Abstract
Many bacteria possess the ability to actively take up DNA from the environment and incorporate it into the chromosome. RecA protein is the key protein achieving homologous recombination. Several of the proteins involved in the transport of DNA across the cell envelope assemble at a single or both cell poles in competent Bacillus subtilis cells. We show that the presumed structure that transports DNA across the cell wall, the pseudopilus, also assembles at a single or both cell poles, while the membrane receptor, ComEA, forms a mobile layer throughout the cell membrane. All other known Com proteins, including the membrane permease, localize again to the cell pole, revealing that the uptake machinery has three distinct layers. In cells having two uptake machineries, one complex is occasionally mobile, with pairs of proteins moving together, suggesting that a complete complex may lose anchoring and become mobile. Overall, the cell pole provides stable anchoring. Only one of two uptake machineries assembles RecA protein, suggesting that only one is competent for DNA transfer. FRAP (fluorescence recovery after photobleaching) analyses show that in contrast to known multiprotein complexes, the DNA uptake machinery forms a highly stable complex, showing little or no exchange with unbound molecules. When cells are converted into round spheroplasts, the structure persists, revealing that the assembly is highly stable and does not require the cell pole for its maintenance. High stability may be important to fulfill the mechanical function in pulling DNA across two cell layers.
Competence refers to the physiological state in which many bacterial species can take up plasmid or chromosomal DNA from the environment, which can be propagated or recombined with their own chromosomal DNA (8). This ability is found in species from a broad spectrum of all bacterial branches, and it is clinically relevant because several human pathogens are naturally competent and can acquire drug resistance. Bacillus subtilis is a model organism for the study of the molecular mechanism of competence. At the transition between exponential growth and stationary phase, B. subtilis cells can enter stationary phase and induce sporulation (to a maximum of 80% of the cells) or competence. Under conditions favoring competence, only up to 20% of the cells become competent. Thus, competence and sporulation are active decisions taken by a subpopulation of cells. These decisions are based on bistable switches, an intriguing phenomenon in bacterial populations (34). Competence is induced through secreted peptide factors that are taken up by the cells in a quorum sensing mechanism and trigger a sophisticated regulatory system, ultimately leading to the stabilization of the otherwise unstable master transcription regulator ComK. In turn, ComK activates transcription of all necessary competence proteins (at least 13: ComGA, -GB, -GC, -GD, -GE, -GF, -GG, -EA, -EB, -EC, -FA, -FB, and -FC), as well as about 100 other proteins (1, 25). ComK is turned off (i.e., targeted for proteolysis) about 2 to 3 h after induction of competence.
It has been proposed that DNA uptake occurs through a three-step mechanism. (i) Transfer of DNA through the cell wall requires the products of the comG operon, which may form a pilus-like structure (5). ComGC is the major constituent of the pilin-like subunits, while ComGA is a cytosolic ATPase required for the assembly of the pseudopilus and also mediates the repression of growth and cell division during competence (11). The other steps are (ii) binding of double-stranded DNA (dsDNA) by the membrane-bound ComEA protein (28) and (iii) cleavage of DNA into shorter fragments by the NucA endonuclease (27), conversion into single-stranded DNA (ssDNA) by an unknown enzyme, and uptake into the cytosol through the ComEC membrane permease (2). ComEC is thought to form an aqueous ssDNA-specific membrane channel, while ComFA is an ATPase that may energize the uptake of DNA (22). Therefore, only one DNA strand enters the cytosol and the other is degraded (2). Single-molecule experiments have revealed that the competence machinery provides strong motor activity, with up to 80 bp/s processively being pulled into the cell. Uptake depends on the membrane proton motive force (24).
Intriguingly, the B. subtilis competence machinery has been shown to be located at one or both cell poles (10), showing that DNA uptake occurs at a specific site in competent cells. Inside the cell, imported DNA is used for homologous recombination (HR) with the chromosome, leading to the transformation of the cell, in case the acquired DNA contains novel or altered genetic information. The ssDNA-binding ATPase RecA is the central player in DNA recombination. It forms right-handed nucleoprotein filaments in vitro and introduces ssDNA into a homologous DNA duplex, thereby mediating DNA strand exchange and extruding one strand of the parental duplex, which is degraded by an unknown factor (6, 17). In vivo, RecA accumulates at the polar DNA uptake machinery and forms dynamic filamentous structures in competent cells, extending from the pole to the chromosome (16). These structures may be the active form of RecA searching for homology.
Alternatively, taken-up DNA can be converted to a circular plasmid, which depends on RecO and RecU proteins, and can be propagated if an autonomous replication origin is present on the plasmid. RecO accumulates at the uptake machinery in response to the addition only of plasmid, but not of chromosomal DNA, while RecU, which is also accumulated at the polar machinery, dissipates from the cell pole upon addition of chromosomal as well as of plasmid DNA (14). Therefore, the recombination machinery is a highly dynamic apparatus that responds differentially to different kinds of incoming DNA. We wished to investigate which proteins are part of the DNA uptake machinery, if this machinery is also a dynamic complex, and how it is maintained at a single cell pole (or both cell poles). We found that the DNA uptake machinery is a very stable protein complex that persists even when cells lose their rod shape. We also show that the pseudopilus structure assembles at a single or both cell poles, showing that DNA uptake across the cell envelope is mediated entirely at the poles by a highly stable molecular machinery.
MATERIALS AND METHODS
Growth conditions.
For vector construction and propagation, Escherichia coli strain XL1-Blue was used. All strains of Bacillus subtilis are derivates of PY79. The strains which are used in this study are listed in Table S1 in the supplemental material. B. subtilis was grown to competence using the two-step protocol, which is described in reference 9. Media were supplemented with antibiotics where appropriate (ampicillin, 100 μg/ml; chloramphenicol [Cm], 5 μg/ml; spectinomycin, 100 μg/ml; tetracycline [Tet], 20 μg/ml; kanamycin, 10 μg/ml). If genes were expressed under the control of a PXyl promoter, an 0.5% final concentration of xylose was added to the medium. For microscopy, cells were mounted on 1% (wt/vol) agarose pads containing the supernatant of cells grown to competence to maintain this developmental state (quorum sensing) under imaging conditions.
Construction of vectors and strains.
