Abstract
Oxidative stress occurs when the generation of reactive oxygen species (ROS) exceeds the capacity of the cell's endogenous systems to neutralize them. Our analyses of the cellular damage and oxidative stress responses of the archaeon Halobacterium salinarum exposed to ionizing radiation (IR) revealed a critical role played by nonenzymatic antioxidant processes in the resistance of H. salinarum to IR. ROS-scavenging enzymes were essential for resistance to chemical oxidants, yet those enzymes were not necessary for H. salinarum's resistance to IR. We found that protein-free cell extracts from H. salinarum provided a high level of protection for protein activity against IR in vitro but did not protect DNA significantly. Compared with cell extracts of radiation-sensitive bacteria, H. salinarum extracts were enriched in manganese, amino acids, and peptides, supporting an essential role in ROS scavenging for those small molecules in vivo. With regard to chemical oxidants, we showed that the damage caused by gamma irradiation was mechanistically different than that produced by hydrogen peroxide or by the superoxide-generating redox-cycling drug paraquat. The data presented support the idea that IR resistance is most likely achieved by a “metabolic route,” with a combination of tightly coordinated physiological processes.
Oxidative stress occurs when the level of reactive oxygen species (ROS) produced in cells by aerobic metabolic activity or environmental challenges overwhelms antioxidant defense mechanisms and damage accumulates (58). ROS include molecules such as hydrogen peroxide (H2O2), superoxide (O2−), and hydroxyl radicals (HO), all of which can be derived from molecular oxygen in metabolically active cells (58). Damage from ROS has also been implicated in a variety of human conditions, including the neurological diseases Alzheimer's and Parkinson's (7), aging, and a wide range of cancers (9, 55, 57). In response to the danger of ROS, organisms have evolved numerous defense and repair mechanisms, including conserved detoxification enzymes (e.g., superoxide dismutases [SOD] and catalases), free radical scavenger systems, and mechanisms to repair DNA damage (28, 30, 58). Among the environmental factors and external sources of toxic oxidants that can increase the production of ROS are starvation, desiccation, heat shock, antibiotics, sunlight, and exposure to ionizing radiation (IR) (14, 30, 67).
Ionizing radiation is of particular interest because most of its deleterious effects are the result of HO, O2−, and H2O2 produced directly via the radiolysis of water, imparting severe oxidative stress to all of the cell's components (54, 63). DNA is associated with water molecules, and the proximity of HO production results in clusters of damage, including strand breaks and oxidative base and sugar damage (27, 47). The most consequential damage by O2− and H2O2 in cells is to proteins that contain exposed iron-sulfur or heme groups (29), resulting in carbonylation of protein residues, formation of protein cross-linkages, oxidation of the protein backbone, and ultimately, protein fragmentation and inactivation (29, 57). In proteins containing exposed Fe2+ groups, O2− and H2O2 can cause the release of ferrous ion and the formation of HO via electron transfer from ferrous iron to H2O2, the so-called Fenton reaction (28, 29). The ability of an organism to prevent specific and nonspecific ROS-mediated cellular damage is therefore key to its survival.
Studies of Deinococcus radiodurans and other radiation-resistant bacteria have demonstrated a strong correlation between high intracellular Mn/Fe concentration ratios and a high level of IR resistance (14, 20, 34). The evidence points to a major role in D. radiodurans for ROS-scavenging orthophosphate and metabolite complexes of Mn2+ that specifically protect proteins from oxidative damage during irradiation (12). A high level of intracellular salts together with a high intracellular Mn/Fe ratio have been implicated in combating ROS in the archaeon Halobacterium salinarum (34). The ability of cells to protect their proteins from oxidation by scavenging IR-induced ROS has been proposed as the key mechanism for survival of IR-resistant microorganisms (12).
H. salinarum is an extreme halophile that experiences a number of oxidative stressors in its natural environment, such as high UV radiation and desiccation/rehydration cycles (15). This microorganism grows optimally at 4.3 M salt and counterbalances the high external osmotic pressure by accumulating up to 4 M intracellular KCl (24). Its extreme resistance to IR and desiccation makes this organism a good model system to investigate the diversity of mechanisms in response to oxidative stress (35, 64).
Here, we characterize the cellular damage and stress responses of H. salinarum exposed to chemical oxidants and to IR. ROS-scavenging enzymes were essential for the survival of H. salinarum exposed to H2O2 or O2−, but those enzymes were not necessary for the survival of H. salinarum following gamma irradiation. Protein-free cell extracts of H. salinarum provided a high level of radioprotection to protein activity but not to DNA in vitro, whereas extracts from radiation-sensitive bacteria did not provide protection to irradiated proteins or DNA. H. salinarum cell extracts were enriched in classes of small molecules that included Mn and peptides also found in high abundance in the protein-free extract of D. radiodurans (12), indicating an essential role of those molecules in ROS scavenging. This study contributes novel findings on the critical role played by nonenzymatic antioxidant systems in IR resistance in H. salinarum, showing that IR resistance is most likely achieved by a “metabolic route” with a combination of tightly coordinated physiological processes.
MATERIALS AND METHODS
Culturing and growth conditions.
Halobacterium salinarum NRC-1 (ATCC 700922) Δura3 and mutant cultures were grown in standard GN101 medium (250 g/liter NaCl, 20 g/liter MgSO4·7H2O, 2 g/liter KCl, 3 g/liter Na citrate, 10 g/liter Oxoid-brand peptone), pH 7.2, with the addition of 1 ml/liter trace element solution (31.5 mg/liter FeSO4·7H2O, 4.4 mg/liter ZnSO4·7H2O, 3.3 mg/liter MnSO4·7H2O, 0.1 mg/liter CuSO4·5H2O) and supplemented with a final concentration of 50 mg/liter uracil (Sigma, St. Louis, MO). Escherichia coli and Pseudomonas putida were grown in LB (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl, pH 7.0) and TGY (10 g/liter Bacto-tryptone, 5 g/liter yeast extract, 1 g/liter glucose, pH 7.0) medium, respectively. Cultures were grown at 37°C for E. coli and P. putida and at 42°C for H. salinarum, with shaking at 220 rpm in a Gyromax 737 shaker (Amerex Instruments, Lafayette, CA). ROS detoxification mutants with Δsod1/2, ΔperA, ΔmsrA, ΔVNG0018H, and ΔVNG0798H mutations have been previously described (32).
Chemical oxidant treatments.
Cells were grown in the appropriate medium to early log phase (optical density at 600 nm [OD600] = 0.4) and treated either with 25 or 30 mM H2O2 (Sigma, St. Louis, MO) or with 4 or 10 mM paraquat (methyl viologen; Sigma, St. Louis, MO), final concentrations, for 2 h at 42°C with shaking at 220 rpm in a Gyromax 737 shaker. Cells were collected by centrifugation and washed once in the appropriate medium. Cells for survival plating and pulsed-field gel electrophoresis (PFGE) analyses were processed immediately; cells for DNA or protein oxidation analyses were stored at −80°C until further processing.
Gamma irradiation.
