, Yersinia enterocolitica, Citrobacter freundii, Pseudomonas aeruginosa, Salmonella enterica serovar Braenderup, Y. pseudotuberculosis, and E. coli CB3537) it became clear that this phenotype did not depend on the prey used.
FIG. 1.
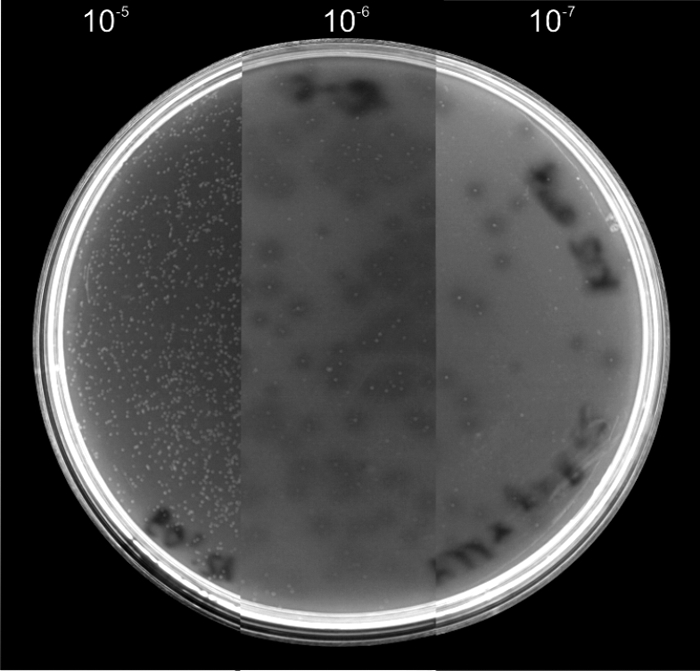
Growth of saprophytic mutant B. bacteriovorus strain M11/Strep on heat-killed E. coli K-12. M11/Strep was cultured on double-layer agar plates containing autoclaved E. coli K-12 within the top layer. Different dilutions of M11/Strep from 10−5 to 10−7 were plated. The facultative predacious strain formed visible colonies which were surrounded by a zone of lysis in the lawn of dead prey cells. In the 10−5 dilution all remains of the heat-killed bacteria were lysed, while in the higher dilutions the remaining dead bacteria were visible as opaque areas.
The saprophytic mutants described so far were also able to grow on live bacteria and form turbid plaques on this prey. When these mutants were streaked on complete bacteriological media such as PYE, colonies developed only when a high density of mutants was plated. This phenotype shows that the saprophytic mutants are an intermediate stage between the prey-dependent and axenic strains.
Three saprophytic mutant strains (M1, M2, and M11/Strep) were chosen to generate axenic mutants. Saprophytic mutants were spread on PYE agar plates, and some of the colonies which developed were cultivated repeatedly until axenic derivatives were obtained (M1.1, M2.1, M11.1, and M11.2; Table 1). Serial dilutions of axenic strains have the same CFU titer on heat-killed prey and on PYE (see Table 2 for summary of growth phenotypes).
TABLE 2.
Phenotype of morphological growth of predatory B. bacteriovorus strains and their saprophytic derivatives on agar plates
| Growth mediuma | Phenotype of B. bacteriovorus strainsb |
||
|---|---|---|---|
| Predatory strains (HD100, HD100-S1) | Saprophytic strains (M1, M2, M11/Strep) | Axenic strains (M1.1, M2.1, M11.1, M11.2) | |
| Live bacteria | Plaques (no colonies) | Turbid plaques | Turbid plaques |
| Heat-killed bacteria | - | Colonies with lysis zones | Colonies with lysis zones |
| PYE | - | - | Colonies |
Either PYE alone or bacterial type on top of agar.
-, few colonies with different phenotypes (see text).
Electron microscopic images of saprophytic and axenic mutants depicted aberrant morphological features, especially an abnormal length (data not shown). This may be the result of cell division hindrances and was also described for other prey-independent mutants of B. bacteriovorus (4, 25, 32).
Analyses of the hit gene in saprophytic derivatives of B. bacteriovorus HD100 and HD100-S1.
It was reported that prey-independent mutants of the strain B. bacteriovorus 109J possess frameshift mutations in a small open reading frame of the hit locus (7). In the B. bacteriovorus HD100 genome sequence, the corresponding ORF is annotated as gene Bd0108. Because in other studies prey-independent mutants derived from strain 109J showed no sequence alterations within this region, the relevance of this locus for prey independence was questioned (4, 23).
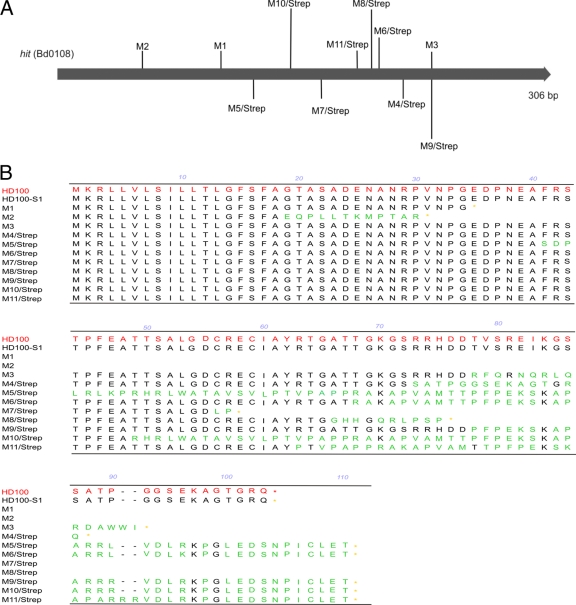
To characterize the sequence of the hit locus in our 11 new saprophytic mutants, we amplified the corresponding region by PCR and sequenced the PCR products. While a mutation could be detected in each Bd0108 ORF of the saprophytic mutants (Fig. 2A), the ORFs of the wild-type strain and HD100-S1 were unaltered. All sequence alterations in the amplified genomic region were within the Bd0108 ORF and changed the primary sequence of the putative gene products considerably (Fig. 2B). These data were in agreement with the findings of Cotter and Thomashow (7), as well as with earlier results of our lab in which another five saprophytic mutants possessed an altered hit gene (D. Schwudke, personal communication).