All strains, oligonucleotides, and plasmids used in this study are listed in Tables S1 and S2 in the supplemental material. C-terminal fusions of fluorescent proteins were constructed by amplifying about 500 bp of the corresponding gene and cloning them into plasmid pSG1164 (21). Competent B. subtilis PY79 wild-type cells were transformed with the generated plasmid, and transformants were selected for chloramphenicol resistance. For the mCherry fusion to comEA, 500 bp of the 3′ gene region was cloned into plasmid JCL259-mCherry and then transformed to competent B. subtilis PY79 cells. The transformants were selected for spectinomycin resistance. To construct the N-terminal fusion to ComEA, plasmid pHJDS1-YFP (7) was used and competent PY79 cells were transformed with plasmid pHJDS1-YFP-ComEA, which integrated at the original gene locus. Transformants were selected for chloramphenicol resistance. In the case of ComGC-FlAsH (fluorescein arsenical hairpin binder), the gene encoding a fusion of the peptide GFLNCCPGCCMEP to the C terminus of ComGC was created by PCR. The resulting gene was inserted into plasmid pSG1164 by eliminating the present fluorescence protein gene, and afterwards, the resulting plasmid was transformed into B. subtilis PY79 wild-type cells.
For colocalization experiments, competent ComEC-yellow fluorescent protein (YFP) cells, competent ComFC-YFP cells, or competent ComGC-FlAsH cells were transformed with chromosomal DNA from ComGA-cyan fluorescent protein (CFP) cells. In the case of colocalization of ComFA-mCherry and ComEC-YFP, the Cm resistance of ComFA-mCherry was exchanged for Tet resistance by transforming competent cells of this strain with pCm::Tet. The resulting strain was transformed with chromosomal DNA from ComEC-YFP cells selecting for Cm and Tet resistance.
Fluorescence microscopy.
The fluorescence microscopy experiments were performed either on an AX70 microscope (Olympus) equipped with a total internal reflection fluorescent (TIRF) objective with a numerical aperture of 1.45 (1.45 NA) and a Photometrics CoolSnapES2 charge-coupled device (CCD) camera (Visitron System GmbH) (images were recorded using VisiView 1.5.8 [Visitron System GmbH]) or on an Axio Observer.Z1 microscope (Zeiss) using a 1.45-NA objective. The images were acquired with a Photometrix Cascade CCD camera (Visitron System GmbH) and recorded using Metamorph 7.5.5.0 software (Universal Imaging Corporation). All images were processed using ImageJ 1.43 (Wayne Rasband, National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/).
Immunofluorescence (IF).
PY79 wild-type cells were grown to competence using the two-step protocol (9). The fixation was done with 500 μl of the bacterial culture as described in reference 26. One difference from this protocol is the use of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8) in contrast to GTE buffer (0.5 M glucose, 10 mM EDTA, 20 mM Tris-HCl [pH 7.5]) and the treatment of the cells with lysozyme for ∼10 min. The cells were treated with 1:5,000-diluted ComGC antiserum from rabbit as the first antibody and with 1:100-diluted Alexa Fluor 488-coupled secondary antibody (Molecular Probes/Invitrogen). A standard fluorescein isothiocyanate (FITC)-GFP filter was used for the detection of fluorescence.
FlAsH.
The cells were grown to competence using the two-step protocol (9). One milliliter of the bacterial culture was centrifuged (13,000 rpm, 1 min, room temperature [RT]), and the pellet was resuspended in 50 μl FlAsH·EDT2 (TC-FlAsH II in-cell tetracysteine tag detection; Molecular Probes/Invitrogen) for 1 h at RT. Afterwards, the cells were washed twice with BAL buffer (2,3-dimercapto-1-propanol) (TC-FlAsH II in-cell tetracysteine tag detection; Molecular Probes/Invitrogen). For microscopy, the cells were resuspended in 50 μl of competence medium. A standard FITC-GFP filter was used for the detection of fluorescence.
FRAP experiments.
For FRAP (fluorescence recovery after photobleaching) analysis, measurements were performed on an Axio Observer.Z1 microscope (Zeiss) equipped with a 1.45-NA objective. Image acquisition was done with a Photometrix Cascade CCD camera. A laser with a 405-nm wavelength was used for bleaching. For image analysis (18), images of a FRAP series were recorded by using the Metamorph 7.5.5.0 program (Universal Imaging Corp.) and were subsequently analyzed by using ImageJ 1.43m (Wayne Rasband, National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/). In order to align a stack of images, the StackReg plug-in (32) was used. Fluorescence intensity of the polar region of interest (ROI) was measured automatically using a custom-written ImageJ plug-in (multimeasure). The continuous bleaching during scanning was compensated by the mean fluorescence of the entire cell in the same image. Background intensity was subtracted. The relative fluorescence intensity was normalized to the relative fluorescence intensity of the ROI before bleaching to facilitate comparison of multiple experiments with different bleaching depths. The average of several experiments was fitted against the time using OriginPro 8.1G (OriginLab Corporation).
RESULTS
Different localization patterns of competence proteins.