Cells grown in the appropriate growth medium to early log phase (OD600 = 0.40) were irradiated using a 60Co gamma source (dose rate = 3.5 kGy/hr; Uniformed Services University of the Health Sciences, Bethesda, MD) to a final dose of 0, 2.5, or 5 kGy. All samples, regardless of the volume of the starting culture, were irradiated in a volume of 1 ml after concentration by centrifugation (8,000 × g for 5 min), resuspension in 1 ml of the appropriate growth medium in a 1.5-ml microcentrifuge tube, and storage on ice until irradiation. Cells for survival plating and PFGE analyses were kept on ice until processing; cells for DNA or protein oxidation analyses were stored at −80°C until further processing.
Survival assays.
Following treatment, H. salinarum cells were serially diluted in basal salt solution (BSS; 250 g/liter NaCl, 20 g/liter MgSO4·7H2O, 2 g/liter KCl, 3 g/liter Na citrate) and plated on GN101 medium supplemented with 50 mg/liter uracil. The plates were incubated at 42°C for 5 to 7 days. Survival was calculated as the number of viable cells following treatment divided by the number of viable untreated cells and graphed with standard error bars.
Genomic DNA extraction and GC-MS analysis.
DNA extractions and gas chromatography-mass spectrometry (GC-MS) with isotope dilution were carried out in triplicate as previously described (34). Briefly, H. salinarum cells were lysed with proteinase K (0.13 mg/ml) (Invitrogen, Carlsbad, CA) in the presence of 2 mM deferoxamine (Desferal; Sigma, St. Louis, MO), the DNA was ethanol precipitated (49), and the resulting DNA pellets were stored under 70% ethanol at −20°C. The quality and quantity of the DNA resuspended in water was determined through absorption spectrophotometry between 200 and 350 nm. Fifty-microgram aliquots of DNA were dried under vacuum and supplemented with an aliquot of [13C,15N2]2,6-diamino-4-hydroxy-5-formamidopyrimidine as an internal standard (Cambridge Isotope Laboratories, Cambridge, MA). These were hydrolyzed for 1 h with 2 μg formamidopyrimidine glycosylase (Fpg) and then analyzed by GC-MS as previously described (46).
Pulsed-field gel electrophoresis.
PFGE was performed as described previously (33), in triplicate. Following treatment, cells were pelleted by centrifugation at 8,000 × g for 5 min and resuspended in room temperature BSS prior to being embedded into InCert agarose plugs (0.8% final concentration prepared in 3:1 BSS-H2O; Bio-Rad, Hercules, CA) at a final cell concentration of 1 × 109 cells/ml. Plugs were lysed in proteinase K solution (0.25 M EDTA [pH 8], 1% N-lauryl sarkosine, and 0.5 mg/ml proteinase K) at 54°C for 1 to 2 days. Plug washes consisted of two washes for 1 h in 20 ml 1× Tris-EDTA (TE) buffer at room temperature, two washes for 1 h in 20 ml 0.5× TE buffer at room temperature, and four washes for 24 h in 0.5× TE buffer at 4°C. Plugs were incubated in Pefabloc (Roche, Indianapolis, IN) solution (10 mM Tris-HCl, 1 mM EDTA, pH 7.0, 1 mM Pefabloc) overnight at 37°C, washed as described above, and stored in 5 ml 0.5× TE buffer at 4°C. Plugs were digested with XbaI (New England BioLabs, Ipswich, MA) for 16 h at 37°C; following equilibration in 2 mM Tris-HCl, 5 mM EDTA, pH 8.0 for 20 min at 4°C, plugs were analyzed using a CHEF DR-III electrophoresis system (Bio-Rad, Hercules, CA) with 1% PFGE certified agarose (Bio-Rad, Hercules, CA) gels and 0.2 5× Tris-borate-EDTA in both the running and gel buffers. The run conditions were 6 V/cm, 10- to 60-s switching times, and a 120° included angle for 24 h at 14°C.
Protein oxidation assays.
Protein oxidation was assessed in triplicate using an OxiSelect protein carbonyl enzyme-linked immunosorbent assay (ELISA) kit (Cell Biolabs, San Diego, CA) and the manufacturer's protocol. Briefly, cell pellets were resuspended in 1 ml cold 1 M salt buffer (50 mM potassium phosphate, pH 7.0, 1 M NaCl, 1% 2-mercaptoethanol) and sonicated for 30 s, followed by 30 s on ice, repeated three times. Cell lysates were then fractionated by centrifugation (12,000 × g, 30 min, 4°C), and the soluble proteins in the supernatant were stored at −20°C. Protein concentration was determined with a Bio-Rad Bradford assay (Hercules, CA) using the manufacturer's protocol. Cell lysates were diluted to 10 μg/ml of protein in 1× PBS, and 1 μg of protein was added to each well in a 96-well protein binding plate. Protein carbonyl-bovine serum albumin (BSA) standards were prepared with concentrations ranging from 0 μg/ml to 7.5 μg/ml, and 1 μg of each was added to the wells of the plate and incubated at 4°C overnight. Three washes with 250 μl of 1× PBS were performed, followed by incubation with 4 μg of DNPH (2,4-dinitrophenylhydrazine) for 45 min at room temperature. Five washes with 250 μl of 1× PBS-ethanol (1:1, vol/vol) and two washes with 250 μl 1× PBS were performed, followed by incubation with blocking buffer for 2 h at room temperature with shaking. Immunodetection was performed using primary (anti-DNP [2,4-dinitrophenol]) and secondary (horseradish peroxidase-conjugated) antibodies provided by the manufacturer. The absorbance of each well was read with a Power Wave 200 microplate spectrophotometer (BioTek Instruments, Winooski, VT) at 450 nm. A standard curve was constructed and used to determine the protein carbonylation levels of the oxidant-treated samples.
Preparation of protein-free UFs.
Protein-free ultrafiltrates (UFs) were prepared for H. salinarum (UFHs), E. coli (UFEc), and P. putida (UFPp). Harvested cells were washed twice with BSS for H. salinarum and with 100 mM Tris-HCl (pH 7.4) for E. coli and P. putida. Amounts of 15 g of wet weight cells were resuspended in distilled and deionized water (ddH2O) and passed through a French press at 900 lb/in2, and the cell extracts were then centrifuged at 12,000 × g (60 min and 4°C). The protein concentration of the supernatants was determined with a Bio-Rad Bradford assay (Hercules, CA) and adjusted to 17 mg/ml with ddH2O. The supernatant was further centrifuged at 190,000 × g (44 h and 4°C) and then subjected to filtration using 3-kDa centrifugal devices (Amicon Ultracel-3 filters; Millipore, Billerca, MA). The resulting UFs were boiled for 30 min and concentrated 5 times in a speed vacuum concentrator (Heto vacuum centrifuge; ATR, Laurel, MD). Samples were aliquoted and stored at −20°C.
DNA protection assay.
pUC19 plasmid DNA (New England BioLabs, Ipswich, MA) was added at a final concentration of 40 ng/μl to UFs diluted 1:5 and to KCl salt solutions at final concentrations of 0.8 and 3.8 M KCl. DNA in 25 mM phosphate buffer, pH 7.0, served as control. These in vitro solutions were irradiated using a 60Co gamma source (dose rate = 3.5 kGy/hr; Uniformed Services University of the Health Sciences, Bethesda, MD) at the following doses: 0, 0.25, 0.5, 1, 2, 4, 6, 8, 10, 12, and 15 kGy. The resulting DNA fragments were electrophoresed on a 0.9% agarose-Tris-borate-EDTA (TBE) gel and visualized using ethidium bromide staining.
Enzyme protection assay.
