Abstract
A number of d-amino acids occur in nature, and there is growing interest in their function and metabolism, as well as in their production and use. Here we use the well-established l-amino-acid-producing bacterium Corynebacterium glutamicum to study whether d-amino acid synthesis is possible and whether mechanisms for the export of these amino acids exist. In contrast to Escherichia coli, C. glutamicum tolerates d-amino acids added extracellularly. Expression of argR (encoding the broad-substrate-specific racemase of Pseudomonas taetrolens) with its signal sequence deleted results in cytosolic localization of ArgR in C. glutamicum. The isolated enzyme has the highest activity with lysine (100%) but also exhibits activity with serine (2%). Upon overexpression of argR in an l-arginine, l-ornithine, or l-lysine producer, equimolar mixtures of the d- and l-enantiomers accumulated extracellularly. Unexpectedly, argR overexpression in an l-serine producer resulted in extracellular accumulation of a surplus of d-serine (81 mM d-serine and 37 mM l-serine) at intracellular concentrations of 125 mM d-serine plus 125 mM l-serine. This points to a nonlimiting ArgR activity for intracellular serine racemization and to the existence of a specific export carrier for d-serine. Export of d-lysine relies fully on the presence of lysE, encoding the exporter for l-lysine, which is apparently promiscuous with respect to the chirality of lysine. These data show that d-amino acids can also be produced with C. glutamicum and that in special cases, due to specific carriers, even a preferential extracellular accumulation of this enantiomer is possible.
Corynebacterium glutamicum is well known for its extraordinary l-amino acid production properties. Its most prominent feature is probably its capacity to produce l-glutamate, 1.8 million tons of which are currently produced per year and used as a sodium salt to be added to food (24). Another amino acid made with C. glutamicum is l-lysine, which is required for animal nutrition. Indeed, over the years, C. glutamicum strains have been developed for the production of almost all the l-amino acids with commercial potential. For instance, our own group has contributed to the development of C. glutamicum strains producing l-isoleucine (7), l-valine (30), l-threonine (5), and l-serine (37).
Obviously, given the success of amino acid production by C. glutamicum and other bacteria, and the fact that sugar is a renewable substrate, bacterial production of further compounds has great appeal. Processes to enable the production of succinate (28), lactate (29), cadaverine (14), putrescine (32), 2-oxoisovalerate (19), and isobutanol (35) by C. glutamicum have been described.
d-Amino acids constitute another interesting group of products, although their direct biotechnological production by bacteria has not yet been investigated. A significant reason is probably that it is unclear whether the cell can tolerate the coexistence of d- and l-amino acids. In fact, the growth of Lactobacillus arabinosus is inhibited by d-leucine (9); Escherichia coli and Staphylococcus aureus are inhibited by d-leucine, d-alanine, or d-valine (17). d-Serine inhibits E. coli at a concentration of just 20 mM (39), and 15 mM d-serine, d-lysine, or d-proline has been shown to amplify growth inhibition under osmotic stress (33). For E. coli K37, d-glutamate, d-threonine, d-lysine, d-valine, d-cysteine, or d-serine at a concentration of 10 mM or less causes a 2-fold decrease in the final optical density (36).
Currently, d-amino acids are produced by different methods and are used mostly as building blocks in chemistry (43). Their production often starts with the l-amino acid and its racemization, due to the unrivaled bacterial production of l-amino acids. In further procedures to derive the d-enantiomer, a number of different physicochemical or biocatalytic treatments are required. Here we present experiments designed to reduce the number of steps required for d-amino acid production by producing racemic mixtures of dl-amino acids directly from C. glutamicum.
MATERIALS AND METHODS
Strains and cultivation.
The wild-type (WT) Corynebacterium glutamicum strain ATCC 13032 or its recombinant derivatives were used throughout. For recombinant work, E. coli DH5α was used; it was grown at 37°C in Luria-Bertani broth (1). All C. glutamicum strains utilized are listed in Table 1 . For Lys, Arg, or Orn production, strains were pregrown overnight at 30°C on the complex medium CGIII (31) and were inoculated into the defined minimal medium CGXII (31) with 4% (wt/vol) glucose as a substrate (CGXII-glucose) to an initial optical density at 600 nm (OD) of 1. Serine formation required an intermediate cultivation on CGXII-glucose to deplete Ser4 and its derivatives of folate. For this purpose, cells were pregrown on CGIII overnight, inoculated into CGXII-glucose to an OD of 1, and grown for 10 h. These cultures were used to inoculate fresh CGXII-glucose medium to an OD of 1, and their production and growth was followed in detail. d-Amino acids and the dipeptide l-Lys-l-Ala were sterile filtered as stock solutions and were added to CGXII after autoclaving.
TABLE 1.
Corynebacterium glutamicum strains and plasmids
| Designation | Features or usea | Source or reference |
|---|---|---|
| Strains | ||
| ATCC 13032 | WT; biotin auxotroph | Culture collection |
| Ser4 | l-Serine producer; WTΔsdaA ΔpabABC (pserABC); Tetr | 37 |
| DM1933 | l-Lysine producer; WTΔpck pyc(P458S) hom(V59A) 2×lysC(T311I) 2×lysE 2×asd 2×dapA 2×dapB 2×ddh 2×lysA | 3 |
| ORN1 | l-Ornithine producer; WTΔargR ΔargF; arginine auxotroph | 13, 32 |
| WTΔargR | Deletion of transcriptional regulator of arginine synthesis | This institute |
| ARG2 | WTΔargR [pVWEx2-argB(A49VM54V)]; Tetr | This study |
| WTΔlysEG | Deletion of lysine exporter and its regulator | 40 |
| Plasmids | ||
| pEKEx2 | Shuttle vector; Kanr Ptac lacIq | 8 |
| pEKEx3 | Derived from pEKEx2; Specr | 11 |
| pVWEx2 | Derived from pEKEx2; Tetr Ptac lacIq | 8 |
| pEKE3-argB(A49VM54V) | With feedback-resistant acetylglutamate kinase; Specr | 13, 32 |
| pEKEx3-ArgR | Carrying the racemase gene of P. taetrolens with its signal sequence deleted | This study |
| pEKEx3-SP-ArgR | Carrying the full-length racemase gene of P. taetrolens | This study |
| pEKEx2-TorA-MalE | Vector providing the torA signal sequence for Tat-dependent expression | F. Lausberg; this institute |
| pEKEx2-TorA-ArgR | Replacement of the signal sequence of P. taetrolens argR by the torA signal sequence | This study |
| pVWEx2-argB(A49VM54V) | With feedback-resistant acetylglutamate kinase; Tetr | 13, 32 |
| pET22b(+)argR | Carrying the racemase gene of P. taetrolens with its signal sequence deleted and with C-terminal His6 | 22 |
WT, wild type; 2×, two copies.
