Abstract
The periplasmic chaperones Skp, SurA, and DegP are implicated in the biogenesis of outer membrane proteins (OMPs) in Escherichia coli. Here, we investigated whether these chaperones exert similar functions in Neisseria meningitidis. Although N. meningitidis does not contain a homolog of the protease/chaperone DegP, it does possess a homolog of another E. coli protein, DegQ, which can functionally replace DegP when overproduced. Hence, we examined whether in N. meningitidis, DegQ acts as a functional homolog of DegP. Single skp, surA, and degQ mutants were easily obtained, showing that none of these chaperones is essential in N. meningitidis. Furthermore, all combinations of double mutants were generated and no synthetic lethality was observed. The absence of SurA or DegQ did not affect OMP biogenesis. In contrast, the absence of Skp resulted in severely lower levels of the porins PorA and PorB but not of other OMPs. These decreased levels were not due to proteolytic activity of DegQ, since porin levels remained low in a skp degQ double mutant, indicating that neisserial DegQ is not a functional homolog of E. coli DegP. The absence of Skp resulted in lower expression of the porB gene, as shown by using a PporB-lacZ fusion. We found no cross-species complementation when Skp of E. coli or N. meningitidis was heterologously expressed in skp mutants, indicating that Skp functions in a species-specific manner. Our results demonstrate an important role for Skp but not for SurA or DegQ in OMP biogenesis in N. meningitidis.
After crossing the inner membrane (IM) and the periplasm, bacterial outer membrane proteins (OMPs) are assembled into the outer membrane (OM) by a machinery that, in Neisseria meningitidis, consists of the central component Omp85 (48), the essential lipoprotein ComL, two nonessential lipoproteins BamC and BamE, and the RmpM protein (47). In Escherichia coli, this machinery was recently renamed Bam (β-barrel assembly machinery) and is similarly composed of BamA (the homolog of Omp85), BamD (the homolog of ComL), BamC, BamE, and additionally, BamB, a lipoprotein that has no homolog in the genus Neisseria (39, 53). E. coli does not contain a homolog of RmpM.
In E. coli, several chaperones have been identified that play a role in the transit of OMPs through the periplasm (25). Among them, Skp, SurA, and DegP appear to be the most prominent ones. Skp was shown to bind unfolded β-barrel OMPs (6, 10), presumably while they emerge from the Sec machinery (13), and to stimulate their release from the IM, resulting in the formation of soluble periplasmic intermediates of these proteins (36). The crystal structure of Skp revealed a homotrimer forming a basket-like shape, which transiently shields substrates from the environment to prevent their aggregation (19, 51, 52). The SurA protein contains two peptidyl-prolyl isomerase (PPIase) domains flanked by N- and C-terminal sequences. The PPIase domains are dispensable for SurA's chaperone activity (1). Its crystal structure shows a core module formed by the N- and C-terminal segments and one PPIase domain, with the other PPIase domain extending away from this core (2). Consistent with its proposed OMP-specific chaperone activity (22, 33), SurA preferentially binds in vitro to peptides containing two consecutive aromatic residues or two aromatic residues separated by one other residue, a motif that is regularly present in OMPs (14).
DegP is thought to function both as a chaperone and as a protease. The protein belongs to the HtrA family of proteases, which is characterized by the presence of one or more PDZ domains and a trypsin-like protease domain (8). E. coli contains three members of this family, DegP, DegS, and DegQ. DegP and DegQ contain two PDZ domains and DegS only one. A variable region, called the Q-linker, is located between the first and second β-strand of the protease domain and is approximately 40 amino acids long in DegP, 20 amino acids in DegQ, and basically absent in DegS. DegQ is a functional substitute for DegP when overexpressed (50). Many other bacteria possess only one HtrA family member, which usually is a DegQ homolog, based on the number of PDZ domains present and the length of the Q-linker (18). Whether this single HtrA family member then fulfils all functions ascribed to DegP, DegS, and DegQ in E. coli is at present unclear.
The roles of these chaperones have been studied almost exclusively in the Enterobacteriaceae. Recently, it was reported that surA in Bordetella pertussis could not be inactivated, suggesting that it is an essential gene in that species (15). Thus, the OMP assembly process in E. coli is not the paradigm for all bacteria. In this respect, N. meningitidis has already revealed some insightful differences with E. coli; for example, in contrast to E. coli, this Gram-negative bacterium can survive and assemble OMPs when the synthesis or transport of lipopolysaccharide (LPS) is disturbed (4, 40). Also, the composition of the Bam complex in N. meningitidis is not identical to that of E. coli (47) and the phenotype of mutants depleted of Bam components is different in each species. In E. coli, the periplasmic accumulation of unfolded OMPs activates the σE response; consequently, unfolded OMPs are degraded by the periplasmic protease DegP and OMP synthesis is downregulated by small RNAs (17, 31). This regulatory pathway is missing in N. meningitidis (5), and unassembled OMPs accumulate in the periplasm when OMP assembly is inhibited (47, 48). The lack of such feedback mechanisms may allow for a clearer interpretation of mutant phenotypes.
Since N. meningitidis has proven to be very informative as a model organism in studies to understand OM biogenesis, we here undertook a systematic study into the role of the periplasmic chaperones Skp, SurA, and DegQ in OMP biogenesis in N. meningitidis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used are described in Table 1. E. coli strains were grown at 37°C either on LB agar plates or in liquid LB medium, supplemented with 25 μg/ml of chloramphenicol or 50 μg/ml kanamycin when appropriate. To analyze the effect of Skp on OmpA levels, BW25113-derived strains were grown at 28°C in M9 medium supplemented with 2 μg/ml thiamine and 0.4% maltose (6) in the absence or presence of 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). N. meningitidis strains were grown at 37°C in candle jars on GC agar plates (Oxoid) supplemented with Vitox (Oxoid) and, when necessary, with 10 μg/ml of chloramphenicol or 80 μg/ml of kanamycin. For liquid cultures, N. meningitidis was grown overnight on plates, from which it was swabbed into tryptic soy broth (TSB) (Becton Dickinson) to an optical density at 550 nm (OD550) of 0.1 and grown for 6 h with shaking.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| TOP10F′ | Cloning strain | Invitrogen |
