Abstract
The cell wall-less prokaryote Mycoplasma pneumoniae causes bronchitis and atypical pneumonia in humans. Mycoplasma attachment and gliding motility are required for colonization of the respiratory epithelium and are mediated largely by a differentiated terminal organelle. P30 is a membrane protein at the distal end of the terminal organelle and is required for cytadherence and gliding motility, but little is known about the functional role of its specific domains. In the current study, domain deletion and substitution derivatives of P30 were engineered and introduced into a P30 null mutant by transposon delivery to assess their ability to rescue P30 function. Domain deletions involving the extracellular region of P30 severely impacted protein stability and adherence and gliding function, as well as the capacity to stabilize terminal organelle protein P65. Amino acid substitutions in the transmembrane domain revealed specific residues uniquely required for P30 stability and function, perhaps to establish correct topography in the membrane for effective alignment with binding partners. Deletions within the predicted cytoplasmic domain did not affect P30 localization or its capacity to stabilize P65 but markedly impaired gliding motility and cytadherence. The larger of two cytoplasmic domain deletions also appeared to remove the P30 signal peptide processing site, suggesting a larger leader peptide than expected. We propose that the P30 cytoplasmic domain may be required to link P30 to the terminal organelle core, to enable the P30 extracellular domain to achieve a functional conformation, or perhaps both.
The cell wall-less prokaryote Mycoplasma pneumoniae is a human pathogen causing community-acquired respiratory disease in persons of all ages but especially older children and young adults (32). Although it is perhaps best known as the etiologic agent of atypical, or “walking,” pneumonia, accounting for up to 20% of all cases of community-acquired pneumonia (41), infections most commonly manifest as tracheobronchitis. Chronic lung disease and extrapulmonary sequelae due to immunopathology or direct invasion can ensue, including a strong correlation with onset and exacerbation of asthma (3, 29, 42). Colonization of the epithelial mucosa of the conducting airways is mediated largely by the terminal organelle, a polar, differentiated, membrane-bound extension of the mycoplasma cell (7, 30). The adhesins P30 and P1 and several adherence accessory proteins localize to this structure (23), which is defined by a complex electron-dense core that is a prominent component of the mycoplasma cytoskeleton (2, 17, 18). In addition to its role in adherence, the terminal organelle is the motor for gliding motility (14), and its duplication precedes cell division (15).
P30 is a membrane protein oriented with the C terminus (domain III) on the mycoplasma cell surface, based on accessibility to antibodies and protease (10, 25) (Fig. 1 A), and the N terminus likely in the cell interior (domain I). Its predicted transmembrane (TM) domain and the region that immediately follows as the beginning of the surface-exposed portion of P30 (domain II) are highly conserved in homologs in Mycoplasma genitalium (33) and Mycoplasma gallisepticum (19) (Fig. 2). Domain III of P30 and its homologs is dominated by P-rich repeats which vary in sequence between species but which occur in M. pneumoniae in three motifs, PGMAPR, PGMPPH, and PGFPPQ (9, 10).
FIG. 1.
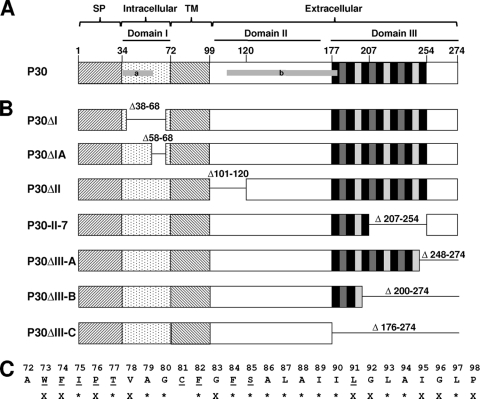
(A) Schematic of wild-type protein P30. Brackets designate the predicted signal peptide (SP), intracellular, transmembrane (TM), and extracellular regions of P30. Domains I, II, and III are indicated by black horizontal lines, while horizontal gray bars marked a and b correspond to antigens used to prepare antisera to the cytoplasmic and extracellular regions of P30, respectively. Numbers indicate amino acid residues. Vertical black and gray bars within domain III correspond to P-rich repeat motifs PGMAPR (black), PGMPPH (dark gray), and PGFPPQ (light gray). (B) Schematic of P30 domain deletion derivatives. Numbers indicate the deleted residues for each. (C) Amino acid residues of the P30 TM domain. Underlining, alanine substitutions generated; X, identical residues in M. pneumoniae, M. genitalium, and M. gallisepticum; *, identical residues in M. pneumoniae and M. genitalium only.
