Abstract
The FsrABDC signal transduction system is a major virulence regulator in Enterococcus faecalis. The FsrC sensor histidine kinase, upon activation by the gelatinase biosynthesis-activating pheromone (GBAP) peptide encoded by the fsrBD genes, phosphorylates the FsrA response regulator required for the transcription of the fsrBDC and the gelE-sprE genes from the fsrB promoter and the gelE promoter, respectively. FsrA belongs to the LytTR family of proteins, which includes other virulence regulators, such as AgrA of Staphylococcus aureus, AlgR of Pseudomonas aeruginosa, and VirR of Clostridium perfringens. The LytTR DNA-binding domain that characterizes these proteins generally binds to two imperfect direct repeats separated by a number of bases that place the repeats on the same face of the DNA helix. In this study, we demonstrated that FsrA also binds to two imperfect direct repeats separated by 13 bp, based on the consensus sequence of FsrA, T/AT/CAA/GGGAA/G, which is consistent with the binding characteristics of LytTR domains.
Enterococcus faecalis is a Gram-positive commensal bacterium that in the last 2 decades has emerged as one of the leading causes of nosocomial infections (12). This opportunistic pathogen has extraordinary capacities to grow under hostile conditions and to colonize and survive in a large range of ecological niches. The mechanisms by which this bacterium is able to cross the barrier from a commensal to a major pathogen are still not well understood. Intrinsic physiological properties of E. faecalis, such as stress response capacities and inherent antibiotic resistance, may also provide an advantage during the infection process (3, 8, 24).
The ability of most bacteria to sense and adapt to changing environmental conditions is mediated largely through two-component signal transduction systems (TCS). TCS generally consist of a sensor histidine kinase and a cognate response regulator. The histidine kinase senses the signal and relays it, through the transfer of a phosphoryl group, to the response regulator, which generally acts as a transcriptional regulator and modulates gene expression (13). A total of 17 TCS and one orphan response regulator have been identified in the genome of Enterococcus faecalis V583 (10). Among these, the Fsr system (also known as RR-HK 15) has been extensively analyzed and shown to be critical for enterococcal virulence (10, 11, 18, 20, 25, 26).
The fsr locus is comprised of 4 genes, fsrA, fsrB, fsrD, and fsrC, whose products form a system that responds to the extracellular accumulation of the gelatinase biosynthesis-activating pheromone (GBAP) peptide encoded by the fsrD gene (19, 21, 26). FsrB acts as a cysteine protease-like processing enzyme involved in the processing of the FsrD peptide (19, 26). Accumulation of this peptide in the extracellular space is sensed by the FsrC membrane histidine kinase, leading to the activation of the response regulator and transcription factor FsrA. Stimulation of FsrC activity in vivo and in vitro by a chemically synthesized GBAP peptide has been demonstrated (16, 20). The FsrABDC proteins are necessary for autoregulation at a promoter located upstream of fsrB and for the expression of two E. faecalis virulence-related proteases, gelatinase (GelE) and serine protease (SprE), from a promoter located upstream of the gelE gene (26). The FsrABCD system and gelatinase have been shown to be relevant for enterococcal virulence in different animal models (9, 18, 25). Furthermore, gelatinase is required for efficient biofilm formation and is a major contributor to the pathogenesis of enterococcal endocarditis (11, 30).
The FsrABDC system is a homologue of the AgrABCD virulence system of Staphylococcus aureus (23). The FsrA response regulator, like AgrA, belongs to the family of proteins characterized by a LytTR DNA-binding domain (22). The LytTR domain is distinct from the more typical helix-turn-helix or winged-helix DNA-binding domains of response regulators and appears to be common in proteins that regulate virulence factors such as toxins, bacteriocins, and extracellular polysaccharide. This family includes, in addition to FsrA and AgrA, the AlgR protein of Pseudomonas aeruginosa, VirR of Clostridium perfringens, BlpR of Streptococcus pneumoniae, PlnC and PlnD of Lactobacillus plantarum, LytT of Bacillus subtilis, and LytR of S. aureus (2, 4, 14, 17, 27).