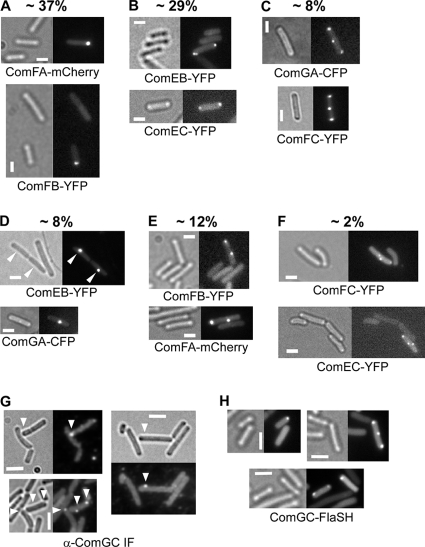
Components of the DNA uptake machinery that mediate the transfer across the cell membrane have been reported to be present at a single cell pole or at both cell poles or distributed throughout the cell membrane in a punctate pattern (10). DNA uptake has been shown to occur with a high force generated close to the cell pole by single-molecule experiments (24). These findings raise several important questions, namely, which proteins other than ComGA and ComFA are part of the polar machinery, what determines if cells have a single DNA uptake machinery or two to several, and where does translocation of external DNA across the cell wall occur. Apparently, the second cell pole is also competent for the assembly of the competence machinery. However, it is possible that single competence proteins may form assemblies on their own, distinct from the main machinery at the pole. In other words, cells may contain a single fully assembled competence machinery and nonassembled subassemblies of single Com proteins. To investigate the nature of the two assemblies at both poles in a large fraction of competent cells, we first set out to obtain a more complete picture of the localization of most proteins known to be involved in competence and subsequently to determine the pattern of proteins involved in competence in more detail. We therefore visualized several Com proteins that have not been observed before and scored the patterns of localization of all Com proteins in detail. All protein fusions except for ComEA are driven by the original promoter, encoded at the original gene locus. We found that ComEB-YFP, ComEC-YFP, ComFB-YFP, and ComFC-YFP all localized to one or both poles or, less frequently, to other places along the cell membrane (Fig. 1A to F). For ComEB, a higher percentage of cells than those for all other fluorescent protein fusions contained single foci and more than one focus per cell was rarely seen, in contrast to all other Com proteins (Table 1). While ComFB-YFP and ComFC-YFP fusions were fully functional in terms of transformation efficiency, the ComEB-YFP fusion showed a reduced transformation rate (∼100-fold), suggesting that the YFP fusion may partially interfere with the function and proper localization of ComEB. Nevertheless, our data suggest that the four Com proteins are also part of the polar competence machinery, with ComEC predicted to be a membrane protein. We scored the number of cells having a particular localization pattern for each competence protein, which is detailed in Table 1. Figure 1 states the average number of cells for all fusions showing a certain localization pattern. It is clear that cells having a single polar competence machinery are predominant (37% of all cells showing foci, not considering ComEB, which behaves abnormally [Fig. 1A]). Cells with two polar assemblies make up the second largest fraction (29% [Fig. 1B]). Further notable patterns are two foci, with one at a pole and one close to the cell center (12% [Fig. 1E]) and three foci, with two at a pole and one close to the center (8% [Fig. 1C]). These two patterns and a further pattern, one focus close to the cell center (8% [Fig. 1D]), may for a large part represent cells that are about to divide, in which a competence machinery assembles at a future division site (membrane staining did not reveal any septation in these cells [data not shown]). Cells having more than four foci or having several foci away from the cell pole represent only a minor subset of all competent cells (residual 4%). Therefore, it is clear that the cell poles, and not the lateral sides, represent major sites for the assembly of the competence machinery.
FIG. 1.
Different localization patterns of known and new competence proteins studied by fluorescence microscopy. (A to F) Localization of the proteins at one cell pole (A), at both cell poles (B), at both cell poles and at midcell (C), only at midcell (D), at one cell pole and at midcell (E), and in two foci at midcell (F). Protein fusions analyzed are stated under the panels. The percent values stated above the panels give the average occurrence of this pattern relative to all fluorescent cells for all fusions. White bars, 2 μm. (G and H) Two different fluorescence methods for localization studies of the pseudopilus protein ComGC: immunofluorescence with anti-ComGC antibody (G) and FlAsH method by C-terminal labeling of ComGC with FlAsH-EDT2 (H). White bars, 2 μm.
TABLE 1.
Quantitative analysis of localization patterns of competent proteins
| Strain | No. of cells | Fluorescent cells (% of cells) | % of fluorescent cells with: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 focus |
2 foci |
3 focia |
4 foci | >4 foci | |||||||
| At the pole | In the cellb | Both at the pole | 1 at the pole, 1 in the cell | Both in the cell | 2 at the pole, 1 in the cell | 1 at the pole, 2 in the cell | |||||
| ComGA-CFP | 1,608 | 16.8 | 33.7 | 5.9 | 26.3 | 16.3 | 0.4 | 10.7 | 1.5 | 3.3 | 1.9 |
| ComFA-mCherry | 1,497 | 9.8 | 37.0 | 7.5 | 28.8 | 8.9 | 0.0 | 13.7 | 0.7 | 2.7 | 0.7 |
| ComGC-FlAsH | 1,117 | 12.5 | 24.3 | 5.0 | 37.1 | 13.6 | 0.0 | 10.0 | 0.0 | 2.9 | 7.1 |
| ComEC-YFP | 573 | 28.4 | 31.9 | 4.3 | 41.1 | 13.5 | 0.0 | 4.9 | 1.8 | 1.8 | 0.6 |
| ComFC-YFP | 689 | 19.9 | 39.4 | 7.3 | 26.3 | 9.5 | 0.0 | 10.9 | 1.5 | 1.5 | 3.6 |
| ComEB-YFPc | 507 | 20.3 | 65.0 | 15.5 | 11.7 | 1.9 | 5.8 | 0.0 | 0.0 | 0.0 | 0.0 |
| ComFB-YFP | 1,861 | 8.0 | 39.6 | 8.1 | 28.8 | 15.3 | 0.0 | 5.4 | 0.9 | 1.8 | 0.0 |
Three foci away from the pole were never observed.
“In the cell” means “not at the pole but toward the cell center.”
Fusion is not fully functional.
To gain further insight into the nature of Com assemblies, we determined the average length of cells showing a certain localization pattern. Indeed, cells having a single assembly were the shortest, whereas cells with more foci were considerably longer (see Table S3 in the supplemental material). Cells containing a central focus were somewhat longer than cells with just polar assemblies, supporting the idea that longer cells assemble additional Com machineries at future division sites. The data also suggest that a second assembly may mature upon cell division to become an active machinery (see below), because cells with two or three foci are larger than cells with a single Com assembly (see Table S3).
Com proteins were visible in somewhat different numbers of cells grown to competence. Most proteins were present in 15 to 20% of the cells, but ComEC was observed in 24%, while ComFB was detectable in only 8% of the cells (Table 1). ComFB-YFP may not be detectable in more cells, because the fluorescence of this fusion construct is relatively faint. We do not know why apparently more cells contain the membrane permease than other Com proteins, but this may also reflect differences due to the abundances of proteins. In toto, these data support the idea that all competence proteins generally form one or two assemblies within competent cells.