FIG. 2.
Location of sequence alterations in Bd0108 (hit) of saprophytic mutants of HD100 and HD100-S1 (A) and resulting amino acid sequences of putative gene products (B).
Generation of saprophytic derivatives of B. bacteriovorus HD100 by transposon mutagenesis.
Mutagenesis by transposon insertion has been previously applied to isolate prey-independent B. bacteriovorus 109J mutants (23, 39). We performed matings between the E. coli donor strain S17-1λ harboring the plasmid pMiniCm and B. bacteriovorus HD100 as the recipient. The suicide plasmid pMiniCm harbors a chloramphenicol resistance (Cmr) gene (10).
To isolate saprophytic growing bdellovibrios, transconjugants were spread on double-layer agar plates with chloramphenicol and heat-killed E. coli in the top layer. The chromosomal DNA of 21 saprophytic strains was isolated and digested with restriction enzymes. The resulting fragments were cloned into the vector pLITMUS38 and introduced into E. coli K-12. Plasmid DNAs of the Cmr transformants were isolated, and the chromosomal insertion site of the Cmr cassette in all 21 cases was verified by sequencing.
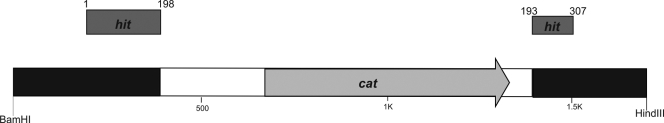
In 20 cases, the inserted DNA from the transconjugants consisted of the Cmr cassette of pMiniCm plasmid flanked on one site by chromosomal DNA of B. bacteriovorus HD100. In only one case, the Cmr cassette was flanked on both sides by chromosomal DNA. This insert shows the resistance cassette positioned in the hit gene which led to a duplication of six nucleotides of the coding region at the integration site (Fig. 3). In the remaining 20 cases, the insertion of the Cmr cassette is in the identical position of the hit gene, either with the 3′ part or with the 5′ part of the hit gene.
FIG. 3.
BamHI-HindIII fragment containing a Cmr cassette integrated into the chromosome of B. bacteriovorus HD100 used for construction of knockout plasmid pNR16. The fragment was obtained by PCR amplification using primers Bd29 and Bd30 from chromosomal DNA of a saprophytic mutant obtained through transposon mutagenesis. The Cmr cassette is indicated by white boxes surrounding the cat gene (light gray) and integrated into the chromosomal DNA (black boxes) interrupting the hit gene (Bd0108) shown above the chromosomal region (dark gray boxes). On each site of the integration six nucleotides are duplicated (positions 193 to 198, GGCGCC).
Knocking out the hit gene leads to saprophytic Bdellovibrio mutants.
The knockout of the Bd0108 gene was done using the plasmid pK18mobsacB (29), a suicide plasmid in B. bacteriovorus carrying the sacB gene for counterselection (37).
In a first attempt we tried to knock out the hit gene by using plasmid constructs containing amplified chromosomal hit regions from our saprophytic mutants (Fig. 2), as these chromosomal regions contain only small insertions or deletions of a few nucleotides in the hit gene. However, we were not able to identify mutants in which an allelic replacement of the wild-type hit gene by an altered hit gene had occurred due to a double crossover, even when we used the counterselection protocol with the addition of 5% sucrose to the medium (37).
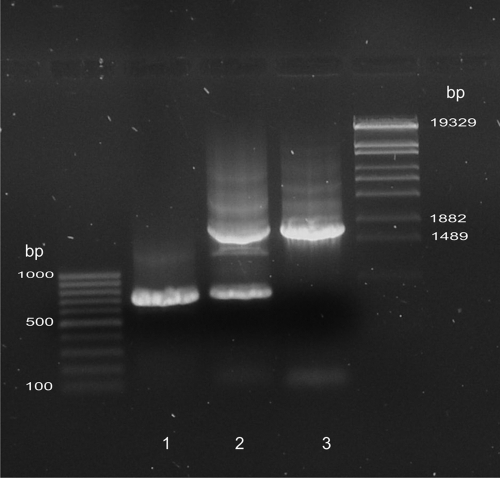
To obtain the knockout in the hit gene, we changed our strategy and amplified the chromosomal region of the saprophytic mutant from the previous transposon mutagenesis experiment which contained the Cmr cassette integrated into the hit gene (Fig. 3). A fragment with the chromosomal region of the Cmr-resistant saprophytic mutant was inserted into the plasmid pK18mobsacB (see Materials and Methods for details) and introduced by mating into B. bacteriovorus HD100. Transconjugants were at first cultured with live E. coli DH5α[pBR329] and then transferred onto double-layer agar plates containing 5% sucrose. Small colonies could finally be observed within the zone of prey lysis. Light microscope examination showed morphological aberrances of isolated bacteria similar to those of saprophytic strains. The colony-forming bacteria were cultivated and purified on autoclaved prey bacteria (see Materials and Methods). PCR amplification clearly revealed that only the knockout construct was present and the small PCR product which is generated by the wild-type hit gene was absent (Fig. 4). The whole hit region was amplified, and the sequence of the PCR products confirmed that the knockout construct had replaced the hit wild-type gene.
