Abstract
YybT family proteins (COG3887) are functionally unknown proteins that are widely distributed among the firmicutes, including the human pathogens Staphylococcus aureus and Listeria monocytogenes. Recent studies suggested that YybT family proteins are crucial for the in vivo survival of bacterial pathogens during host infection. YybT family proteins contain an N-terminal domain that shares minimum sequence homology with Per-ARNT-Sim (PAS) domains. Despite the lack of an apparent residue for heme coordination, the putative PAS domains of BsYybT and GtYybT, two representative members of the YybT family proteins from Bacillus subtilis and Geobacillus thermodenitrificans, respectively, are found to bind b-type heme with 1:1 stoichiometry. Heme binding suppresses the catalytic activity of the DHH/DHHA1 phosphodiesterase domain and the degenerate GGDEF domain. Absorption spectroscopic studies indicate that YybT proteins do not form stable oxyferrous complexes due to the rapid oxidation of the ferrous iron upon O2 binding. The ferrous heme, however, forms a hexacoordinated complex with carbon monoxide (CO) and a pentacoordinated complex with nitric oxide (NO). The coordination of NO, but not CO, to the heme stimulates the phosphodiesterase activity. These results suggest that YybT family proteins function as stress-signaling proteins for monitoring cellular heme or the NO level by using a heme-binding PAS domain that features an unconventional heme coordination environment.
COG3887 or YybT family proteins are widely distributed among the firmicutes phylum, including such pathogens as Staphylococcus aureus, Streptococcus mutans, and Listeria monocytogenes. COG3887 family proteins contain two N-terminal transmembrane helices and three predicted protein domains: a putative Per-ARNT-SIM (PAS) domain, a highly degenerate GGDEF domain, and a DHH/DHHA1 domain (Fig. 1). Although the biological function of this family of proteins remains to be fully unveiled, genetic studies have revealed the connection of COG3887 family proteins with several phenotypes in various bacterial strains. The disruption of the llmg1816 gene, which encodes LlYybT by transposon insertion, rendered Lactococcus lactis more tolerant to acid stress (28). The knockout of the gcp gene, which encodes SmYybT, led to abnormal biofilm formation in Streptococcus mutans (44). In Staphylococcus aureus, the disruption of the SA0013 gene, which encodes SaYybT, abolished the secretion of the virulence factor hemolysin for iron acquisition, suggesting that SaYybT is crucial for the in vivo survival of S. aureus (4). Another study also found that the deletion of SA0013 seems to attenuate virulence during murine infection (2). The latest study from our laboratory demonstrated that the ΔBsYybT strain of Bacillus subtilis becomes more resistant to acid and nalidixic acid-caused DNA damage (31).
FIG. 1.
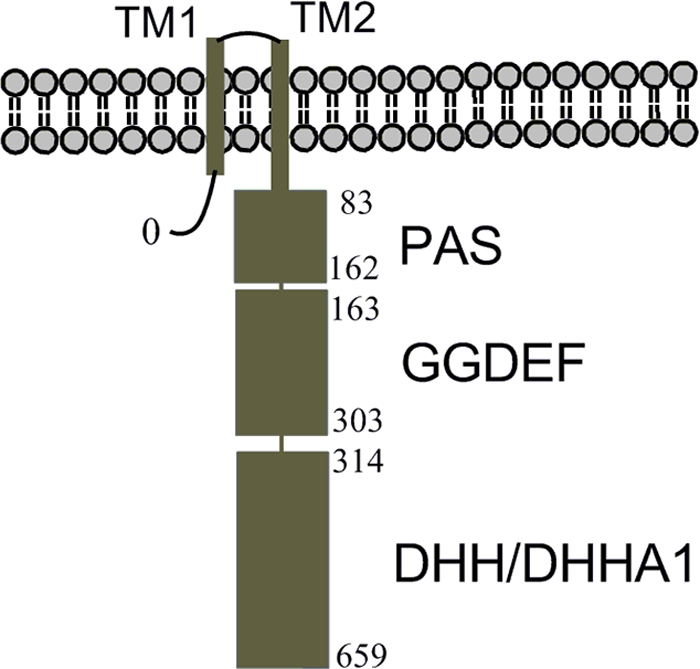
Domain organization of YybT family proteins (COG3887). The residue numbers are shown to indicate the domain boundaries of BsYybT.
The enzymatic activities of the two putative catalytic domains of BsYybT have been examined by us recently. Among a collection of potential substrates, the C-terminal DHH/DHHA1 domain exhibits phosphodiesterase activity toward the cyclic dinucleotides cyclic (c)-di-AMP and c-di-GMP, with a micromolar Michaelis-Menten constant (Km) for c-di-AMP (31). At this moment, however, it is not known whether c-di-AMP is truly the physiological substrate of the DHH/DHHA1 domain despite the potent enzymatic activity. The GGDEF domain possesses weak ATPase activity instead of the diguanylate cyclase (DGC) activity associated with orthodox GGDEF domains. COG3887 family proteins also contain a putative PAS domain immediately following the second transmembrane helix (Fig. 1). The prevalent PAS domain usually is comprised of a five-strand anti-parallel β-sheet and three or four flanking α-helices (17, 40). Many PAS domains function as sensor domains by binding ligand or cofactors such as heme, flavin, and para-hydroxcinnamate (19, 22, 24, 27, 33). The best-characterized PAS domains include the ones that bind heme for sensing changes in oxygen concentration and redox state (13, 16, 22). Given the low sequence identity shared with other characterized PAS domains, little information about the function of the PAS domain of COG3887 family proteins can be derived from protein sequences.
In this report, we present results to demonstrate that the PAS domains of two YybT proteins are able to bind b-type heme despite the lack of apparent residue(s) for heme coordination. The heme-bound BsYybT and its thermophilic homolog (GtYybT) in their oxidized, reduced, and nitric oxide (NO)-, carbon monoxide (CO)-, and cyanide (CN−)-ligated forms were characterized by absorption spectroscopy. Enzymatic activity measurement showed that the enzymatic activity of the DHH/DHHA1 domain of BsYybT is affected by the binding of the heme and coordination of cyanide and NO to the heme iron. The results from the in vitro studies suggest that the PAS domain of COG3887 family proteins represent a unique subfamily of a heme-binding PAS domain involved in heme or NO sensing.
