Abstract
Most iron in mammals is found within the heme prosthetic group. Consequently, many bacterial pathogens possess heme acquisition systems to utilize iron from the host. Here, we demonstrate that Mycobacterium tuberculosis can utilize heme as an iron source, suggesting that M. tuberculosis possesses a yet-unknown heme acquisition system.
Owing to its versatile redox potential under physiological conditions, iron is an essential nutrient for the vast majority of organisms (6, 31). Organisms tightly sequester and regulate their iron supplies to limit the toxicity of the ferrous ion and to deal with the insolubility of the ferric ion (2). In the mammalian host, these mechanisms create an iron-scarce environment for bacterial pathogens. As a result, all bacterial pathogens have evolved specialized iron acquisition systems to utilize host iron (30). Many bacteria produce siderophores, small high-affinity iron chelators, to acquire iron from their environments (13). However, 70% of total iron in an adult mammal is bound within the heme prosthetic group (1), and as a consequence many bacterial pathogens also use heme acquisition systems in addition to siderophores (29).
Mycobacterium tuberculosis, the causative agent of tuberculosis, is a global health problem. In 2008, M. tuberculosis caused more deaths than any other bacterial pathogen (32). A hallmark of M. tuberculosis virulence is its capability to proliferate within arrested phagosomes of macrophages. This growth is dependent on siderophore biosynthesis (11, 16). However, M. tuberculosis mutants deficient in siderophore transport are not completely attenuated following an aerosol infection in mice, suggesting that M. tuberculosis might possess alternative iron acquisition systems (25). Recently, a novel heme-degrading enzyme in M. tuberculosis (Rv3592, MhuD) was characterized (5). Furthermore, the slow-growing Mycobacterium haemophilum displays a strict heme requirement for growth in vitro (9, 28). These reports suggest that M. tuberculosis and other mycobacteria might be capable of utilizing heme as an iron source.
Construction of a siderophore-deficient strain of M. tuberculosis.
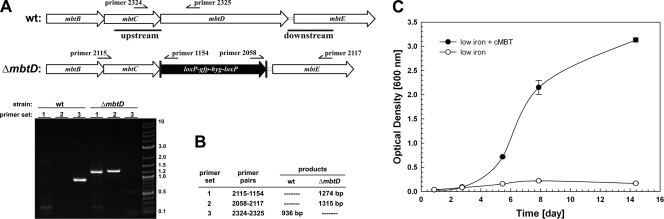
The siderophores of M. tuberculosis are called mycobactins and chelate iron from a core unit assembled by nonribosomal peptide synthases and polyketide synthases, encoded by the genes mbtA to mbtH (7). In order to unambiguously determine if M. tuberculosis can utilize heme as an iron source, it is necessary to have a siderophore-deficient mutant. Deletion of the mbtD gene encoding the putative polyketide synthase completely eliminated mycobactin production in Mycobacterium smegmatis (27). Therefore, we deleted the mbtD gene by allelic exchange in the avirulent M. tuberculosis strain mc26230 (ΔRD1 ΔpanCD; referred to here as the wild type [wt]) (26) (Fig. 1A) using the plasmid pML1816 (Table 1), which contained 1,000-bp regions upstream and downstream of mbtD. The analysis of double-crossover candidate 1 by PCR of chromosomal DNA demonstrated the absence of the mbtD gene (Fig. 1B). This ΔmbtD mutant was named M. tuberculosis ML1600 and was used in further experiments. To test whether M. tuberculosis ML1600 displayed a low-iron growth defect, we prepared a low-iron minimal medium consisting of 500 μM MgCl2·6H2O, 7 μM CaCl2·2H2O, 1 μM NaMoO4·2H2O, 2 μM CoCl2·6H2O, 6 μM MnCl2·4H2O, 7 μM ZnSO4·7H2O, 1 μM CuSO4·5H2O, 15 mM (NH4)2SO4, 12 mM KH2PO4 (pH 6.8), and 1% (wt/vol) glucose, which was supplemented with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC; Middlebrook), and 0.2% (wt/vol) Casamino Acids. The iron content of this medium was determined to be approximately 500 nM by inductively coupled plasma-mass spectrometry (ICP-MS) (not shown). We inoculated this low-iron minimal medium and low-iron minimal medium supplemented with purified ferri-carboxymycobactin from Mycobacterium bovis BCG (cMBT) with M. tuberculosis ML1600. Figure 1C shows that M. tuberculosis ML1600 does indeed display a growth defect in low-iron minimal medium and that this growth defect is rescued by the addition of purified cMBT. This biochemical complementation experiment provides evidence that the growth defect of M. tuberculosis ML1600 was caused by the elimination of siderophores and not by secondary mutations in siderophore acquisition or an incomplete medium.
FIG. 1.