FIG. 4.
Identification of B. bacteriovorus HD100 knockout mutants. By using primers Bd29 and Bd30, PCR amplification of genomic DNA of wild-type stains yields a PCR product of 696 bp, while using genomic DNA of the saprophytic mutant isolated through transposon mutagenesis yields a PCR product of 1695 bp. Lane 1 shows the hit-containing PCR fragment of the wild type; in lane 2, the merodiploid state is visible, and lane 3 shows the PCR product from the saprophytic knockout mutant after allelic exchange.
As this knockout strain grew on heat-killed bacteria, the approach confirmed the role of the hit gene in the development of saprophytic growth.
Whole-genome sequencing of different saprophytic and axenic mutant strains.
The above-described experiments focused on the investigation of the involvement of the hit locus in the development of the saprophytic lifestyle.
However, none of the approaches ruled out the possibility of the involvement of more genetic factors in the development of the prey-independent lifestyle. Therefore, we decided to analyze complete genomes of saprophytic mutants to determine whether more mutations had taken place during development of the saprophytic mutants. For this purpose, whole-genome sequencing of three saprophytic (M1, M2, and M11/Strep) was performed. Furthermore, we analyzed two axenically growing derivatives of M1 and M2 (mutants M1.1 and M2.1; Table 1) in order to clarify which additional mutations might have taken place to achieve complete prey independency.
Alignment of the sequencing data to the already known B. bacteriovorus HD100 sequence, published by Rendulic et al. (accession NC_005363 [26]) revealed genomic sequence differences between the wild-type HD100 strain we used and the published strain. Therefore, we decided to sequence the genome of our wild-type strain. All of the differences between the two wild-type strains are shown in Table S1 in the supplemental material. These sequence differences were not connected to the lifestyle, as both HD100 strains are obligately dependent on prey bacteria. Compared to the already known HD100 sequence (NC_005363) the sequence of our HD100 lab strain possesses modifications at 33 positions. Ten of the detected modifications reside in the noncoding region of the DNA, and the other mutations lead to modifications of 12 genes (see Table S1 in the supplemental material). Sequence translations into proteins indicated that most of the altered DNA sequences are responsible for minor changes of the gene products, which means that the amino acid sequence of the respective protein shows changes of only the last few amino acids (Bd0001, Bd0062, Bd1824, Bd1873, Bd2717, and Bd3497). Only three putative gene products show changes which result in a major alteration of the amino acid composition (Bd0538, Bd1200, and Bd3469). Database search via the BIOCYC database collection (www.biocys.org) revealed that two of the three putative gene products provide an unknown function, whereas the protein of the Bd1200 gene is a nucleotide utilizing enzyme (CinA). The sequence differences between the predatory wild types may be due to changes during growth and maintenance in different laboratories.
To summarize, all sequence alignments of saprophytic and axenic mutants were performed in comparison to the genome sequence of the wild-type HD100 strain used in our laboratory. The results of the genomic analyses are shown in Table 3.
TABLE 3.
Sequence alterations of saprophytic and axenic B. bacteriovorus HD100 mutants
| Position | Gene or proteina | Amino acid change (position in gene product) | Sequence alterationc |
|||||
|---|---|---|---|---|---|---|---|---|
| HD100-wtb | M1 | M1.1 | M2 | M2.1 | M11/Strep | |||
| 35,765 | Bd0040 | Unmodified protein | C | T | T | |||
| 96,997 | hit [Bd0108] | Frameshift, C-terminal amino acids modified (64 to 103) | GTAGG | |||||
| 97,076 | hit [Bd0108] | Stop codon (37) | C | A | A | |||
| 97,125 | hit [Bd0108] | Frameshift, C-terminal amino acids modified (19 to 100) | C | * | * | |||
| 170,857 | Bd0183 | H to N (160) | G | T | T | |||
| 521,234 | citZ [Bd0562] | Frameshift, change of amino acids 261 to 382 (end) | T | * | ||||
| 707,368 | Bd0745 | P to S (87) | C | T | T | |||
| 1,331,024 | Bd1408 | V to A (1) | A | G | G | |||
| 1,723,594 | Bd1797 | N to K (288) | T | A | ||||
| 2,317,782 | cheY [Bd2409] | D to G (12) | C | A | ||||
| 2,318,005 | cheY [Bd2409] | L to F (86) | T | C | C | |||
| 2,567,148 | Bd2644 | T to I (420) | C | T | ||||
| 2,876,738 | rpsL [Bd2981] | K to R (43) | T | C | ||||
| 2,880,920 | rpoC [Bd2983] | E to K (86) | C | T | ||||
| 2,974,067 | nifA [Bd3063] | G to D (160) | C | T | ||||
| 3,370,837 | rhlB [Bd3461] | Stop codon (461) | G | A | ||||
| 3,374,202 | pcnB [Bd3464] | Stop codon (328) | C | T | ||||
| 3,617,768 | Intergenic region between Bd3743 and Bd3745 | C | A | A | ||||
| 3,704,211 3,704,212 | Bd3838 | Frameshift, C-terminal amino acids modified (92 to 120) | AA | * | ||||
hit, hypothetical protein associated with the host (prey) interaction, unknown function; citZ, citrate synthase hypothetical protein; cheY, chemotaxis response regulator; rpsl, 30S ribosomal protein S12; rpoC, DNA-directed RNA polymerase, beta subunit; nifA, transcriptional regulator; rhlB, ATP-dependent RNA helicase; pcnB, poly(A) polymerase; Bd0040, putative radical activating enzyme; Bd1408, outer membrane protein; Bd0183, Bd0745, Bd1797, and Bd2644, hypothetical proteins.