Molecular work.
The primers used in this study are listed in Table 2 . For the construction of pEKEx3-ArgR, arg was amplified from pET11b-argR (22) using primers argR-PstI (fw) and argR-BamHI (rev). The product was treated with PstI/BamHI and was ligated with similarly treated pEKEx3. The construction of pEKEx3-SP-ArgR was similar, but primers argR-SP-PstI (fw) and argR-BamHI (rev) were used. For the construction of pEKEx3-TorA-ArgR, primers argR-Esp3I (fw) and argR-Esp3I (rev) were used with pEKEx3-ArgR as the template. The resulting fragment was treated with Esp3I and was ligated with EcoRI-cleaved pEKEx2-TorA-MalE. To enable the isolation of ArgR, primers argR-NdeI (Fw) and argR-XhoI (rev), with pEKEx3-ArgR as the template, served for argR amplification. The product was treated with NdeI/XhoI and was ligated with similarly treated pET22b(+). To construct pVWEx2-argB(A49VM54V), mutated argB was amplified from pEKEx3-argB(A49VM54V) (32) using primers argB-Cg-SalI (fw) and argB-Cg-BamHI (rev). The product was treated with SalI/BamHI, ligated with similarly treated pVWEx2, and used to transform E. coli DH5α to tetracycline resistance (15 μg ml−1). The inserts of all plasmids made were confirmed by sequencing. Plasmids were introduced into C. glutamicum by electroporation. This technique, as well as other methods specific for C. glutamicum, such as plasmid isolation, was used as described previously (31).
TABLE 2.
Primers used in this study
| Primer | Sequence | Feature(s) |
|---|---|---|
| argR-PstI (fw) | TTTCTGCAGAAGGAGATATAGATATGGCGCCACCC | RBS,a PstI |
| argR-SP-PstI (fw) | TTTCTGCAGAAGGAGATATAGATATGCCCTTCTC | RBS, PstI |
| argR-BamHI (rev) | GCAGGATCCTCGTTCAATATACGG | BamHI |
| argR-Esp3I (fw) | ATTCGTCTCGAATTCGATGCGCCACCCCTGTCGATGAC | Esp3I |
| argR-Esp3I (rev) | CGGCGTCTCGAATTCTCATTACTGATCTTTCAGGATTTTAGG | Esp3I |
| argR-NdeI (fw) | CAACATATGGCGCCACCCCTGTCG | NdeI |
| argR-XhoI (rev) | CCCCTCGAGCTGATCTTTCAGGATTTT | XhoI |
| argB-Cg-SalI (fw) | GCTTAAGTCGACAAGGAGATATAGATGAATGACTTGATCAAAGAT | RBS, SalI |
| argB-Cg-BamHI (rev) | ATATGTGGATCCTTACAGTTCCCCATCCTTGTCGTC | BamHI |
RBS, ribosome binding site.
Localization studies.
Cells were grown overnight on CGIII with antibiotics (50 μg kanamycin [Kan] ml−1 and/or 250 μg spectinomycin [Spec] ml−1), and corresponding cultures were used to inoculate the main culture with an antibiotic to an OD of 1. At the same time, 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added. After further cultivation for 24 h, 4-ml cultures were harvested by centrifugation (10 min, 16,000 × g). The proteins in the supernatant were precipitated with trichloroacetic acid (final concentration, 10%), harvested by centrifugation, subsequently washed with 100% and 80% acetone, and dissolved in 80 μl of 50 mM Tris-HCl (pH 7.5) at 37°C. The pelleted cells were dissolved in 200 μl lysis buffer (10 mM Tris-HCl [pH 8.0], 25 mM MgCl2, 200 mM NaCl), and cells were disrupted by bead beating for 3 min using a SpeedMill P12 system (Analytik Jena). The supernatant obtained after centrifugation served as the extract that was analyzed by polyacrylamide gel electrophoresis (PAGE). The volume of extract (Fig. 1, E) used was equivalent to an OD of 0.5, whereas that of the supernatant (Fig. 1, S) corresponded to an OD of 1.5.
FIG. 1.
Localization of ArgR and derivatives in C. glutamicum. Shown are extracts (E) and supernatants (S) applied to an SDS gel for analysis of recombinants containing either an empty vector (pEKEx3-), argR with the signal sequence (SP-ArgR), argR with the signal sequence deleted (ArgR), or argR in which the signal sequence was replaced by the TorA signal peptide (TorA-ArgR). Gels were stained with Coomassie blue, and the ArgR protein is boxed.
Protein work.
To obtain ArgR protein, E. coli C41(DE3) pET22b(+)argR was grown in 100 ml LB at 37°C. After initial growth to an OD of 0.4 to 0.5, IPTG was added to give a concentration of 0.4 mM, and the cells were further cultivated at 30°C for 4 to 6 h. Cells were harvested, disrupted by sonication, and centrifuged, and the clear supernatant was applied to a 1-ml Ni2+-nitrilotriacetic acid (NTA)-agarose column. After a wash with a buffer consisting of 50 mM NaH2PO4·H2O, 300 mM NaCl, and 10 mM imidazole (pH 8), unspecific proteins were eluted in two additional wash steps with the same buffer but containing increased concentrations of 60 and 100 mM imidazole. ArgR was eluted with the buffer containing 250 mM imidazole.