| DH5α | Cloning strain | Laboratory collection |
| BW25113 | Parent strain of the Keio collection | NBRPa |
| JW0173 | BW25113 with skp replaced by a kan cassette | NBRP |
| JW0173(pSkpEc) | JW0173 containing pFP10-SkpEc | This study |
| JW0173(pSkpNm) | JW0173 containing pFP10-SkpNm | This study |
| N. meningitidis strains | ||
| HB-1 | Serogroup B strain H44/76 with the capsule locus replaced by an erythromycin resistance gene cassette | 3 |
| HB-1Δskp1 | HB-1 with skp replaced by a kan cassette | This study |
| HB-1skp2 | HB-1 with a kan cassette inserted in skp | This study |
| HB-1ΔsurA | HB-1 with surA replaced by a kan cassette | This study |
| HB-1ΔsurA-1 | HB-1 with surA replaced by a cat cassette | This study |
| HB-1ΔdegQ | HB-1 with degQ replaced by a kan cassette | This study |
| HB-1ΔdegQ(pDegQ) | HB-1ΔdegQ containing pEN11-DegQ | This study |
| HB-1Δskp1(pSkp) | HB-1Δskp1containing pEN11-Skp | This study |
| HB-1skp2(pSkp) | HB-1skp2 containing pEN11-Skp | This study |
| HB-1skp2(pSkpEc) | HB-1skp2 containing pFP10-SkpEc | This study |
| HB-1skp2ΔsurA | HB-1skp2 with surA replaced by a cat cassette | This study |
| HB-1skp2ΔdegQ | HB-1skp2 with degQ replaced by a cat cassette | This study |
| HB-1ΔsurAΔdegQ | HB-1ΔsurA-1 with degQ replaced by a kan cassette | This study |
| HB-1(pNhhA) | HB-1 containing pEN11-NhhA | This study |
| HB-1skp2(pNhhA) | HB-1(pNhhA) with a kan cassette inserted in skp | This study |
| HB-1ΔsurA(pNhhA) | HB-1(pNhhA) with surA replaced by a kan cassette | This study |
| HB-1ΔdegQ(pNhhA) | HB-1(pNhhA) with degQ replaced by a kan cassette | This study |
| HB-1(pFrpB) | HB-1 containing pFP10-c-frpB | This study |
| HB-1skp2(pFrpB) | HB-1skp2 containing pFP10-c-frpB | This study |
| HB-1ΔsurA(pFrpB) | HB-1ΔsurA containing pFP10-c-frpB | This study |
| HB-1ΔdegQ(pFrpB) | HB-1ΔdegQ containing pFP10-c-frpB | This study |
| HB-1skp2ΔNMB1332 | HB-1skp2 with NMB1332 replaced by a cat cassette | This study |
| HB-1skp2ΔNMB1433 | HB-1skp2 with NMB1433 replaced by a cat cassette | This study |
| HB-1lacZ | HB-1 with promoterless lacZ gene inserted in the hrtA locus | This study |
| HB-1porB-lacZ | HB-1 with PporB-lacZ fusion inserted in the hrtA locus | This study |
| HB-1porB-lacZ skp2 | HB-1 with PporB-lacZ fusion inserted in the hrtA locus and a cat cassette inserted in skp | This study |
| Plasmids | ||
| pCRII-TOPO | TA-cloning vector for PCR products | Invitrogen |
| pMB25 | pCRII-TOPO with imp inactivated by a kan cassette | 4 |
| pCRII-cat | pCRII-TOPO with a cat cassette flanked by AccI sites | 47 |
| pCRII-Δskp1 | skp deletion plasmid containing a kan cassette | This study |
| pCRII-skp2 | Plasmid containing kan cassette insertion in skp | This study |
| pCRII-skp2-cat | Plasmid containing cat cassette insertion in skp | This study |
| pCRII-ΔsurA | surA deletion plasmid containing a kan cassette | This study |
| pCRII-ΔsurA-1 | surA deletion plasmid containing a cat cassette | This study |
| pCRII-ΔdegQ | degQ deletion plasmid containing a kan cassette | This study |
| pCRII-ΔdegQ-1 | degQ deletion plasmid containing a cat cassette | This study |
| pCRII-ΔNMB1332 | NMB1332 deletion plasmid containing a cat cassette | This study |
| pCRII-ΔNMB1433 | NMB1433 deletion plasmid containing a cat cassette | This study |
| pSMS1 | Plasmid containing promoterless lacZ gene inserted into the neisserial hrtA locus | 6 |
| pSMS-1235porB | pSMS1 with porB promoter region, including the first 18 bp of the ORF, cloned in front of lacZ | This study |
| pEN11-Imp | Neisseria replicative plasmid containing H44/76-derived imp under lac promoter control | 4 |
| pEN11-Skp | pEN11-imp with imp replaced by N. meningitidis skp | This study |
| pEN11-NhhA | pEN11-imp with imp replaced by N. meningitidis nhhA | This study |
| pFP10-c-lbpA | Neisseria replicative plasmid containing the lbpA gene of H44/76 under lac promoter control | 27 |
| pFP10-c-frpB | Neisseria replicative plasmid containing the frpB gene of H44/76 under lac promoter control | 20 |
| pFP10-SkpEc | pFP10-c-lbpA with the mature LbpA-encoding part replaced by that of E. coli Skp | This study |
| pFP10-SkpNm | pFP10-c-lbpA with the mature LbpA-encoding part replaced by that of N. meningitidis Skp | This study |
| pPU100 | pET11a containing nhhA | 44 |
NBRP, National Bioresource Project (NIG, Japan): E. coli.
Plasmid and mutant constructions.
The plasmids and primers used in this study are summarized in Tables 1 and 2, respectively. All N. meningitidis DNA fragments were obtained by PCR using genomic DNA from strain HB-1 as the template. Deletion constructs of skp, surA, degQ, NMB1332 (locus tag), and NMB1433 were obtained by amplifying DNA fragments upstream and downstream of these genes by PCR using the primers indicated with Up-For and Up-Rev and Down-For and Down-Rev in Table 2. The fragments were cloned into pCRII-TOPO. Next, the upstream and downstream fragments of each gene were joined together in one plasmid using the AccI sites that were introduced via the primers and the XbaI site in the vector. A kanamycin resistance gene (kan) cassette that includes the neisserial DNA uptake sequence was obtained from pMB25 and inserted in some plasmids after AccI restriction, yielding pCRII-Δskp1, pCRII-ΔsurA, and pCRII-ΔdegQ. Alternatively, a chloramphenicol-resistance gene (cat) cassette was obtained from pCRII-cat and inserted after AccI restriction, resulting in pCRII-ΔsurA-1, pCRII-ΔdegQ-1, pCRII-ΔNMB1332, and pCRII-ΔNMB1433. For allelic replacements, constructs containing the antibiotic-resistance cassette in the same transcriptional direction as the gene to be replaced were used.
TABLE 2.