The sequence of wild-type B. bacteriovorus HD100 used in this study.
*, deletion.
With regard to the first step of the change from prey-dependent to saprophytic growth, it is remarkable that the hit gene (Bd0108) is the only gene which possessed sequence differences in each mutant strain. All other genomic regions showing sequence deviations from the wild-type sequence are unique in the three saprophytic strains and are therefore unlikely to account for the change of the life cycle. The sequence analysis of the M11/Strep mutant was especially remarkable, as the data reveal only two mutations within the whole genome compared to its wild-type ancestor. One mutation was a nucleotide exchange in the coding sequence of the rpsL gene, which caused streptomycin resistance and was already present in the predatory predecessor HD100-S1, and the second mutation was an insertion of five nucleotides in the hit coding region (Fig. 2).
In addition to the mutation of the hit gene, the saprophytic strain M1 had three and M2 had four nucleotide sequence differences compared to the wild type. Most of these (Table 3) led to single amino acid substitutions in the respective gene products and one nucleotide exchange was in a noncoding region (Table 3).
Furthermore, the data shown in Table 3 gave a clear indication regarding the development from saprophytic to completely prey-independent (axenic) growth. The axenic strains M1.1 and M2.1 showed more mutations than their predecessors M1 and M2, respectively (with the exception of gene Bd1797 in M1.1). The strain M1.1 has three and M2.1 has five additional sequence differences compared to M1 and M2, respectively.
Identification of genes involved in axenic growth.
Comparison of the sequences of the two axenically growing mutants M1.1 and M2.1 did not clearly point to one gene which may be involved in the development of axenic growth. However, the two adjacent genes, Bd3461—coding for the ATP-dependent RNA helicase RhlB—in strain M2.1 and the gene for the poly(A) polymerase PcnB (Bd3464) in strain M1.1, are potential candidates. Nonsense mutations in the coding regions of the two genes led to premature terminations of the proteins. Mutant M2.1 possessed two more, genes citZ (Bd0562) and Bd3838 (unknown function), whose putative gene products were largely modified by frameshift mutations.
To obtain more comprehensive information, new axenically growing mutant strain were selected from the strain B. bacteriovorus M11/Strep, which possessed only the mutation in the hit gene. We isolated two new axenic strains, M11.1 and M11.2, and sequenced eight of the genes which were detected in the previously described genomic analysis (Table 4). From these genes only gene Bd3461, coding for an RNA helicase, RhlB, was mutated in both axenic mutants. This result indicated a correlation between RNA processing and the ability for axenic growth, as in all axenic strains (M1.1, M2.1, M11.1, and M11.2) mutations affected genes involved in RNA modification. This hypothesis was tested in further experiments by complementation (see below).
TABLE 4.
Identification of genes involved in axenic growth of B. bacteriovorusa
| Gene | Mutation detected in strainb |
|
|---|---|---|
| M11.1 | M11.2 | |
| Bd0562 [citZ] | - | - |
| Bd2409 [cheY] | - | - |
| Bd2644 | - | - |
| Bd2983 [rpoC] | - | - |
| Bd3063 [nifA] | - | - |
| Bd3461 [rhlB] | Deletion of 630 bp between position 205 and 837 | Insertion of T at position 744 |
| Bd3464 [pcnB] | - | - |
| Bd3838 | - | - |
Eight genes were selected (Table 3) to analyze axenically growing strains B. bacteriovorus M11.1 and M11.2.
-, no mutation detected.
Saprophytic mutants return to a prey-dependent lifestyle after complementation with the wild-type hit gene.
The restoration of the predatory phenotype by complementation with the wild-type hit gene is indicative of the importance of mutations in the hit gene for the change from a prey-dependent to a saprophytic lifestyle. For this purpose, the wild-type allele of the hit gene was amplified by PCR and inserted into the plasmid pSUP404.2, which replicates stably in bdellovibrios (27).
The recombinant plasmid pNR22 (Table 1) was tested in two saprophytic mutants: the streptomycin-resistant strain M11/Strep, which possessed only a mutation of the hit gene, and the strain M1, which has three additional mutations. After matings with the E. coli strain S17-1λ[pNR22] as the donor, transconjugants were spread on double-layer agar plates containing live E. coli cells within the top layer. After approximately 3 days of incubation, plaques became visible. Single plaques were isolated, and the presence of the plasmid was tested by plasmid extraction. The presence of two alleles of the hit sequence was demonstrated via Southern blot followed by a DNA-DNA hybridization using a HD100-hit probe (data not shown).