In the present study, FsrA and FsrC were purified, and by means of electrophoretic mobility shift assays (EMSA) and DNase I protection footprinting, we identified the DNA promoter regions in fsrB and gelE that are recognized by FsrA. The consensus sequence appeared to be an imperfect direct repeat of 8 bp separated by 13 bases and located immediately upstream of the −35 promoter region. This consensus sequence is consistent with FsrA being a member of the LytTR family of DNA-binding proteins.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The E. faecalis strain used in this study was the clinical isolate V583 (28). Plasmids and oligonucleotide primers used in this study are listed in Tables 1 and 2. Strains were cultured in Todd-Hewitt broth (THB). Escherichia coli DH5α and TB1 were used for plasmid constructions and propagation. Strains were cultivated in Luria-Bertani broth. Antibiotics used for selection in E. coli and E. faecalis were spectinomycin (150 and 750 μg/ml, respectively), ampicillin (100 μg/ml E. coli only), and tetracycline (15 μg/ml). Electroporation of E. faecalis and E. coli was carried out as described previously (6, 7).
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pET28a | Expression vector | Novagen |
| pET-FsrA | pET28 containing E. faecalis fsrA | |
| pML28 | pAT28 plus the aphA3 promoter on a 369-bp EcoRI/BamHI fragment | |
| pDII32 | pML28 expressing E. faecalis FsrA-His6 | |
| pWH1520 | E. coli/B. megaterium shuttle vector | MoBiTec |
| pWH-FsrA | pWH1520 expressing E. faecalis FsrA-His6 | |
| pMAL-c2E | Expression vector | New England Biolabs |
| pMAL-FsrC | pMAL-c2E containing the E. faecalis FsrC cytosolic region | |
| pBluescript IIKS | Phagemid from pUC19 | Stratagene |
| pBlue-fsrA | pBluescript IIKS containing the fsrB promoter region (−351 to +33) | |
| pBlue-fsrB | pBluescript IIKS containing the fsrB promoter region (−283 to +14) | |
| PBlue-gelE | pBluescript IIKS containing the gelE promoter region (−307 to +52) | |
| pCR2.1-TOPO | Cloning vector | Invitrogen |
| pTOPO-fsrB | pCR2.1-TOPO containing the fsrB promoter region (−283 to +14) | |
| pTOPO-gelE | pCR2.1-TOPO containing the gelE promoter region (−305 to +49) |
TABLE 2.
Oligonucleotides primers used in this study
| Primer | Sequencea |
|---|---|
| EffsrA5′BspH | 5′-GAAAGTCATGAGTGAACAAATGGCTA-3′ |
| EffsrA3′XhoI | 5′-ATCCCCTCGAGAGTAAGAAATAGTGCCTTG-3′ |
| EffsrA5′Bam | 5′-ATAGGATCCGAAAGGGATGAGTGA-3′ |
| pET283′Sal | 5′-AATTAGTCGACCTTCCTTTCGGGCTTTGTTAGC-3′ |
| EffsrC3′Bam | 5′-GACATGGATCCCTGTATTGCCCCTC-3′ |
| EffsrC5′Kpn | 5′-GCTTAGGTACCAGAACAACGTATCAACACTC-3′ |
| FsrA5′MfeI | 5′-TTAACCCAATTGGACCAATGAATTGATTTTGTC-3′ |
| FsrA3′EcoRI | 5′-ATTTGGAATTCTAATATATAAATAGCC-3′ |
| FsrB5′MfeI | 5′-ATTATACAATTGAATTTTATGGAACGTTATCA-3′ |
| FsrB3′EcoRI | 5′-TTTTAGAATTCAATCGATTAGCATATCG-3′ |
| GelE5′MfeI | 5′-ATTTAACAATTGAAAAAAATCATAACA-3′ |
| GelE3′EcoRI | 5′-CCTAAGAATTCTAAAATTTTATTTCCCTTC-3′ |
| GelE5′Mfe2 | 5′-GTCCTCAATTGAAGGAGCGGTCACTCAAC-3′ |
| GelE3′Eco2 | 5′-ACATGGAATTCCAACAAAGATGCCTGTAC-3′ |
| FsrB3′EcoSeq | 5′-AATTCAATCGATTAGCA-3′ |
| GelE3′EcoSeq | 5′-AATTCTAAAATTTTATT-3′ |
Restriction sites are in boldface.
Expression and purification of soluble FsrA-His6.