Primers used in this study
| Primer name | Sequence (5′-3′) | Underlined restriction site |
|---|---|---|
| Skp-Up-For | GTGGCAGAACACCTGACC | |
| Skp-Up-Rev | ATGTCGACCTGAAGGGCTTCAGACGGCATT | AccI |
| Skp-Down-For | ATGTCGACTTCAGACGGCATACCGAA | AccI |
| Skp-Down-Rev | ATGACGGTGTGCGAACCGATT | |
| Skp-For | ATCATATGACCCGTTTGACCC | NdeI |
| Skp-For2 | ATCATATGCCGACACCTTCCAAAAAATCG | NdeI |
| Skp-Rev | ATGACGTCTCATCAGCGGGCGT | AatII |
| SurA-Up-For | TACGGCAACGACAGGATTA | |
| SurA-Up-Rev | ATGTCGACACGGTGCTCCTGCCAGGTT | AccI |
| SurA-Down-For | ATGTCGACGAGCAGGCGGGAATCCGGTT | AccI |
| SurA-Down-Rev | ATGACGTCGGCAACTTCTGAATCGTC | |
| SurA-For | ATCATATGATGAAAATCAAAGCCCTG | NdeI |
| SurA-Rev | ATGACGTCTTAGCGGATGTCGACATACGCGC | AatII |
| DegQ-Up-For | ATCCGACCACCGAGCTGAATTTC | |
| DegQ-Up-Rev | ATGTCGACATTCTACAACGTCCGTCC | AccI |
| DegQ-Down-For | ATGTCGACTCCGACGCGGCAGAACGCG | AccI |
| DegQ-dOwn-Rev | GCCTAAAGACAGCAGTACGC | |
| DegQ-for | ATCATATGTTCAAAAAATACCAATACC | NdeI |
| DegQ-for1 | AATCCGATGTCGCCCTTCTG | |
| DegQ-rev | ATGACGTCTTATTGCAGGTTTAATGC | AatII |
| 1332-up-for | ATCGGAATTCGGTTATGGGTATCGGCAG | |
| 1332-up-rev | AGTCGTCGACGGCCACGCCGCTGATTG | AccI |
| 1332-down-for | ATCGGTCGACTCGCAGGTGCATTGCAGG | AccI |
| 1332-down-rev | CTTCTGCCACTGCTCGGGCG | |
| 1332-int | CAAGCCGATAGTCGTCAACCTG | |
| 1433-up-for | ATGCGAATTCTGACCAACTCGCTGCAG | |
| 1433-up-rev | ATCGGTCGACTCGTGCCGCATGAGGCG | AccI |
| 1433-down-for | ATCGGTCGACCTCTCCACACCGTTTTAC | AccI |
| 1433-down-rev | CTGAAAGCTTGCACGTCGAAGGCGATGC | |
| 1433-int | GCAACCGGCTTCGATTGCAG | |
| A | ATGCCGTCTGAACGCCGAAATCGAA | |
| B | TTTGGACTAGGTGTCGACGTGCGCGC | AccI |
| C | ACGTCGACACCTAGTCCAAAAAATCG | AccI |
| D | ATGCCGTCTGAAAACAGGCGGGCGACTTTGG | |
| Skp-Eco-For | ATCATATGCTGACAAAATTGCA | NdeI |
| Skp-Eco-Rev | ATGACGTCTTATTTAACCTGTTTC | AatII |
| lac-cass | TCTGGATAATGTTTTTTGCGCCGAC | |
| rmpMF | CAGGCTCCGCAATATGTTGA | |
| rmpMR | GTTGTCTTGAGCTTCGGCG | |
| lpxDF | GGACATTTCCGTTACCGCC | |
| lpxDR | CTGTCGTGGACTTCGGCTTT | |
| fabZF | ATCCAAAAACTCATCCCCCAC | |
| fabZR | GGTGACGTTTTTAATCGCGGT | |
| SMS-1 | GCGCGCGGATCCAGGGCAATCAGGGATTTTTTC | BamHI |
| SMS-7 | GCGCGCGGATCCAATGACGGGATTTTAGGTTTC | BamHI |
To create an insertional skp mutant, a fragment consisting of the 3′ region of omp85 and the 5′ region of skp was amplified using primers A and B, and another fragment consisting of the 3′ region of skp and the 5′ region of lpxD was amplified using primers C and D. The fragments were cloned into pCRII-TOPO, yielding pCRII-skp-AB and pCRII-skp-CD, respectively. The AccI-SpeI fragment of pCRII-skp-CD was then ligated into AccI/SpeI-restricted pCRII-skp-AB together with the AccI-restricted kan cassette released from pMB25, yielding pCRII-skp2. The final construct contained a mutant skp allele with a kan cassette inserted after nucleotide 68 in the same transcriptional direction as skp and a 2-nucleotide insertion directly downstream of the cassette. The kan cassette was replaced by the cat cassette from pCRII-cat, using AccI restriction and ligation, resulting in pCRII-skp2-cat.
For complementation experiments, N. meningitidis skp was obtained by PCR using primers Skp-For and Skp-Rev and cloned into pCRII-TOPO. From there, the gene was excised with NdeI and AatII and subcloned into NdeI/AatII-digested pEN11-Imp, producing pEN11-Skp. A similar strategy was used to construct a degQ complementation plasmid, using primers DegQ-for and DegQ-rev, resulting in pEN11-DegQ. E. coli skp was amplified by PCR from genomic DNA of strain DH5α using primers Skp-Eco-For and Skp-Eco-Rev and cloned into pCRII-TOPO. From there, the gene was excised with NdeI and AatII and subcloned into NdeI/AatII-digested pFP10-c-lbpA, producing pFP10-SkpEc. As a result of the cloning strategy, the signal sequence of the E. coli Skp was replaced with that of the N. meningitidis lactoferrin-binding protein A (LbpA) (MNKKHSFPLTLTALAIATAFPSYA). For comparison, pFP10-SkpNm was constructed in a similar fashion, using primers Skp-For2 and Skp-Rev, resulting in a construct encoding the neisserial Skp with the signal sequence of LbpA. Plasmid pEN11-NhhA was constructed by subcloning nhhA (locus tag NMB0992) from pPU100 into pEN11-Imp using NdeI/AatII restriction and ligation.
Plasmid pSMS1 contains a promoterless lacZ gene inserted into the hrtA (high rate of transformation) locus of N. meningitidis (7). A 1,235-bp fragment of the porB promoter region, including its ribosome-binding site and the first 18 bp of the open reading frame (ORF), was obtained by PCR amplification using primers SMS-1 and SMS-7 and chromosomal DNA of strain HB-1 as the template. The PCR product was digested with BamHI, and the released insert was cloned into the BglII site of pSMS1 to generate a translational fusion of the 5′ end of porB to the lacZ gene. Correct orientation of the promoter relative to the lacZ gene was confirmed by PCR. The resulting plasmid, pSMS-1235porB, was verified by sequencing and used to transform strain HB-1.
Meningococci were transformed on plates by adding 1 μg of PCR product or plasmid in 10 mM MgCl2 to a few freshly restreaked colonies for 6 h. Then, bacteria were plated on GC agar plates containing appropriate antibiotics for mutant selection. The transformants were restreaked on similar selection plates to ensure their antibiotic-resistant phenotype and to prevent contamination of any remaining DNA used for the transformation in subsequent PCRs. To verify the mutants, a few colonies of each transformant were resuspended in H2O, boiled, and centrifuged for 5 min at 13,000 rpm in a microcentrifuge. The resultant supernatant was used as template in a series of PCRs. The knockout mutants were tested for the presence of the mutant allele by using the relevant Up-For and Down-Rev primers and for the absence of the wild-type allele by using internal primers annealing within the removed coding sequence combined with appropriate Up-For or Down-Rev primers. These internal primers were for the following constructs: for the ΔdegQ mutant, DegQ-for and DegQ-for1; for the ΔsurA mutant, SurA-for and SurA-rev; for the Δskp1 mutant, Skp-for; for the ΔNMB1332 mutant, 1332-int; and for the ΔNMB1433 mutant, 1433-int. The presence of pEN11-Skp, pFP10-SkpNm, or pFP10-SkpEc was tested by PCR using a primer annealing within the lac/tac promoter region (primer lac-cass) combined with Skp-Rev or Skp-Eco-Rev. For all PCRs, wild-type bacteria were used as controls.
Antibiotic sensitivity.
Meningococci grown overnight on GC agar plates were resuspended in 100 μl of TSB to an OD550 of 0.2 and plated on GC agar plates. Paper discs containing 30 μg of vancomycin (BD Biosciences) were placed on top of the agar. The plates were incubated at 37°C for 24 h, after which growth inhibition zones around the discs were measured in mm from the rim of the disk. All tests were repeated at least three times.
SDS-PAGE and immunoblot analysis.
Denaturing and seminative SDS-polyacrylamide electrophoresis (SDS-PAGE) and immunoblotting procedures were used as described previously (47). Protein bands in gels were stained with Coomassie brilliant blue. To enhance epitope recognition on immunoblots, native proteins were denatured within the seminative SDS-PAGE gels by leaving the gels in steam for 20 min prior to blotting.