To compare the growth characteristics of M11/Strep and M11/Strep[pNR22] as well as those of M1 and M1[pNR22], different dilution steps of each culture were spotted on double-layer agar plates with live E. coli cells in the top layer (Fig. 5).
FIG. 5.

Plaque-forming ability of wild-type, saprophytic strains (M11/Strep, M1/Strep) and transconjugant derivatives containing pNR22. The plaque-forming ability of the complemented strain and its respective mutant was checked by spotting dilutions of 10-μl liquid cultures from 10° on top of the plates (undiluted, top of the plate) to 10−6 (third line, last spot) (see diagram at top of figure). (A to E) Double-layer agar plates containing living prey: A, B. bacteriovorus HD100 wild type (control); B, M11/Strep; C, M11/Strep[pNR22]; D, M1; E, M1[pNR22]. (F to I) Double-layer agar plates containing autoclaved prey: F, M11/Strep; G, M11/Strep[pNR22]; H, M1; I, M1[pNR22].
Figure 5 shows that the plaque-forming ability on live bacteria of the two transconjugant strains recovered when the wild-type HD100 hit gene resided within the bacteria (Fig. 5C and E). While M1 and M11/Strep did not form plaques in the lawn of live prey (Fig. 5B and D), the pNR22-containing transconjugants were able to do so, and plaque formation up to the 10−6 dilution was clearly visible. It is remarkable that the complemented strains lacked the ability to grow on autoclaved prey (Fig. 5G and I), whereas their saprophytic predecessors formed colonies surrounded by the typical zone of lysis for all of the tested dilution steps (Fig. 5F and H). This observation proved the importance of the hit gene in respect to the predatory lifestyle and confirmed the observation made by Cotter and Thomashow, who discovered that the hit locus enhanced plaque-forming ability. (7).
An axenic mutant showed reduced colony-forming ability on PYE medium after complementation with the wild-type rhlB gene.
To test whether the axenically growing derivative M11.1 that had a mutation in the rhlB gene (Bd3461) could be altered in its growth characteristics, a complementation experiment with the wild-type allele of Bd3461 was carried out. For this reason, Bd3461 was amplified from chromosomal DNA of HD100 and cloned into the plasmid pSUP404.2 via the restriction sites NheI and HindIII. The resulting recombinant plasmid, pNR29, was transferred into the axenic mutant strain M11.1, which had a deletion of 631 bp between positions 205 and 837 in the coding region of Bd3461 (Table 4). Transconjugants showing plasmid-encoded resistance against chloramphenicol were tested for their growth abilities by plating on double-layer plates with live or killed prey bacteria and PYE medium (Fig. 6).
FIG. 6.
Growth of M11.1[pNR29] on live or dead prey and PYE agar. B. bacteriovorus strain M11.1 containing the plasmid pNR29 was diluted up to 10−6, and 10 μl of each dilution was spotted on top of the agar plates. (A) Living E. coli JC3272[pBR329]. (B) Autoclaved E. coli K-12. (C) PYE agar. For details of the spotting scheme, see Fig. 5.
As seen in Fig. 6, the introduction of the wild-type gene Bd3461 into the axenic B. bacteriovorus HD100 strain M11.1 had a major impact on the prey-independent cultivation. The number of colonies of M11.1[pNR29] was significantly reduced when they were plated on PYE medium compared to the number of colonies after plating on plates with heat-killed or living prey. The axenic predecessor strain M11.1 was able to form typical isolated colonies on PYE agar plates up to high dilution steps. The transconjugant strain M11.1[pNR29], however, formed colonies only to a 10−2 or 10−3 dilution on PYE medium. When grown on double-layer agar plates containing heat-killed E. coli, colony formation was observed up to a 10−6 or 10−7 dilution.
While the colony-forming ability was reduced by the introduction of plasmid pNR29, no effect was found regarding the ability to grow on either autoclaved or live prey. When grown on autoclaved prey, the transconjugant strain still formed the typical colonies with a zone of lysis. When grown on live prey, M11.1[pNR29] caused turbid plaques.
These experiments were carried out three times with similar results. Therefore, it can be assumed that the gene Bd3461 is of major importance for axenic growth yet does not influence the ability of plaque formation.
DISCUSSION
The first genetic studies with B. bacteriovorus were carried out to analyze the change from the predatory lifestyle to prey-independent growth and resulted in the identification of the hit locus (7). The hit locus described had a size of 959 bp and contained only one complete ORF (Bd0108). The biological role of this genetic locus has remained obscure. Later studies reported that prey independency was found without alterations of the hit locus (4, 23). Also Southern hybridization experiments indicated that the hit locus is present only in B. bacteriovorus strains (18, 30), although related genetic sequences which are too different to be detected by hybridization possibly exist in other predators. The putative gene product of the hit gene of strain HD114 shows a similarity of only 54% to the protein of Bd0108, although all other gene products of HD100 and HD114 in this region possess similarities of 80 to 95% (31). A BLAST search against the complete genome of Bacteriovorax marinus (FQ312005) also did not reveal any positive result. This could indicate a high level of diversity of proteins with similar functions in predatory strains.
The complete genome analysis of the B. bacteriovorus HD100 strain showed that the hit locus lies in a section of a type IV pilus cluster and might be transcriptionally coupled to it (26) The same genetic localization of a related Hit protein was also found in the predatory strain B. bacteriovorus HD114 (31). In a previous approach we tried to determine whether a gene product of the hit locus can be detected in synchronized cultures of the predatory strain by a proteomic approach; this, however, failed. Nevertheless, prey-independent mutants were isolated and all were mutated in the hit locus (D. Schwudke, unpublished results).