The fsrA coding region was amplified from E. faecalis V583 using primers EffsrA5′BspH and EffsrA3′XhoI. The resulting PCR product digested with BspHI and XhoI was cloned into pET28b(+) cut with NcoI/XhoI, thus generating a fusion of 6 His codons to the 3′ end of the gene. All cloning steps involving bacterial growth were carried out at room temperature.
This construct was used as a PCR template with primers EffsrA5′Bam and pET283′Sal. The PCR product was cut with the BamHI and SalI enzymes and ligated into pML28 (11), which was similarly digested, yielding plasmid pDII32. This plasmid was cut with BamHI and SphI, and the fragment carrying the fsrA coding sequence and the His tag at the 3′ end was ligated into pWH1520 (MoBiTec), which was similarly digested, obtaining plasmid pWH-FsrA. All constructs were verified by sequence analysis. Plasmid pWH-FsrA was used to transform protoplasts of Bacillus megaterium according to the protocol recommended by the supplier (MoBiTec).
Expression of FsrA-His6 was induced by the addition of 0.5% d-xylose, and growth was carried out overnight at 28°C. Cells were then harvested, washed in buffer B (Tris-HCl, 50 mM, pH 8.0, 100 mM NaCl, 1 mM phenylmethylsulfonyl fluoride [PMSF]), resuspended in buffer B with 10 mg/ml lysozyme, incubated for 30 min at 4°C, and lysed by sonication. The FsrA-His6 protein was then purified by immobilized-metal affinity chromatography using the Ni-nitrilotriacetic acid (NTA) resin (Qiagen). FsrA-His6 was eluted from the column by a linear gradient of 30 to 300 mM imidazole in buffer B. The FsrA-His6-containing fractions were pooled, dialyzed, and then loaded onto a 5-ml hydroxylapatite column that had been preequilibrated with buffer H (40 mM K2HPO4, pH 7.5, 2 mM dithiothreitol [DTT], and 0.2 mM PMSF). The column was washed with 30 ml of buffer H, and FsrA-His6 was eluted with a 50-ml linear gradient of 40 to 400 mM K2HPO4, pH 7.5. The FsrA-His6-containing fractions were pooled and dialyzed against 50 mM Tris, pH 8.0, 100 mM NaCl, 20% glycerol. The protein was concentrated and stored at −20°C.
Expression and purification of MBP-FsrC.
The coding region of the cytosolic portion of FsrC (27 kDa, from amino acid 192 to the stop codon) was amplified from E. faecalis V583 using primers EffsrC5′KpnI and EffsrC3′BamHI. The resulting PCR product was cloned into pMAL-c2E (New England BioLabs) digested with KpnI/BamHI, generating a fusion to the coding region of the maltose-binding protein (MBP). The construct was verified by sequence analysis. The plasmid pMAL-FsrC was transformed into the E. coli expression strain BL21(DE3)pLyS (Novagen). A culture grown in LB with ampicillin and 0.2% glucose was induced at an optical density at 600 nm (OD600) of 0.5 with 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and allowed to grow for 4 h at 16°C. Cells were harvested by centrifugation, and the pellet was resuspended in lysis buffer C (20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM EDTA). Cells were lysed by sonication, and the debris was spun down by a 30-min centrifugation at 20,000 × g at 4°C. The supernatant was diluted 5 times in lysis buffer and then loaded onto amylose resin (New England BioLabs). Purification was as described by the manufacturer, with 10 mM maltose used for protein elution. Fractions were collected and analyzed by sodium dodecyl sulfate (SDS)-PAGE. The protein was dialyzed against the lysis buffer, concentrated, and stored in 40% glycerol at −20°C.
Protein phosphorylation assays.
The FsrC phosphorylation kinetic and phosphoryl transfer reactions were carried out at 30°C in the presence of 5 μCi of [γ-32P]ATP (specific activity, 3,000 Ci/mmol) in buffer A (20 mM HEPES, pH 7.0, 50 mM KCl, 2 mM MgCl2, 5 mM CaCl2, 10 mM DTT, 10 μM bovine serum albumin [BSA]) with 5 μM purified MBP-FsrC and 2.5 μM FsrA-His6. The reactions were initiated by the addition of [γ-32P]ATP into a 15-μl reaction mixture and terminated by the addition of 5× SDS sample buffer, and the reaction mixtures were subjected to SDS-PAGE on 15% polyacrylamide gels (15). Gels were dried, and labeled proteins were detected by a PhosphorImager screen (Amersham-Molecular Dynamics).