Cellular fractions.
Cell envelopes were prepared as described previously (47). Extracellular media were collected from cultures grown for 6 h in TSB. Bacteria were removed by centrifugation (6,000 × g for 10 min), and proteins were precipitated from the supernatant with 10% trichloroacetic acid (TCA).
Urea solubilization.
Cell envelopes were incubated in 20 mM Tris-HCl, 100 mM glycine, 6 M urea (pH 7.6) for 1 h at room temperature. Nonsoluble material was separated from soluble material by ultracentrifugation (170,000 × g for 1 h). The resulting pellet was dissolved in 2 mM Tris-HCl (pH 7.6). Proteins in the supernatant were precipitated by 10% TCA.
Protease treatment.
Intact cells were suspended in phosphate-buffered saline supplemented with 1 mM MgCl2 and 0.5 mM CaCl2 (pH 7.6) to an OD550 of 1 and treated with 50 μg/ml proteinase K (Merck) for 15 min at room temperature. After the addition of 1 mM phenylmethanesulfonyl fluoride (Sigma), cells were boiled in SDS-PAGE sample buffer. Cell envelopes in 2 mM Tris-HCl (pH 7.6) were treated with 50 μg/ml trypsin (Sigma) for 16 h at room temperature and subsequently boiled in SDS-PAGE sample buffer.
Immunofluorescence microscopy.
Labeling and immunofluorescence microscopy analysis of formaldehyde-fixed bacteria were performed as described previously (48).
qRT-PCR.
Quantitative reverse transcription-PCR (qRT-PCR) was performed as described previously (42). Primer couples lpxDF/lpxDR, fabZF/fabZR, and rmpMF/rmpMR (Table 2) were used to amplify the cDNAs of lpxD, fabZ, and rmpM, respectively. The rmpM transcript was used to normalize all data.
Antisera.
Monoclonal antibodies (MAbs) against PorA and PorB were provided by the Netherlands Vaccine Institute (Bilthoven). The anti-PilE MAb was a generous gift from John Heckels (University of Southampton Medical School, United Kingdom). Mouse antisera directed against FrpB, PilQ, and fHbp and MAbs against Omp85, NspA, and NhhA were provided by GlaxoSmithKline Biologicals (Rixensart, Belgium). Mouse antiserum against N. meningitidis Skp (26) was generously provided by Gerardo Guillen Nieto (Center for Genetic Engineering and Biotechnology, Havana, Cuba), and rabbit antiserum against E. coli Skp was a kind gift from Mathias Müller (Universität Freiburg, Germany). Rabbit antisera against N. meningitidis Imp/LptD and E. coli SecB and a MAb against E. coli OmpA came from our laboratory stocks.
β-Galactosidase assays.
Standard β-galactosidase assays were conducted as described previously (24). Data are presented as the mean results ± standard errors of the means from one representative experiment performed in triplicate.
RESULTS
Construction of skp, surA, and degQ mutants in N. meningitidis.
Homology searches using E. coli Skp, SurA, and DegP sequences yielded locus tags NMB0181, NMB0281, and NMB0532, respectively, in the sequenced genome of N. meningitidis strain MC58. The Skp and SurA homologs were found in genetic locations similar to those in E. coli, i.e., downstream of the genes encoding Omp85/BamA and Imp/LptD, respectively (see Fig. S1 in the supplemental material). Imp is an OMP that is required for transport of lipopolysaccharide (LPS) to the cell surface (4) and was recently renamed LptD in E. coli (5). A reciprocal search using the amino acid sequence of NMB0532 on E. coli genome sequences showed that NMB0532 is more similar to DegQ than to DegP (the percentages of identity and similarity at the amino acid level to DegQ are 39% and 59% and to DegP are 34% and 56%). Also, the Gln-rich region called the Q-linker, characteristic of DegP, is absent in NMB0532. Therefore, we will refer to NMB0532 as DegQ. No other significant hits came up in searching the MC58 genome using DegP, DegQ, DegS, or COG0265, the conserved domain of the HtrA protein family, as queries. Thus, N. meningitidis appears to contain only one member of the HtrA protein family. To investigate the functions of the Skp, SurA, and DegQ proteins in N. meningitidis, mutants were constructed by replacing the main part of their coding sequences in strain HB-1 with antibiotic resistance cassettes (see Fig. S1 in the supplemental material). Correct mutants were easily obtained, demonstrating that none of these genes is essential. We found no growth defects for the ΔsurA mutant in liquid medium, whereas the ΔdegQ mutant demonstrated an enhanced lag time which was restored when a complementing copy of degQ was expressed from plasmid (see Fig. S2A and B in the supplemental material). In contrast, the Δskp mutant, designated HB-1Δskp1, grew significantly more slowly than any of the other strains (see Fig. S2A in the supplemental material). This defect in growth was not restored when skp was expressed from a complementing plasmid (data not shown), indicating that the Δskp1 mutation could be polar. Indeed, qRT-PCR analysis revealed that the transcript levels of lpxD and fabZ, which are located downstream of skp in the same operon (11), were ∼30 and ∼9 times lower in HB-1Δskp1 than in the parent strain. Therefore, we constructed an alternative skp mutant by the insertion of a kan cassette into the 5′ part of the skp gene without a concomitant deletion of skp sequences (see Fig. S1 in the supplemental material). This mutant, designated HB-1skp2, did not demonstrate any growth defect (see Fig. S2A in the supplemental material). The lpxD transcript level in the skp2 mutant was ∼5 times lower than that of the parent strain, indicating that, indeed, the polar effect of the skp2 mutation is much smaller than that of the Δskp1 mutation.
OMP assembly in the chaperone deletion mutants.
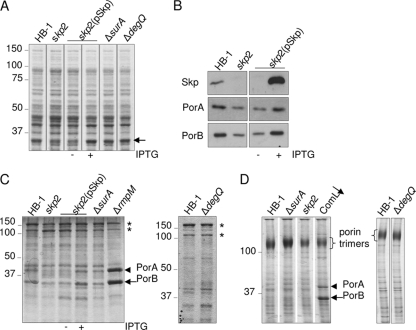
The protein profiles of the cells and culture supernatants were analyzed by SDS-PAGE. The comparison of cell lysates yielded one obvious difference: the skp2 mutant contained lower levels of a protein migrating at 35 kDa. This defect was indeed due to the absence of Skp, since it was restored upon the expression of skp from plasmid (Fig. 1 A, compare lanes without and with [− and +] IPTG). The absence and presence of the Skp protein was confirmed by immunoblotting (Fig. 1B, upper panel). As the 35-kDa band represented one of the most abundant cellular proteins, we reasoned that it might be PorB, i.e., one of the two major porins present in this strain. This notion was confirmed by immunoblotting (Fig. 1B). Similar blots using an antibody against the other porin, PorA, showed that the levels of this protein also were affected by the absence of Skp. Extracellular protein profiles showed the expected presence of two processed forms of IgA protease (45) in the parent strain (Fig. 1C), which were similar in the mutants, demonstrating that none of the three chaperones is essential for the correct processing and secretion of this autotransporter. Neisserial strains are known to shed OM blebs spontaneously, a process that is considerably enhanced in mutants lacking the RmpM protein, which anchors the OM to the peptidoglycan (41). As a consequence, high levels of porins are present in spent media of such mutants (Fig. 1C). The porin levels in the extracellular media of the chaperone mutants showed no evidence for enhanced blebbing for any of these mutants (Fig. 1C). In fact, the levels of porins in the spent media of the Skp variants reflected the differential cellular porin levels. Thus, the lower porin levels in the skp2 mutant are not due to increased blebbing.