Selection for prey-independent growth requires some rounds of cultivation; thus, it is possible that during the selection process several genomic changes take place. This was already described by Thomashow and Cotter (38), who summarized several reports from early studies of prey-independent mutants and differentiated between type I and type II mutants. Type I mutants show growth only on rich medium and only when prey extracts were added. When type I mutants were plated on prey-free complete medium, a density-dependent growth was observed. This means that only in parts of the plate where at least 104 colonies were plated could colonies be found. In contrast to this, type II mutants isolated with a lower frequency can form colonies on prey-free complete medium. In a recent paper, a modified selection technique was described, which strongly enhanced the frequency of prey-independent mutants (8). No genetic analysis of these mutants was performed, and the authors did not explain why the new selection technique yielded higher mutation rates than previous procedures (32, 35). The rationale for this observation remains therefore unclear.
To isolate type I prey-independent mutants, we selected on media with prey extracts (autoclaved E. coli strains). We isolated 11 prey-independent mutants which all showed mutations in the hit locus. All of the mutants were facultatively predatory and did not grow axenically on complete medium. We identified all mutations as nonsense or frameshift mutations affecting the putative gene products of a small ORF (Bd0108, termed hit gene) of the hit locus significantly (Fig. 2).
Transposon mutagenesis has been employed to identify genes involved in the interactions between predator and prey, indicating that mutations in motility, penetration, and chemotaxis are involved in the interaction between predator and prey (23, 39). Under our selection conditions, we identified an integration of the transposon only into the hit gene, and by performing a knockout of the hit gene (Bd0108) a prey-independent growth was also achieved. However, it is possible that during the selection procedure additional mutations may have emerged. The selection for prey-independent growth is bound to generate more mutations, as each round of growth may favor mutations conferring improved abilities to grow in the absence of live prey.
Whole-genome sequencing of mutants is a new advanced tool to find unequivocal proof for the molecular changes taking place. We performed the analysis of three mutants (M1, M2, M11/Strep) which were saprophytic but unable to grow axenically. The result of the genomic analysis revealed that in two mutants, four or five mutations (including the hit gene) had occurred. However, one saprophytic strain (M11/Strep) derived from a streptomycin-resistant predatory predecessor had only one frameshift mutation in the hit gene, thus proving that this mutation alone is sufficient for the switch from the predatory to the saprophytic lifestyle. We could confirm this assumption by complementation with the wild-type hit gene using a plasmid vector which replicates stably in Bdellovibrio with a copy number of approximately seven per cell (27). The introduction of the wild-type hit gene significantly enhanced the plaque formation on live prey cells of M11/Strep and of M1. The latter experiments also demonstrated that other mutations outside the hit locus which were present in the saprophytic strain M1 were not involved in the development of prey-independent growth.
The hit gene appears to be transcriptionally coupled to a type IV pilus cluster (26). Database analyses of the Hit protein sequence indicated that the Hit protein shows an identity of 34% (52% positive) to a hypothetical protein (RC1_3569) of Rhodospirillum centenum. Compared to the fact that the Hit proteins of B. bacteriovorus HD100 and HD114 had an identity of just 54% (63% positive) (31), this similarity could be significant. The RC1_3569 protein has a motif that can be found in VirB5 type IV secretion system (T4SS) proteins which are involved in the assembly of the T4 pilus (1). It is an intriguing idea that the Hit protein could be a component of an adhesive pilus and thus be involved in the process of penetration and invasion of prey bacteria.
Type II mutants were obtained from the saprophytic mutants by repeated cultivation on complete medium without addition of prey extracts. The genome analysis of two axenically growing derivatives, M1.1 and M1.2, showed additional information related to the ability of forming colonies. The comparison of the two genome sequences did not reveal only one gene responsible for the axenic development; instead, two adjacent arranged genes (Bd3461 and Bd3464) involved in RNA processing showed mutations (Table 3). With the M11/Strep mutant at hand we selected axenically growing derivatives, analyzed all potentially mutated loci (Table 4), and performed a complementation experiment. Thus, it was revealed that the wild-type Bd3461 gene coding for the ATP-dependent RNA helicase RhlB reduced the colony formation on PYE in the mutants M11.1 and M11.2.
It is not clear why the colony-forming ability is enhanced by the loss of functions in RNA processing. Interestingly, Bd3461 (rhlB) is one part of the degradosome, a large multiprotein complex involved in RNA degradation and decay in Escherichia coli (2). The interaction of different proteins of the degradosome is required for efficient degradation of RNA, and the disruption of the complex has stabilizing effects on bacterial transcripts (19). No further experimental evidence was found for a role of Bd3464 coding for the poly(A) polymerase PcnB; however, it is intriguing that this gene also encodes an enzyme that catalyzes RNA polyadenylation at the 3′ end, which leads to its faster degradation (24). Inactivation of pcnB significantly increased the half-lives of various specific transcripts in E. coli. The connective link between the two genes might be indicated by the observation that the addition of a 3′ poly(A) tail stimulated the degradation of the mRNA by the degradosome (5). The finding that Bd3461 and Bd3164 were mutated in axenically growing mutants could indicate that specific transcripts were increased in stability, thus enabling colony formation. However, more research is necessary to test this hypothesis.