In order to quantify racemase activity, the extracts were first passed through PD-10 columns (Amersham Biosciences, Freiburg, Germany) equilibrated with 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) (pH 10). In the assay, 30 to 800 μg total-cell protein or 2 to 200 μg isolated ArgR protein was used. Besides protein, the assay mixture contained 10 mM CAPS (pH 10), 250 nM pyridoxal-5′-phosphate, and 50 mM substrate (either l-arginine, l-lysine, l-ornithine, or l-serine). The assay mixture was incubated at 37°C, and at different intervals, 150-μl aliquots were removed, mixed with 50 μl of 10% trichloroacetic acid, and either frozen prior to analysis or directly subjected to reversed-phase high-performance liquid chromatography (RP-HPLC) for amino acid quantification.
Amino acid determination.
Intracellular amino acids were quantified using silica oil centrifugation (16). In brief, 400-μl Beckman tubes (Beckman Instruments GmbH, Munich, Germany) containing 30 μl of 20% (vol/vol) perchloric acid with an overlay of 65 μl silica oil (ρ, 1.04 g cm−3) were prepared. A 200-μl sample of the cell suspension was added to the tube, which was immediately centrifuged (for 45 s at 13,750 rpm in a Microfuge E centrifuge [Beckman Instruments GmbH, Munich, Germany]) to separate the medium from the cells. Cells were pelleted and inactivated in the bottom perchloric acid phase, whereas the medium was found on top of the oil. The cellular fraction was further treated by sonication, neutralized with 5 M KOH-1 M triethanolamine, and centrifuged as described previously (12). The resulting supernatant was analyzed by RP-HPLC for amino acids resulting from the cytosol, as was the fraction containing amino acids in the medium. For calculation purposes, it was assumed that an OD of 1 corresponded to 0.3 mg (dry weight) ml−1 and that the cell volume was 1.6 μl mg (dry weight)−1 (12).
Quantification of d- and l-amino acids was performed by automatic precolumn derivatization using the chiral thiol butyloxycarbonyl (Boc)-l-cysteine (10), which enables the formation of strongly fluorescent diastereomeric isoindole derivatives. The derivatization reagent was prepared by mixing 5 ml solution A (0.4 M borate buffer [pH 10.4]) with 0.1 ml solution B (75 mg ml−1 o-phthaldialdehyde and 75 mg ml−1 Boc-l-cysteine dissolved in methanol). Amino acid derivatives were separated by RP-HPLC (LiChrospher 100 RP-18 end-capped [EC] column; particle size, 5 μm; dimensions, 125 by 4 mm) using a gradient of 0.1 M sodium acetate (pH 7.2) with increasing methanol concentrations. Detection was performed at 450 nm, with an excitation wavelength of 230 nm. The entire analysis was conducted with an Agilent 1100 ChemStation.
RESULTS
Influence of d-amino acids on bacterial growth.
Corynebacterium glutamicum was grown on the minimal medium CGXII-glucose, to which individual d-amino acids were added to give a final concentration of 100 mM. As can be seen in Table 3, d-Thr had no influence on growth. The amino acids d-Arg, d-Lys, d-Ser, and d-Ala had a slight influence; d-Ser resulted in the strongest reduction in the growth rate, from 0.38 h−1 to 0.28 h−1. A rather strong influence on growth was observed for d-Met (μ, 0.20 h−1) and d-Asn. In all cases, the maximal OD reached in the stationary phase was not significantly affected.
TABLE 3.
Growth response of C. glutamicum and utilization of added d-amino acids
| d-Amino acid | μ (h−1) | Concn (mM)a |
|---|---|---|
| None | 0.38 ± 0.02 | |
| d-Thr | 0.39 ± 0.03 | 89 ± 7 |
| d-Lys | 0.33 ± 0.01 | 95 ± 8 |
| d-Arg | 0.29 ± 0.02 | 102 ± 4 |
| d-Ala | 0.29 ± 0.03 | 3 ± 2 |
| d-Ser | 0.28 ± 0.01 | 77 ± 8 |
| d-Asn | 0.24 ± 0.03 | 92 ± 4 |
| d-Met | 0.20 ± 0.04 | 97 ± 12 |
Concentration of the d-amino acid remaining after 48 h of cultivation, when growth was completed. Amino acids had been added at a concentration of 100 mM.
Determination of the levels of d-amino acids in the culture supernatant after 48 h revealed that d-Thr, d-Arg, d-Lys, and d-Met concentrations were not significantly reduced in the three biological replicates analyzed (Table 3). In contrast, d-Ser was reduced to a concentration of 77 ± 8 mM and thus was apparently partly metabolized. d-Ala, with a remaining concentration of 3 ± 1 mM, was almost fully consumed. Since the l-Ala concentration was also low (≤4 mM), it is most likely that d-Ala was converted to pyruvate via ubiquitous alanine racemase (27) and transaminase activities (20). Direct incorporation of d-Ala into peptidoglycan is also possible (38).
Racemase selection and localization.
Like almost every bacterium, C. glutamicum has alanine and glutamate racemase for cell wall synthesis. These enzymes are substrate specific (27, 38) and consequently are not suited for the racemization of other amino acids. We therefore searched for other racemases and selected the broad-substrate-specific arginine racemase of Pseudomonas taetrolens for the catabolism of d-amino acids (44). However, this racemase possesses a signal peptide resulting in translocation via the Sec system and has been demonstrated to be present in the periplasmic space in both P. taetrolens and E. coli (22). We therefore first studied arginine racemase and its derivatives for their localization in C. glutamicum.
The argR gene (GenBank accession no. AB096176) encoding the racemase was cloned into pEKEx3 to yield pEKEx3-SP-ArgR and was also cloned without the 5′ end, coding for a signal sequence of 23 amino acids, to yield pEKEx3-ArgR. In addition, plasmid pEKEx3-TorA-ArgR was constructed, where the signal sequence was replaced by the signal peptide sequence of the trimethylamine N-oxide (TMAO) reductase (TorA) of E. coli, which enables Tat-dependent translocation of folded proteins together with their cofactor (2). C. glutamicum was transformed with either of these plasmids or with pEKEx3 as a control. The recombinant strains were grown in the complex medium CGIII in the presence of 0.1 mM IPTG for 24 h, and subsequently, proteins from crude extracts and culture supernatants were prepared for sodium dodecyl sulfate (SDS)-PAGE analysis (Fig. 1).