Gel retardation assay.
Labeled fsrA, fsrB, and gelE promoter probes were generated by digesting plasmids pBlue-fsrA, pBlue-fsrB, and pBlue-gelE, respectively, with EcoRI, followed by a fill-in reaction with Klenow polymerase (New England BioLabs) in the presence of [α-32P]dATP. The fragment was then released by MfeI digestion and purification from 5% acrylamide gel by electroelution. The labeled fragments were ethanol precipitated and resuspended in TE buffer (Tris-HCl, 10 mM, pH 8.0, EDTA, 1 mM). Binding of FsrA-His6 to the DNA fragment was performed at room temperature in buffer E (20 mM HEPES, pH 7.5, 1 mM EDTA, 50 mM KCl, 2 mM DTT, 100 μg/ml BSA, 8% glycerol). Phosphorylated FsrA-His6 (FsrA-His6∼P) was prepared as described above, and the reaction mixture was added, with the final concentrations of response regulator indicated in Fig. 2. The reaction mixtures were incubated for 30 min after the addition of the fragment DNA (0.05 ng/μl) and applied to a 5% acrylamide gel running at 200 V in Tris-borate-EDTA (TBE) buffer. Specific or nonspecific DNA (unlabeled fsrB or gelE promoter fragments or salmon sperm DNA, respectively) was used at a 100-fold excess. Electrophoresis was continued at 15 mA for approximately 15 min. The gels were dried and exposed to a PhosphorImager screen (Amersham-Molecular Dynamics).
DNase I footprinting assays.
Labeled probes were generated by PCR amplification using the oligonucleotide primer pairs FsrB5′MfeI-FsrB3′EcoRI and GelE5′Mfe2-GelE3′EcoRI2. Prior to PCR amplification, primers FsrB3′EcoRI and GelE5′Mfe2 were labeled with [γ-32P]ATP and polynucleotide kinase as recommended by the supplier (New England BioLabs). Probes were purified from agarose gels using the Invitrogen PureLink gel extraction kit. The labeled probes (20,000 cpm) were incubated in the presence of FsrA-His6 or its storage buffer at the concentrations indicated in the legend to Fig. 3 in a l5-μl reaction mixture containing 1× DNase I buffer (New England BioLabs). Reaction mixtures were incubated for 3 min at room temperature before the addition of DNase I and further incubation for 1 min. The reactions were stopped by the addition of 6 μl of stop solution (0.1 M Na2-EDTA, 0.5% SDS, 0.4 mg/ml sheared salmon sperm DNA). The DNA was precipitated with 500 μl ethanol. The dried pellet was resuspended in 6 μl of loading dye (95% deionized formamide, 10 mM Na2-EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol) and heated for 2 min at 90°C before it was loaded onto a 5% polyacrylamide gel containing 6 M urea in 1× TBE.
The primers FsrB3′EcoRI and GelE5′Mfe2 were used for dideoxy chain termination sequencing of plasmids pTOPO-fsrB and pTOPO-gelE, respectively, with the Sequenase version 2.0 DNA sequencing kit (USB). The sequencing reactions were run on the gel alongside the footprinting reactions. The gels were dried and exposed to an X-ray film.
RESULTS
Overexpression and purification of FsrA and FsrC.
Several attempts to clone the fsrA gene in different E. coli expression plasmids never resulted in viable transformants. Attempts made at overexpressing FsrA in B. subtilis or E. faecalis resulted in viable colonies that nevertheless did not overproduce the protein. However, the fsrA gene modified to carry 6 histidine codons at the 3′ end could be cloned in plasmid pWH1520 using E. coli DH5α as the host strain. Plasmid pWH1520 allows heterologous gene expression for protein overproduction in B. megaterium by utilizing the xylose-inducible xylA promoter from this organism. After transformation of plasmid pWH-FsrA in protoplasts of B. megaterium, viable colonies were obtained; thus, the resulting strain was tested for FsrA production upon induction with xylose at 37, 28, and 16°C for 2-h or 12-h induction times. In all cases, the expression was very low. Nevertheless, the protein was purified from cells grown at 28°C as described in Materials and Methods with highly variable yields ranging from 30 to 100 μg/liter of culture. The purified product was found to migrate to the expected molecular mass (29 kDa) by Coomassie blue-strained SDS-PAGE and Western blot analysis using an anti-His tag monoclonal antibody (data not shown).