The restriction enzyme DdeI was added at a final concentration of 1 unit/μl to UFs diluted 1:5, to 25 mM phosphate buffer (PiB), pH 7.0, and to a solution of 0.8 M KCl, final concentration. The solutions were irradiated using a 60Co gamma source (dose rate = 3.5 kGy/hr; Uniformed Services University of the Health Sciences, Bethesda, MD) at the following doses: 0, 0.5, 1, 2, 4, 6, 8, 10, 12, and 15 kGy. Samples were kept on ice until digestion of 1 μg of pUC19 DNA using 1 U of enzyme from each irradiated solution at 37°C for 1 h. The resulting pUC19 DNA fragments were separated by electrophoresis on 1% agarose-TBE gels and visualized with ethidium bromide staining.
Determination of amino acid concentration.
Free and total amino acid concentrations in the UFs of H. salinarum, E. coli, and P. putida were determined by using a ninhydrin assay (10). Briefly, tryptophan standard solutions were prepared at concentrations of from 0 to 200 nmol tryptophan. Ninhydrin reagent (Sigma-Aldrich, St. Louis, MO) was added to the UFs and to the standards and boiled for 20 min. Isopropanol was added to 30% final concentration, and the absorbance was read at 570 nm. A standard curve was constructed based on tryptophan standards to determine the free amino acid concentrations in the UFs. Determination of the total amino acid concentration was performed with an acid hydrolysis as described in reference 60 before assaying the free amine concentration with the ninhydrin assay. In short, the UFs were diluted 1:10 in ddH2O and an equal amount of 10.5 N HCl was added. The mixture was flushed with nitrogen, sealed in a glass ampoule, and incubated at 110°C for 24 h. Ninhydrin reagent (Sigma-Aldrich, St. Louis, MO) was added to the resulting digestions, and amino acid concentrations were measured as described above.
ICP-MS and ion chromatography.
The Mn, Fe, and PO4 concentrations in H. salinarum, E. coli, and P. putida UFs and cells (Mn and Fe) were determined using induced coupled plasma (ICP)-MS (Mn and Fe) and ion chromatography (PO4) at the Division of Environmental Health Engineering, Johns Hopkins University School of Public Health. For ICP-MS analysis, 50 μl of UF was transferred to a precleaned 15-ml polystyrene tube and diluted to a final volume of 1.5 ml with 1% HNO3 plus 0.5% 1 N HCl. Cells were prepared by adding 1.5 ml of concentrated HNO3 to a pellet of 1012 cells, vortexing, and diluting 50 μl of the digest into 4.95 ml of H2O, yielding a 1% final concentration of HNO3. Internal standards (Mn or Fe) were added to each sample to monitor for sample matrix effects of the plasma. Analysis was performed with an Agilent 7500ce induced coupled plasma-mass spectrometer (Agilent Technologies, Santa Rosa, CA). A standard calibration curve was generated from multielement standards (Elements, Inc., Shasta Lake, CA) at the following concentrations: 0, 1, 5, 10, 50, 100, 500, and 1,000 μg/liter. The reported sample concentrations of Mn and Fe were blank and dilution corrected. Standard Reference Material 1643e (trace elements in water; National Institute of Standards and Technology, Gaithersburg, MD) was used to test the accuracy of sample preparation and was prepared in the same manner as the samples. For ion chromatography analysis, 25 μl of UF was transferred into a precleaned Dionex IC vial (Dionex Corp., Sunnyvale, CA), MilliQ water was added up to a 1.5-ml final volume, and the sample was vortexed to ensure thorough mixing. Analysis was performed using a Dionex DX600 ion chromatograph (Dionex Corp., Sunnyvale, CA). A standard calibration curve was generated from a multianion solution (Elements, Inc., Shasta Lake, CA) containing the anion of interest (PO4). The concentrations of the calibration curve were as follows: 0, 1, 2, 4, 6, 12, 16, and 20 μg/ml. Samples were run on an IonPac AS14A anion exchange column (4- by 250-mm; Dionex Corp., Sunnyvale, CA) and AS14A guard column (3- by 150-mm; Dionex Corp., Sunnyvale, CA) using 1.08 mM Na2CO3 and 1.02 mM NaHCO3 as the eluent. Samples were suppressed using an ASRS 4-mm suppressor (Dionex Corp., Sunnyvale, CA) with a current of 100 mA. Samples were eluted for 30 min to ensure complete anion exchange. Anion retention times (±5%) were determined based upon the certificate of analysis for the column. The sample concentrations of PO4 were reported as the average of the two replicates after blank and dilution correction.
LC-MS analysis.
The nucleoside and nucleotide composition of the UFs was determined by LC-MS. Twenty microliters of ultrafiltrates was injected onto an Agilent prep C18 column (LiChrosphere 125-mm by 4-mm, 5-mm bead size RP-18; Agilent, Santa Clara, CA) at 45°C and subjected to a 0.9-ml/min isocratic elution with 0.1 M triethanolamine acetate, pH 6.5, using an Agilent 1100 high-performance liquid chromatograph (HPLC) (Agilent 1100 LC-MS system; Agilent, Santa Clara, CA). UV peaks were detected based on their UV absorbance at 254 and 270 nm. For the MS analysis, the flow from the HPLC (0.9 ml/min) was pumped into the MS electrospray chamber with the addition of 0.1 ml/min of 1% formic acid in methanol. The MS was set up for optimal nucleotide/nucleoside ionization by using a fragmentor voltage of 350 V and a capillary voltage of 4,000 V. At these settings, doubly charged ions were minimized and the total ion abundance of the singly charged parent was at a maximum. Standards (uracil, inosine, uridine, adenosine, cytidine, thymidine, guanidine, 2′-deoxyadenosine, 2′-deoxycytidine, 2′-deoxyguanosine, 2′-deoxyuridine, and 2′-deoxythymidine) purchased from Sigma (Sigma, St. Louis, MO) were run under the same conditions for each UF analysis. Peak areas of the UV spectra were used to calculate base and nucleoside concentrations with standard curves for each standard.
RESULTS
Cellular lesions following oxidative stress.
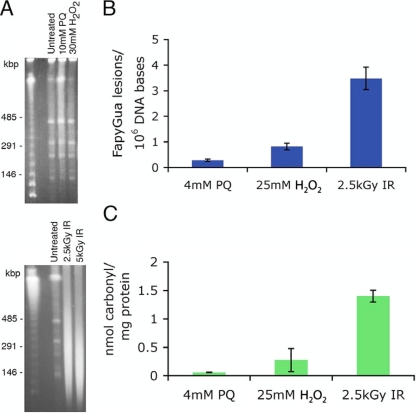
The cellular effects of oxidative stress from exposure to chemical oxidants and to IR were determined for H. salinarum cells exposed to H2O2, paraquat (an O2− generator), and 60Co gamma radiation. Oxidative damage to DNA and proteins was measured at doses that yielded 80% survival of the cells (32, 64). Hydrogen peroxide and IR were applied directly to H. salinarum cells, whereas O2− radicals were produced indirectly via exposure to paraquat (25). DNA double-strand breaks (DSBs) were visualized using PFGE; extensive DNA fragmentation was observed when H. salinarum cells were exposed to 2.5 kGy IR but not in cells exposed to H2O2 or O2−, even at doses higher than those yielding 80% survival (Fig. 1A). The modified DNA base 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua) was quantified using GC-MS with isotope dilution as a measure of the oxidation level of the DNA (34). Consistent with the PFGE results, the number of FapyGua lesions accumulated per 106 DNA bases was significantly higher in cells exposed to IR than in cells exposed to H2O2 or O2− (Fig. 1B). We also measured the level of 8-hydroxyguanine in cells using the same method. No statistically significant increase was observed in the level of this product by any treatment used in this work (data not shown). Thus, for H. salinarum cells treated with H2O2 or O2−, cell killing was not explained by the levels of DNA damage. Protein oxidation was determined by immunodetection of carbonyl groups in H. salinarum protein extracts (57); we found a significantly higher level of protein oxidation in H. salinarum cells exposed to IR than in cells exposed to H2O2 or O2− (Fig. 1C), although the pattern of oxidized proteins visualized by immunoblotting was comparable for the three treatments (data not shown).