FIG. 2.
Boxshade of M. pneumoniae P30 (MPN P30), M. genitalium P32 (MGN P32), and M. gallisepticum MGC2 (MGA MGC2). Black and gray blocks indicate identical and similar amino acids, respectively. The solid bar is above the predicted P30 TM domain, and the double line is above the conserved region between P30 and P32. The alignment of sequences was analyzed by the “ClustalW2” program on http://www.ebi.ac.uk/ and boxshaded by BOXSHADE 3.21 on http://www.ch.embnet.org.
Mutant analysis has begun to elucidate the role of P30 in terminal organelle function (16, 22, 34). Loss of P30 due to a frameshift in its corresponding open reading frame (ORF), MPN453 (mutant II-3), is accompanied by reduced levels of the terminal organelle protein P65 (1, 20, 31, 34). Mutant II-3 is nonmotile, noncytadherent, and avirulent and has an ovoid, branched shape rather than the tapered appearance characteristic of wild-type cells (22, 34). A second site mutation in mutant II-3 generated mutant II-3R, having a wild-type reading frame for all but 17 residues within domain II of P30 and restoring cytadherence to a level approximately 60% of the wild-type level but gliding to a level only 5% of that of the wild type (21, 34). Mutant II-7 produces a truncated but stable P30 derivative due to a 144-bp internal deletion near the 3′ end of MPN453 (1, 25) and has a morphology similar to that of the wild type but exhibits markedly reduced gliding velocity and frequency (16, 34). P65 stability is reduced in mutant II-7 but not II-3R (20, 31), suggesting that the P-rich repeats of domain III are required to stabilize P65. By definition, adherence to surfaces is required for gliding motility, but the gliding defects in these P30 mutants do not correlate strictly with their cytadherence phenotypes, indicating that these functions are distinguishable (16). Finally, introduction of the recombinant wild-type allele by transposon delivery restores normal cytadherence, gliding motility, and cell morphology to mutants II-3 and II-7 (34), setting the stage for further analysis of P30 functional domains. Here we engineered P30 domain deletion and amino acid substitution derivatives and introduced these into mutant II-3 to explore further their specific contributions to P30 function in cytadherence and gliding motility. Deletion in extracellular domain II, truncation of extracellular domain III, and amino acid substitutions at specific sites in the TM domain severely impacted protein stability and function. Surprisingly, deletions in the putative cytoplasmic domain I had little effect on P30 stability, localization, or capacity to stabilize P65 but resulted in failure to rescue motility or cytadherence. The larger of these deletions also appeared to remove the P30 signal peptide processing site, suggesting a larger leader peptide than expected. We propose that this domain may link P30 to the terminal organelle core and/or direct folding of the extracellular domain to a functional conformation.
MATERIALS AND METHODS
Mycoplasma strains and culture media.
M. pneumoniae M129 (17th broth passage [26]), spontaneously arising M129 noncytadherent mutants II-3 and II-7, and mutant II-3 transformed with the recombinant wild-type allele encoding P30 (22, 34) were cultured in SP4 medium (28) and on enriched pleuropneumonialike organism (PPLO) agar plates with gentamicin, as appropriate (13, 16). See Fig. S1 in the supplemental material for details on the construction of deletion and substitution derivatives, as well as primers and plasmids. All derivatives were sequenced through their entire length (Georgia Genomics Facility, University of Georgia). Standard protocols were used for all DNA manipulations and electroporation of M. pneumoniae cells (34). Multiple M. pneumoniae transformants were isolated and characterized in each case to control for possible positional effects of the transposon insertion.
Western immunoblotting.