The coding sequence for the cytoplasmic portion of FsrC was cloned downstream of the maltose-binding protein, and expression of soluble MBP-FsrC fusion protein was found to be optimal when the E. coli cells were grown at 16°C. The yield was ∼10 g/liter of culture.
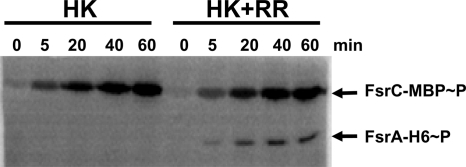
To confirm the functionality of the FsrA and FsrC proteins, a phosphorylation assay was carried out as described in Materials and Methods. Autophosphorylation of MBP-FsrC occurred even in the absence of the GBAP-activating pheromone, and this was followed by phosphoryl transfer to FsrA-His6 (Fig. 1). FsrA-His6 did not phosphorylate when incubated with a noncognate enterococcal sensor histidine kinase, thus indicating that specificity existed between FsrA and FsrC (data not shown).
FIG. 1.
Phosphorylation assay of FsrA-His6. MBP-FsrC was incubated with [γ-32P]ATP in the absence and presence of FsrA-His6 for the indicated times. The samples were separated by 15% SDS-PAGE and exposed to a PhosphorImager screen.
These results indicated that both proteins were functionally active and suitable for further in vitro analysis.
FsrA binds to the fsrB and gelE promoters.
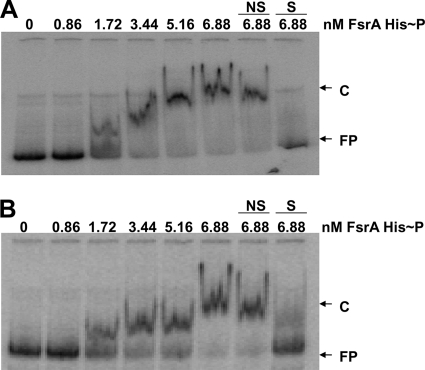
To test the ability of the FsrA response regulator to bind to the fsrB and gelE promoter regions, EMSA were performed using purified FsrA-His6 protein that had previously been incubated with ATP and MBP-FsrC, to generate FsrA-His6∼P. Radioactively labeled fragments containing the fsrA, fsrB, and gelE promoter regions were obtained as described in Materials and Methods. The results shown in Fig. 2 indicated that binding of phosphorylated FsrA-His6 to the fsrB and gelE promoter regions occurred, as retarded mobility of the DNA fragment was observed. An approximate equilibrium dissociation constant (KD) of 1.7 nM was deduced from the protein concentration needed to retard 50% of the DNA. Notably, no specific binding of FsrA was observed with the fsrA promoter fragment (data not shown), as was expected from previous in vivo analyses (26).
FIG. 2.
Gel mobility shift assays. The 297-bp (positions −283 to +14) end-labeled fsrB promoter fragment (A) or the 359-bp (positions −307 to + 52) end-labeled gelE fragment (B) was incubated with FsrA-His6 at the concentrations indicated in the figure. C, complex; FP, free probe; NS, nonspecific DNA; S, specific DNA.
In order to determine specificity of binding, the assays were also carried out in the presence of an excess of nonspecific DNA (salmon sperm DNA), which did not affect DNA mobility, or specific DNA (the unlabeled fsrB or gelE DNA promoter), which significantly reduced the mobility of the corresponding labeled fragment.
Unphosphorylated FsrA was also used in the gel retardation assay, and 50% retardation of the DNA fragments was obtained with 5.16 nM protein, although specificity of binding was unaffected (data not shown).
The results indicated that FsrA binds specifically to the fsrB and gelE promoters and that DNA-binding efficiency was stimulated by phosphorylation.
DNase I protection assay of fsrB and gelE promoters.