Construction of a mycobactin-deficient M. tuberculosis strain. (A) Scheme of the mycobactin biosynthesis locus of M. tuberculosis encompassing mbtD. Genomic deletion of the mbtD gene was obtained by allelic exchange as described in the text. (B) Verification of the ΔmbtD mutant ML1600 by colony PCR. Bands in the DNA marker are denoted with their lengths in kilobase pairs. (C) Rescue of the ML1600 by purified ferri-carboxymycobactin (cMBT) from M. bovis BCG. Error bars represent standard deviations from the mean of results from biological duplicates.
TABLE 1.
Plasmids used in this work
| Plasmid | Parent vector, relevant genotype, and propertiesa | Source or reference |
|---|---|---|
| pML523 | pUC origin, PAL5000 origin; sacB xylE loxP-gfp2+m-HygR-loxP; 9,845 bp | This study |
| pML1815 | pML523; mbtDuploxP-gfp2+m-hyg-loxP; 10,766 bp | This study |
| pML1816 | pML1815; loxP-gfp2+m-hyg-loxP mbtDdown; 11,825 bp | This study |
Up- and downstream homologous sequences of genes are noted with a subscript “up” and “down,” respectively. Origin, origin of replication. The annotation HygR indicates that the plasmid confers resistance to hygromycin.
Utilization of heme by the M. tuberculosis ΔmbtD mutant.
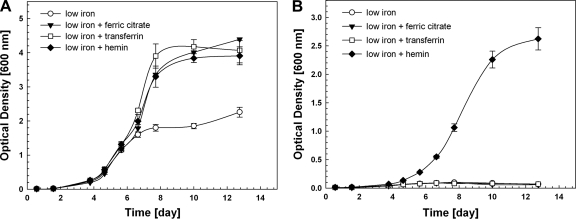
To test whether M. tuberculosis can utilize heme as an iron source, we measured growth of wt M. tuberculosis and the siderophore-deficient M. tuberculosis mutant ML1600 in low-iron medium and low-iron medium supplemented with 10 μM ferric citrate (prepared in a 1:200 iron-to-citrate ratio), 10 μM holo-transferrin (Sigma), and 10 μM hemin (Sigma). M. tuberculosis ML1600 displayed a severe growth defect in the low-iron medium which cannot be restored with 10 μM ferric citrate, indicating that in the absence of siderophores, M. tuberculosis cannot utilize ferric citrate under these conditions (Fig. 2B). Similar results were obtained with ferric chloride (not shown). Additionally, the low-iron growth defect of ML1600 was not rescued by 10 μM holo-transferrin (Fig. 2B), suggesting that M. tuberculosis does not possess a mechanism to liberate iron from transferrin in the absence of siderophores in contrast to Haemophilus influenzae and Neisseria gonorrhoeae (3). However, the growth defect of ML1600 under these conditions was rescued by hemin at a concentration of either 1 μM (not shown) or 10 μM (Fig. 2B), demonstrating that M. tuberculosis ML1600 can utilize hemin as an iron source. The growth rate of the ΔmbtD mutant grown with 1 μM hemin (doubling time ± standard deviation, 31.3 ± 0.1 h) was significantly lower than the growth rate of the mutant grown with 1 μM cMBT (doubling time of 27.1 ± 0.5 h; P = 0.01, two-tailed Student's t test) (not shown). In addition, stationary-phase optical densities were lower in the presence of hemin than in the presence of cMBT. In contrast, the growth rates of wt M. tuberculosis were similar in the presence of hemin or cMBT (not shown). Taken together, these data indicate that heme utilization is not as efficient as siderophore utilization under the conditions tested. It was reported that M. haemophilum requires heme concentrations greater than 7 μM in order to grow to visible colonies on solid medium, whereas heme concentrations of 2 μM did not yield any colonies (28). We observed that M. tuberculosis grows well with 1 μM hemin, suggesting that heme utilization in M. tuberculosis is more efficient than in M. haemophilum.
FIG. 2.
M. tuberculosis can utilize heme as an iron source. (A) Growth of wt M. tuberculosis in low-iron medium or low-iron medium supplemented with 10 μM ferric citrate, 10 μM holo-transferrin, or 10 μM hemin. (B) Growth of M. tuberculosis ML1600 in low-iron medium or low-iron medium supplemented with 10 μM ferric citrate, 10 μM holo-transferrin, or 10 μM hemin. The media for both these growth experiments were identical. Error bars represent standard deviations from the mean of results from biological triplicates.