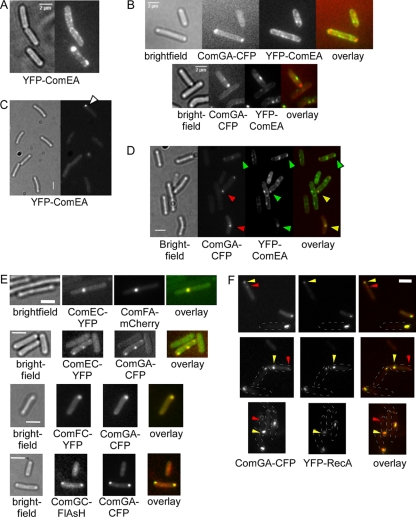
ComEA is the sole exception to the polarity rule for all other Com proteins investigated: ComEA has been reported to be present at several positions within the whole-cell membrane, in a seemingly helical pattern, using immunofluorescence (IF) (10). As IF can occasionally lead to artifacts, we generated a fluorescent protein fusion for ComEA, YFP-ComEA (the N terminus is supposed to be within the cytosol), which is fully functional (data not shown) and is expressed from the original gene locus. YFP-ComEA localized throughout the membrane in a punctate pattern in all cells grown to competence (Fig. 2A), because it is driven by a xylose promoter and not by the original ComK promoter. In some cells, YFP-ComEA formed polar accumulations (Fig. 2A, lower cell), possibly getting recruited to the competence machinery. To determine if these cells correspond to competent cells, we colocalized YFP-ComEA and ComGA-CFP. There was little correlation between ComGA-CFP foci and YFP-ComEA accumulation, and in the case that cells contained both kinds of accumulations, these rarely colocalized (Fig. 2B). These results show that ComEA does not specifically gather at the competence machinery but reveals individual accumulation patterns. To rule out the chance that the localization pattern is caused by protein overproduction, we grew cells with lower concentrations of the inducer for yfp-comEA transcription. Under conditions of a 50-fold-reduced amount of inducer, the foci along the lateral side were much fainter than under full induction but still visible (Fig. 1C). Polar accumulations were more apparent than they were under high-xylose conditions, and these often (but not always) colocalized with ComGA-CFP (Fig. 2D). These data suggest that the localization pattern of YFP-ComEA is not caused by an overproduction of the protein, and additionally, the pattern of localization is quite similar to that seen by the Dubnau laboratory through the use of IF. Thus, in contrast to all other Com proteins, ComEA localizes to many sites along the cell membrane and occasionally accumulates at the polar DNA uptake machinery but is not exclusively part of the competence machinery.
FIG. 2.
Colocalization of competence proteins in B. subtilis cells grown to competence. (A) Localization of YFP-ComEA. (B) YFP-ComEA does not accumulate at the polar competence machinery, visualized with ComGA-CFP. Lower panels show distinct locations of YFP-ComEA and ComGA-CFP foci. (C) Localization of YFP-ComEA under lower inducer levels (0.01 instead of 0.5%); the white triangle indicates polar accumulation of YFP-ComEA. (D) Colocalization of ComGA-CFP and of YFP-ComEA under lower inducer levels (0.01%); green triangles indicate polar YFP-ComEA accumulations, red triangles indicate polar ComGA-CFP foci, and yellow triangles indicate colocalization of the two proteins. (E) Examples of different pairs of competence proteins, as stated underneath the panels. (F) Localization of ComGA-CFP and YFP-RecA. Outlines of cells are indicated by dashed ovals. Yellow triangles indicate polar ComGA-CFP foci that colocalize with YFP-RecA foci, and red triangles indicate additional ComGA-CFP foci lacking any YFP-RecA signal. White bars, 2 μm.
It was important to verify that all competence proteins are present in the same structure. We therefore colocalized many different pairs of competence proteins, with the rationale that if all combinations of proteins colocalize in cells containing a single assembly, it follows that the whole machinery must be present at the same location. Alternatively, if a considerable number of noncolocalizing pairs of proteins are detected, different subcomplexes or partially assembled machineries most likely exist. We colocalized almost all possible combinations of pairs of Com proteins. In Fig. 2E, different pairs with distinct localization patterns are shown as examples. In 84% of all cases, protein pairs colocalized, as single or two polar foci or, more rarely, as foci at the cell center or at random positions in the cell. In about 13% of all cells analyzed (>500), only one of the two fusions but not the other (usually the fainter one) was visible. Only 3% of the cells showed noncolocalizing foci, which, however, were juxtaposed in all cases. We presume that these cases represent Com assemblies that are mobile (see below) and that filter changes resulted in a shift of the second acquired focus.
In toto, these experiments do not provide any evidence for Com proteins forming an assembly independently of other Com proteins. Therefore, it is reasonable to assume that foci of competence proteins represent complete competence machineries and that no subcomplexes lacking any known protein exist to a notable extent. This important finding leads to the conclusion that the Com complex assembles as a whole entity at the cell pole and is not composed of dynamically interacting subcomplexes or is set up from preexisting subcomplexes.
The pseudopilus structure assembles at a single cell pole or at both cell poles.
We used three different strategies to visualize the major component ComGC of the pseudopilus, which is required to allow the passage of DNA to the ComEA receptor, located at the cell membrane (4). ComGC belongs to the family of pilins and has been shown to be part of a multimeric structure (3). The addition of mCherry to ComGC rendered cells nontransformable, showing that a large tag interferes with the function of the pseudopilus. We therefore used two different strategies to visualize ComGC, namely, IF and the fluorescence arsenical hairpin (FlAsH) tag, a small 10-amino-acid (aa) extension that allows coupling to a chromophore through high-affinity binding in vivo. A ComGC-FlAsH fusion was almost fully competent for transformation (85% transformation efficiency), showing that a small tag does not interfere with the assembly and function of the pseudopilus. Addition of the FlAsH substrate revealed discrete foci, which in 24% of the competent cells (13% of all cells grown to competence showed fluorescence) were present at a single cell pole and in 37% were present at both cell poles (Fig. 1H). Faint foci could be seen along the longitudinal side in 10% of the cells.
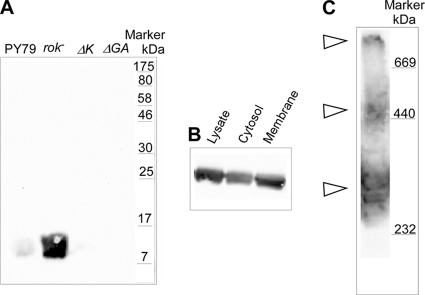
We also generated an immunoserum against the last 10 aa of ComGC, which recognized a single protein in Western blotting, corresponding to the size of ComGC (Fig. 3A), which is absent in comK or comGA mutant cells (note that a rok deletion increases the expression of ComK and thus of Com proteins). IF microscopy supported the findings of the FlAsH tag, namely, predominant localization to a single cell pole in competent cells but also localization to both poles in a substantial fraction of cells (Fig. 1G). Interestingly, ComGC-mCherry showed a similar pattern of localization (data not shown), indicating that the pseudopilus may in fact assemble with the fusion protein but is not functional, maybe because the mCherry moiety obstructs the passage of the DNA or interferes with assembly dynamics.