FsrA belongs to the AlgR/AgrA/LytR family of transcriptional regulators characterized by the LytTR DNA-binding domain. In all cases studied, LytTR domains bind to two imperfect direct 9-bp repeats separated by 12 bp and located just upstream of the promoter −35 consensus sequence.
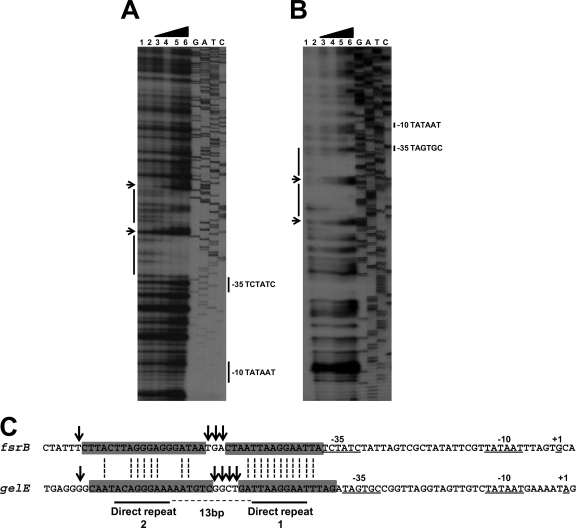
In order to precisely identify the binding site of FsrA on the fsrB and gelE promoters and pinpoint the recognition sequence, DNase I protection experiments were carried out, and the results are shown in Fig. 3. For both promoters, the region protected by FsrA was immediately upstream of the −35 recognition sequence for RNA polymerase. A single discrete area of protection spanning 37 to 38 bp was seen in both promoters, indicating the presence of a single binding site for FsrA. Within this area, 3 to 4 bases demonstrated increased susceptibility to DNase I. Some homology between the protected sequences upstream and downstream of the hypersusceptible sites is consistent with the presence of imperfect direct repeats separated by 12 bases, typical of LytTR proteins (Fig. 3C). Protected sequences at the 3′ end of the hypersusceptible bases (direct repeat 1) contained 11 bases that were identical in the two promoters. Base conservation was less extensive in the protected area at the 5′ end of the hypersusceptible residues (direct repeat 2). Consistently, direct repeat 1 in both promoters seemed to be protected from DNase I more efficiently than direct repeat 2, with the effect being more prominent with the fsrB promoter.
FIG. 3.
DNase I footprinting analysis of the promoter-regulatory regions of fsrB (A) and gelE (B). The labeled DNA fragments were obtained as described in Materials and Methods. The fragments were incubated with different amounts of FsrA-His6 before DNase I treatment. Protein concentrations used in each reaction mixture were 0 (lanes 1 and 2), 0.66 μM (lane 3), 1.3 μM (lane 4), 2.6 μM (lane 5), and 5.3 μM (lane 6). Dideoxy sequencing reactions of pTOPO-fsrB (A) and pTOPO-gelE (B) are also shown. (C). Alignment of the nucleotide sequence of the fsrB and gelE promoters identifying the regions protected by FsrA and the −10 and −35 promoter elements (26). Regions protected from DNase I digestion are indicated by gray boxes. Arrows show nucleotides hypersensitive to DNase I digestion. The direct repeats shown are based on the consensus sequence identified in Fig. 4.
FsrA consensus site.
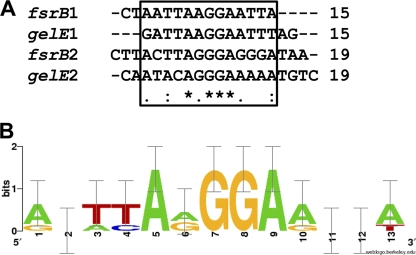
By generating an alignment of the fsrB and gelE protected regions using Clustal W, the FsrA consensus site was identified and is shown, by use of Web Logo (5), in Fig. 4. The consensus site is characterized by a central conserved sequence (T/AT/CAA/GGGAA/G) located in the middle of the two regions that flank the hypersusceptible sites and protected by FsrA from DNase I digestion. The 8-bp consensus sequence of direct repeat 1 and direct repeat 2 for fsrB and gelE are separated by 13 bases, which, given the imperfect nature of the upstream repeat, is consistent with the binding characteristics of other LytTR domain-containing proteins.
FIG. 4.