FIG. 1.
Quantification of cellular lesions in H. salinarum Δura3 cells exposed to H2O2, paraquat, and IR. (A) DNA DSBs were visualized by PFGE; samples were taken before (untreated) and following treatments with H2O2, paraquat, and IR and then embedded in InCert agarose plugs at a final density of 1× 109 cells/ml; the plugs were digested with XbaI prior to gel electrophoresis. The first lanes are molecular size ladders. (B) The level of modified base FapyGua was measured using GC-MS with isotope dilution; the numbers of FapyGua lesions per 106 DNA bases were background subtracted. (C) Protein carbonyl residues were quantified by immunodetection; the nmol of carbonyl residues per mg of proteins was background subtracted. All data shown are the average results of at least three replicates; the uncertainties are standard errors.
Survival and cellular oxidation in ROS detoxification mutants.
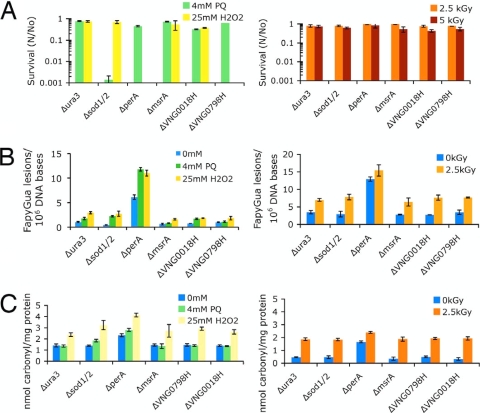
In-frame knockout mutants for major oxygen detoxification enzymes were previously tested for their survival following exposure to H2O2 and O2− and to IR (Fig. 2A). The survival of the superoxide dismutase 1 and 2 double mutant, the Δsod1/2 strain, was decreased by three orders of magnitude compared to the survival of the control Δura3 strain under O2− but not H2O2 stress. In contrast, less than 0.001% survivors were found for the peroxidase mutant, the ΔperA strain (VNG6294G), and the putative Dyp-type peroxidase mutant, the ΔVNG0798H strain, with H2O2 but not with O2−. The strain with a mutation of MsrA, a methionine sulfoxide reductase involved in the removal of methionine sulfoxide residues, did not show any loss in survival compared to the survival of the control strain for either treatment. Remarkably, all the mutants tested showed the same rate of survival as the control strain, H. salinarum Δura3, when exposed to 2.5 or 5 kGy of IR (Fig. 2A).
FIG. 2.
Survival and quantification of cellular lesions of H. salinarum Δura3 and mutant strains exposed to H2O2, paraquat, and IR. (A) Survival was calculated as the average ratio (N/No) of surviving CFU from treated (N) compared to untreated (No) cultures. (B) Modified base FapyGua was quantified using GC-MS with isotope dilution. (C) Protein carbonyl residues were quantified by immunodetection. All data shown are the average results of at least three replicates; the uncertainties are standard errors.
The levels of DNA and protein oxidation were assessed in the ROS detoxification mutants exposed to H2O2, O2−, and IR, using GC-MS to quantify FapyGua lesions and immunodetection to quantify carbonyl residues (Fig. 2B and C). We did not find a significant correlation between the level of DNA and protein oxidation and the level of survival for the ROS detoxification mutants we tested. The number of FapyGua lesions was similar in cells treated with H2O2 and O2−, whereas we found large differences (≥3 logs) in survival between the two treatments, in particular for the mutants with Δsod1/2, ΔperA, ΔVNG0018H (a catalase), and ΔVNG0798H mutations (Fig. 2A and B). A similar level of DNA oxidation was found for the ΔVNG0018H and ΔVNG0798H mutants treated with H2O2, but the survival of the ΔVNG0798H mutant decreased by more than 99.9% when treated with H2O2, while the survival of the ΔVNG0018H mutant only decreased by ∼60% compared to that of the control Δura3 strain. Likewise, the levels of DNA oxidation in the Δsod1/2 and ΔVNG0798H mutants or the ΔVNG0018H mutant were comparable, but the survival of the mutants exposed to H2O2 or O2− was very different. Notably, the level of protein oxidation was higher in all the mutants exposed to H2O2, and yet, we did not find a correlation between protein oxidation and survival for these chemical oxidants (Fig. 2A and C). For example, an increase in protein oxidation in the Δsod1/2 mutant exposed to H2O2 relative to the level in this mutant exposed to O2− did not result in decreased survival of this mutant under H2O2 treatment.
The levels of oxidatively induced damage to DNA and protein in cells exposed to IR showed a different picture. Comparable yields of FapyGua and carbonyl residues were found in the mutants and the control strain (Δura3), which displayed similar levels of survival with exposure to 2.5 kGy and 5.0 kGy (Fig. 2A). An exception was the ΔperA mutant, which accumulated high levels of oxidative damage even without treatment. This strain grew very slowly under any condition tested and showed high levels of background oxidative stress.
Protection against IR by nonenzymatic processes.
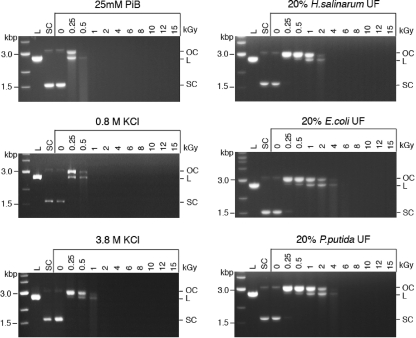
While ROS detoxification enzymes are required for the survival of H. salinarum cells treated with chemical oxidants, we showed that the same enzymes were dispensable for survival following IR. To further investigate the molecular basis of IR protection in H. salinarum, we prepared an ultrafiltered, protein-free cell extract of H. salinarum (the IR dose at which 10% of the cells survive [D10] is 5.0 kGy) (64) and tested its ability to protect DNA and enzyme activity from IR in vitro. Ultrafiltrates (UFs) (ultrafiltered, protein-free cell extracts) from two radiation-sensitive bacteria, E. coli (D10 = 0.7 kGy) and P. putida (D10 = 0.25 KGy) (14), were prepared and used in those assays for comparison with H. salinarum UF. Plasmid DNA was irradiated at increasing doses of IR in phosphate buffer (PiB), in UFs (diluted 1:5) prepared from H. salinarum (UFHs), E. coli (UFEc), and P. putida (UFPp), in 0.8 M KCl, which corresponds to the salt concentration in the 1:5-diluted UFHs, or in 3.8 M KCl, which is the intracellular concentration of KCl in H. salinarum (44, 48) (Fig. 3). Following IR, the damage to the supercoiled (SC) plasmid was analyzed by agarose gel electrophoresis as described previously (34). The plasmid DNA damage assay is extremely sensitive to HO generated by IR, where one single-strand break (SSB) in an SC plasmid molecule yields an open circular (OC) form that is readily distinguished from the SC form (Fig. 3). In this assay, the SC form was progressively broken into an OC form, then into a linear form (L), and finally completely degraded as the IR doses increased (Fig. 3, 25 mM PiB). While the high salt (0.8 M KCl) provided some protection to the plasmid DNA, up to 0.5 kGy, compared to the protection provided by the phosphate buffer (PiB), the UFs from H. salinarum and the two bacteria protected the plasmid to 2 kGy (Fig. 3). However, we did not find a significant difference in the protection afforded by H. salinarum UF compared to the protection conferred by the UFs of E. coli and P. putida, which are IR sensitive.