FIG. 1.
Phenotypes of chaperone mutants. (A) Protein profiles of cell lysates observed by SDS-PAGE followed by Coomassie brilliant blue staining. An arrow indicates the band with the most variable intensity. (B) Identical amounts of cell lysates, based on the OD550 of the cultures, were immunoblotted with antibodies against the proteins indicated on the left. (C) Protein profiles of extracellular media. The bands indicated with asterisks represent two different secreted forms of IgA protease (45). (D) Assembly of porin trimers observed by seminative SDS-PAGE of cell envelopes followed by Coomassie brilliant blue staining. The lane indicated by ComL↓ contains cell envelopes of a ComL-/BamD-depleted strain and was included to show the profile of a strain with a porin assembly defect (47). In panels C and D, monomeric forms of PorA and PorB are indicated by arrowheads and full arrows, respectively. When relevant, the presence or absence of 1 mM IPTG during growth is indicated. The positions of molecular mass standard proteins are shown in kDa at the left side of the gels.
Porins are present as trimers in the OM, which can be visualized by seminative SDS-PAGE, where they migrate as high molecular weight (HMW) complexes. Porin assembly defects are manifested by the appearance of porins migrating at their denatured monomeric position, as demonstrated in Fig. 1D for a strain depleted of ComL/BamD (47). No unfolded porins were detected in seminative SDS-PAGE analysis of cell envelopes of the various chaperone mutants (Fig. 1D). Consistent with its overall lower cellular porin levels, the amount of trimers was decreased in the skp2 mutant, without concomitant accumulation of monomeric forms (Fig. 1D).
Lack of synthetic lethality of skp, surA, and degQ mutations in N. meningitidis.
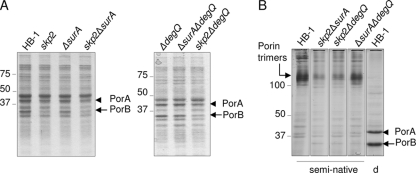
In E. coli, the simultaneous absence of any two of the chaperones Skp, SurA, and DegP leads to synthetic defects, often resulting in lethality (32, 36, 38). To analyze the potential existence of similar synthetic phenotypes in N. meningitidis, we constructed skp2 ΔsurA, skp2 ΔdegQ, and ΔsurA ΔdegQ double mutants. These mutants were all easily obtained, showing that none of the combinations is synthetically lethal. Protein profiles of cell lysates (Fig. 2 A) and extracellular media (data not shown) of the skp2 ΔsurA and skp2 ΔdegQ double mutants revealed no other or stronger defects than those already observed in the skp2 single mutant, i.e., lower levels of porins, most significantly of PorB. The porins were completely assembled into trimers, since we did not observe any unassembled monomeric porins in seminative SDS-PAGE (Fig. 2B). Of note, these results also demonstrate that the decrease in porin levels in the absence of Skp is not due to proteolytic degradation of unassembled forms by DegQ, as the levels of the porins were very similar in the skp2 single and skp2 ΔdegQ double mutants (Fig. 2A) and no unassembled forms were detected in the double mutant (Fig. 2B).
FIG. 2.
Protein profiles of chaperone double mutants. (A) Cell lysates were analyzed by SDS-PAGE followed by Coomassie brilliant blue staining. (B) Cell envelopes prepared from the indicated strains were subjected to seminative or denaturing (d) SDS-PAGE followed by Coomassie brilliant blue staining. Molecular weight, in thousands, is shown beside the gels.
In E. coli, the absence of periplasmic chaperones results in an increased sensitivity to antimicrobial agents, presumably because of a compromised OM integrity. We tested the sensitivity of the neisserial mutants to vancomycin in a disk diffusion assay. The parent strain, the skp2, ΔsurA, and ΔdegQ single mutants and the ΔsurA ΔdegQ double mutant were completely resistant to this antibiotic. Only the skp2 ΔdegQ and skp2 ΔsurA double mutants were slightly sensitive, as seen from the appearance of a clearing zone of 1.5 mm around the vancomycin-containing disk. For comparison, strains lacking BamE yielded clearing zones of 6 mm (47).
Assembly of OMPs other than porins.
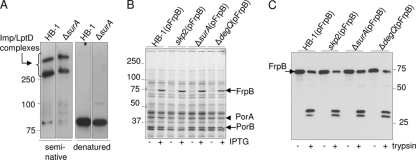
To test whether any of the chaperones is involved in the assembly of only a limited set of proteins, we investigated the levels and assembly of several OMPs and surface-associated proteins in the various mutants. We were particularly interested to see whether the absence of SurA would affect LptD assembly, as the LptD protein was recently reported to be one of the few “true” SurA substrates in E. coli (46). The cellular levels of LptD were not changed in the ΔsurA mutant (see Fig. S3A in the supplemental material). The LptD protein migrates as two HMW bands in seminative SDS-PAGE (Fig. 3 A). These two HMW forms of LptD appeared not to be affected in the ΔsurA mutant (Fig. 3A) or in any of the other mutants (data not shown), suggesting that none of the chaperones is essential for LptD biogenesis. Next, we assessed the levels of FrpB (FetA), a TonB-dependent receptor that can be as highly expressed as the porins (20). To that end, a plasmid containing frpB under IPTG control was introduced into the various mutants. High levels of FrpB were detected after growth in the presence of IPTG in all strains (Fig. 3B). All FrpB produced was correctly assembled in the OM, as deduced from the lack of urea extractability of the protein (data not shown) and from the similar tryptic profiles of FrpB in the various mutants (Fig. 3C). Also, the levels and assembly (tested as described in reference 47) of the secretin PilQ, the OMP assembly factor Omp85/BamA, and the small 8-stranded β-barrel OMP NspA (43) were unaffected, as were the levels of the pilin subunit PilE (data not shown and Fig. S3A and B in the supplemental material, respectively).
FIG. 3.
Levels and assembly of OMPs in chaperone mutants. (A) Cell envelopes of the indicated strains were subjected to seminative or denaturing SDS-PAGE followed by immunoblotting with anti-LptD antiserum. (B) Cell lysates of the indicated strains grown with or without 1 mM IPTG were subjected to SDS-PAGE and Coomassie brilliant blue staining. (C) Cell envelopes of the indicated strains grown in the presence of IPTG were treated with urea and subsequently treated or not treated with trypsin, as indicated, and analyzed by immunoblotting with an anti-FrpB antiserum.