Sequence logo of the repeats of 8 nucleotides that form the FsrA consensus site. (A) Sequence alignment of the regions protected by FsrA in the fsrB and gelE promoters obtained with Clustal W. The sequences in the box were used to generate the logo. An asterisk indicates identical bases, a colon indicates a different nucleotide base group, and a period indicates the same nucleotide base group. (B) The height of each letter is proportional to the frequency of the base, and the height of the letter stack is the conservation in bits at that position. Error bars are shown at the tops of the stacks. The logo was obtained with the WebLogo server (http://weblogo.berkeley.edu) (5).
DISCUSSION
In this study, electrophoretic mobility shift assays and DNase I protection footprinting were used to identify the binding site of the E. faecalis FsrA response regulator and transcription factor to its target promoters, fsrB and gelE.
Previous in vivo studies established that FsrA is required for the transcription of the fsrB and gelE promoters (26). By means of DNA sequence alignments, those studies identified two conserved direct repeats required for activation of these promoters, and deletion of either repeat abolished fsrB or gelE expression. With a consensus sequence deduced from those studies and in silico as well as microarray gene expression analyses, it was also shown that a third gene, EF1097, was dependent on FsrA for transcription and that its promoter contained two direct repeats with similarities to the ones present in the fsrB and gelE promoters (1). The present studies used in vitro approaches to identify and confirm the direct repeats in the fsrB and gelE promoters as the target of FsrA and allowed better definition of the DNA consensus sequence recognized by this transcription factor. These results were obtained through the use of a Bacillus megaterium protein expression system that overcame the lethal effect of expressing FsrA in E. coli.
Footprinting experiments revealed that FsrA binds a region immediately upstream of the −35 consensus sequences of both promoters. Within this region, two major areas of protection from DNase I digestion were observed, and these areas were separated by 3 to 4 hypersensitive bases. These protected areas included imperfect direct repeats of 8 bp separated by 13 nucleotides. Alignments of the repeat sequences allowed us to propose the following consensus sequence for FsrA binding to its target promoters: T/AT/CAA/GGGAA/G. The downstream direct repeats (DR1) in fsrB and gelE are identical, with the identity extending for a stretch of 11 bases. The upstream direct repeats (DR2) are less conserved between fsrB and gelE (5-bp identity out of 8 bp) and also vary in 3 out of 8 positions with respect to their corresponding DR1. DR1 is likely to be an optimal binding site as, for both promoters, protection appeared to be more efficient in this region than in DR2.
The binding characteristics of FsrA to its target promoters are highly reminiscent of the behavior of the other proteins in the LytTR family. These all bind to a region that includes 2 imperfect direct repeats of 8 to 9 bases separated by a 12- to 13-bp spacer. In the spacer, 2 to 3 bp is generally DNase I hypersensitive, usually indicative of DNA bending. Structures and modeling of the LytTR domain of the staphylococcal AgrA regulator (AgrAC) bound to its DNA consensus site suggest that two protein molecules interact, each with one of the direct repeats, in a tandem orientation along the same face of the DNA. The length of the space between the two direct repeat sequences is then critical for the positioning of the binding sites on the same face of the DNA helix.
It was shown that truncation of any of the two direct repeats in fsrB and gelE abolished promoter activity completely, suggesting that the two sites are active only in a tandem arrangement, and thus FsrA likely binds as a dimer to the target promoters (26).
The observation that the DNA within the AgrAC-DNA complex adopts the B form suggests that the DNA is subject to a degree of bending in order to adapt to the binding surface of the regulator protein, and this may explain the hypersensitivity to DNase I digestion by the central base pairs of the spacer (29).
The sequence of the 8-bp FsrA consensus site (T/AT/CAA/GGGAA/G) proposed in this study differs from the previously identified LytTR consensus site because of the absence of the central TT base pairs that appeared to be absolutely conserved in this family of proteins. Based on the structure of the DNA-bound AgrA protein, these two residues may act as a hinge that enables the DNA to adapt to the surface of the protein. If a change in DNA conformation is induced by FsrA as well, as suggested by the DNase I-hypersensitive sites, clearly the pair of TT residues does not appear to be a requirement for these enterococcal promoters.
Acknowledgments
We thank M. A. Strauch (University of Maryland, Baltimore, MD) and Ignacio E. Sánchez (University of Buenos Aires, Argentina) for helpful suggestions and Mike Green for protein purification.