FIG. 3.
Blue native gel electrophoresis of B. subtilis cells grown to competence. (A) Western blot of whole-cell extracts separated by SDS-PAGE of strains as indicated using ComGC antiserum. (B) Western blot of ComGC in cytosolic and membrane fractions relative to the whole-cell lysate. (C) Western blot of ComGC from 5 to 15% blue-native PAGE of isolated membrane fractions. Membranes were solubilized in Triton X-100 (3%). Triangles indicate ComGC-containing complexes that have entered the gradient gel.
To verify that the pseudopilus assembles at the site where the DNA uptake machinery is present, we generated a strain expressing ComGA-CFP and ComGC-FlAsH, because ComGA has been shown to colocalize with several other proteins involved in DNA uptake. ComGA-CFP and ComGC-FlAsH colocalized in 96% of the cells when they were at a single cell pole or at both cell poles (Fig. 2E, fourth row). A considerable number of cells contained ComGC-FlAsH at sites along the lateral membrane, not colocalizing with a ComGA-CFP focus, suggesting that nonpolar pseudopilus structures are generally not associated with the rest of the uptake machinery. Thus, the ComGC pseudopilus structure is also present at the cell poles, where the rest of the competence machinery assembles, apparently ensuring that uptake across the cell wall is directly coupled to transport across the cell membrane.
ComGC forms distinct-sized membrane-associated complexes.
We aimed at obtaining further information on the organization of the presumed pseudopilus formed by the proteins of the comG operon. We used ComGC antiserum to detect the main structural component of the pseudopilus. ComGC was found in the cytosolic fraction as well as in the membrane (Fig. 3B), in accordance with the pilin-like nature of the protein. To identify ComGC-containing membrane-associated protein complexes in a native form and in the absence of any fixing agent, we performed blue native gel electrophoresis. Figure 3C shows that ComGC is present in several high-molecular-weight complexes in Triton-solubilized membrane fractions. The smallest metastable subcomplex identified migrates with an apparent molecular mass of 250 to 300 kDa, and a second subcomplex runs as a 450- to 500-kDa band on blue native gradient gels (Fig. 3C). The small and medium-size complexes were observed with a variety of different detergents tested (see Fig. S1 in the supplemental material) and are thus not induced by special detergent conditions. The largest complex is found in the megadalton range. As this complex species has clearly entered the gradient gel, it is most likely a native form and not the result of protein precipitation (Fig. 3C). The high-molecular-weight complex is also observed with all tested detergents (see Fig. S1), in agreement with ComGC building up a large pilus-like structure. Our data suggest that a ComGC-containing 250- to 300-kDa complex may represent the building block of this structure. The diffuse migration behavior of the detected membrane-bound subcomplexes indicates that they may not be completely uniform in their protein composition or association with membrane lipids, pointing to a dynamic assembly mechanism for the pseudopilus that, once assembled, can span the cell wall.
RecA assembles only at a single competence machinery.
In previous work, we have stated that the RecA protein almost exclusively localizes to a single cell pole in cells grown to competence, in a manner dependent on the DNA uptake machinery, and not to both cell poles. To clarify if we had overlooked cells having two YFP-RecA foci (the fusion is fully functional [15, 16]) or if only one of two polar uptake machineries is favored by RecA (and thus is active for transformation), we scored cells expressing YFP-RecA and ComGA-CFP in detail. Out of 120 cells having ComGA-CFP foci analyzed, we never found a cell that showed both two ComGA-CFP and two YFP-RecA assemblies. In the cells having two ComGA foci, YFP-RecA localized invariably to the brighter ComGA-CFP focus. Figure 2F shows two cells with bipolar ComGA-CFP signals and one with a polar focus and one away from a pole. In these cells, RecA assembles only at the brighter polar ComGA-CFP focus. These data suggest that only a single DNA uptake machinery can recruit RecA and that additional DNA uptake machineries are unlikely to be active for transformation.
Different dynamics of competence assemblies.
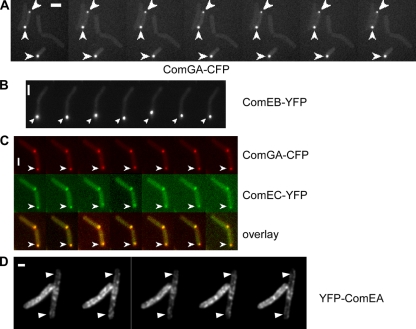
We investigated if the polar and the lateral DNA uptake complexes are mobile or are statically anchored at their position. We therefore performed time-lapse experiments with most of the fluorescent Com protein fusions that we generated. Interestingly, polar competence protein foci were predominantly static and did not move within a time scale of minutes. Figure 4A shows an example of a cell containing a single ComGA-CFP focus (lower cell) and one cell with two foci (upper cell), all of which remain static for 1 min. In most cells, foci remained static for more than 2 min (55 cells analyzed). However, in 10 to 15% of the cells a single focus was mobile and moved around the cell pole (Fig. 4B; see also Movie S1 in the supplemental material). In all cases of moving signals, foci moved away and back to the cell pole, or moved around the cell center, and did not stay away from the pole or the center for an extended period of time. Mobile foci could move about 0.4 μm in 10 s and thus were moderately dynamic. In cells containing two foci (29% of all cells with foci), one focus was invariantly static, while the other was mobile in 35% of these cases (data not shown). In 80% of the cases, the movable machinery was less bright than the other, static assembly; in 20%, the two foci were equally bright (data not shown). Thus, a minority of competence assemblies are mobile and they mostly contain fewer molecules than the second, static complex, while most complexes are static and anchored at the cell pole.
FIG. 4.
Dynamics of competence proteins. Time-lapse microscopy with pictures acquired every 10 s. (A) Static ComGA-CFP foci. (B) Mobile ComEB-YFP focus. (C) Colocalization of ComGA-CFP and ComEC-YFP over time (images were acquired with a time delay of 2 s). In the panels, arrowheads show the localization of the proteins at the beginning of the experiment. (D) YFP-ComEA. Arrowheads indicate quickly remodeling areas. White bars, 2 μm.