FIG. 3.
Protection of DNA integrity. Plasmid pUC19 DNA was irradiated up to 15 kGy in 25 mM PiB, in 0.8 and 3.8 M KCl, and in protein-free UFs of H. salinarum, E. coli, and P. putida (diluted 1:5). DNA topology was analyzed by agarose gel electrophoresis. The first lanes are molecular size ladders. L, linear; SC, supercoiled; OC, open circular plasmid.
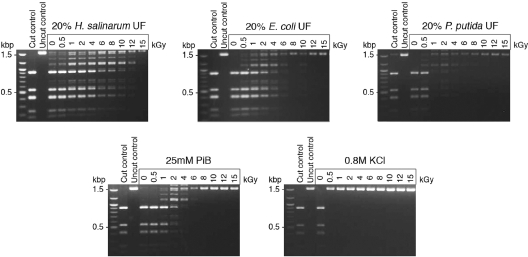
We next tested the ability of the UFs to protect the activity of a purified enzyme irradiated in aqueous solution, the restriction enzyme DdeI. The residual activity of the enzyme was measured after IR by restriction digest of plasmid DNA and analysis of the fragments by agarose gel electrophoresis (Fig. 4). We demonstrated extensive protection of the irradiated enzyme using H. salinarum UF, up to 12 kGy, but the UFs of E. coli and P. putida were much less protective (Fig. 4).
FIG. 4.
Protection of enzyme activity. The restriction enzyme DdeI was irradiated up to 15 kGy in 25 mM PiB, protein-free UFs of H. salinarum, E. coli, and P. putida (diluted 1:5), and 0.8 M KCl. Residual restriction enzyme activity was assayed by the digestion of pUC19 plasmid DNA; fragments were analyzed by agarose gel electrophoresis. The first lanes are molecular size ladders.
Composition of the protein-free cell extracts.
Orthophosphate-Mn complexes have been found to play a major role in vivo and in vitro in resistance to oxidative stress, radiation resistance, and the scavenging of ROS (13, 39). To determine a potential role of orthophosphate complexes with Mn in the radioprotection by H. salinarum UF, we measured the concentrations of Mn, Fe, and phosphate in the UFs of H. salinarum, E. coli, and P. putida, using ICP-MS and ion chromatography (Table 1). Compared to the UFs of E. coli and P. putida, the H. salinarum UF was highly enriched in Mn (∼100 times) and also enriched in orthophosphate (Pi) (4 times). Whole-cell analysis showed that H. salinarum accumulated 10 times more Mn than E. coli and P. putida (Table 1). Analysis of the Mn concentration during the preparation of the UFs revealed that 30% of the Mn was retained in the UF of H. salinarum, whereas only 2% was carried through the UFs of E. coli and P. putida.
TABLE 1.
Ultrafiltrate and whole-cell concentrations of Mn, Fe, and PO4
| Organism | Concn in: |
||||
|---|---|---|---|---|---|
| Ultrafiltrate |
Whole cells |
||||
| Mn (μM) | Fe (μM) | PO4 (mM) | Mn (ng/109 cells) | Fe (ng/109 cells) | |
| H. salinarum | 87 | 8.9 | 22 | 155 | 818 |
| E. coli | 0.6 | 3.5 | 5.9 | 14 | 645 |
| P. putida | 0.9 | 6.1 | 4.5 | 18 | 1045 |
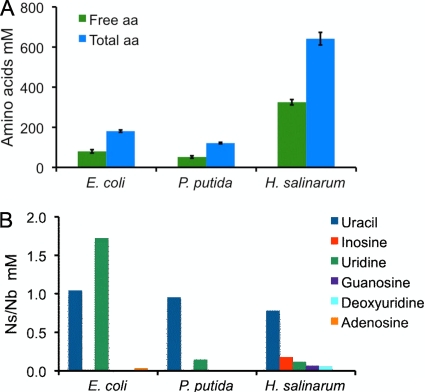
We also found that total amino acids and peptides were significantly overrepresented in UFHs compared to their levels in UFEc and UFPp (Fig. 5A). The amounts of nucleotides, nucleosides, and bases in the UFs were measured using LC-MS. Uracil and uridine were the most abundant species detected in all the UFs, and inosine, guanosine, and deoxyuridine were only present in the H. salinarum UF. Relative to their levels in UFEc and UFPp, we did not find any large increase of nucleobases or nucleosides in UFHs (Fig. 5B).
FIG. 5.
Total and free amino acids (aa) (A) and nucleosides (Ns) and nucleobases (Nb) (B) measured in the protein-free UFs of H. salinarum, E. coli, and P. putida.
DISCUSSION
The molecular response of H. salinarum cells exposed to IR was dramatically different from the response of cells exposed to chemical oxidants, as determined by DNA and protein damage assays. This is the first time that such a detailed analysis of DNA and protein lesions has been carried out, comparing oxidative stress from chemicals and from IR. At doses that yielded similar levels of survival, we found a significantly higher level of cellular damage in H. salinarum cells exposed to IR than to H2O2 and O2−, underlining cellular targets for the toxicity of IR that are different from those damaged by chemical oxidants. In this work, O2− was generated by exposure of the cells to paraquat (N,N′-dimethyl-4,4′-bipyridinium dichloride), which produces O2− from molecular oxygen by redox cycling (5, 19). This reaction occurs principally in the cell membrane, where a localized production of O2− directly interferes with energy transduction systems, thereby disrupting the cell's redox homeostasis (5, 61). It is also known that O2− generation by paraquat redox cycling results in the depletion of intracellular reducing equivalents, such as NADPH (5, 23, 25, 59). The highly localized production of O2− by paraquat and the O2−-mediated insults to the cells represent primary toxic events leading to the disruption of specific energy metabolism pathways and, ultimately, cell death. In concurrence, it has been reported that the exposure of E. coli cells to paraquat selectively caused the oxidation of the β-subunit of F0F1-ATPase, resulting in early depletion of ATP levels and loss of membrane potential (6, 61). While paraquat damage is localized to a subcellular compartment, H2O2 is readily diffusible throughout the cell. Exogenous H2O2 rapidly diffuses across the cellular membrane, and in the presence of transition metal ions, oxidizes methionine and cysteine residues and attacks [4Fe-4S] clusters of labile dehydratases by direct oxidation of their solvent-exposed clusters (30). A Fenton-like reaction produces a catalytically inactive cluster, resulting in enzyme inactivation, pathway failure, and the proliferation of ROS via the release of Fe2+ (29, 31). In E. coli, H2O2 has been found to preferentially target dehydratases, such as aconitase and fumarase (tricarboxylic acid [TCA] cycle), and isopropylmalate isomerase (leucine biosynthesis), interfering directly with essential metabolic pathways in the cell (8, 21, 31, 62). Measurement of the level of protein carbonylation in E. coli also revealed that exposure to H2O2 resulted in specific oxidation of elongation factor G (protein synthesis), heat shock protein DNA K (chaperone function), enolase (glucose catabolism), and ATPase subunits (6, 61). The data presented herein support the conclusion that the exposure of H. salinarum to paraquat-generated O2− and exogenously administered H2O2 affects specific cellular pathways and processes that cause cell death before extensive and generalized cellular damage occurs. In contrast, high levels of DNA and protein damage in H. salinarum cells exposed to IR support a mechanism of oxidation that is more generalized, targeting all cellular compartments and macromolecules.