MATERIALS AND METHODS
Materials.
c-di-AMP was purchased from Biolog. The NO donors MAHMA NONOate, DEA-NONOate, sodium nitroprusside (SNP), the CO donor tricarbonyldichlororuthenium (II) dimer (CORM2), potassium cyanide and potassium ferricyanide, hemin chloride, and thrombin were purchased from Sigma. Spermine NONOate was from Caymen Chemical. Strains and plasmids are described in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant phenotype | Description and/or reference |
|---|---|---|
| Strain | ||
| E. coli BL21(DE3) | Camr | Cells enable high-level expression of heterologous proteins in E. coli (Stratagene) |
| Bacillus subtilis 168 | Wild-type Marburg strain; trpC2 | 45 |
| Plasmid | ||
| pET28(a+) | Knr | Plasmid for the overexpression of His6-tagged proteins in E. coli (Novagen) |
| pET28-BsYybT84-659 | Knr | Plasmid for the overexpression of the PAS-GGDEF-DHH/DHHA1 fragment of BsYybT; this work |
| pET28-BsYybT150-659 | Knr | Plasmid for the overexpression of the GGDEF-DHH/DHHA1 fragment of BsYybT; this work |
| pET28-BsYybT84-303 | Knr | Plasmid for the overexpression of the PAS-GGDEF fragment of BsYybT; this work |
| pET-BsYybT55-162 | Knr | Plasmid for the overexpression of the PAS domain fragment of BsYybT; this work |
| pET28-GtYybT55-658 | Knr | Plasmid for the overexpression of the PAS-GGDEF-DHH/DHHA1 fragment of GtYybT; this work |
| pET28-GtYybT55-162 | Knr | Plasmid for the overexpression of the stand-alone PAS domain of GtYybT; this work |
| pET28-GtYybT55-304 | Knr | Plasmid for the overexpression of the PAS-GGDEF fragment of GtYybT; this work |
Protein expression and purification.
The preparation of BsYybT constructs has been described previously (31). Site-directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Stratagene). The gene encoding GtYybT from Geobacillus thermodenitrificans (GtYybT55-658) was cloned into pET28(a+) (Novagen) between the NdeI and XhoI restriction sites. Escherichia coli strain BL21(DE3) was transformed with the plasmid for the expression of the N-terminal His6-tagged recombinant construct GtYybT55-658. The shorter constructs (GtYybT55-162 and GtYybT55-304) were prepared by PCR cloning and ligated into pET28(a+) vector. The procedures for the expression and purification of BsYybT and GtYybT proteins are similar. Briefly, 1 liter of bacterial culture (in LB medium) was grown to an optical density of 0.8 before induction with 0.8 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The culture was shaken at 16°C for ∼12 h before it was pelleted by centrifugation. The cells were lysed in 20 ml lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 5% glycerol, 0.1% β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). After centrifugation at 25,000 rpm for 30 min, the supernatant was filtered and then incubated with 2 ml of nickel nitrilotriacetic acid (Ni-NTA) resin (Qiagen) for 1 h at 4°C. The resin was washed with 50 ml of W1 buffer (lysis buffer with 20 mM imidazole) and 20 ml of W2 buffer (lysis buffer with 50 mM imidazole). The proteins were eluted using a step gradient method, with the elution buffer containing 50 mM Tris (pH 8.0); 150 mM NaCl; 5% glycerol; and 200, 300, or 500 mM imidazole. After being analyzed by SDS-PAGE, fractions with purity higher than 95% were pooled together. Size-exclusion chromatography was carried out at 4°C using the AKTA fast protein liquid chromatography (FPLC) system equipped with a Superdex 200 HR 16/60 column (Amersham Biosciences). The buffer used for gel filtration was comprised of 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 5% glycerol. The proteins were stored at −80°C after being flash frozen, and concentrations were measured by the Bradford assay method.
Pyridine hemochrome assay.
To determine the identity of the heme bound by the recombinant BsYybT and GtYybT, a pyridine hemochrome assay was performed according to the method of Berry and Trumpower (3). Briefly, 0.5-ml aliquots of the sample were added to the stock solution [200 mM NaOH, 40% (vol/vol) pyridine, 3 μl of 0.1 M K3Fe(CN4)], followed by thorough mixing. The oxidized spectrum was recorded several times using a Shimadzu UV-1700 spectrophotometer. Sodium dithionite (2 to 5 mg) was added to the cuvette, and the spectrum of the reduced pyridine hemochrome was recorded. To estimate the percentage of heme incorporation, the absorbance difference at the selected wavelength between the reduced and oxidized spectra was used to calculate the concentration of heme based on the extinction coefficients as described previously (3).
Heme reconstitution.
The heme chloride stock (50 mM) in 0.1 M NaOH was slowly titrated into the protein solution (50 μM) with gentle agitation for ∼4 h (4°C), with a final heme/protein ratio of 5:1. Free heme was separated from the protein using size-exclusion chromatography with an AKTA FPLC system (31). The fractions containing the holo protein were pooled and dialyzed into pH 5.5 phosphate-buffered saline (PBS) buffer gradually. The protein then was exchanged back to the original buffer (50 mM Tris [pH 8.0], 150 mM NaCl, and 5% glycerol) by dialysis and concentrated for storage.
UV-vis spectroscopy.
Unless otherwise noted, all absorption spectra were measured with 200 μl holo protein (10 to 20 μM) in 50 mM Tris (pH 8.0), 150 mM NaCl at 23°C in a sealed quartz cuvette. Ferrous deoxy-BsYybT was obtained after the addition of 1 mM sodium dithionite (DTH). The NO donor stocks (100 mM) were made in nitrogen-saturated 0.01 M NaOH in an anaerobic chamber and stored at −80°C in aliquots. The CO donor (CORM2) was prepared in dimethylsulfoxide (DMSO) as a 100 mM stock and stored at −80°C. Following the addition of KCN (1 mM), NO donor (100 μM), CO donor (100 μM), or oxygen (by air bubbling), the absorption spectra of the various forms of BsYybT were recorded with the UV-vis spectrophotometer.
Enzymatic activity assays.
Enzymatic reactions were performed in a Coy anaerobic chamber. The enzymatic assay conditions and steady-state kinetic measurements for apo-BsYybT were reported previously (31). Assays for the ferric holo protein in the presence and absence of potassium cyanide (1 mM) were performed under similar conditions. The assays for the reduced protein form (ferrous state) were performed in the reaction buffer containing 100 mM Tris-HCl (pH 7.3), 20 mM KCl, and 0.5 mM MnCl2. One hundred μl protein (0.5 mg/ml) was split into two vials, followed by the addition of 0.5 μl of 100 mM sodium dithionite to one vial and 0.5 μl of 100 mM NaCl solution to the other. To remove residual dithionite that may interfere with the reaction, sodium dithionite was removed by buffer exchange through the use of desalting spin columns (Pierce) inside the anaerobic chamber. Five μl of the reduced and oxidized proteins was added to 195 μl reaction buffer containing 5 μM c-di-AMP. The reaction was stopped at various time points by the addition of a 1/10 volume of 0.5 M EDTA. The progress of c-di-AMP hydrolysis was monitored using the same Agilent LC1200 system as that described before (31). For the NO- and CO-ligated protein forms, the reaction buffer was first saturated with N2 or CO gas through extensive bubbling. The effect of NO was evaluated by the addition of 100 μM DEA-NONOate solution prepared in situ. The high-performance liquid chromatography (HPLC) conditions for product analysis and quantification have been described previously (27, 29-31).