Whereas the extracts (E) and supernatants (S) of C. glutamicum(pEKEx3) and of C. glutamicum(pEKEx3-SP-ArgR) did not reveal a dominant band that could be due to ArgR, a band of ∼42 kDa was present in the extract of C. glutamicum(pEKEx3-ArgR) and was verified as ArgR protein by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (data not shown). Also, the relatively weak band present in the supernatant, possibly due to lysis of cells, was identified as ArgR. For pEKEx3-TorA-ArgR, ArgR was found to be localized largely in the supernatant, and to a minor extent also in the extract, again as verified by MALDI-TOF analyses. With pEKEx3-TorA-ArgR, ArgR has an increased molecular mass of ∼43 kDa due to 11 additional amino acids resulting from a linker and TorA sequences. The data obtained with the TorA signal sequence suggest that extracellularly produced ArgR protein could possibly be used. However, we focused on the application of cytosolic ArgR, produced with pEKEx3-ArgR, which has no signal sequence.
Comparison of ArgR activities.
To assay for specific racemase activities, argR without the signal sequence was cloned into pET22b(+), expressed in E. coli C41(DE3), and isolated as ArgR-His6 protein via affinity chromatography. From a 500-ml culture, about 7.5 mg ArgR-His6 was isolated; it exhibited the expected size of ∼42 kDa and no significant impurities upon SDS-PAGE analysis (data not shown). Racemase activity was assayed with the purified ArgR-His6 protein and with a crude extract prepared from C. glutamicum(pEKEx3-ArgR) grown in the presence of 0.1 mM IPTG. The resulting specific activities are shown in Table 4 . Isolated racemase exhibited the highest activity with lysine, followed by ornithine and arginine, findings that largely agree with previous data (22, 44). Interestingly, the enzyme had significant racemase activity with serine as a substrate, which was previously unknown. With extracts of C. glutamicum(pEKEx3-ArgR), the specific activities based on total protein were naturally lower, but the activity ratios among the four amino acids were almost identical. In controls with extracts prepared from WT C. glutamicum, no racemase activity with any of the four amino acids was detectable. These promising data prompted us to assay pEKEx3-ArgR for its applicability to d-amino acid formation by C. glutamicum strains producing l-amino acids.
TABLE 4.
Comparison of ArgR racemase activities with different substrates
| Substrate | Sp act (μmol min−1 mg−1 [%]) of: |
|
|---|---|---|
| ArgR-His6 protein | Crude extract of cells expressing argRa | |
| l-Lysine | 1,260 ± 22 (100) | 119 ± 10 (100) |
| l-Ornithine | 1,135 ± 20 (90) | 96 ± 7 (81) |
| l-Arginine | 879 ± 47 (70) | 86 ± 2 (72) |
| l-Serine | 34 ± 2 (3) | 3 ± 1 (2) |
Crude extract from C. glutamicum pEKEx3-ArgR. With a crude extract from WT C. glutamicum, no activity (<0.01 μmol min−1 mg−1) was detectable with any of the four amino acids.
Production of d-serine.
In earlier work, we engineered C. glutamicum to derive the l-serine producer C. glutamicum Ser4, whose folate auxotrophy is key to triggering l-serine accumulation (37). This strain was transformed with pEKEx3-ArgR. The resulting strain and the control strains Ser4 and Ser4(pEKEx3) were assayed in minimal medium cultures for serine formation; the results are shown in Fig. 2. As expected, the parent strain Ser4 accumulated 81 mM l-Ser. The presence of the empty plasmid pEKEx3 (in the absence of spectinomycin) apparently stressed the cell, resulting in growth retardation and altered l-Ser accumulation. Whereas the control strains formed no d-Ser, d-Ser accumulated extracellularly with strain Ser4(pEKEx3-ArgR), thus demonstrating the function of the engineered broad-substrate-specific racemase of Pseudomonas taetrolens in l-Ser-producing C. glutamicum. Interestingly, the d-enantiomer was even present in excess. At a fermentation time of 120 h, d-Ser was present at a concentration as high as 81 mM and l-Ser at a concentration of 37 mM.
FIG. 2.
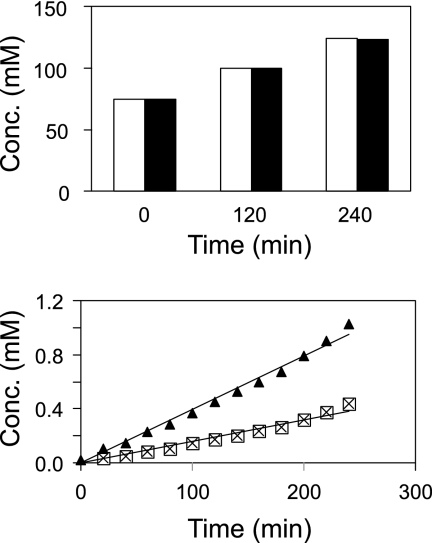
Extracellular accumulation of d-Ser and l-Ser. Shown are growth (⊠) and the accumulation of l-Ser (▴) and d-Ser (▵). In each graph, the Ser concentration (mM) is shown along the left y axis, growth (OD) along the right y axis, and time (h) along the x axis.
d-Serine export.
The preferential extracellular accumulation of d-Ser is surprising, since racemization is expected to occur within the cytosol, resulting in an equimolar mixture of the two Ser enantiomers. At most, an excess of l-Ser would be conceivable if the in vivo racemase activity were too low. In order to obtain information on this subject, we determined intracellular concentrations and excretion rates in a separate assay. For this purpose, Ser4(pEKEx3-ArgR) was grown as described above, but cells were removed after 15 h of cultivation and were transferred to an OD of 1 into new, prewarmed minimal medium cultures. This enabled extracellular Ser, which otherwise would have interfered with the determination of intracellular Ser levels, to be present at very low concentrations. The sample taken immediately after inoculation and subjected to silica oil centrifugation revealed that both Ser enantiomers were present at high cytosolic concentrations of about 75 mM each (Fig. 3). Equimolar concentrations of the enantiomers were also present in samples taken 2 h and 4 h later, while the total intracellular Ser concentration increased from 150 mM to 250 mM. These data indicate that the in vivo ArgR activity of Ser4(pEKEx3-ArgR) is sufficient to obtain full racemization of the l-Ser produced within the cell.