The management of oxidative stress in microorganisms is attributed to specific enzymatic detoxification systems, including superoxide dismutases (SOD), catalases, and peroxidases, and antioxidants, such as glutathione (58). In cells exposed to chemical oxidants, such as H2O2 and O2−, those enzymes were found to be essential for survival (30). We show here that the deletion of SODs, catalases, and peroxidases renders H. salinarum highly sensitive to H2O2 and O2− but not IR. These results are substantiated by the results of our functional genomic studies of H. salinarum exposed to IR, H2O2, and O2−, where we found an increase in mRNA levels for SODs, catalases, and peroxidases with chemical oxidant stressors but not with IR (32, 64). In previous work on D. radiodurans exposed to high doses of IR, SOD and catalase mutants showed almost no increase in IR sensitivity compared to that of the wild type (14, 37). These findings support a key role for nonenzymatic antioxidant processes in the survival of H. salinarum exposed to IR and underline a common mechanism for radiation resistance in Bacteria and Archaea.
We report an increase in H. salinarum oxidative lesions in DNA and proteins in vivo at increasing IR doses. Previous studies showed that IR-resistant organisms had significantly lower levels of protein damage than IR-sensitive microorganisms for the same IR doses, whereas the DNA was equally damaged (13, 34, 36), suggesting that, in H. salinarum, proteins were protected from extensive damage by IR, allowing for DNA repair and survival. This is supported by our in vitro data, where we showed radioprotection of protein activity by H. salinarum UF, while the DNA was not significantly protected. The results from our studies with H. salinarum (34) and those of others with D. radiodurans (12, 13, 36) demonstrate that the radiation resistance of an organism stems from its ability to protect its proteins from extensive oxidative damage.
The results of whole-cell analysis showed that H. salinarum has a Mn/Fe concentration ratio (0.19) that is of the same order of magnitude as the cellular Mn/Fe ratio of D. radiodurans (0.24) and much higher than those of radiation-sensitive bacteria (0.0072 for E. coli and <0.0001 for P. putida) (14, 34). Here, we showed that H. salinarum UF was highly enriched in Mn and that 30% of the cellular Mn was retained in the UF. Although Mn2+ is most commonly associated with a role as a catalytic cofactor of proteins, a significant fraction of the total Mn content of H. salinarum appears not to be bound to proteins and is presumably present as small Mn2+ complexes. A strong mechanistic link has been demonstrated between high Mn/Fe concentration ratios and radiation resistance in both Bacteria and Archaea (14, 34), and IR-resistant bacteria have been shown to accumulate low-molecular-weight Mn2+ complexes (12). Antioxidant properties of Mn have been demonstrated in vitro by the scavenging of HO and O2− by Mn2+, Mn-PO4 complexes, and Mn associated with metabolic intermediates, such as lactate and malate (1-3, 17, 18, 56). In particular, Mn2+ and orthophosphate, which do not significantly scavenge HO, can form complexes that catalytically remove O2− in vitro via a disproportionation mechanism (1, 2). Whereas HO generated during IR reacts indiscriminately with all of the cell's macromolecules, the most severe damage by O2− and H2O2 is to proteins that contain iron-sulfur groups, cysteine residues, and other sites where iron-catalyzed oxidation takes place (29). Those proteins are at high risk for inactivation unless ROS are catalytically removed by Mn-orthophosphate complexes, preventing site-specific oxidative protein damage (11, 14). Measurements of Mn speciation in yeast using 1H and 31P electron-nuclear double resonance (ENDOR) provided evidence for a major in vivo role of orthophosphate-Mn2+ complexes in the resistance to oxidative stress of this organism (39). Recent work with D. radiodurans also demonstrated a critical role for Mn2+ complexes, in the scavenging of ROS, in the extreme radiation resistance of this organism (11, 14). In this work, the high level of protection of protein against IR afforded by H. salinarum UF combined with the high Mn and orthophosphate concentrations in the UF—and in the cells of the organism—clearly demonstrate that the scavenging of IR-produced ROS by Mn complexes is also key for H. salinarum protein protection and its survival of exposure to IR.
The increased accumulation of amino acids and small peptides (<3,000 Da, ∼20 amino acids in length) in H. salinarum UF compared to their accumulation in the UFs of E. coli and P. putida also suggest a ROS-scavenging activity of those small molecules, resulting in protection of the cell's macromolecules against IR. The antioxidant properties of free amino acids and small peptides have also been reported in various protein hydrolysates (40, 43, 52, 65). In mackerel protein hydrolysate, peptides of 1,400 Da in size had the strongest in vitro antioxidant activity (65), and a potent antioxidant peptide isolated from algae was shown to scavenge HO, O2−, peroxyl radicals, and other free radicals in vitro (52). The amino acid composition, structure, and solvent accessibility of the amino acids for this 11-residue polypeptide were essential for its antioxidant activity, with the most reactive amino acids being those with nucleophilic sulfur-containing side chains, aromatic side chains, or imidazole-containing side chains (16, 52). In addition to Mn2+, phosphate, and small peptides, D. radiodurans also accumulates large amounts of uridine, adenosine, and uracil, and in vitro experiments to analyze protein protection against IR have confirmed the ROS-scavenging properties of uridine in the presence of Mn and phosphate (12). The UF of H. salinarum was not enriched in nucleosides or bases, illustrating that organisms can adopt diverse strategies for scavenging ROS. Cyanobacteria, fungi, microalgae, and small invertebrates were found to accumulate mycosporines and mycosporine-like amino acids as defense against oxidative stress (41, 66); spores of Bacillus subtilis have high intracellular levels of both Mn2+ and dipicolinic acid (50, 51), making those spores highly resistant to IR, and radiation-resistant cyanobacteria accumulate nonreducing disaccharides, such as trehalose, together with Mn2+ (4, 53).
The results from this work clearly show that large amounts of Mn, orthophosphate, and other small ROS-scavenging molecules represent a metabolic route to achieve a high level of resistance to IR in H. salinarum. This is a nonenzymatic route that is not inducible—preconditioning of H. salinarum with a low IR dose did not increase resistance (35)—and represents a first line of response to neutralizing the high levels of ROS produced by IR. This immediate response does not require a role for detoxification enzymes, and indeed, we showed that, within 30 min of IR exposure, the mRNA levels for SODs, catalases, and peroxidases did not increase, in contrast to the results of H2O2 and O2− exposure (32, 64). The high levels of survival of the detoxification enzyme mutants also showed that those nonenzymatic antioxidant defenses were sufficient to eliminate most of the deleterious ROS from IR exposure in H. salinarum and promote cell survival.