Heme utilization often involves lysins and proteases to make host heme available (12). Contact-dependent hemolytic activity has been reported in Mycobacterium avium (24) and M. tuberculosis (10). However, the molecular determinants of the M. tuberculosis hemolytic activity are not entirely clear. A putative hemolysin in M. tuberculosis was first identified based on sequence homology to the hemolysin TlyA from the porcine pathogen Serpula hyodysenteriae (21). Expression of the M. tuberculosis tlyA homolog in Escherichia coli JM109 and M. smegmatis conferred contact-dependent hemolytic activity to these otherwise nonhemolytic strains (33). However, the tlyA gene product was later shown to possess 2′-O-methyltransferase activity that methylates the 16S and 23S rRNA subunits of both M. tuberculosis and M. smegmatis (15). The 2′-O-methyltransferase activity of TlyA renders these strains sensitive to the antibiotic capreomycin (20). Most recently, both in vitro hemolytic activity and in vitro methyltransferase activity were demonstrated for recombinant TlyA (23). Those authors argue that TlyA has evolved functions as a hemolysin and an rRNA 2′-O-methyltransferase. However, this notion is challenged by the fact that mutations in M. smegmatis tlyA result in capreomycin resistance, while no contact-dependent hemolytic activity has been observed in this organism. Additionally, TlyA-dependent hemolytic activity has not been reported in M. tuberculosis, only in E. coli and M. smegmatis overexpressing M. tuberculosis tlyA. Further research is necessary to determine if TlyA is indeed a bona fide hemolysin of M. tuberculosis.
It is unknown how heme is taken up by M. tuberculosis. In Gram-negative bacteria, heme uptake is mediated by TonB-dependent high-affinity receptors in the outer membrane, periplasmic binding proteins, and inner membrane ABC transporters to facilitate passage into the cytosol (17). In M. tuberculosis, heme uptake would also require transport across the outer and inner membranes (14, 22). However, bioinformatic analysis of the M. tuberculosis genome did not reveal homologs to TonB or TonB-dependent receptors in Gram-negative bacteria. Homology searches for heme acquisition systems using the heme uptake operon phuRSTUVW and hemophore export system hasRADEF from Pseudomonas aeruginosa as models revealed only Rv1747 of M. tuberculosis (49% similar to the heme permease PhuV) and Rv0194 of M. tuberculosis (54% similar to the hemophore transporter HasD). However, Rv0194 was recently shown to be a multidrug efflux protein of M. tuberculosis (8), making it unlikely that it is part of a hemophore export system. Furthermore, a sequence similarity of 43% between M. tuberculosis FecB and the Staphylococcus aureus heme binding protein HtsA (43%) was observed but not any other proteins. A possible explanation for the apparent absence of sequence similarity of M. tuberculosis proteins to known heme uptake systems is that heme uptake in M. tuberculosis occurs nonspecifically, such as that seen in Escherichia coli K-12, where heme uptake across the inner membrane is dependent on the dipeptide transporter DppBCDF (18).
The molecular weight of heme exceeds the diffusion limit of the outer membrane in Gram-negative bacteria (4). Thus, acquisition is mediated by dedicated uptake machinery. However, heme has been shown to diffuse through liposome membranes (19), suggesting that it could passively diffuse through the mycobacterial outer membrane. We find this unlikely due to the fact that free heme is toxic and is always bound by proteins (12). Any bacterial uptake mechanism would then have to compete for heme binding, whether it is a secreted hemophore or a cell surface receptor. Therefore, M. tuberculosis likely has evolved a novel mechanism for the utilization of heme.
Conclusions.
Heme uptake by bacterial pathogens is a common mechanism for iron acquisition in the host. Here, we demonstrate that M. tuberculosis can utilize heme as an iron source, suggesting that it has evolved a heme acquisition system like many other bacterial pathogens to exploit the most abundant iron source inside the human body.
Acknowledgments
This work was supported by a fellowship from training grant T32 AI007493 to C.M.J. and by the grants R01 AI063432 and R01 AI083632 to M.N. from the National Institutes of Health.
We are thankful to Colin Ratledge for providing purified ferri-carboxymycobactin from M. bovis BCG.
Footnotes
Published ahead of print on 4 February 2011.
REFERENCES
- 1.Andrews, N. C. 2003. Disorders of iron metabolism, p. 1401-1433. In S. E. Lux, R. I. Handin, and T. P. Stossel (ed.), Blood: principles and practice of hematology, 2nd ed. Lippincott Williams and Walkins, Philadelphia, PA.