NhhA is a trimeric autotransporter (44) whose membrane-embedded translocator domain is formed by three monomers, each donating four β-strands to the 12-stranded β-barrel, resulting in a heat- and SDS-resistant molecule migrating at a much higher apparent molecular weight than expected from the predicted size of a monomer with a molecular weight of 60,000 (9, 44). We anticipated that chaperones might be required for the biogenesis of such a complex molecule. As natural levels of NhhA could not be detected in HB-1, we introduced a plasmid containing nhhA under IPTG control in HB-1 and subsequently constructed skp2, ΔsurA, and ΔdegQ mutations in this background. Growth of the strains in the presence of IPTG resulted in comparable, huge levels of NhhA migrating in a HMW position in SDS-PAGE, indicating that the translocator domain is assembled correctly in all strains (see Fig. S3C and D in the supplemental material). Also, immunofluorescence experiments using an antibody against the passenger domain of NhhA showed similar positive staining of parent and mutant strains (data not shown), indicating that the chaperone mutants correctly assemble this protein. Factor H-binding protein (fHbp) is a surface-exposed lipoprotein (23) whose presence at the cell surface can be determined by using its high sensitivity to extracellular proteases. Both the levels and protease accessibility of fHbp were unaffected in the mutants (see Fig. S3E in the supplemental material). Thus, none of the chaperones Skp, SurA, and DegQ are required for the biogenesis of the cell surface-exposed lipoprotein fHbp, the OMPs PilQ, Omp85/BamA, Imp/LptD, NspA, and FrpB, or the trimeric autotransporter NhhA, even when this protein is highly overexpressed.
The absence of Skp affects porB expression.
The lower levels of PorB in the skp2 mutant were not due to proteolytic activity of DegQ, as shown by the results described above. Similarly, we found no increase in porin levels or the accumulation of unfolded porins when we inactivated in the skp2 mutant the genes for two other periplasmic proteases, i.e., NMB1332, which is a prc/tsp homolog (37), and NMB1433, an spr homolog (12) (data not shown). To test whether the expression of porB might be affected by the absence of Skp, we constructed a single-copy PporB-lacZ translational fusion in the chromosomal hrtA locus in strain HB-1 and generated a skp2 mutation in this strain. A control strain containing a promoterless lacZ gene did not generate detectable β-galactosidase activity. In contrast, the PporB-lacZ parent strain produced high levels of β-galactosidase activity: 7,299 ± 244 Miller units. In cells lacking Skp, the β-galactosidase activity dropped by 44%, to 4,154 ± 38 Miller units, demonstrating that the lower PorB levels in the skp2 mutant are caused at least in part by decreased production of the protein.
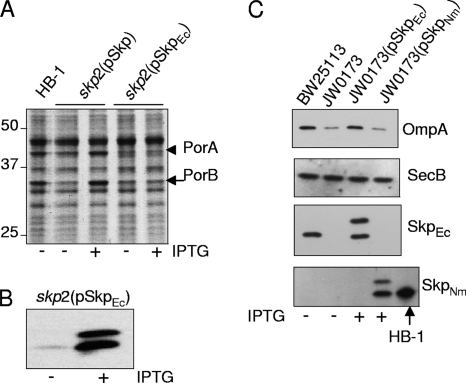
Cross-complementation analysis.
E. coli-derived Skp was reported to bind the neisserial autotransporter NalP with high affinity in vitro (29). To determine whether E. coli Skp can substitute for N. meningitidis Skp in vivo, we introduced a copy of E. coli skp under IPTG control on plasmid pFP10-SkpEc into the skp2 mutant. Porin levels were not restored upon induction of the expression of the E. coli skp gene, whereas this was clearly the case when neisserial skp was present on the plasmid (Fig. 4 A). Immunoblot analysis of cell envelopes confirmed that E. coli Skp was indeed produced upon IPTG induction, albeit with somewhat incomplete processing as suggested by the two bands detected with the antiserum (Fig. 4B). These bands probably correspond to the precursor and mature forms of Skp. Thus, E. coli Skp is not able to functionally replace N. meningitidis Skp. For the reverse experiment, we introduced pFP10-SkpNm and pFP10-SkpEc into an E. coli skp mutant. As reported before (6), OmpA levels decreased in the absence of Skp and were restored by expressing a plasmid copy of the E. coli-derived skp (Fig. 4C, top panel). However, the expression of neisserial skp did not restore OmpA levels (Fig. 4C, top panel), even though the Skp protein was produced and for the most part processed (Fig. 4C, bottom panel). Thus, the Skp proteins of E. coli and N. meningitidis cannot replace each other in vivo, at least not in maintaining wild-type OMP levels.
FIG. 4.
Lack of cross-complementation of E. coli and N. meningitidis Skp. (A) The indicated N. meningitidis strains were grown in the presence or absence of 1 mM IPTG as specified below the gel and analyzed as cell lysates by denaturing SDS-PAGE and Coomassie brilliant blue staining. The positions of PorA and PorB are indicated. (B) Synthesis of E. coli Skp in N. meningitidis. Cell envelopes of HB-1 carrying pFP10-SkpEc grown in the presence or absence of 1 mM IPTG were analyzed by denaturing SDS-PAGE and immunoblotting with antiserum directed against E. coli Skp. (C) The indicated E. coli strains were grown in M9 medium with or without 0.5 mM IPTG as indicated. Cell lysates were processed for immunoblots and probed with antisera against the proteins indicated on the right. SecB levels served as loading controls. The lower panel shows an extra lane containing cell envelopes from HB-1 to indicate the position in the gel of fully processed Skp.
DISCUSSION
In this work, we investigated the effect of Skp, SurA, and DegQ deficiencies on OMP biogenesis in N. meningitidis. Individual and double deficiencies had surprisingly little effect on OMP biogenesis. Only the absence of Skp resulted in a clear defect, i.e., lower cellular levels of the major OMPs, the porins PorA and PorB. The levels of other, minor OMPs, such as PilQ, NspA, Omp85/BamA, and Imp/LptD, were not affected by the absence of Skp. As the levels of another highly expressed OMP, FrpB, were also not diminished, it appears that the absence of Skp specifically affected the porins. In E. coli, the absence of Skp also results in diminished cellular levels of major OMPs, such as OmpA (Fig. 4C) (6), which is, at least in part, due to DegP-mediated degradation of unfolded OmpA, since aggregates of unfolded OmpA accumulate in the periplasm of an skp degP double mutant (36). N. meningitidis, which naturally lacks DegP, is apparently different in this respect, since we did not find any evidence for the presence of unfolded porins in the skp mutant. We also found no indications that other periplasmic proteases, i.e., DegQ, NMB1332, or NMB1433, were responsible for the removal of unfolded porins. However, we cannot exclude a possible role for other periplasmic proteases that we did not investigate.
Using a PporB-lacZ translational fusion, we observed that the expression of porB was reduced at the transcriptional and/or translational level in the skp mutant, suggesting that a feedback system is operating when Skp is absent. In E. coli, a number of signaling systems exist that respond to extracytoplasmic stress (31). The σE pathway that responds to the accumulation of unfolded OMPs in the periplasm of E. coli is absent in N. meningitidis (5). Possibly other pathways, such as the Cpx or Bae two-component systems, which were shown to respond to disorders in OMP biogenesis in E. coli (30), play similar roles in N. meningitidis. Meningococci contain only four two-component systems, which are not yet well characterized; one of them may be functionally homologous to the Cpx or Bae system in sensing OMP assembly defects and may respond by repressing the transcription of porin genes. Of note, however, unfolded OMPs accumulate in meningococcal strains with a defective Bam complex (47, 48), suggesting that under those conditions, the feedback inhibition system is not triggered. In E. coli, Skp was reported to bind OMPs while they are still engaged with the Sec machinery (13) and to stimulate their release from the IM (36). Possibly, the occupancy of the Sec system is sensed in a skp mutant of N. meningitidis, resulting in the feedback inhibition of the synthesis of the major exported proteins, the porins.