Our data confirm the considerations of Thomashow and Cotter (38) expressed in a short review paper in which the results concerning the hit locus were discussed in relation to a model based on earlier Bdellovibrio research. In this model it was suggested that two major signals are necessary for the predatory life cycle (15). One signal triggers the differentiation of attack-phase bdellovibrios into growth-phase cells, and the second initiates DNA replication. Both signals initiate two distinct waves of protein synthesis. The hit locus was suggested to be responsible for the first step in these processes. The whole-genome sequence analysis of mutant strain M11/Strep, together with the results of the restoration of predatory lifestyle by complementation with the wild-type hit gene, clearly indicates that the Hit protein is involved in the switch from attack to growth phase. The potential transcriptional coupling of the hit gene to a pilus cluster (26) suggests that the gene product is involved in the penetration/invasion process. Lack of the Hit protein substitutes the signal for the predator to be inside the prey and to start growth. The mutations in genes involved in RNA processing indicate an involvement of the bacterial degradosome in processes preceding DNA replication. Mutations in parts of the degradosome potentially increase the half-life of specific RNAs, which could be involved in the initiation of DNA replication and thus deliver the signal for formation of colonies on complete medium. Interestingly the complementation experiments demonstrated that plaque formation and colony formation are two independent phenotypes.
All of our data were obtained with the reference strain B. bacteriovorus HD100, whose genome sequence has been previously published (26). The study of Cotter and Thomashow was done with another reference strain, B. bacteriovorus 109J, which is closely related. Two later studies reported the isolation of prey-independent mutants derived from B. bacteriovorus 109J which were not mutated at hit (18, 23); however, new investigations revealed that the majority of prey-independent mutants of this strain carried mutations in the hit gene (40). Thus, if pathways to prey-independent growth circumventing the hit gene are possible, they are likely to be of minor relevance. Another possibility is that there are strain-specific differences between HD100 and 109J in this respect.
The vast changes that occur in predatory bacteria by switching from attack phase to predatory phase have been studied by proteomic (11) and transcriptional profiling (22). If a single mutation is able to allow for prey-independent growth, it is likely that a protein derived from the affected gene plays a key regulatory role in the life cycle. We have begun to address the possible function of the Hit protein in these processes by proteomic investigations.
Supplementary Material
Acknowledgments
We thank Lothar Beutin, Anne Mayer-Scholl, and Sebastian Beck for critical reading of the manuscript.
N.R. was supported by a grant from DFG (STR 1034/1-1).
Footnotes
Published ahead of print on 28 January 2011.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aly, K. A., and C. Baron. 2007. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens. Microbiology 153:3766-3775. [DOI] [PubMed] [Google Scholar]
- 2.Arraiano, C. M., et al. 2010. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 34:883-923. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., et al. 1987. Current protocols in molecular biology. Wiley Interscience, New York, NY.
- 4.Barel, G., and E. Jurkevitch. 2001. Analysis of phenotypic diversity among host-independent mutants of Bdellovibrio bacteriovorus 109J. Arch. Microbiol. 176:211-216. [DOI] [PubMed] [Google Scholar]
- 5.Blum, E., A. J. Carpousis, and C. F. Higgins. 1999. Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J. Biol. Chem. 274:4009-4016. [DOI] [PubMed] [Google Scholar]
- 6.Cotter, T. W., and M. F. Thomashow. 1992. A conjugation procedure for Bdellovibrio bacteriovorus and its use to identify DNA sequences that enhance the plaque-forming ability of a spontaneous host-independent mutant. J. Bacteriol. 174:6011-6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter, T. W., and M. F. Thomashow. 1992. Identification of a Bdellovibrio bacteriovorus genetic locus, hit, associated with the host-independent phenotype. J. Bacteriol. 174:6018-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Covarrubias, L., and F. Bolivar. 1982. Construction and characterisation of new cloning vehicles. VI. Plasmid pBR329, a new derivative of pBR328 lacking the 428-base-pair inverted duplication. Gene 17:79-89. [DOI] [PubMed] [Google Scholar]
- 8.Dashiff, A., and D. E. Kadouri. 2009. A new method for isolating host-independent variants of Bdellovibrio bacteriovorus using E. coli auxotrophs. Open Microbiol. J. 3:87-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta, N., R. W. Hedges, E. J. Shaw, R. B. Sykes, and M. H. Richmond. 1971. Properties of an R factor from Pseudomonas aeruginosa. J. Bacteriol. 108:1244-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dori-Bachash, M., B. Dassa, S. Pietrovski, and E. Jurkevitch. 2008. Proteome-based comparative analyses of growth stages reveal new cell cycle-dependent functions in the predatory bacterium Bdellovibrio bacteriovorus. Appl. Environ. Microbiol. 74:7152-7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flannagan, R. S., M. A. Valvano, and S. F. Koval. 2004. Downregulation of the motA gene delays the escape of the obligate predator Bdellovibrio bacteriovorus 109J from bdelloplasts of bacterial prey cells. Microbiology 150:649-656. [DOI] [PubMed] [Google Scholar]
- 13.Friedberg, D. 1978. Growth of host dependent Bdellovibrio in host cell free systems. Arch. Microbiol. 116:185-190. [DOI] [PubMed] [Google Scholar]
- 14.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 15.Gray, K. M., and E. G. Ruby. 1991. Intercellular signalling in the Bdellovibrio developmental cycle, p. 333-366. In M. Dworkin (ed.), Microbial cell-cell interactions. American Society for Microbiology, Washington, DC.