The ATPase assay was performed as described previously (31). For the HPLC method with an Eclipse XDB-C18 reverse-phase column (4.6 by 150 mm), the flow rate was maintained at 1.5 ml/min, with the mobile phase being comprised of 100 mM KH2PO4 (pH 6.5), 10 mM tetrabutyl ammonium bromide (TBAB), and 5.5% acetonitrile. The mobile phase allowed us to differentiate the reactant and product readily, with a retention time of 5.0 and 12.0 min for ADP and ATP, respectively.
Structural modeling.
The structural model of the PAS domain was constructed by using the program I-Tasser (35) and manually aligned with the structures of the PAS domain of EcDOS and BjFixL for comparison.
RESULTS
Overexpression of heme-bound BsYybT and GtYybT proteins.
In a previous study, we cloned three BsYybT constructs of different lengths for enzymatic activity assays (31). It was noticed that the concentrated BsYybT84-303 protein solution had a faint brownish appearance. Absorption spectroscopy revealed an absorbance band at 412 nm and two other weaker bands at 533 and 566 nm (Fig. 2A). Although the spectrum is reminiscent of heme proteins, it was attributed to a copuri- fied E. coli heme protein. Later, when the homologous GtYybT55-658 and GtYybT55-304 from the thermophilic firmicutes Geobacillus thermodenitrificans were expressed and purified, the proteins obtained after the metal affinity and gel-exclusion chromatographic steps are noticeably brownish in color. The A412/A278 ratio did not decrease with increased protein homogeneity, suggesting that putative heme ligand is associated with the recombinant proteins. GtYybT55-658 and GtYybT55-304 exhibit absorption bands similar to those of BsYybT84-303 with the characteristic Soret and α/β bands (Fig. 2B). The A412/A278 ratio for the purified GtYybT55-304 is 10 times greater than that of BsYybT84-303, indicating higher heme occupancy for GtYybT55-304.
FIG. 2.
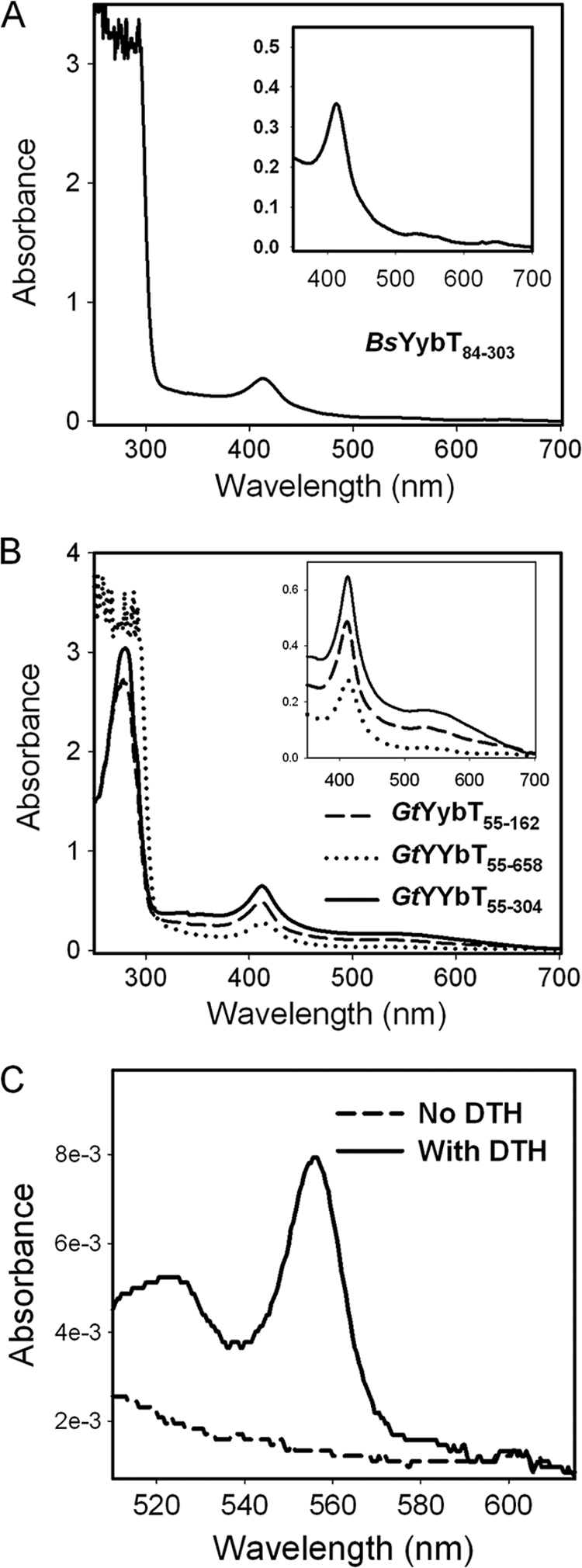
Absorption spectra of recombinant YybT proteins expressed from E. coli. (A) Absorption spectrum of BsYybT84-303. (B) Absorption spectra of the three GtYybT protein constructs. (C) Identification of type b heme by the pyridine hemochrome method.
Although many PAS domains are known to bind heme for O2 and redox sensing, the PAS domain of YybT family proteins was not expected to be a heme-binding domain because of the lack of a conserved histidine or cysteine residue for heme coordination. To verify that the heme is indeed bound by the PAS domain, we cloned and expressed the PAS domains of BsYybT and GtYybT as stand-alone proteins. While the insolubility of the BsYybT55-162 construct prevented further study, the PAS domain of GtYybT (GtYybT55-162) was expressed and purified with partial heme occupancy. The A412/A278 ratio for GtYybT55-162 is comparable to that of the longer construct, GtYybT55-304 (Fig. 2B). The heme bound by GtYybT55-162 was confirmed to be type b heme by the pyridine hemochrome method (3), with the diagnostic absorption bands at 557 and 525 nm upon reduction (Fig. 2C). As quantified by the pyridine hemochrome assay, the heme occupancy is ca. 5% for GtYybT55-162, which is comparable to that of GtYybT55-304. Despite the substoichiometric heme to protein ratio, the results clearly demonstrate the binding capability of the PAS domain of YybT family proteins for type b heme.