FIG. 3.
Intracellular Ser concentrations in C. glutamicum Ser4(pEKEx3-ArgR) (top) and extracellular accumulations from the same experiment (bottom). Open bars, l-Ser; filled bars, d-Ser. Symbols: ▴, d-Ser; ⊠, l-Ser.
The experiment described above also served to determine Ser excretion rates. Aliquots of the culture supernatant were removed at 20-min intervals and were analyzed for dl-Ser. The results are shown at the bottom of Fig. 3. The accumulation of d-Ser was significantly faster than that of l-Ser. The calculated d-Ser export rate was 16.20 ± 1.19 nmol min−1 (mg [dry weight])−1, more than twice the rate of l-Ser export, which was 6.69 ± 0.51 nmol min−1 (mg [dry weight])−1. In view of this result, it is very unlikely that the high extracellular d-Ser accumulation (higher than that of l-Ser [Fig. 2]) is due to reuptake of l-Ser and its metabolism (26); rather, our finding suggests active export of d-Ser.
Production of d-lysine.
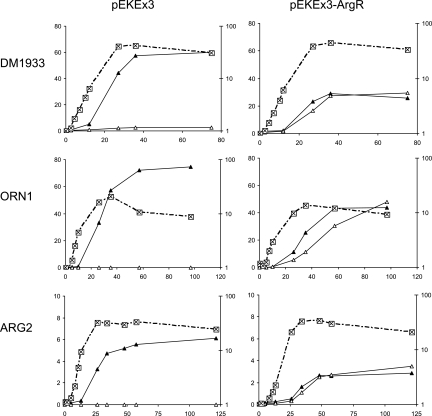
C. glutamicum DM1933 is an l-Lys producer that carries 3 point mutations and 1 deletion and has duplications of genes coding for l-lysine biosynthesis and l-lysine export (3). We transformed this strain with pEKEx3-ArgR and with pEKEx3 as a control, and we assayed for growth and product formation in CGXII-glucose (Fig. 4, top). Whereas the control accumulated 62 mM l-Lys in the culture supernatant, the strain expressing argR accumulated 26 mM l-Lys plus 29 mM d-Lys. In contrast to the situation with extracellular Ser accumulation (Fig. 2), there was no preferential extracellular accumulation of one of the Lys enantiomers. Furthermore, the comparable growth of DM1933 with and without ArgR means that intracellular d-Lys does not have an unfavorable effect on the metabolism of C. glutamicum, as already concluded from the experiment with extracellular d-Lys addition (Table 3).
FIG. 4.
Extracellular ArgR-dependent amino acid accumulation with C. glutamicum. The growth (⊠) and l-amino acid (▴) and d-amino acid (▵) accumulation of the lysine producer DM1933, the ornithine producer ORN1, and the arginine producer ARG2, without ArgR (pEKEx3) and with ArgR (pEKEx3-ArgR), are shown. In each graph, the amino acid concentration (mM) is shown along the left y axis, growth (OD) along the right y axis, and time (h) along the x axis.
d-Lysine export.
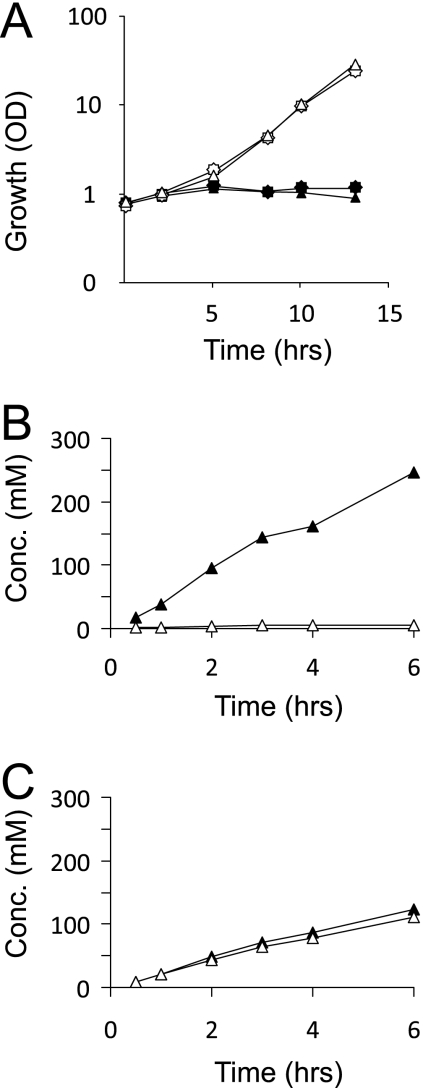
There is apparently no preferred export of d- or l-Lys. Translocation of charged amino acids through lipid bilayers requires active export (18, 23), and we have demonstrated that l-Lys export with C. glutamicum is fully dependent on the lysine exporter LysE (40). We hypothesize here that d-Lys is also exported actively and that LysE might possibly translocate d-Lys, too. To assay for this, we used the dipeptide l-Lys-l-Ala, which is rapidly taken up by C. glutamicum and is hydrolyzed intracellularly, resulting in an increased cytosolic l-Lys concentration even in the wild type. When this experiment was carried out with C. glutamicum ΔlysEG, in which the l-Lys exporter was deleted together with its transcriptional regulator, the strain was unable to excrete l-Lys (40). As a consequence, l-Lys piled up intracellularly to reach concentrations exceeding 300 mM, resulting in growth arrest.