M.F.D.P. is a member of CONICET-Argentina. The present work was supported in part by NIH, U.S. PHS, grants AI052289 to M.P. and GM19416 to J. A. Hoch.
This is manuscript no. 21012 from the Department of Molecular and Experimental Medicine, The Scripps Research Institute.
Footnotes
Published ahead of print on 21 January 2011.
REFERENCES
- 1.Bourgogne, A., S. G. Hilsenbeck, G. M. Dunny, and B. E. Murray. 2006. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J. Bacteriol. 188:2875-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunskill, E. W., and K. W. Bayles. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chavers, L. S., et al. 2003. Vancomycin-resistant enterococci: 15 years and counting. J. Hosp. Infect. 53:159-171. [DOI] [PubMed] [Google Scholar]
- 4.Cheung, J. K., and J. I. Rood. 2000. The VirR response regulator from Clostridium perfringens binds independently to two imperfect direct repeats located upstream of the pfoA promoter. J. Bacteriol. 182:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruz-Rodz, A. L., and M. S. Gilmore. 1990. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol. Gen. Genet. 224:152-154. [DOI] [PubMed] [Google Scholar]
- 7.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flahaut, S., et al. 1996. Relationship between stress response toward bile salts, acid and heat treatment in Enterococcus faecalis. FEMS Microbiol. Lett. 138:49-54. [DOI] [PubMed] [Google Scholar]
- 9.Garsin, D. A., et al. 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. U. S. A. 98:10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock, L. E., and M. Perego. 2002. Two-component signal transduction in Enterococcus faecalis. J. Bacteriol. 184:5819-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock, L. E., and M. Perego. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 186:5629-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hidron, A. I., et al. 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect. Control Hosp. Epidemiol. 29:996-1011. [DOI] [PubMed] [Google Scholar]
- 13.Hoch, J. A., and T. J. Silhavy. 1995. Two-component signal transduction. American Society for Microbiology, Washington, DC.
- 14.Koenig, R. L., J. L. Ray, S. J. Maleki, M. S. Smeltzer, and B. K. Hurlburt. 2004. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J. Bacteriol. 186:7549-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Ma, P., et al. 2008. Expression, purification and activities of the entire family of intact membrane sensor kinases from Enterococcus faecalis. Mol. Membr. Biol. 25:449-473. [DOI] [PubMed] [Google Scholar]
- 17.Mohr, C. D., J. H. J. Leveau, D. P. Krieg, N. S. Hibler, and V. Deretic. 1992. AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J. Bacteriol. 174:6624-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mylonakis, E., et al. 2002. The Enterococcus faecalis fsrB gene, a key component of the fsr quorum-sensing system, is associated with virulence in the rabbit endophthalmitis model. Infect. Immun. 70:4678-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakayama, J., et al. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145-154. [DOI] [PubMed] [Google Scholar]
- 20.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, and H. Nagasawa. 2001. Chemical synthesis and biological activity of the gelatinase biosynthesis-activating pheromone of Enterococcus faecalis and its analogs. Biosci. Biotechnol. Biochem. 65:2322-2325. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama, J., et al. 2006. Revised model for Enterococcus faecalis fsr quorum-sensing system: the small open reading frame fsrD encodes the gelatinase biosynthesis-activating pheromone propeptide corresponding to staphylococcal agrD. J. Bacteriol. 188:8321-8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikolskaya, A. N., and M. Y. Galperin. 2002. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 30:2453-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 24.Palmer, K. L., V. N. Kos, and M. S. Gilmore. 2010. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol. 13:632-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183:3372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Risoen, P. A., et al. 2001. Regulation of bacteriocin production in Lactobacillus plantarum depends on a conserved promoter arrangement with consensus binding sequence. Mol. Genet. Genomics 265:198-206. [DOI] [PubMed] [Google Scholar]
- 28.Sahm, D. F., et al. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sidote, D. J., C. M. Barbieri, T. Wu, and A. M. Stock. 2008. Structure of the Staphylococcus aureus AgrA LytTR domain bound to DNA reveals a beta fold with an unusual mode of binding. Structure 16:727-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thurlow, L. R., et al. 2010. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect. Immun. 78:4936-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]