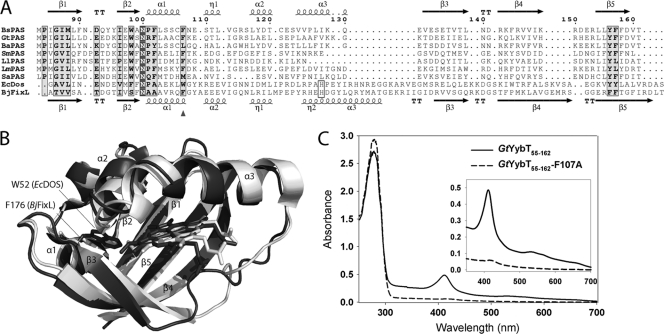
The sequence alignment of the YybT homologs with other heme PAS domains did not reveal any potential proximal ligand for the coordination of heme iron (Fig. 3A). The binding mode of heme thus is likely to be different from the canonical heme-binding PAS domains in EcDOS and BjFixL. The heme-binding pocket of EcDOS and BjFixL lies in a well-defined hydrophobic cavity flanked by a twisted β-sheet and a few short helices (Fig. 3B) (15, 22). Secondary structure prediction and structural modeling suggest that the same topology is conserved in the PASYybT domain despite the noticeable shortening of the α3 helix and β4 strand (Fig. 3A). Based on sequence alignment and structural modeling studies, a few bulky hydrophobic residues (Phe107, Tyr156, and Phe157 of BsYybT) seem to be conserved in EcDOS, BjFixL, and YybT family proteins. The corresponding residue of Phe107 (BsYybT) in EcDOS (Trp52) or BjFixL (Phe176) is located in the heme-binding pocket and is in direct contact with the heme cofactor. If we assume that PASYybT binds heme in a fashion similar to that of EcDOS or BjFixL, the side chain of Phe107 would be involved in protein-heme interaction as well. To test whether Phe107 is involved in protein-heme interaction, the F107A mutant of the GtYybT55-162 protein was prepared to analyze the heme occupancy. The F107A mutant exhibits a similar expression yield and size-exclusion chromatogram but with a remarkable 90% reduction in heme content based on the heme-to-protein ratio (Fig. 3C). Despite the inherent inaccuracy associated with protein structure prediction, this observation underpins the importance of Phe107 for heme binding and provides some circumstantial evidence supporting the idea that the heme is bound in the well-defined binding pocket rather than at protein surface in a nonspecific manner.
FIG. 3.
Sequence and modeling analysis of heme binding. (A) Sequence alignment of the PAS domains of YybT family proteins with the heme-binding PAS domain of BjFixL and EcDOS. Residue numbering is based on BsYybT. The position of residue Phe107 is highlighted by the triangle, and the conserved histidine residues for heme coordination in BjFixL and EcDOS are boxed. The predicted secondary structure for PASBsYybT is shown on the top, and the secondary structure for BjFixL is shown at the bottom. Sequences were aligned by using the program ClustalW, and the figure was generated by using the program ESPript. (BsPAS is from Bacillus subtilis; BaPAS is from Bacillus anthracis; GtPAS is from Geobacillus thermodenitrificans; LlPAS is from Lactooccus lactis; LmPAS is from Listeria monocytogene; SaPAS is from Staphylococcus aureus; SmPAS is from Streptococcus mutans). (B) Overlaid structures of the PAS domains of EcDOS (black structure) and BjFixL (gray structure) with the heme and the corresponding residue of Phe107 shown as sticks. (C) Absorption spectra of the wild-type GtYybT55-162 and F107A mutant.
Heme reconstitution of YybT proteins.
Given the low heme-to-protein ratio for the recombinant proteins, the reconstitution method described in the experimental section was used to prepare holo proteins for further study. The reconstituted proteins exhibit spectra similar to that of the purified BsYybT84-303 and GtYybT55-162, with 412-, 533-, and 566-nm absorbance maxima (Fig. 4). The heme-to-protein ratio that indicates the effectiveness of reconstitution varies by protein construct. The heme-to-protein ratio is close to 1:1 for the reconstituted full-length constructs BsYybT84-659 and GtYybT55-658 according to pyridine hemochrome assay, whereas the ratio is significantly lower for the shorter construct GtYybT55-162, indicating that the heme-binding pocket is perturbed in GtYybT55-162. The His6 tag-free GtYybT55-162, prepared by thrombin cleavage, still is capable of binding heme, suggesting that the histidine residues of the His6 tag are not involved in heme coordination. In addition, the BsYybT150-659 construct, which lacks the PAS domain, failed to bind heme through reconstitution, confirming that the binding of heme is indeed through the PAS domain.
FIG. 4.
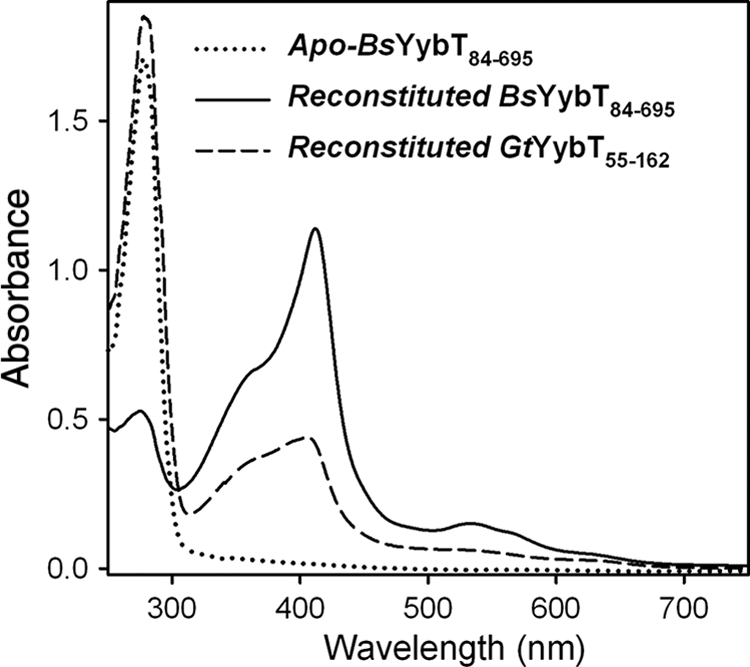
UV-vis absorption spectra of apo-BsYybT84-659 and reconstituted BsYybT84-659 and GtYybT55-162. The proteins were reconstituted with ferric heme in the absence of reducing agent. The spectra were taken in 50 mM Tris (pH 8.0), 150 mM NaCl at 23°C.
Spectroscopic characterization of holo-BsYybT.
The absorption spectrum of heme proteins is sensitive to the change of redox, spin state, and coordination environment of the heme iron. To investigate whether the heme-binding PAS domain could act as a sensor domain for redox signal or the gaseous ligands (O2, NO, CO), the reduced, oxidized, and ligated forms of BsYybT84-659 were characterized by absorption spectroscopy as described below.