We grew C. glutamicum ΔlysEG(pEKEx3-ArgR) and C. glutamicum ΔlysEG(pEKEx3) overnight on the complex medium CGIII and transferred the cells into CGXII-glucose alone and in parallel into CGXII-glucose to which 3 mM l-Lys-l-Ala was added. Under the latter conditions, the growth of the strains was severely inhibited (Fig. 5 A), and neither l-Lys nor d-Lys was found extracellularly (≤0.05 mM). Together with the fact that, due to pEKEx3-ArgR, d-Lys accumulates extracellularly in a background with lysEG present (Fig. 4), this finding suggests that LysE is responsible for d-Lys export. For further verification, we grew the strains under conditions identical to those applied previously and quantified intracellular dl-Lys in the initial 6 h of cultivation. As can be seen in Fig. 5B, the control strain C. glutamicum ΔlysEG(pEKEx3) accumulated l-Lys in its cytosol up to a bacteriostatic concentration of about 250 mM, whereas C. glutamicum ΔlysEG(pEKEx3-ArgR) (Fig. 5C) accumulated intracellularly an enantiomeric mixture reaching a similar concentration of 233 mM Lys (122 mM l-Lys plus 111 mM d-Lys). These results demonstrate the presence of a high intracellular d-Lys concentration, and they confirm that the mutant in which the lysine exporter is deleted is unable to export d-Lys. We thus conclude that LysE is promiscuous with respect to the d- and l-enantiomers of lysine.
FIG. 5.
Effect of lysEG deletion on lysine export with C. glutamicum. (A) Growth of C. glutamicum ΔlysEG(pEKEx3-ArgR) (star) and C. glutamicum ΔlysEG(pEKEx3) (triangle) without l-Lys-l-Ala (open symbols) or with the addition of 3 mM l-Lys-l-Ala (solid symbols). (B and C) Intracellular Lys concentrations in C. glutamicum ΔlysEG(pEKEx3) (B) and C. glutamicum ΔlysEG(pEKEx3-ArgR) (C) plus 3 mM l-Lys-l-Ala. ▵, d-Lys; ▴, l-Lys.
Production of d-ornithine and d-arginine.
To further extend the spectrum of cellular d-amino acid production, we took the C. glutamicum l-ornithine producer ORN1 (32). We also deleted argR, which bears the same name as the racemase gene from P. taetrolens but in C. glutamicum encodes the transcriptional regulator of arginine synthesis, to yield C. glutamicum WTΔargR. Furthermore, we cloned a gene encoding a feedback-resistant N-acetylglutamate kinase [argB(A49V M54V)] (13, 32) to yield pVWEx2-argB(A49VM54V). This vector was used to transform WTΔargR to yield the l-Arg producer ARG2. Both ORN1 and ARG2 were transformed with pEKEx3-ArgR. Their product accumulations are shown in Fig. 4 (center and bottom). In both cases, roughly equimolar concentrations of the corresponding enantiomers accumulated. The final concentrations were 48 mM d-Orn plus 44 mM l-Orn and 3 mM d-Arg plus 3 mM l-Arg, respectively. The total concentrations were clearly dependent on the production capacities of the control strains without ArgR, which accumulated 75 mM l-Orn and 6 mM l-Arg, respectively.
DISCUSSION
As outlined in the introduction, there are numerous examples illustrating the sensitivity of E. coli (17, 33, 36) and other bacteria (9, 17) to added d-amino acids. In contrast, C. glutamicum is rather insensitive to d-amino acid addition. Even more interestingly, this is also the case when d-amino acids are formed intracellularly, which was observed for the first time in the present work. d-Ser is tolerated in C. glutamicum at cytosolic concentrations exceeding 75 mM without any strong influence on growth. It is known that the presence of a d-amino acid can result in its transfer onto tRNA(s) by aminoacyl-tRNA synthetases, as demonstrated for E. coli tyrosyl-, tryptophanyl-, aspartyl-, lysyl-, and histidyl-tRNA synthetases (4). This harmful reaction is counteracted by the enzyme d-aminoacyl-tRNA deacylase, encoded by dtd in E. coli. The absence of dtd increases sensitivity to an added d-amino acid, and this is similarly the case in the absence of the deacylase in Saccharomyces cerevisiae (4, 36). A gene encoding d-aminoacyl-tRNA deacylase, NCgl1843, is also present in C. glutamicum. It is possible that high deacylase activity in this organism is one reason for its tolerance of d-amino acids.
The P. taetrolens ArgR racemase is apparently very suitable for providing C. glutamicum with new racemase activity. Although the in vivo specific activity with l-Ser is very low, at 3 μmol min−1 (mg of protein)−1, compared to the activity with l-Lys, this level is quite sufficient for cellular racemization. The excretion rate for l-Ser plus d-Ser, based on the dry weight of cells, is 22.89 nmol min−1 (mg [dry weight])−1. Since only about half of the dry weight of cells is protein, 45.78 nmol of Ser min−1 (mg of protein)−1 is excreted, which is still 2 orders of magnitude lower than the in vivo racemase activity. Therefore, it is export rather than racemase activity that is limiting, and the high cytosolic concentrations determined agree with this view. Export is recognized as a major issue in biotechnological production processes (reviewed in reference 6), although it is seldom considered. Moreover, the conclusion that weak in vivo racemase activity is sufficient to turn C. glutamicum into a d-amino acid producer offers the prospect of using ArgR or related enzymes to broaden the spectrum of d-amino acid production with C. glutamicum even further. For instance, it would be worthwhile to assay C. glutamicum l-Trp producers together with the mutated racemase BAR from Pseudomonas putida IFO 12996, which displays a specific activity of 52.8 nmol min−1 (mg of protein)−1 with l-Trp as a substrate (15), in order to produce d-Trp, an interesting d-amino acid, as well.