To shed light onto the nature of the mobile foci, we tested if possibly single protein fusions fall apart from a static machinery and move or if a whole machinery is mobile. We performed fast time-lapse experiments with two colors, in which the fusions are imaged every 10 s, with a delay of 2 s between the fusions. We found that ComGA-CFP and ComEC-YFP invariably moved together during 10-s intervals for the duration of 2 min. Figure 4C (see also Movies S2, S3, and S4 in the supplemental material) shows an example where one static focus is present in the upper cell, while the lower cell contains a focus that moves away from and back to a cell pole during the course of the experiment. Although it is possible that mobile assemblies lack a single component, our data suggest that a whole DNA uptake machinery can become mobile and detached from the cell pole, given that, except for ComEA, all Com proteins colocalize in 97% of the competent cells.
The rather static behavior of the Com proteins was not seen for YFP-ComEA. This protein fusion showed a different localization pattern between 10-s intervals, which is most easily seen in the lighter parts of the cells shown in Fig. 4D, suggesting that the distinct assemblies of ComEA are not static but are mobile. The mobility of YFP-ComEA foci is apparent from Movie S5 in the supplemental material and also in Movie S6, showing stream acquisition of YFP-ComEA with 10 frames/s in TIRF microscopy. In all cells, several accumulations of YFP-ComEA are stationary for an extended period of time, while other foci appear and disappear within intervals of a second. Thus, YFP-ComEA shows considerable dynamics within the membrane.
The DNA uptake machinery is a static complex.
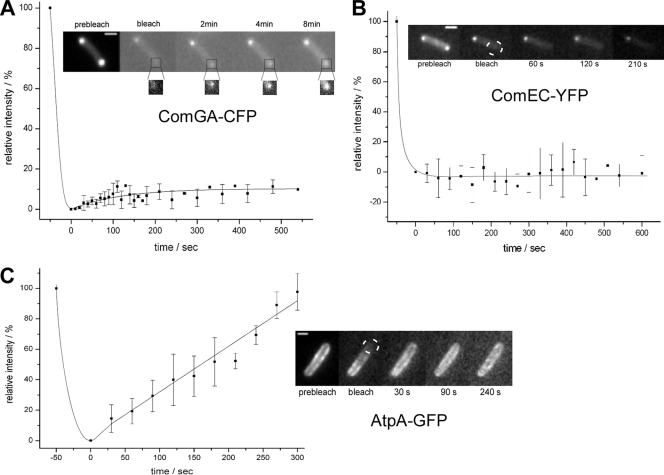
A fundamental biological question is how protein complexes are set up and are maintained. FRAP is a powerful technique to analyze if a protein complex shows high or low turnover, i.e., exchange of bound subunits with freely diffusing nonbound subunits. We bleached a small area close to a cell pole, which contained a DNA uptake assembly, and monitored the recovery of fluorescence of different components (ComGA, ComEA, ComEC, ComEB, and ComFC) of the polar uptake machinery. Figure 5A shows an example of a FRAP experiment, in which one ComGA-CFP focus is bleached in a cell containing two foci. At different time intervals after the bleach, a low degree of recovery of fluorescence can be observed. The enlargements show an increase of fluorescence; however, the images are highly increased for fluorescence intensity. The other nonbleached focus stays relatively constant for its fluorescence, i.e., it does not lose considerable fluorescence other than a small amount due to the bleaching during image acquisition. Although some fluorescence recovery is apparent and could be observed in many FRAP experiments, overall, fluorescence recovery was very low, with less than 10% recovery on average (15 experiments were scored [Fig. 5A]). Even more strikingly, fluorescence recovery for ComEC was close to 0% (Fig. 5B), revealing an entirely static behavior. The same observations were made for ComFC and ComEB fusions (data not shown). It should be noted that cells were fully competent for transformation (except that of ComEB; see above), even after FRAP experiments, showing that cells were fully viable. To rule out the possibility that all proteins become static within the membrane of cells grown to competence, we performed FRAP analysis on ATP synthase (AtpA-GFP), which shows recovery within 6 min in exponentially growing cells (13). In all cells grown to competence that were analyzed, ATP synthase recovered within 150 s for 50% fluorescence and within 300 s for full recovery (Fig. 5C), showing that ATP synthase remains mobile within the membrane of competent cells. These experiments show that the DNA uptake machinery is highly stable and to our knowledge the most stable complex found in a biological system so far.
FIG. 5.
FRAP experiments of the competence proteins ComGA-CFP (A) and ComEC-YFP (B). Diagrams show the relative intensities of the fluorescent area before and after bleaching over time, normalized against gradual bleaching of the images (between 12 and 15 experiments were analyzed). Pictures give examples of the microscopic acquisitions. The bleached area in panel A is highly increased in intensity in the inset, to reveal a low degree of fluorescence recovery—insets are equally scaled relative to each other. (C) Equivalent experiments with ATP synthase subunit AtpA-GFP in cells grown to competence. White bars, 1 μm.
The assembly of the uptake machinery persists even after a loss of rod shape.
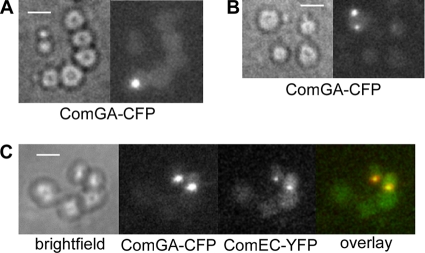
We wished to know if the competence machinery remains assembled in cells after a loss of their rod shape, i.e., in round cells. We therefore treated competent cells with lysozyme, after addition of a high concentration of sucrose to the medium to stabilize the cells. Intriguingly, 15% of the cells grown to competence (i.e., all competent cells) retained fluorescent foci (85% of the fluorescent cells had a single focus, 12% had two foci, and 3% had three or more foci), showing that the assembly of ComGA molecules persists even after the loss of rod shape (Fig. 6A and B). Capturing of Z-stacks through the cells revealed that there are one or two foci only (data not shown). Time-lapse microscopy showed little to no mobility of ComGA-CFP foci within the membrane of round cells (data not shown). To find out if this is also true for other components of the machinery and if the machinery remains assembled, we monitored the localization of ComGA-CFP and ComEC-YFP in the same cells. In all cases (93 cells analyzed), ComGA and ComEC signals were coincident at the cell membrane in the round cells (Fig. 6C). Again, 89% of the cells showing fluorescent foci contained a single focus, and 11% contained two foci, all of which coincided. Fluorescent lectin staining revealed that less than 50% of all spheroplasts retained residual cell wall material (data not shown), suggesting that in at least half of the spheroplasts containing a Com assembly, no more cell wall material is present. We infer from these findings that the competence machinery not only shows very little exchange with nonbound subunits but also remains intact when the physical structure of the cell pole is lost.