- 16.Horowitz, A. T., M. Kessel, and M. Shilo. 1974. Growth cycle of predacious bdellovibrios in a host-free extract system and some properties of the host extract. J. Bacteriol. 117:270-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reference deleted.
- 18.Jurkevitch, E., D. Minz, B. Ramati, and G. Barel. 2000. Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl. Environ. Microbiol. 66:2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaberdin, V. R., and S. Lin-Chao. 2009. Unraveling new roles for minor components of the E. coli RNA degradosome. RNA Biol. 6:402-405. [DOI] [PubMed] [Google Scholar]
- 20.Lambert, C., M. C. Smith, and R. E. Sockett. 2003. A novel assay to monitor predator-prey interactions for Bdellovibrio bacteriovorus 109J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ. Microbiol. 5:127-132. [DOI] [PubMed] [Google Scholar]
- 21.Lambert, C., et al. 2006. Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus. Mol. Microbiol. 60:274-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert, C., C. Y. Chang, M. J. Capeness, and R. E. Sockett. 2010. The first bite—profiling the predatosome in bacterial pathogen Bdellovibrio. PLoS One 5:e8599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Maidhof, H., B. Guerra, S. Abbas, H. M. Elsheikha, T. S. Whittam, and L. Beutin. 2002. A multiresistant clone of Shiga toxin-producing Escherichia coli O118:[H16] is spread in cattle and humans over different European countries. Appl. Environ. Microbiol. 68:5834-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medina, A. A., R. M. Shanks, and D. E. Kadouri. 2008. Development of a novel system for isolating genes involved in predator-prey interactions using host independent derivatives of Bdellovibrio bacteriovorus 109J. BMC Microbiol. 8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadratowska-Wesołowska, B., et al. 2010. Transcription regulation of the Escherichia coli pcnB gene coding for poly(A) polymerase I: roles of ppGpp, DksA and sigma factors. Mol. Genet. Genomics 284:289-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nunez, M. E., M. O. Martin, L. K. Duong, E. Ly, and E. M. Spain. 2003. Investigations into the life cycle of the bacterial predator Bdellovibrio bacteriovorus 109J at an interface by atomic force microscopy. Biophys. J. 84:3379-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Priefer, U. B., R. Simon, and A. Pühler. 1985. Extension of the host range of Escherichia coli vectors by incorporation of RSF1010 replication and mobilization functions. J. Bacteriol. 163:324-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rendulic, S., et al. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689-692. [DOI] [PubMed] [Google Scholar]
- 27.Roschanski, N., and E. Strauch. 2010. Assessment of the mobilizable vector plasmids pSUP202 and pSUP404.2 as genetic tools for the predatory bacterium Bdellovibrio bacteriovorus. Curr. Microbiol. 62:589-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Schäfer, A., et al. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 30.Schwudke, D., E. Strauch, M. Krueger, and B. Appel. 2001. Taxonomic studies of predatory bdellovibrios based on 16S rRNA analysis, ribotyping and the hit locus and characterization of isolates from the gut of animals. Syst. Appl. Microbiol. 24:385-394. [DOI] [PubMed] [Google Scholar]
- 31.Schwudke, D., et al. 2005. Transcriptional activity of the host-interaction locus and a putative pilin gene of Bdellovibrio bacteriovorus in the predatory life cycle. Curr. Microbiol. 51:310-316. [DOI] [PubMed] [Google Scholar]
- 32.Seidler, R. J., and M. P. Starr. 1969. Factors affecting the intracellular parasitic growth of Bdellovibrio bacteriovorus developing within Escherichia coli. J. Bacteriol. 97:912-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selbitschka, W., S. Niemann, and A. Pühler. 1993. Construction of gene replacement vectors for gram-negative bacteria using a genetically modified sacB gene as a positive selection marker. Appl. Microbiol. Biotechnol. 38:615-618. [Google Scholar]
- 34.Sharma, D., A. R. Cukras, E. J. Rogers, D. R. Southworth, and R. Green. 2007. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J. Mol. Biol. 374:1065-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shilo, M., and B. Bruff. 1965. Lysis of Gram-negative bacteria by host-independent ectoparasitic Bdellovibrio isolates. J. Gen. Microbiol. 40:317-328. [DOI] [PubMed] [Google Scholar]
- 36.Sockett, R. E. 2009. Predatory lifestyle of Bdellovibrio bacteriovorus. Annu. Rev. Microbiol. 63:523-539. [DOI] [PubMed] [Google Scholar]
- 37.Steyert, S. R., and S. A. Pineiro. 2007. Development of a novel genetic system to create markerless deletion mutants of Bdellovibrio bacteriovorus. J. Bacteriol. 73:4717-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomashow, M. F., and T. W. Cotter. 1992. Bdellovibrio host dependence: the search for signal molecules and genes that regulate the intraperiplasmic growth cycle. J. Bacteriol. 174:5767-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tudor, J. J., J. J. Davis, M. Panichella, and A. Zwolak. 2008. Isolation of predation-deficient mutants of Bdellovibrio bacteriovorus by using transposon mutagenesis. Appl. Environ. Microbiol. 74:5436-5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wurtzel, O., M. Dori-Bachash, S. Pietrokovski, E. Jurkevitch, and T. R. Sorek. 2010. Mutation detection with next-generation resequencing through a mediator genome. PLoS One 5:e15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.