We previously showed that the high halide concentration in the cytoplasm of H. salinarum was a major factor in protecting its macromolecules against the oxidative effects of IR (34) and resulted directly from its adaptation to a high-salt environment. H. salinarum is found in hypersaline pools, where it is subjected to cycles of desiccation and rehydration (15), and halophiles have been shown to survive extended periods of time encased inside salt crystals (42). The adaptation of H. salinarum to desiccation is another example of the link between IR resistance and desiccation that has been reported for D. radiodurans and other dry-climate-adapted bacteria (20, 38, 45) and for some lower eukaryotes, such as rotifers and tardigrades (22, 26). It is still too early to conclude whether there is a universal role for Mn in IR protection, but it is important to recognize the extreme diversity of microorganisms and their adaptation to a wide range of environmental conditions, and therefore, the great potential for novel mechanisms for radioprotection.
Acknowledgments
This work was supported by the AFOSR (grant FA95500710158 to J.D.) and NIH (grants P50GM076547 and 1R01GM077398-01A2), and portions conducted by ENIGMA were supported by the Office of Biological and Environmental Research of the Office of Science, U.S. Department of Energy, under contract no. DE-AC02-05CH11231 to N.S.B. The ICP-MS work was supported in part by the Maryland Cigarette Restitution Fund Program at Johns Hopkins and the NIEHS Center (grant P30 ES00319).
We thank Elena Gaidamakova and Vera Matrosova at the Uniformed Services University of the Health Sciences (USUHS), Bethesda, MD, for helpful discussions and for their technical support using the gamma source at USUHS and M. J. Daly for his advice and support.
Certain commercial equipment or materials are identified in this paper in order to specify adequately the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
Footnotes
Published ahead of print on 28 January 2011.
REFERENCES
- 1.Archibald, F. S., and I. Fridovich. 1982. The scavenging of superoxide radical by manganous complexes: in vitro. Arch. Biochem. Biophys. 214:452-463. [DOI] [PubMed] [Google Scholar]
- 2.Barnese, K., E. B. Gralla, D. E. Cabelli, and J. S. Valentine. 2008. Manganous phosphate acts as a superoxide dismutase. J. Am. Chem. Soc. 130:4604-4606. [DOI] [PubMed] [Google Scholar]
- 3.Berlett, B. S., P. B. Chock, M. B. Yim, and E. R. Stadtman. 1990. Manganese(II) catalyzes the bicarbonate-dependent oxidation of amino acids by hydrogen peroxide and the amino acid-facilitated dismutation of hydrogen peroxide. Proc. Natl. Acad. Sci. U. S. A. 87:389-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billi, D., E. I. Friedmann, K. G. Hofer, M. Grilli Caiola, and R. Ocampo-Friedmann. 2000. Ionizing-radiation resistance in the desiccation-tolerant cyanobacterium Chroococcidiopsis. Appl. Environ. Microbiol. 66:1489-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bus, J. S., and J. E. Gibson. 1984. Paraquat: model for oxidant-initiated toxicity. Environ. Health Perspect. 55:37-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabiscol, E., J. Tamarit, and J. Ros. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3:3-8. [PubMed] [Google Scholar]
- 7.Calabrese, V., et al. 2005. Oxidative stress, mitochondrial dysfunction and cellular stress response in Friedreich's ataxia. J. Neurol. Sci. 233:145-162. [DOI] [PubMed] [Google Scholar]
- 8.Carlioz, A., and D. Touati. 1986. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 5:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chobotova, K. 2009. Aging and cancer: converging routes to disease prevention. Integr. Cancer Ther. 8:115-122. [DOI] [PubMed] [Google Scholar]
- 10.Clark, J. M. 1964. Experimental biochemistry. W. H. Freeman and Co, New York, NY.
- 11.Daly, M. J. 2009. A new perspective on radiation resistance based on Deinococcus radiodurans. Nat. Rev. Microbiol. 7:237-245. [DOI] [PubMed] [Google Scholar]
- 12.Daly, M. J., et al. 2010. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS One 5:e12570. doi: 10.1371/journal.pone.0012570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daly, M. J., et al. 2007. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 5:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daly, M. J., et al. 2004. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306:1025-1028. [DOI] [PubMed] [Google Scholar]
- 15.DasSarma, S., and P. Arora. 2001. Halophiles, p. 1-9, Encyclopedia of life sciences. Nature Publishing Group, New York, NY.
- 16.Elias, R. J., S. S. Kellerby, and E. A. Decker. 2008. Antioxidant activity of proteins and peptides. Crit. Rev. Food Sci. Nutr. 48:430-441. [DOI] [PubMed] [Google Scholar]
- 17.Ezra, F. S., D. S. Lucas, R. V. Mustacich, and A. F. Russell. 1983. Phosphorus-31 and carbon-13 nuclear magnetic resonance studies of anaerobic glucose metabolism and lactate transport in Staphylococcus aureus cells. Biochemistry 22:3841-3849. [DOI] [PubMed] [Google Scholar]
- 18.Ezra, F. S., D. S. Lucas, and A. F. Russell. 1984. 31P-NMR and ESR studies of the oxidation states of manganese in Staphylococcus aureus. Biochim. Biophys. Acta 803:90-94. [DOI] [PubMed] [Google Scholar]
- 19.Fisher, M. T. 1993. On the assembly of dodecameric glutamine synthethase from stable chaperonin complexes. J. Biol. Chem. 268:13777-13779. [PubMed] [Google Scholar]
- 20.Fredrickson, J. K., et al. 2008. Protein oxidation: key to bacterial desiccation resistance? ISME J. 2:393-403. [DOI] [PubMed] [Google Scholar]
- 21.Gardner, P. R., and I. Fridovich. 1991. Superoxide sensitivity of the Escherichia coli aconitase. J. Biol. Chem. 266:19328-19333. [PubMed] [Google Scholar]
- 22.Gladyshev, E., and M. Meselson. 2008. Extreme resistance of bdelloid rotifers to ionizing radiation. Proc. Natl. Acad. Sci. U. S. A. 105:5139-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gram, T. E. 1997. Chemically reactive intermediates and pulmonary xenobiotic toxicity. Pharmacol. Rev. 49:297-341. [PubMed] [Google Scholar]
- 24.Grant, W. D. 2004. Life at low water activity. Philos. Trans. R. Soc. B Biol. Sci. 359:1249-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassan, H. M., and I. Fridovich. 1979. Paraquat and Escherichia coli. Mechanism of production of extracellular superoxide radical. J. Biol. Chem. 254:10846-10852. [PubMed] [Google Scholar]
- 26.Horikawa, D. D., et al. 2006. Radiation tolerance in the tardigrade Milnesium tardigradum. Int. J. Radiat. Biol. 82:843-848. [DOI] [PubMed] [Google Scholar]
- 27.Hutchinson, F. 1985. Chemical changes induced in DNA by ionizing radiation. Prog. Nucleic Acid Res. Mol. Biol. 32:115-154. [DOI] [PubMed] [Google Scholar]
- 28.Imlay, J. A. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77:755-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imlay, J. A. 2006. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59:1073-1082. [DOI] [PubMed] [Google Scholar]