- 2.Andrews, N. C. 2000. Iron homeostasis: insights from genetics and animal models. Nat. Rev. Genet. 1:208-217. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 4.Braun, V., and H. Killmann. 1999. Bacterial solutions to the iron-supply problem. Trends Biochem. Sci. 24:104-109. [DOI] [PubMed] [Google Scholar]
- 5.Chim, N., A. Iniguez, T. Q. Nguyen, and C. W. Goulding. 2010. Unusual diheme conformation of the heme-degrading protein from Mycobacterium tuberculosis. J. Mol. Biol. 395:595-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crichton, R. R., and J. L. Pierre. 2001. Old iron, young copper: from Mars to Venus. Biometals 14:99-112. [DOI] [PubMed] [Google Scholar]
- 7.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danilchanka, O., C. Mailaender, and M. Niederweis. 2008. Identification of a novel multidrug efflux pump of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 52:2503-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawson, D. J., and F. Jennis. 1980. Mycobacteria with a growth requirement for ferric ammonium citrate, identified as Mycobacterium haemophilum. J. Clin. Microbiol. 11:190-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshpande, R. G., M. B. Khan, D. A. Bhat, and R. G. Navalkar. 1997. Isolation of a contact-dependent haemolysin from Mycobacterium tuberculosis. J. Med. Microbiol. 46:233-238. [DOI] [PubMed] [Google Scholar]
- 11.de Voss, J. J., et al. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. U. S. A. 97:1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genco, C. A., and D. W. Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 13.Hider, R. C., and X. Kong. 2010. Chemistry and biology of siderophores. Nat. Prod. Rep. 27:637-657. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann, C., A. Leis, M. Niederweis, J. M. Plitzko, and H. Engelhardt. 2008. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. U. S. A. 105:3963-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansen, S. K., C. E. Maus, B. B. Plikaytis, and S. Douthwaite. 2006. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol. Cell 23:173-182. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, E. E., et al. 2010. Siderocalin inhibits the intracellular replication of Mycobacterium tuberculosis in macrophages. FEMS Immunol. Med. Microbiol. 58:138-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krewulak, K. D., and H. J. Vogel. 2008. Structural biology of bacterial iron uptake. Biochim. Biophys. Acta 1778:1781-1804. [DOI] [PubMed] [Google Scholar]
- 18.Letoffe, S., P. Delepelaire, and C. Wandersman. 2006. The housekeeping dipeptide permease is the Escherichia coli heme transporter and functions with two optional peptide binding proteins. Proc. Natl. Acad. Sci. U. S. A. 103:12891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Light, W. R., III, and J. S. Olson. 1990. Transmembrane movement of heme. J. Biol. Chem. 265:15623-15631. [PubMed] [Google Scholar]
- 20.Maus, C. E., B. B. Plikaytis, and T. M. Shinnick. 2005. Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muir, S., et al. 1992. Cloning and expression of a Serpula (Treponema) hyodysenteriae hemolysin gene. Infect. Immun. 60:529-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niederweis, M., O. Danilchanka, J. Huff, C. Hoffmann, and H. Engelhardt. 2010. Mycobacterial outer membranes: in search of proteins. Trends Microbiol. 18:109-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman, A., S. S. Srivastava, A. Sneh, N. Ahmed, and M. V. Krishnasastry. 2010. Molecular characterization of tlyA gene product, Rv1694 of Mycobacterium tuberculosis: a non-conventional hemolysin and a ribosomal RNA methyl transferase. BMC Biochem. 11:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rindi, L., D. Bonanni, N. Lari, and C. Garzelli. 2003. Most human isolates of Mycobacterium avium Mav-A and Mav-B are strong producers of hemolysin, a putative virulence factor. J. Clin. Microbiol. 41:5738-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez, G. M., and I. Smith. 2006. Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J. Bacteriol. 188:424-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambandamurthy, V. K., et al. 2006. Mycobacterium tuberculosis ΔRD1 ΔpanCD: a safe and limited replicating mutant strain that protects immunocompetent and immunocompromised mice against experimental tuberculosis. Vaccine 24:6309-6320. [DOI] [PubMed] [Google Scholar]
- 27.Siegrist, M. S., et al. 2009. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc. Natl. Acad. Sci. U. S. A. 106:18792-18797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sompolinsky, D., A. Lagziel, D. Naveh, and T. Yankilewitz. 1978. Mycobacterium haemophilum sp. nov., a new pathogen of humans. Int. J. Syst. Bacteriol. 28:67-75. [Google Scholar]
- 29.Tong, Y., and M. Guo. 2009. Bacterial heme-transport proteins and their heme-coordination modes. Arch. Biochem. Biophys. 481:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinberg, E. D. 1974. Iron and susceptibility to infectious disease. Science 184:952-956. [DOI] [PubMed] [Google Scholar]
- 31.Weinberg, E. D. 1997. The Lactobacillus anomaly: total iron abstinence. Perspect. Biol. Med. 40:578-583. [DOI] [PubMed] [Google Scholar]
- 32.WHO. 2010. World health statistics 2010. World Health Organization, Geneva, Switzerland.
- 33.Wren, B. W., et al. 1998. Characterization of a haemolysin from Mycobacterium tuberculosis with homology to a virulence factor of Serpulina hyodysenteriae. Microbiology 144(Pt. 5):1205-1211. [DOI] [PubMed] [Google Scholar]