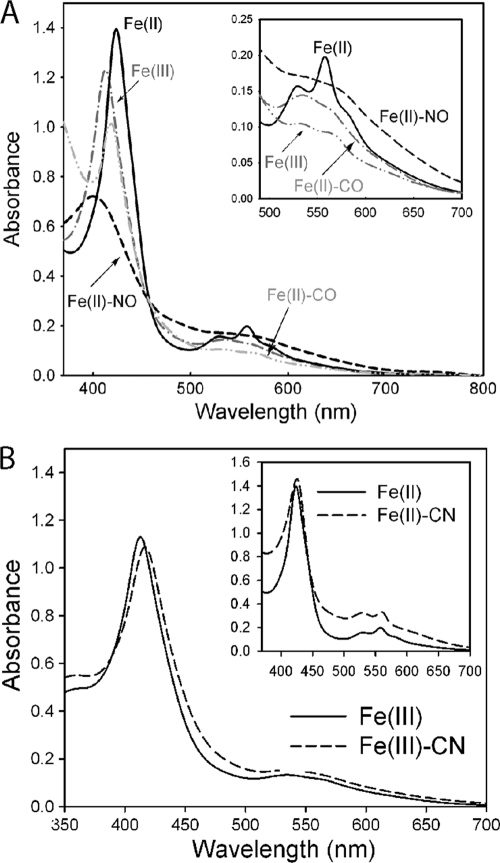
Since penta- and hexacoordinated hemes usually exhibit a distinctive Soret band with the pentacoordinated heme in the region of 350 to ∼400 nm and the hexacoordinated heme above 400 nm (37), the 412-nm Soret band of the ferric BsYybT84-659 is consistent with a hexacoordinated heme. The treatment of BsYybT84-659 with the reducing agent dithiothreitol (DTT) readily generated the ferrous form, as evidenced by the spectral changes. Upon reduction, the Soret band is red shifted to 423 nm and the β and α bands are blue shifted to 528 and 557 nm, respectively (Fig. 5A, Table 2). The relative intensity of the β and α bands also changes upon reduction, with the α band at 557 nm becoming more intense than the β band at 528 nm. The shape and wavelength of the absorption bands for the ferrous form are similar to those of some of the hexacoordinated low-spin heme proteins. When the ferrous BsYybT55-659 protein was exposed to air or oxygen gas, the absorption spectrum rapidly reverted back to that of the ferric form within seconds. The rapid oxidation suggests that the PAS domain of BsYybT is very different from those of some of the O2-sensing heme-PAS sensors that feature a long-lasting Fe(II)-O2 species.
FIG. 5.
Absorption spectra of BsYybT84-659 and its adducts. (A) Spectra of BsYybT84-659 in its ferric, ferrous, Fe(II)-CO, and Fe(II)-NO forms. (B) Spectra of BsYybT84-659 in its Fe(II)-CN− and Fe(III)-CN− forms.
TABLE 2.
UV-vis absorption maxima of BsYybT84-659 and its adducts
| Heme state | Soret band (nm) | α/β band (nm) | Others (nm) |
|---|---|---|---|
| Fe(III) | 412 | 566/533 | 639 (small) |
| Fe(II) | 423 | 557/528 | |
| Fe(II)-NO | 398 | 568/532 | |
| Fe(II)-CO | 418 | 567/536 | |
| Fe(II)-CN− | 426 | 561/529 | |
| Fe(III)-CN− | 417 | 536 |
The addition of KCN to ferric BsYybT84-659 caused a 5-nm red shift of the Soret peak to 417 nm and the merging of α and β bands into a single band at 536 nm (Fig. 5B). Although cyanide binding often is observed only for ferric heme proteins (7, 21), the addition of KCN to ferrous BsYybT84-659 also resulted in small shifts in the Soret band (from 423 to 426 nm) and the β and α bands (from 557 and 528 nm to 561 and 529 nm, respectively) (Fig. 5B), suggesting that ferrous heme iron also forms a complex with CN−. Little spectral change in the spectrum was observed when the small-molecule NO or CO donor was added to the ferric protein solution, which suggests that CO and NO do not form adducts with Fe(III)-BsYybT. In contrast, the addition of NO and CO donors to the ferrous protein solution immediately caused significant changes to the Soret and α/β bands, likely due to the formation of Fe(II)-NO and Fe(II)-CO adducts (Fig. 5A, Table 2). The formed adducts are remarkably stable and resistant to O2 oxidation, as the spectra remained unchanged after being exposed to air for more than 60 min. The spectral changes caused by CO and NO binding differ drastically. The binding of CO shifts the Soret peak from 423 to 418 nm and resulted in less intense α/β bands, indicating that the Fe(II)-CO adduct still contains a hexacoordinated low-spin heme. In comparison, the binding of NO resulted in a broad and less intense Soret band at 398 nm as well as ill-defined α and β bands. The 398-nm Soret band of the Fe(II)-NO complex is indicative of a pentacoordinated heme that resembles those of the NO-sensing H-NOX domain of the soluble guanylate cyclase (sGC), nitrate respiration regulator (DNR), and nuclear receptor E75 (12, 26, 32).
Taken together, the spectroscopic observations suggest that Fe(II)-BsYybT binds NO, CO, and CN−, whereas Fe(III)-BsYybT binds CN− but not NO and CO. Although Fe(II)-BsYybT is able to bind O2, the oxyferrous species undergoes rapid oxidation to yield Fe(III). Similar spectroscopic changes also were observed with GtYybT55-162 (data now shown), indicating a similar heme coordination environment in the PAS domain of GtYybT.
Effect of heme and ligand binding on catalytic activity.
The DHH/DHHA1 and GGDEF domains of BsYybT were found to possess phosphodiesterase (PDE) and weak ATPase activity, respectively (31). We previously noticed a difference in PDE activity between apo-BsYybT84-659 and the BsYybT150-659 construct, which lacks the PAS domain (Table 3) (31). In light of the heme-binding capability of the PAS domain, we further examined the effect of heme and ligand binding on the catalytic activities of the DHH/DHHA1 and GGDEF domains.
TABLE 3.