- 30.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 31.Jang, S., and J. A. Imlay. 2007. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 282:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaur, A., et al. 2010. Coordination of frontline defense mechanisms under severe oxidative stress. Mol. Syst. Biol. 6:393. doi: 10.1038/msb.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kish, A., and J. DiRuggiero. 2008. Rad50 is not essential for the Mre11-dependent repair of DNA double strand breaks in Halobacterium sp. str. NRC-1. J. Bacteriol. 190:5210-5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kish, A., et al. 2009. Salt shield: intracellular salts provide protection against ionizing radiation in the halophilic archaeon, Halobacterium salinarum NRC-1. Environ. Microbiol. 11:1066-1078. [DOI] [PubMed] [Google Scholar]
- 35.Kottemann, M., A. Kish, C. Iloanusi, S. Bjork, and J. DiRuggiero. 2005. Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation. Extremophiles 9:219-227. [DOI] [PubMed] [Google Scholar]
- 36.Kriško, A., and M. Radman. 2010. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc. Natl. Acad. Sci. U. S. A. 107:14373-14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markillie, L. M., S. M. Varnum, P. Hradecky, and K. K. Wong. 1999. Targeted mutagenesis by duplication insertion in the radioresistant bacterium Deinococcus radiodurans: radiation sensitivities of catalase (katA) and superoxide dismutase (sodA) mutants. J. Bacteriol. 181:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattimore, V., K. S. Udupa, G. A. Berne, and J. R. Battista. 1995. Genetic characterization of forty ionizing radiation-sensitive strains of Deinococcus radiodurans: linkage information from transformation. J. Bacteriol. 177:5232-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McNaughton, R. L., et al. 2010. Probing in vivo Mn2+ speciation and oxidative stress resistance in yeast cells with electron-nuclear double resonance spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 107:15335-15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendis, E., N. Rajapakse, and S. K. Kim. 2005. Antioxidant properties of a radical-scavenging peptide purified from enzymatically prepared fish skin gelatin hydrolysate. J. Agric. Food Chem. 53:581-587. [DOI] [PubMed] [Google Scholar]
- 41.Oren, A., and N. Gunde-Cimerman. 2007. Mycosporines and mycosporine-like amino acids: UV protectants or multipurpose secondary metabolites? FEMS Microbiol. Lett. 269:1-10. [DOI] [PubMed] [Google Scholar]
- 42.Park, J. S., et al. 2009. Haloarchaeal diversity in 23, 121 and 419 MYA salts. Geobiology 7:515-523. [DOI] [PubMed] [Google Scholar]
- 43.Peng, X., Y. L. Xiong, and B. Kong. 2009. Antioxidant activity of peptide fractions from whey protein hydrolysates as measured by electron spin resonance. Food Chem. 113:196-201. [Google Scholar]
- 44.Pérez-Fillol, M., and F. Rodríguez-Valera. 1986. Potassium ion accumulation in cells of different halobacteria. Microbiologia 2:73-80. [PubMed] [Google Scholar]
- 45.Rainey, F. A., et al. 2005. Extensive diversity of ionizing-radiation-resistant bacteria recovered from Sonoran desert soil and description of nine new species of the genus Deinococcus obtained from a single soil sample. Appl. Environ. Microbiol. 71:5225-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddy, P., P. Jaruga, T. O'Connor, H. Rodriguez, and M. Dizdaroglu. 2004. Overexpression and rapid purification of Escherichia coli formamidopyrimidine-DNA glycosylase. Protein Expr. Purif. 34:126-133. [DOI] [PubMed] [Google Scholar]
- 47.Riley, P. A. 1994. Free radicals in biology: oxidative stress and the effects of ionizing radiation. Int. J. Radiat. Biol. 65:27-33. [DOI] [PubMed] [Google Scholar]
- 48.Roberts, M. F. 2005. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Systems 4:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 50.Setlow, B., S. Atluri, R. Kitchel, K. Koziol-Dube, and P. Setlow. 2006. Role of dipicolinic acid in resistance and stability of spores of Bacillus subtilis with or without DNA-protective alpha/beta-type small acid-soluble proteins. J. Bacteriol. 188:3740-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Setlow, P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101:514-525. [DOI] [PubMed] [Google Scholar]
- 52.Sheih, I. C., T. K. Wu, and T. J. Fang. 2009. Antioxidant properties of a new antioxidative peptide from algae protein waste hydrolysate in different oxidation systems. Bioresource Technol. 100:3419-3425. [DOI] [PubMed] [Google Scholar]
- 53.Shirkey, B., et al. 2003. Genomic DNA of Nostoc commune (Cyanobacteria) becomes covalently modified during long-term (decades) desiccation but is protected from oxidative damage and degradation. Nucleic Acids Res. 31:2995-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stadtman, E. R. 1993. Oxidation of free amino acids and amino acid residues in proteins by radiolysis and by metal-catalyzed reactions. Annu. Rev. Biochem. 62:797-821. [DOI] [PubMed] [Google Scholar]
- 55.Stadtman, E. R. 1992. Protein oxidation and aging. Science 257:1220-1224. [DOI] [PubMed] [Google Scholar]
- 56.Stadtman, E. R., B. S. Berlett, and P. B. Chock. 1990. Manganese-dependent disproportionation of hydrogen peroxide in bicarbonate buffer. Proc. Natl. Acad. Sci. U. S. A. 87:384-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stadtman, E. R., and R. L. Levine. 2003. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids 25:207-218. [DOI] [PubMed] [Google Scholar]
- 58.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 59.Suntres, Z. E. 2002. Role of antioxidants in paraquat toxicity. Toxicology 180:65-77. [DOI] [PubMed] [Google Scholar]
- 60.Svensson, E., A. Skoog, and J. P. Amend. 2004. Concentration and distribution of dissolved amino acids in a shallow hydrothermal vent system, Vulcano Island (Italy). Organic Geochem. 35:1001-1014. [Google Scholar]
- 61.Tamarit, J., E. Cabiscol, J. Aguilar, and J. Ros. 1997. Differential inactivation of alcohol dehydrogenase isoenzymes in Zymomonas mobilis by oxygen. J. Bacteriol. 179:1102-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Varghese, S., Y. Tang, and J. A. Imlay. 2003. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J. Bacteriol. 185:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.von Sonntag, C. 1987. The chemical basis of radiation biology. Taylor and Francis, London, United Kingdom.
- 64.Whitehead, K., et al. 2006. An integrated systems approach for understanding cellular responses to gamma radiation. Mol. Syst. Biol. 2:47. doi: 10.1038/msb4100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, H. C., H. M. Chen, and C. Y. Shiau. 2003. Free amino acids and peptides as related to antioxidant properties in protein hydrolysates of mackerel (Scomber austriasicus). Food Res. Int. 36:949-957. [Google Scholar]
- 66.Yakovleva, I., R. Bhagooli, A. Takemura, and M. Hidaka. 2004. Differential susceptibility to oxidative stress of two scleractinian corals: antioxidant functioning of mycosporine-glycine. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 139:721-730. [DOI] [PubMed] [Google Scholar]
- 67.Zuber, P. 2009. Management of oxidative stress in Bacillus. Annu. Rev. Microbiol. 63:575-597. [DOI] [PubMed] [Google Scholar]