FIG. 6.
Localization of competence proteins in spheroplasts. (A) Single ComGA-CFP focus. (B) Cells with two ComGA-CFP foci. (C) Colocalization of ComGA-CFP and ComEC-YFP. The cells were grown to competence using the two-step protocol and were treated with 1 mg/ml (final concentration) lysozyme for 10 min. White bars, 2 μm.
DISCUSSION
Our work establishes that DNA uptake in competent Bacillus subtilis cells occurs via three differently organized layers of proteins. An important finding is the demonstration that ComGC, a pilin-like component of the subcomplex that is required for the passage of DNA across the cell wall, localizes to a single cell pole or, less frequently, to both cell poles (and very rarely to the lateral cell membrane). ComGC colocalizes with the DNA transfer proteins situated in and at the cell membrane, even in cells with one polar assembly, revealing that uptake of DNA across the cell wall and that across the membrane occur at the same polar position. ComGC is part of a several-MDa structure that can be isolated under native conditions and likely forms two subcomplexes of 250 and 400 kDa, respectively, as smaller units. These data suggest that the pseudopilus is indeed a large structure that can extend through the cell wall and assembles at a cell pole in competent cells. Our data are compatible with the idea that the pseudopilus may extend and retract, i.e., it may be a dynamic structure with the potential to pull on extracellular DNA for uptake across the cell wall. The second layer is ComEA, the membrane receptor for incoming double-stranded DNA (dsDNA) (28). ComEA is present at many sites within the membrane and can also bind to DNA, which may cross the cell wall but diffuse away from the pole in the space between wall and membrane. The third layer, the membrane translocase ComEC and associated ATPase ComFA, is again localized at a single pole or, less frequently, at both poles. Therefore, external DNA traverses the wall at a pole to be directly further processed and transported into the cell as ssDNA. Given a jam of DNA and a delay in membrane transport, we expect the additional ComEA proteins to sequester DNA, which can later be translocated into the cytoplasm. This is likely, because we show that ComEA moves within the entire membrane and can deliver dsDNA to the polar uptake channel. Interestingly, even if a cell has two DNA uptake machineries, transport of ssDNA to the chromosome and homology search by RecA appear to occur only from one pole at a time, because RecA never assembles at more than one uptake complex in the same cell. This spatial arrangement yields a vectoral DNA transport process from a single cell pole to the chromosome, which may render transformation much more efficient than would many randomly localized subcomplexes, which would attempt homology search from many directions onto the nucleoid.
We found that proteins of unknown functions, ComFB, ComFC, and ComEB, coassemble with the polar Com machinery, suggesting that all Com proteins are involved in DNA uptake. All protein combinations analyzed showed colocalization, suggesting that all Com proteins are present at the cell pole and that no partially assembled or distinct subcomplexes exist.
Several proteins as well as large complexes of multiple proteins in bacterial cells have been reported to be located at specific sites within the cytosol or the cell membrane (12, 23). It is still a mystery how specific subcellular localization of proteins in noncompartmentalized cells is achieved. The polar DNA uptake machinery in competent B. subtilis cells does not require a physical cell pole for its maintenance, as it assembles within long extended cells that do not contain proper rings of FtsZ (10), the first known protein to mark a future division site (and thus a new cell pole). This has also been shown for IcsA protein from Shigella pathogens (12) and for the chemosensory system (33). Thus, localization/assembly without a known physical structure corresponding to a future pole is a property shared by many bacterial membrane proteins. A remarkable finding is the fact that the competence machinery remains intact when rod shape is lost, i.e., when the physical structure of the pole is destroyed. Thus, the Com complex and possibly many other protein complexes must possess an autonomous factor that ensures the maintenance of assembly, independent of membrane curvature. This factor could be a single component or several components, whose main function is the bridging between the complex subunits (i.e., acting as molecular clamps), or could be an inherent property of the whole assemblage of Com proteins. However, there must be something special about the cell pole, because although nonpolar Com assemblies do exist, these are very rare and thus the cell pole must present something that highly favors the positioning of the complex. We have found that the DNA uptake machinery is generally not mobile, i.e., it is anchored at the pole, even in round cells. However, a low number of Com assemblies do move around a cell pole within a time frame of a few seconds, suggesting that the static position of most of the Com assemblies is not an intrinsic property of the complex but a feature of the cell pole or a combination of the pole and a feature of the complex. In this context, it is interesting that DivIVa has been shown to localize in response to high negative membrane curvature, i.e., at the cell pole and the newly invaginating septum (20, 29).
A further key finding is that the DNA uptake machinery has—to our knowledge—unprecedentedly low exchange with nonbound proteins. FRAP analysis has shown that all components of the complex analyzed show very little to no exchange, revealing that the assembly is highly stable. This is in contrast to the MotB subunit of the flagellar rotor, which is exchanged with nonbound proteins in a time frame of half a second for each of the 22 copies within the complex (19), and different from the chemosensory machinery at the cell pole in E. coli, which shows different degrees of exchange with nonbound subunits, from a few seconds (CheR and CheB) to 30 min (the Tar receptor core) (30). Thus, the competence machinery has amazingly dynamic interactions at its periphery, i.e., different recombination proteins are associated with the machinery or leave the machinery upon addition of different kinds of DNA (14, 16, 31), but the core complex is highly static and stable. It is possible that a complex that exerts high force against an object, such as the Com complex pulling DNA into the cell, must possess stable mechanical properties to achieve its motor function, while a sensory system must be flexible to respond to changing cues and environmental conditions. Our analysis rules out the notion that specific subcellular localization of the Com complex depends on diffusion/capture kinetics that are most stable at a cell pole but reveals that a multiprotein complex can assemble and remain inert for a long period of time.