Of course, the most ambitious endeavor would be to produce just the d-amino acid directly, and not the racemic mixture. In this regard, the example of d-Ser and l-Ser formation with C. glutamicum is of particular interest, since the excretion and accumulation of d-Ser, rather than l-Ser, are preferred. This suggests the existence of an exporter with a preference for the d-enantiomer in C. glutamicum. In general, the export of bacterial metabolites has been little investigated, and the stereospecificity of transporters even less. One instance in which the components of metabolite export have been partially deciphered is l-Thr export by C. glutamicum (34). This occurs by (i) passive diffusion, (ii) active export catalyzed by the threonine exporter ThrE (34), and (iii) further active export by unknown carriers. Due to its related structure, Ser is also expected to be exported partly by diffusion (18, 23), and this physical process, of course, relates to both enantiomers. Since ThrE also utilizes l-Ser as a substrate, it would be interesting to test the consequences of ThrE deletion for Ser accumulation. An example of the stereospecificity of transporters is the rabbit Na+/glucose cotransporter, which favors the recognition of d-glucose over that of l-glucose (41). Other examples relate to the specificity of drugs accepted by transporters, such as the proton-coupled folate transporter PCFT of Mammalia, which has 40-fold greater affinity for the folate analogue l-amethopterin than for d-amethopterin (25). The indications obtained in the present work point to the presence of an enantiomer-specific exporter in C. glutamicum with a preference for accepting d-Ser, which has to be identified among the roughly 180 candidate exporter genes in the C. glutamicum genome (21, 42).
In the case of d-Lys export by C. glutamicum, the data allow us to conclude that LysE accepts d-Lys just as efficiently as l-Lys. The broad substrate specificity of LysE with respect to the compounds accepted has already been recognized: LysE has been demonstrated to accept l-Lys, l-Arg, and l-His (40). Cadaverine is also a substrate (our unpublished data), and canavanine could be a substrate, too. The present data show that LysE has even broader substrate specificity with respect to the stereochemistry of substrates, and this is the first example of the promiscuousness of an amino acid exporter in this regard, illustrating the potential of cells for the excretion of metabolites.
Acknowledgments
We thank Amino GmbH, 38373 Frellstedt, Germany, for supporting this work.
We are also grateful to M. Broker for MALDI-TOF analyses, to R. Freudl and F. Lausberg for advice on protein export, and to D. Matsui for providing pEKEx3-ArgR.
Footnotes
Published ahead of print on 21 January 2011.
REFERENCES
- 1.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaudeck, N., G. A. Sprenger, R. Freudl, and T. Wiegert. 2001. Specificity of signal peptide recognition in tat-dependent bacterial protein translocation. J. Bacteriol. 183:604-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blombach, B., S. Hans, B. Bathe, and B. J. Eikmanns. 2009. Acetohydroxyacid synthase, a novel target for improvement of l-lysine production by Corynebacterium glutamicum. Appl. Environ. Microbiol. 75:419-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calendar, R., and P. Berg. 1967. d-Tyrosyl RNA: formation, hydrolysis and utilization for protein synthesis. J. Mol. Biol. 26:39-54. [DOI] [PubMed] [Google Scholar]
- 5.Diesveld, R., et al. 2009. Activity of exporters of Escherichia coli in Corynebacterium glutamicum, and their use to increase l-threonine production. J. Mol. Microbiol. Biotechnol. 16:198-207. [DOI] [PubMed] [Google Scholar]
- 6.Eggeling, L. 15 April 2010. Microbial metabolite export, p. 1-10. In M. C. Flickinger (ed.), Encyclopedia of industrial biotechnology: bioprocess, bioseparation, and cell technology. John Wiley & Sons, Inc., New York, NY. doi: 10.1002/9780470054581.eib426. [DOI]
- 7.Eggeling, L., S. Morbach, and H. Sahm. 1997. The fruits of molecular physiology: engineering the l-isoleucine biosynthesis pathway in Corynebacterium glutamicum. J. Biotechnol. 56:167-182. [Google Scholar]
- 8.Eikmanns, B. J., E. Kleinertz, W. Liebl, and H. Sahm. 1991. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene 102:93-98. [DOI] [PubMed] [Google Scholar]
- 9.Fox, S. W., M. Fling, and G. N. Bollenback. 1944. Inhibition of bacterial growth by d-leucine. J. Biol. Chem. 155:465-468. [Google Scholar]
- 10.Hashimoto, A., T. Nishikawa, T. Oka, K. Takahashi, and T. Hayashi. 1992. Determination of free amino acid enantiomers in rat brain and serum by high-performance liquid chromatography after derivatization with N-tert.-butyloxycarbonyl- l-cysteine and o-phthaldialdehyde. J. Chromatogr. 582:41-48. [DOI] [PubMed] [Google Scholar]
- 11.Hoffelder, M., K. Raasch, J. van Ooyen, and L. Eggeling. 2010. Pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase: the E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl residues of the acetyltransferase AceF. J. Bacteriol. 192:5203-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoischen, C., and R. Krämer. 1990. Membrane alteration is necessary but not sufficient for effective glutamate secretion in Corynebacterium glutamicum. J. Bacteriol. 172:3409-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda, M., S. Mitsuhashi, K. Tanaka, and M. Hayashi. 2009. Reengineering of a Corynebacterium glutamicum l-arginine and l-citrulline producer. Appl. Environ. Microbiol. 75:1635-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kind, S., W. K. Jeong, H. Schröder, O. Zelder, and C. Wittmann. 2010. Identification and elimination of the competing N-acetyldiaminopentane pathway for improved production of diaminopentane by Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 76:5175-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kino, K., M. Sato, M. Yoneyama, and K. Kirimura. 2007. Synthesis of dl-tryptophan by modified broad specificity amino acid racemase from Pseudomonas putida IFO 12996. Appl. Microbiol. Biotechnol. 73:1299-1305. [DOI] [PubMed] [Google Scholar]
- 16.Klingenberg, M., and E. Pfaff. 1967. Means of terminating reactions. Methods Enzymol. 10:680-684. [Google Scholar]
- 17.Kobayashi, Y., M. Fling, and S. W. Fox. 1948. Antipodal specificity in the inhibition of growth of Escherichia coli by amino acids. J. Biol. Chem. 174:391-398. [PubMed] [Google Scholar]
- 18.Krämer, R. 1994. Systems and mechanisms of amino acid uptake and excretion in prokaryotes. Arch. Microbiol. 162:1-13. [DOI] [PubMed] [Google Scholar]
- 19.Krause, F. S., B. Blombach, and B. J. Eikmanns. 2010. Metabolic engineering of Corynebacterium glutamicum for 2-ketoisovalerate production. Appl. Environ. Microbiol. 76:8053-8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marienhagen, J., N. Kennerknecht, H. Sahm, and L. Eggeling. 2005. Functional analysis of all aminotransferase proteins inferred from the genome sequence of Corynebacterium glutamicum. J. Bacteriol. 187:7639-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marin, K., and R. Krämer. 2007. Amino acid transport systems in biotechnologically relevant bacteria, p. 289-326. In V. F. Wendisch (ed.), Amino acid biosynthesis: pathways, regulation and metabolic engineering. Microbiology Monographs no. 5. Springer, Berlin, Germany.