A surprising result of our study is the apparent absence of a role for SurA in OMP biogenesis. SurA was proposed to be the major periplasmic chaperone involved in OMP biogenesis in E. coli, whereas Skp would act in a parallel rescue pathway that deals only with substrates that fall off from the SurA pathway (38). Also, the absence of synthetic lethality of the skp and surA mutations in N. meningitidis constitutes another difference from the situation in E. coli but is consistent with the fact that we did not observe any phenotype of the surA mutation. The different function of SurA in N. meningitidis compared to that in E. coli might be reflected in its different structure. N. meningitidis SurA lacks one of the two PPIase domains found in E. coli SurA. Based on sequence alignments (data not shown), the PPIase 1 domain, which together with the N- and C-terminal parts forms the core of the protein in the crystal structure of E. coli SurA (2), is missing. Although the PPIase domains are dispensable for function (1), the PPIase 1 domain of E. coli SurA was shown to selectively bind peptides that are rich in aromatic residues and characteristic for OMPs (14, 54). Thus, SurA of N. meningitidis may fail to specifically recognize OMP substrates because it lacks the PPIase 1 domain. Interestingly, the SurA proteins of several other bacteria, such as those of Haemophilus influenzae and Pasteurella multocida, also lack the PPIase 1 domain (2), and we speculate that in these bacteria also, SurA has no general role in OMP biogenesis.
We found no evidence for a DegP-like function of neisserial DegQ as a chaperone or as a protease in the degradation of unassembled OMPs, although the E. coli DegQ can functionally replace DegP when overproduced (50). Recent structural studies have shed new light on the functional mechanisms of DegP. DegP appears to exist in several different conformations: in solution, inactive hexameric forms can transform into huge, cagelike 12- and 24-mers upon binding of substrates, which then become enclosed within the cage, where they are stabilized in a folded conformation or are degraded (16, 21). Interestingly, the neisserial DegQ protein is likely anchored to the inner or outer membrane by a lipid tail, since the C terminus of its signal sequence shows a characteristic lipobox sequence, LAGC. It seems impossible that a membrane-anchored lipoprotein could form cagelike assemblies similar to those of DegP, reinforcing the notion that neisserial DegQ is not a functional homolog of E. coli DegP.
The role of periplasmic chaperones in autotransporter biogenesis has been explored in only a few studies. The autotransporters IcsA of Shigella flexneri and EspP of E. coli require DegP, Skp, and SurA for proper cell surface presentation (28, 34, 49), whereas the biogenesis of Hbp in E. coli is affected by the absence of SurA but not by that of DegP or Skp (35). We found no defects in the assembly of the trimeric autotransporter NhhA or in the levels of the secreted monomeric autotransporter IgA protease in the neisserial chaperone mutants; apparently, chaperone requirements may be species and autotransporter specific.
Outer membrane-destined lipoproteins are chaperoned through the periplasm by the LolA protein (55). In E. coli, these lipoproteins are anchored in the inner leaflet of the OM; however, in N. meningitidis and in many other pathogens, lipoproteins anchored in the outer leaflet of the OM are also found. It is completely unknown how these lipoproteins reach the cell surface. As we did not find any defect in the cell surface exposure of the lipoprotein fHbp in our chaperone mutants, we conclude that SurA, Skp, and DegQ are required neither directly for the biogenesis of such lipoproteins nor indirectly by playing an essential role in the assembly of a putative lipoprotein transfer system.
Interestingly, we found no cross-species complementation of E. coli and N. meningitidis skp. Skp does not demonstrate much species specificity in vitro, as E. coli-derived Skp was reported to bind a neisserial OMP (29). Since, nevertheless, E. coli Skp could not functionally replace its homolog in N. meningitidis and vice versa, it appears that Skp interacts with another component of the OMP biogenesis machinery and that such interaction possibly happens in a species-specific manner.
In conclusion, we found no general role for Skp, SurA, or DegQ as an OMP chaperone in N. meningitidis. However, Skp is involved in the biogenesis of porins by affecting protein production, presumably through a feedback system that remains to be identified.
Supplementary Material
Acknowledgments
We thank Heike Claus and Ulrich Vogel (University of Würzburg, Germany) for donation of the pSMS1 plasmid and the National Bioresource Project (NIG, Japan): E. coli for providing strains.
This work was supported by the Netherlands Research Councils for Chemical Sciences (CW) and Earth and Life Sciences (ALW) of the Netherlands Organization for Scientific Research (NWO) and by GlaxoSmithKline Biologicals.
Published ahead of print on 4 February 2011.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Behrens, S., R. Maier, H. de Cock, F. X. Schmid, and C. A. Gross. 2001. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J. 20:285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bitto, E., and D. B. McKay. 2002. Crystallographic structure of SurA, a molecular chaperone that facilitates folding of outer membrane porins. Structure 10:1489-1498. [DOI] [PubMed] [Google Scholar]
- 3.Bos, M. P., and J. Tommassen. 2005. Viability of a capsule- and lipopolysaccharide-deficient mutant of Neisseria meningitidis. Infect. Immun. 73:6194-6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos, M. P., B. Tefsen, J. Geurtsen, and J. Tommassen. 2004. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl. Acad. Sci. U. S. A. 101:9417-9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bos, M. P., V. Robert, and J. Tommassen. 2007. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61:191-214. [DOI] [PubMed] [Google Scholar]
- 6.Chen, R., and U. Henning. 1996. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol. Microbiol. 19:1287-1294. [DOI] [PubMed] [Google Scholar]
- 7.Claus, H., M. Frosch, and U. Vogel. 1998. Identification of a hotspot for transformation of Neisseria meningitidis by shuttle mutagenesis using signature-tagged transposons. Mol. Gen. Genet. 259:363-371. [DOI] [PubMed] [Google Scholar]
- 8.Clausen, T., C. Southan, and M. Ehrmann. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10:443-455. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, S. E., N. K. Surana, and J. W. St. Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13:199-205. [DOI] [PubMed] [Google Scholar]
- 10.de Cock, H., et al. 1999. Affinity of the periplasmic chaperone Skp of Escherichia coli for phospholipids, lipopolysaccharides and non-native outer membrane proteins. Role of Skp in the biogenesis of outer membrane protein. Eur. J. Biochem. 259:96-103. [DOI] [PubMed] [Google Scholar]
- 11.Genevrois, S., L. Steeghs, P. Roholl, J. Letesson, and P. van der Ley. 2003. The Omp85 protein of Neisseria meningitidis is required for lipid export to the outer membrane. EMBO J. 22:1780-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hara, H., N. Abe, M. Nakakouji, Y. Nishimura, and K. Horiuchi. 1996. Overproduction of penicillin-binding protein 7 suppresses thermosensitive growth defect at low osmolarity due to an spr mutation of Escherichia coli. Microb. Drug Resist. 2:63-72. [DOI] [PubMed] [Google Scholar]
- 13.Harms, N., et al. 2001. The early interaction of the outer membrane protein PhoE with the periplasmic chaperone Skp occurs at the cytoplasmic membrane. J. Biol. Chem. 276:18804-18811. [DOI] [PubMed] [Google Scholar]
- 14.Hennecke, G., J. Nolte, R. Volkmer-Engert, J. Schneider-Mergener, and S. Behrens. 2005. The periplasmic chaperone SurA exploits two features characteristic of integral outer membrane proteins for selective substrate recognition. J. Biol. Chem. 280:23540-23548. [DOI] [PubMed] [Google Scholar]