Kinetic parameters for c-di-AMP hydrolysis by BsYybT proteins
| Protein | kcat (s−1) | Km (μM) | kcat/Km (s−1 μM−1) | Reference or source |
|---|---|---|---|---|
| BsYybT150-659 | 0.074 ± 0.002 | 3.0 ± 0.22 | 0.024 ± 0.002 | 31 |
| Apo-BsYybT84-659 | 0.55 ± 0.02 | 1.3 ± 0.24 | 0.42 ± 0.08 | 31 |
| Holo-BsYybT84-659 | 0.024 ± 0.003 | 16 ± 4.0 | 0.0015 ± 0.0004 | This work |
| CN−-BsYybT84-659 | 0.083 ± 0.003 | 8.9 ± 1.0 | 0.009 ± 0.001 | This work |
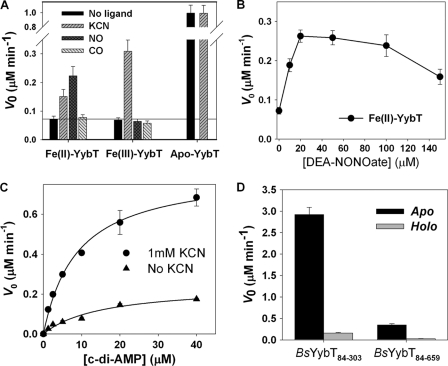
Whether c-di-AMP is the physiological substrate of the DHH/DHHA1 domain remains to be established by in vivo experimental evidence. Here, we use c-di-AMP as the substrate for enzymatic assay given the efficient turnover of c-di-AMP to 5′-pApA, which is catalyzed by the DHH/DHHA1 domain. Compared to that of apo-YybT84-659, a reduction of 15-fold in initial velocity (ν0) was observed for holo-YybT84-659 at a 5 μM substrate concentration (Fig. 6A). Steady-state kinetic measurement revealed a kcat of 0.024 ± 0.003 s−1 for holo-BsYybT, which is 23-fold lower than that of apo-BsYybT84-659. With a concomitant 12-fold increase in Km, holo-BsYybT84-659 exhibits a catalytic efficiency (kcat/Km) that is 276-fold lower than that of the apo protein (Table 3). Hence, heme binding in the PAS domain seems to exert an inhibitory effect on the DHH/DHHA1 domain. Size-exclusion chromatography indicates that BsYybT84-659 exists as heterogeneous high-order oligomers in solution (data now shown). Although the elution profile does not seem to differ significantly upon heme reconstitution, it is not known whether the binding of heme affects the catalytic activity through a conformational change within the individual protein subunit or an alteration of protein oligomeric state.
FIG. 6.
Regulation of the activities of the catalytic domains by the PAS domain. (A) The effect of heme binding and ligand coordination on PDE activity. (B) Dependence of the PDE activity of ferrous BsYybT84-659 on the concentration of NO donor. (C) Steady-state kinetic measurement of PDE activity of ferric BsYybT84-659 in the presence and absence of KCN. (D) Effect of heme binding on ATP hydrolysis catalyzed by the GGDEF domain of BsYybT84-659.
No statistically significant difference in PDE activity was observed between Fe(II)- and Fe(III)-BsYybT84-659, suggesting that the redox state does not regulate the catalytic activity. The binding of NO and CN− to Fe(II)-BsYybT84-659 caused a 3- and 2-fold increase in initial velocity, respectively, at saturating substrate concentration, whereas the binding of CO had a negligible effect. The stimulatory effect of NO was observed for all three small-molecule NO donors tested. In contrast to Fe(II)-BsYybT84-659, no significant effect of CO and NO on Fe(III)-BsYybT84-659 was observed, which is consistent with the spectroscopic observation that CO and NO do not bind to Fe(III)-BsYybT84-659. The stimulatory effect of NO is dependent on the concentration of the NO donor, with a saturating concentration of 20 μM observed for DEA-NONOate (Fig. 6B). Considering that NO also does not affect the PDE activity of apo- and Fe(III)-BsYybT84-659, the result strongly suggests that NO exerts its effect through the ferrous heme iron. CN− is the only ligand that forms an adduct with Fe(III)-YybT84-659, and the CN− adduct exhibits a v0 (5 μM c-di-AMP) that is 5-fold greater and a kcat that is 3.5-fold greater than those of the unligated form (Fig. 6C, Table 3).
Unlike the orthodox GGDEF domains, the defective GGDEF domain of BsYybT lacks a GGDEF motif and does not possess diguanylate cyclase (DGC) activity. We previously found that the GGDEF domain of apo-BsYybT84-303 exhibits unexpected ATPase activity, with a kcat of 0.59 min−1. The binding of heme to both BsYybT84-303 and BsYybT84-659 further suppresses the weak ATPase activity to a residual level that is close to the detection limit of the HPLC method, whereas the binding of the ligands CO, NO, and CN− to the heme iron did not seem to stimulate the ATPase activity (31) (Fig. 6D).
DISCUSSION
Heme-containing PAS domains are best known as sensor domains for perceiving redox signal and the gaseous ligands such as O2 and CO (17, 24, 40). Redox- and O2-sensing PAS domains are exemplified by the proteins FixL, AxPDEA1, and EcDOS (14, 16, 41), whereas the CO sensors are represented by the transcription factors RocM and NPAS2 (11, 20). High-resolution crystal structures of the PAS domains of EcDOS and FixL have revealed an invariant histidine as the proximal ligand for the heme iron (15, 22). The use of histidine for heme coordination is also seen in other non-PAS fold sensor proteins, such as H-NOX, Dos-S and CooA (6, 23, 26). The capability of the PASYybT domain to bind heme is unexpected given the lack of a conserved histidine (or cysteine) in the PAS domain. Intriguingly, the UV-vis spectra of the ferric and ferrous BsYybT84-659 closely resemble those of the heme-binding SOUL protein that is known to coordinate the heme iron with a histidine residue (36). The rapid oxidation of the ferrous form by O2 prevents the formation of stable oxyferrous complex, indicating that the heme coordination environment is different from those of AxPDEA-1 and EcDOS. Crane and coworkers suggested that the binding of heme by certain proteins, such as the PAS domain-containing PER2, may be biologically irrelevant because the heme is bound on the surface of the protein in a nonspecific manner (1). Several observations indicate that the binding of heme by PASYybT is unlikely to be caused by the nonspecific interaction between heme and surface residues. First, even the heme-to-protein ratio is substoichiometric; a portion of the recombinant BsYybT or GtYybT was expressed and purified as heme-bound protein. This is in sharp contrast to the PER2 protein, which can be expressed and isolated only as a heme-free protein (1). Second, it was suggested that PER2 binds heme by using surface histidine or cysteine residues. The PAS domain of BsYybT does not contain any histidine, and the Ala substitution of Cys106 did not seem to perturb heme binding (data not shown). It is unlikely that PASYybT binds heme in a fashion similar to that of PER2. Third, an Ala replacement of the Phe107 residue that putatively is located in the heme-binding pocket reduces the occupancy of heme significantly, hinting that the residues in the hydrophobic pocket are involved in heme binding. These considerations led us to suggest that the binding of heme by the PAS domain of YybT proteins could be a biologically relevant observation, with the PAS domain containing a well-defined heme-binding pocket or cleft. There are examples of proteins that bind heme without using histidine or cysteine (10). However, it remains to be seen whether the heme-binding mode of PASYbbT resembles any of these proteins.