As stated above, a small number of cells contain competence assemblies that are mobile. Most of these mobile complexes represent a second assembly in a cell having two assemblies, usually a larger (as deduced from fluorescence intensity) static complex and a second mobile and smaller one. We have found that the mobile assemblies contain at least two proteins moving together. Because all protein fusions analyzed show some mobile foci and because all analyzed competence proteins colocalize, it is most likely that the mobile assemblies contain most, if not all, Com proteins. Interestingly, we have found that RecA protein (the key protein mediating homologous recombination with the chromosome) assembles only at a single (usually the brighter) Com assembly in cells having two assemblies, indicating that only one machinery is the fully “competent” assembly, while the other is not, although it appears to contain all or the vast majority of Com proteins. Possibly, the first complex to be assembled becomes active for RecA recruitment and the second remains inactive, although it also appears to comprise all known Com proteins. Thus, assembly could be stochastic, with a pole providing a key for assembly but not being required for maintenance of assembly. It will be interesting to find out the molecular basis of this phenomenon.
In toto, our results reveal an astonishing robustness for the maintenance of the large competence protein complex, which comprises a mobile DNA-binding layer in the membrane and two polar layers, one in the cell wall and one in the membrane for DNA transport. It remains a key question which factors are responsible for the assembly of the large machinery at the cell pole.
Supplementary Material
Acknowledgments
We thank Andrea Zimmermann for technical help.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 746).
Footnotes
Published ahead of print on 28 January 2011.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Berka, R. M., et al. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43:1331-1345. [DOI] [PubMed] [Google Scholar]
- 2.Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241-249. [DOI] [PubMed] [Google Scholar]
- 3.Chen, I., R. Provvedi, and D. Dubnau. 2006. A macromolecular complex formed by a pilin-like protein in competent Bacillus subtilis. J. Biol. Chem. 281:21720-21727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung, Y. S., F. Breidt, and D. Dubnau. 1998. Cell surface localization and processing of the ComG proteins, required for DNA binding during transformation of Bacillus subtilis. Mol. Microbiol. 29:905-913. [DOI] [PubMed] [Google Scholar]
- 5.Chung, Y. S., and D. Dubnau. 1998. All seven comG open reading frames are required for DNA binding during transformation of competent Bacillus subtilis. J. Bacteriol. 180:41-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox, M. M. 2003. The bacterial RecA protein as a motor protein. Annu. Rev. Microbiol. 57:551-577. [DOI] [PubMed] [Google Scholar]
- 7.Defeu Soufo, H. J., and P. L. Graumann. 2006. Dynamic localization and interaction with other Bacillus subtilis actin-like proteins are important for the function of MreB. Mol. Microbiol. 62:1340-1356. [DOI] [PubMed] [Google Scholar]
- 8.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 9.Dubnau, D., and R. Davidoff-Abelson. 1971. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J. Mol. Biol. 56:209-221. [DOI] [PubMed] [Google Scholar]
- 10.Hahn, J., B. Maier, B. J. Haijema, M. Sheetz, and D. Dubnau. 2005. Transformation proteins and DNA uptake localize to the cell poles in Bacillus subtilis. Cell 122:59-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haijema, B. J., J. Hahn, J. Haynes, and D. Dubnau. 2001. A ComGA-dependent checkpoint limits growth during the escape from competence. Mol. Microbiol. 40:52-64. [DOI] [PubMed] [Google Scholar]
- 12.Janakiraman, A., and M. B. Goldberg. 2004. Evidence for polar positional information independent of cell division and nucleoid occlusion. Proc. Natl. Acad. Sci. U. S. A. 101:835-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, A. S., S. van Horck, and P. J. Lewis. 2004. Dynamic localization of membrane proteins in Bacillus subtilis. Microbiology 150:2815-2824. [DOI] [PubMed] [Google Scholar]
- 14.Kidane, D., et al. 2009. Evidence for different pathways during horizontal gene transfer in competent Bacillus subtilis cells. PLoS Genet. 5:e1000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kidane, D., and P. L. Graumann. 2005. Dynamic formation of RecA filaments at DNA double strand break repair centers in live cells. J. Cell Biol. 170:357-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kidane, D., and P. L. Graumann. 2005. Intracellular protein and DNA dynamics in competent Bacillus subtilis cells. Cell 122:73-84. [DOI] [PubMed] [Google Scholar]
- 17.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, M., M. S. Mommer, and V. Sourjik. 2010. Mobility of cytoplasmic, membrane, and DNA-binding proteins in Escherichia coli. Biophys. J. 98:552-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leake, M. C., et al. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443:355-358. [DOI] [PubMed] [Google Scholar]
- 20.Lenarcic, R., et al. 2009. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 28:2272-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, P. J., and A. L. Marston. 1999. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227:101-110. [DOI] [PubMed] [Google Scholar]
- 22.Londono-Vallejo, J. A., and D. Dubnau. 1994. Mutation of the putative nucleotide binding site of the Bacillus subtilis membrane protein ComFA abolishes the uptake of DNA during transformation. J. Bacteriol. 176:4642-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 24.Maier, B., I. Chen, D. Dubnau, and M. P. Sheetz. 2004. DNA transport into Bacillus subtilis requires proton motive force to generate large molecular forces. Nat. Struct. Mol. Biol. 11:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogura, M., et al. 2002. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J. Bacteriol. 184:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pogliano, K., E. Harry, and R. Losick. 1995. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol. Microbiol. 18:459-470. [DOI] [PubMed] [Google Scholar]
- 27.Provvedi, R., I. Chen, and D. Dubnau. 2001. NucA is required for DNA cleavage during transformation of Bacillus subtilis. Mol. Microbiol. 40:634-644. [DOI] [PubMed] [Google Scholar]
- 28.Provvedi, R., and D. Dubnau. 1999. ComEA is a DNA receptor for transformation of competent Bacillus subtilis. Mol. Microbiol. 31:271-280. [DOI] [PubMed] [Google Scholar]
- 29.Ramamurthi, K. S., and R. Losick. 2009. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc. Natl. Acad. Sci. U. S. A. 106:13541-13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulmeister, S., et al. 2008. Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105:6403-6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tadesse, S., and P. L. Graumann. 2007. DprA/Smf protein localizes at the DNA uptake machinery in competent Bacillus subtilis cells. BMC Microbiol. 7:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thevenaz, P., U. E. Ruttimann, and M. Unser. 1998. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7:27-41. [DOI] [PubMed] [Google Scholar]
- 33.Thiem, S., D. Kentner, and V. Sourjik. 2007. Positioning of chemosensory clusters in E. coli and its relation to cell division. EMBO J. 26:1615-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veening, J.-W., W. K. Smits, and O. P. Kuipers. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62:193-210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.