- 15.Hodak, H., et al. 2008. The peptidyl-prolyl isomerase and chaperone Par27 of Bordetella pertussis as the prototype for a new group of parvulins. J. Mol. Biol. 376:414-426. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, J., et al. 2008. Activation of DegP chaperone-protease via formation of large cage-like oligomers upon binding to substrate proteins. Proc. Natl. Acad. Sci. U. S. A. 105:11939-11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansen, J., A. A. Rasmussen, M. Overgaard, and P. Valentin-Hansen. 2006. Conserved small non-coding RNAs that belong to the σE regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol. 364:1-8. [DOI] [PubMed] [Google Scholar]
- 18.Kim, D. Y., and K. K. Kim. 2005. Structure and function of HtrA family proteins, the key players in protein quality control. J. Biochem. Mol. Biol. 38:266-274. [DOI] [PubMed] [Google Scholar]
- 19.Korndörfer, I. P., M. K. Dommel, and A. Skerra. 2004. Structure of the periplasmic chaperone Skp suggests functional similarity with cytosolic chaperones despite differing architecture. Nat. Struct. Mol. Biol. 11:1015-1020. [DOI] [PubMed] [Google Scholar]
- 20.Kortekaas, J., et al. 2007. Shielding of immunogenic domains in Neisseria meningitidis FrpB (FetA) by the major variable region. Vaccine 25:72-84. [DOI] [PubMed] [Google Scholar]
- 21.Krojer, T., et al. 2008. Structural basis for the regulated protease and chaperone function of DegP. Nature 453:885-890. [DOI] [PubMed] [Google Scholar]
- 22.Lazar, S. W., and R. Kolter. 1996. SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 178:1770-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madico, G., et al. 2006. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J. Immunol. 177:501-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Mogensen, J. E., and D. E. Otzen. 2005. Interactions between folding factors and bacterial outer membrane proteins. Mol. Microbiol. 57:326-346. [DOI] [PubMed] [Google Scholar]
- 26.Pajon, R., et al. 2009. Identification of new meningococcal serogroup B surface antigens through a systematic analysis of neisserial genomes. Vaccine 28:532-541. [DOI] [PubMed] [Google Scholar]
- 27.Pettersson, A., et al. 2006. Vaccine potential of the Neisseria meningitidis lactoferrin-binding proteins LbpA and LbpB. Vaccine 24:3545-3557. [DOI] [PubMed] [Google Scholar]
- 28.Purdy, G. E., C. R. Fisher, and S. M. Payne. 2007. IcsA surface presentation in Shigella flexneri requires the periplasmic chaperones DegP, Skp, and SurA. J. Bacteriol. 189:5566-5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu, J., C. Mayer, S. Behrens, O. Holst, and J. H. Kleinschmidt. 2007. The trimeric periplasmic chaperone Skp of Escherichia coli forms 1:1 complexes with outer membrane proteins via hydrophobic and electrostatic interactions. J. Mol. Biol. 374:91-105. [DOI] [PubMed] [Google Scholar]
- 30.Raivio, T. L. 2005. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol. Microbiol. 56:1119-1128. [DOI] [PubMed] [Google Scholar]
- 31.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 32.Rizzitello, A. E., J. R. Harper, and T. J. Silhavy. 2001. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J. Bacteriol. 183:6794-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rouvière, P. E., and C. A. Gross. 1996. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 10:3170-3182. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz-Perez, F., et al. 2009. Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J. Bacteriol. 191:6571-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sauri, A., et al. 2009. The Bam (Omp85) complex is involved in secretion of the autotransporter haemoglobin protease. Microbiology 155:3982-3991. [DOI] [PubMed] [Google Scholar]
- 36.Schäfer, U., K. Beck, and M. Müller. 1999. Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 274:24567-24574. [DOI] [PubMed] [Google Scholar]
- 37.Silber, K. R., K. C. Keiler, and R. T. Sauer. 1992. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proc. Natl. Acad. Sci. U. S. A. 89:295-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sklar, J. G., T. Wu, D. Kahne, and T. J. Silhavy. 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 21:2473-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sklar, J. G., et al. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 104:6400-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steeghs, L., et al. 1998. Meningitis bacterium is viable without endotoxin. Nature 392:449-450. [DOI] [PubMed] [Google Scholar]
- 41.Steeghs, L., et al. 2002. Expression of foreign LpxA acyltransferases in Neisseria meningitidis results in modified lipid A with reduced toxicity and retained adjuvant activity. Cell. Microbiol. 4:599-611. [DOI] [PubMed] [Google Scholar]
- 42.Stork, M., et al. 2010. An outer membrane receptor of Neisseria meningitidis involved in zinc acquisition with vaccine potential. PLoS Pathog. 6:e1000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vandeputte-Rutten, L., M. P. Bos, J. Tommassen, and P. Gros. 2003. Crystal structure of Neisserial surface protein A (NspA), a conserved outer membrane protein with vaccine potential. J. Biol. Chem. 278:24825-24830. [DOI] [PubMed] [Google Scholar]
- 44.van Ulsen, P., L. van Alphen, C. T. Hopman, A. van der Ende, and J. Tommassen. 2001. In vivo expression of Neisseria meningitidis proteins homologous to the Haemophilus influenzae Hap and Hia autotransporters. FEMS Immunol. Med. Microbiol. 32:53-64. [DOI] [PubMed] [Google Scholar]
- 45.van Ulsen, P., et al. 2003. A neisserial autotransporter NalP modulating the processing of other autotransporters. Mol. Microbiol. 50:1017-1030. [DOI] [PubMed] [Google Scholar]
- 46.Vertommen, D., N. Ruiz, P. Leverrier, T. J. Silhavy, and J. F. Collet. 2009. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics 9:2432-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volokhina, E. B., F. Beckers, J. Tommassen, and M. P. Bos. 2009. The β-barrel outer membrane protein assembly complex of Neisseria meningitidis. J. Bacteriol. 191:7074-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voulhoux, R., M. P. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262-265. [DOI] [PubMed] [Google Scholar]
- 49.Wagner, J. K., J. E. Heindl, A. N. Gray, S. Jain, and M. B. Goldberg. 2009. Contribution of the periplasmic chaperone Skp to efficient presentation of the autotransporter IcsA on the surface of Shigella flexneri. J. Bacteriol. 191:815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waller, P. R., and R. T. Sauer. 1996. Characterization of degQ and degS, Escherichia coli genes encoding homologs of the DegP protease. J. Bacteriol. 178:1146-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walton, T. A., and M. C. Sousa. 2004. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol. Cell 15:367-374. [DOI] [PubMed] [Google Scholar]
- 52.Walton, T. A., C. M. Sandoval, C. A. Fowler, A. Pardi, and M. C. Sousa. 2009. The cavity-chaperone Skp protects its substrate from aggregation but allows independent folding of substrate domains. Proc. Natl. Acad. Sci. U. S. A. 106:1772-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, T., et al. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235-245. [DOI] [PubMed] [Google Scholar]
- 54.Xu, X., S. Wang, Y.-X. Hu, and D. B. McKay. 2007. The periplasmic bacterial molecular chaperone SurA adapts its structure to bind peptides in different conformations to assert a sequence preference for aromatic residues. J. Mol. Biol. 373:367-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yokota, N., T. Kuroda, S. Matsuyama, and H. Tokuda. 1999. Characterization of the LolA-LolB system as the general lipoprotein localization mechanism of Escherichia coli. J. Biol. Chem. 274:30995-30999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.