Heme binding exerts a strong inhibitory effect on the PDE activity of BsYybT84-659 by reducing kcat and kcat/Km, which raises the possibility that the PAS domain functions as a direct heme sensor. Sensor proteins for monitoring extra and intracellular heme in pathogenic bacteria such as S. aureus have been reported, but none of them adopts the PAS fold (38, 42). Intriguingly, a link between heme intake and YybT proteins has been indicated by an earlier in vivo study. Burnside et al. found that the knockout of the SA0013 gene that encodes SaYybT disrupts hemolysin secretion in S. aureus (4). Hemolysin is an essential virulence factor or exotoxin for acquiring nutrients (e.g., heme iron) from the host by lysing the host blood cells. Given the heme-binding capability of the PAS domain, YybT family proteins may play an important role in mediating hemolysin secretion in response to low cellular heme levels.
As evidenced by the spectral changes, the binding of NO, CO, and CN− to BsYybT84-659 led to the formation of Fe(II)-NO, Fe(II)-CO, and Fe(II/III)-CN− complexes. The binding of CO and CN− resulted in spectra that are consistent with a hexacoordinated iron heme; whereas the characteristic 398-nm Soret band for the Fe(II)-NO complex is indicative of a pentacoordinated heme that often is observed for the NO sensors, such as H-NOX, DNR, and E75 (5, 9, 18, 32). The transition from hexa- to pentacoordinated heme iron upon NO binding suggests that NO triggers the dissociation of the unidentified proximal ligand from the iron center. Such change in coordination number has been reported for NO sensor proteins as well as proteins such as CooA (5, 8, 34). The coordination of NO or CN−, but not CO, to the ferrous heme iron stimulates the catalytic activity of the PDE domain. Although CN− is unlikely to be a physiologically relevant ligand molecule, NO has been known as a signaling molecule and a reactive nitrogen species (RNS) released by host immune cells for bacteria killing. Although no heme-binding PAS domain has been reported to be an NO sensor so far, at least four protein domains that use heme cofactor for NO sensing have been proposed. The best-studied NO sensors are the H-NOX domain of mammalian sGC and the N-terminal domain of the transcription factor DNR from Pseudomonas aeruginosa (5, 12, 25, 26). The other examples include the sensor domain of the Drosophila nuclear receptor E75 and the N-terminal domain of the heme-regulated eukaryotic initiation factor 2α kinase (HRI-ND) (18, 32, 43). Compared to the 3-fold increase in PDE activity upon NO binding, the guanylate cyclase activity of sGC was stimulated by several hundredfold in the presence of NO (39), and the kinase activity of HRI is increased by 5-fold. The transcription activation function of DNR is augmented by ca. 4-fold (5), while the transcription repressor activity of E75 is significantly reduced in the presence of NO (32). As an alternative to the heme-sensing function, the formation of a stable Fe(II)-NO adduct and the effect of NO on PDE activity raises the possibility that the PAS domain functions as an NO sensor.
Studies have implicated YybT family proteins in the regulation of hemolysin secretion, biofilm formation, and acid and DNA damage resistance in several bacterial strains. Hemolysin is crucial for heme iron acquisition during host infection, whereas biofilm formation and acid/DNA damage resistance could affect the in vivo survival rate of the pathogen. Hence, although YybT family proteins are not essential for the in vitro survival of the pathogens, they may be crucial for the in vivo survival in the host environment. With the aim of uncovering the biological function of the YybT family proteins, the studies described here establish the unexpected heme-binding capability of the PAS domain of two YybT family proteins. The lack of conserved histidine or cysteine ligand for heme coordination indicates an uncommon heme-binding mode that entails further investigation. Based on the observations, we postulate that the PAS domain plays a role in sensing cellular heme or NO. Heme and NO may serve as the molecular marker of mammalian host environment for triggering cellular responses in the invading bacterial pathogens. The significance of monitoring cellular heme or NO level in pathogenic bacteria is apparent, given that heme or iron is essential for the biogenesis of many bacterial heme proteins, and that the defense against host immune cells is vital for the in vivo survival of the bacterial pathogens. In vivo studies in the future are needed to validate the role of the PAS domain and the correlation between heme/NO sensing and bacterial pathogenesis.
Acknowledgments
This work is supported by a Biomedical Research Council (BMRC) grant (06/1/22/19/464) from the Agency of Science and Technology and an ARC Tier I grant (RG/35/08) from the Ministry of Education of Singapore.
Footnotes
Published ahead of print on 21 January 2011.
REFERENCES
- 1.Airola, M. V., J. Du, J. H. Dawson, and B. R. Crane. 2010. Heme binding to the mammalian circadian clock protein period 2 is nonspecific. Biochemistry 49:4327-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benton, B. M., et al. 2004. Large-scale identification of genes required for full virulence of Staphylococcus aureus. J. Bacteriol. 186:8478-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry, E. A., and B. L. Trumpower. 1987. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal. Biochem. 161:1-15. [DOI] [PubMed] [Google Scholar]
- 4.Burnside, K., et al. 2010. Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS One 5:e11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castiglione, N., S. Rinaldo, G. Giardina, and F. Cutruzzola. 2009. The transcription factor DNR from Pseudomonas aeruginosa specifically requires nitric oxide and haem for the activation of a target promoter in Escherichia coli. Microbiology 155:2838-2844. [DOI] [PubMed] [Google Scholar]
- 6.Cho, H. Y., H. J. Cho, Y. M. Kim, J. I. Oh, and B. S. Kang. 2009. Structural insight into the heme-based redox sensing by DosS from Mycobacterium tuberculosis. J. Biol. Chem. 284:13057-13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai, Z., and E. M. Boon. 2010. Engineering of the heme pocket of an H-NOX domain for direct cyanide detection and quantification. J. Am. Chem. Soc. 132:11794-11803. [DOI] [PubMed] [Google Scholar]
- 8.Deinum, G., J. R. Stone, G. T. Babcock, and M. A. Marletta. 1996. Binding of nitric oxide and carbon monoxide to soluble guanylate cyclase as observed with resonance Raman spectroscopy. Biochemistry 35:1540-1547. [DOI] [PubMed] [Google Scholar]
- 9.Derbyshire, E. R., and M. A. Marletta. 2009. Biochemistry of soluble guanylate cyclase. Handb. Exp. Pharmacol. 191:17-31. [DOI] [PubMed] [Google Scholar]