- 22.Matsui, D., et al. 2009. A periplasmic, pyridoxal-5′-phosphate-dependent amino acid racemase in Pseudomonas taetrolens. Appl. Microbiol. Biotechnol. 83:1045-1054. [DOI] [PubMed] [Google Scholar]
- 23.Milner, J. L., B. Vink, and J. M. Wood. 1987. Transmembrane amino acid flux in bacterial cells. Crit. Rev. Biotechnol. 5:1-47. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura, J., S. Hirano, H. Ito, and M. Wachi. 2007. Mutations of the Corynebacterium glutamicum NCgl1221 gene, encoding a mechanosensitive channel homolog, induce l-glutamic acid production. Appl. Environ. Microbiol. 73:4491-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narawa, T., and T. Itoh. 2010. Stereoselective transport of amethopterin enantiomers by the proton-coupled folate transporter. Drug Metab. Pharmacokinet. 25:283-289. [DOI] [PubMed] [Google Scholar]
- 26.Netzer, R., P. Peters-Wendisch, L. Eggeling, and H. Sahm. 2004. Cometabolism of a nongrowth substrate: l-serine utilization by Corynebacterium glutamicum. Appl. Environ. Microbiol. 70:7148-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oikawa, T., A. Tauch, S. Schaffer, and T. Fujioka. 2006. Expression of alr gene from Corynebacterium glutamicum ATCC 13032 in Escherichia coli and molecular characterization of the recombinant alanine racemase. J. Biotechnol. 125:503-512. [DOI] [PubMed] [Google Scholar]
- 28.Okino, S., et al. 2008. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl. Microbiol. Biotechnol. 81:459-464. [DOI] [PubMed] [Google Scholar]
- 29.Okino, S., M. Suda, K. T. Fujikura, M. Inui, and H. Yukawa. 2008. Production of d-lactic acid by Corynebacterium glutamicum under oxygen deprivation. Appl. Microbiol. Biotechnol. 78:449-454. [DOI] [PubMed] [Google Scholar]
- 30.Radmacher, E., et al. 2002. Linking central metabolism with increased pathway flux: l-valine accumulation by Corynebacterium glutamicum. Appl. Environ. Microbiol. 68:2246-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyes, O., and L. Eggeling. 2005. Experiments, p. 535-566. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. Taylor and Francis, Boca Raton, FL.
- 32.Schneider, J., and V. F. Wendisch. 2010. Putrescine production by engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 88:859-868. [DOI] [PubMed]
- 33.Shahjee, H. M., K. Banerjee, and F. Ahmad. 2002. Comparative analysis of naturally occurring l-amino acid osmolytes and their d-isomers on protection of Escherichia coli against environmental stresses. J. Biosci. 27:515-520. [DOI] [PubMed] [Google Scholar]
- 34.Simic, P., H. Sahm, and L. Eggeling. 2001. l-threonine export: use of peptides to identify a new translocator from Corynebacterium glutamicum. J. Bacteriol. 183:5317-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, K. M., K. M. Cho, and J. C. Liao. 2010. Engineering Corynebacterium glutamicum for isobutanol production. Appl. Microbiol. Biotechnol. 87:1045-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soutourina, J., P. Plateau, and S. Blanquet. 2000. Metabolism of d-aminoacyl-tRNAs in Escherichia coli and Saccharomyces cerevisiae cells. J. Biol. Chem. 275:32535-32542. [DOI] [PubMed] [Google Scholar]
- 37.Stolz, M., et al. 2007. Reduced folate supply as a key to enhanced l-serine production by Corynebacterium glutamicum. Appl. Environ. Microbiol. 73:750-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tauch, A., S. Götker, A. Pühler, J. Kalinowski, and G. Thierbach. 2002. The alanine racemase gene alr is an alternative to antibiotic resistance genes in cloning systems for industrial Corynebacterium glutamicum strains. J. Biotechnol. 99:79-91. [DOI] [PubMed] [Google Scholar]
- 39.Trippen, B., W. P. Hammes, K.-H. Schleifer, and O. Kandler. 1976. Die Wirkung von d-Aminosäuren auf die Struktur und Biosynthese des Peptidoglycans. Arch. Mikrobiol. 109:247-261. [DOI] [PubMed] [Google Scholar]
- 40.Vrljic, M., H. Sahm, and L. Eggeling. 1996. A new type of transporter with a new type of cellular function: l-lysine export from Corynebacterium glutamicum. Mol. Microbiol. 22:815-826. [DOI] [PubMed] [Google Scholar]
- 41.Wimmer, B., M. Raja, P. Hinterdorfer, H. J. Gruber, and R. K. Kinne. 2009. C-terminal loop 13 of Na+/glucose cotransporter 1 contains both stereospecific and non-stereospecific sugar interaction sites. J. Biol. Chem. 284:983-991. [DOI] [PubMed] [Google Scholar]
- 42.Winnen, B., J. Felce, and M. H. Saier, Jr. 2005. Genomic analyses of transporter proteins in Corynebacterium glutamicum and Corynebacterium efficiens, p. 149-186. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. Taylor and Francis, Boca Raton, FL.
- 43.Yagasaki, M., and A. Ozaki. 1998. Industrial biotransformations for the production of d-amino acids. J. Mol. Catalysis B Enzymatic 4:1-11. [Google Scholar]
- 44.Yorifuji, T., H. Misono, and K. Soda. 1971. Arginine racemase of Pseudomonas graveolens. II. Racemization and transamination of ornithine catalyzed by arginine racemase. J. Biol. Chem. 246:5093-5101. [PubMed] [Google Scholar]