- 10.Dias, J. S., et al. 2006. The first structure from the SOUL/HBP family of heme-binding proteins, murine P22HBP. J. Biol. Chem. 281:31553-31561. [DOI] [PubMed] [Google Scholar]
- 11.Dioum, E. M., et al. 2002. NPAS2: a gas-responsive transcription factor. Science 298:2385-2387. [DOI] [PubMed] [Google Scholar]
- 12.Giardina, G., et al. 2008. NO sensing in Pseudomonas aeruginosa: structure of the transcriptional regulator DNR. J. Mol. Biol. 378:1002-1015. [DOI] [PubMed] [Google Scholar]
- 13.Gilles-Gonzalez, M.-A., and G. Gonzalez. 2004. Signal transduction by heme-containing PAS-domain proteins. J. Appl. Physiol. 96:774-783. [DOI] [PubMed] [Google Scholar]
- 14.Gilles-Gonzalez, M. A., et al. 1994. Heme-based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autoxidation. Biochemistry 33:8067-8073. [DOI] [PubMed] [Google Scholar]
- 15.Gong, W., et al. 1998. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. U. S. A. 95:15177-15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez, G., et al. 2002. Nature of the displaceable heme-axial residue in the EcDos protein, a heme-based sensor from Escherichia coli. Biochemistry 41:8414-8421. [DOI] [PubMed] [Google Scholar]
- 17.Hefti, M. H., K. J. Francoijs, S. C. de Vries, R. Dixon, and J. Vervoort. 2004. The PAS fold-A redefinition of the PAS domain based upon structural prediction. Eur. J. Biochem. 271:1198-1208. [DOI] [PubMed] [Google Scholar]
- 18.Igarashi, J., et al. 2004. Activation of heme-regulated eukaryotic initiation factor 2α kinase by nitric oxide is induced by the formation of a five-coordinate NO-heme complex: optical absorption, electron spin resonance, and resonance raman spectral studies. J. Biol. Chem. 279:15752-15762. [DOI] [PubMed] [Google Scholar]
- 19.Ihee, H., et al. 2005. Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds. Proc. Natl. Acad. Sci. U. S. A. 102:7145-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerby, R. L., H. Youn, and G. P. Roberts. 2008. RcoM: a new single-component transcriptional regulator of CO metabolism in bacteria. J. Bacteriol. 190:3336-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, A., J. C. Toledo, R. P. Patel, J. R. Lancaster, Jr., and A. J. Steyn. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. U. S. A. 104:11568-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurokawa, H., et al. 2004. A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor. J. Biol. Chem. 279:20186-20193. [DOI] [PubMed] [Google Scholar]
- 23.Lanzilotta, W. N., et al. 2000. Structure of the CO sensing transcription activator CooA. Nat. Struct. Biol. 7:876-880. [DOI] [PubMed] [Google Scholar]
- 24.Möglich, A., R. A. Ayers, and K. Moffat. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17:1282-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olea, C. Jr., M. A. Herzik, Jr., J. Kuriyan, and M. A. Marletta. 2010. Structural insights into the molecular mechanism of H-NOX activation. Protein Sci. 19:881-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pellicena, P., D. S. Karow, E. M. Boon, M. A. Marletta, and J. Kuriyan. 2004. Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases. Proc. Natl. Acad. Sci. U. S. A. 101:12854-12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi, Y., F. Rao, Z. Luo, and Z. X. Liang. 2009. A flavin cofactor-binding PAS domain regulates c-di-GMP synthesis in AxDGC2 from Acetobacter xylinum. Biochemistry 48:10275-10285. [DOI] [PubMed] [Google Scholar]
- 28.Rallu, F., A. Gruss, S. D. Ehrilich, and E. Maguin. 2000. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol. Microbiol. 35:517-528. [DOI] [PubMed] [Google Scholar]
- 29.Rao, F., et al. 2009. Enzymatic synthesis of c-di-GMP using a thermophilic diguanylate cyclase. Anal. Biochem. 389:138-142. [DOI] [PubMed] [Google Scholar]
- 30.Rao, F., et al. 2009. The functional role of a conserved loop in EAL domain-based c-di-GMP specific phosphodiesterase. J. Bacteriol. 191:4722-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao, F., et al. 2010. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J. Biol. Chem. 285:473-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinking, J., et al. 2005. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell 122:195-207. [DOI] [PubMed] [Google Scholar]
- 33.Repik, A., et al. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds, M. F., et al. 2000. Electronic absorption, EPR, and resonance Raman spectroscopy of CooA, a CO-sensing transcription activator from R. rubrum, reveals a five-coordinate NO-heme. Biochemistry 39:388-396. [DOI] [PubMed] [Google Scholar]
- 35.Roy, A., A. Kucukural, and Y. Zhang. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato, E., et al. 2004. SOUL in mouse eyes is a new hexameric heme-binding protein with characteristic optical absorption, resonance raman spectral, and heme-binding properties. Biochemistry 43:14189-14198. [DOI] [PubMed] [Google Scholar]
- 37.Smulevich, G., F. Neri, M. P. Marzocchi, and K. G. Welinder. 1996. Versatility of heme coordination demonstrated in a fungal peroxidase. Absorption and resonance Raman studies of Coprinus cinereus peroxidase and the Asp245→Asn mutant at various pH values. Biochemistry 35:10576-10585. [DOI] [PubMed] [Google Scholar]
- 38.Stauff, D. L., and E. P. Skaar. 2009. Bacillus anthracis HssRS signaling to HrtAB regulates heme resistance during infection. Mol. Microbiol. 72:763-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stone, J. R., and M. A. Marletta. 1994. Soluble guanylate cyclase from bovine lung: activation with nitric oxide and carbon monoxide and spectral characterization of the ferrous and ferric states. Biochemistry 33:5636-5640. [DOI] [PubMed] [Google Scholar]
- 40.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomita, T., G. Gonzalez, A. L. Chang, M. Ikeda-Saito, and M.-A. Gilles-Gonzalez. 2002. A comparative resonance raman analysis of heme-binding PAS domains: heme iron coordination structures of the BjFixL, AxPDEA1, EcDOS, and MtDOS proteins. Biochemistry 41:4819-4826. [DOI] [PubMed] [Google Scholar]
- 42.Torres, V. J., et al. 2007. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1:109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uma, S., B. G. Yun, and R. L. Matts. 2001. The heme-regulated eukaryotic initiation factor 2alpha kinase. A potential regulatory target for control of protein synthesis by diffusible gases. J. Biol. Chem. 276:14875-14883. [DOI] [PubMed] [Google Scholar]
- 44.Yan, W., et al. 2010. The effect of c-di-GMP (3′-5′-cyclic diguanylic acid) on the biofilm formation and adherence of Streptococcus mutans. Microbiol. Res. 165:87-96. [DOI] [PubMed] [Google Scholar]
- 45.Zeigler, D. R., et al. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J. Bacteriol. 190:6983-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]