Samples were analyzed by Western immunoblotting as described previously (24, 40), except that membranes were blocked in 5% skim milk in 20 mM Tris-buffered saline (pH 7.5) and probed with alkaline phosphatase-conjugated secondary antibody (Promega, Madison, WI). Rabbit polyclonal antisera to the cytoplasmic (residues 36 to 55) and extracellular (residues 108 to 181) regions of P30 (Fig. 1A) were used at 1:250 and 1:3,000, respectively (34), while rabbit polyclonal anti-P65 antiserum was used at 1:5,000 (31).
Microscopy, cytadherence, and gliding motility.
Mycoplasmas were examined by immunofluorescence microscopy as described previously (6), except that mycoplasma cells were grown on poly-l-lysine-coated coverslips in 24-well dishes. Mouse monoclonal anti-P30 antibody was used at 1:50, and rabbit polyclonal anti-P65 antibody was used at 1:500. Images were captured with OpenLab Imaging software v5.01 (Improvision, Lexington, MA) as described previously (16) and processed with Adobe Photoshop CS3 v10 (Adobe Systems, Inc., San Jose, CA). Mycoplasma hemadsorption (HA) was assessed as an indicator of cytadherence as described previously (22, 38) but using sheep blood cells. Mycoplasmas grown in borosilicate glass chamber slides (Nunc Nalgene, Naperville, IL) were examined at 24-h intervals for satellite growth as a qualitative indicator of gliding motility or at 37°C for 18 h for quantitative assessment of gliding parameters for individual cells (16), with at least 1,000 cells observed per strain for gliding frequency and at least 38 cells per strain for mean and corrected gliding velocities (16).
RESULTS
P30 domain deletion derivatives differ in stability and capacity to stabilize P65.
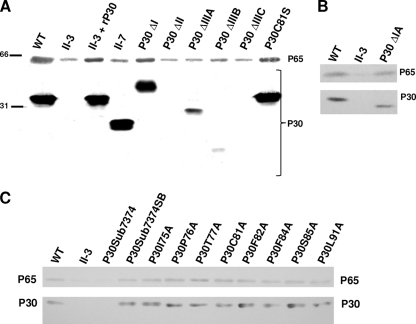
We targeted three regions of the extracellular domain of P30 for analysis of function in adherence, gliding motility, and P65 stabilization, i.e., the extreme C-terminal end (P30ΔIIIA) and P-rich repeat region (P30ΔIIIB and P30ΔIIIC) by truncation and a highly conserved portion of domain II (P30ΔII) by in-frame deletion. Each was engineered downstream of its normal promoter and transformed into P30 null mutant II-3 by recombinant transposon delivery (34). As expression levels vary with chromosome insertion site (34), we examined multiple transformants for each by Western immunoblotting, with representative transformants having the highest levels of each derivative shown here (Fig. 3; summarized in Table 1). P30 derivatives truncated at the C terminus appeared to be less stable than wild-type P30 in a manner proportional to protein length, with P30ΔIIIA the most abundant and P30ΔIIIB and P30ΔIIIC barely detectable and not detected, respectively. The deletion in domain II likewise rendered P30 undetectable in immunoblots, presumably due to instability.
FIG. 3.
Western immunoblot analysis of P30 and P65 in domain deletion mutants (A and B) and A-scanning TM domain substitution derivatives (C). Total mycoplasma protein loaded per lane for polyacrylamide gel electrophoresis was 100 μg (A) or 40 μg (B and C). P30 C-terminus-specific antibodies were used for results shown in panels A and B, and N-terminus-specific antibodies were used for results shown in panel C, but comparable results were obtained with both antibody specificities for all P30 derivatives tested here (data not shown). P65 and P30 or derivatives thereof are indicated on the right (all panels), and size standards in kilodaltons are indicated on the left (A). WT, wild-type M. pneumoniae; rP30, recombinant P30.
TABLE 1.
Summary of phenotypes conferred by P30 deletion and substitution derivatives
| P30 derivative(s) | Phenotypea |
|||||
|---|---|---|---|---|---|---|
| P30 stability | P30 localization | P65 stability | P65 localization | HA | Gliding | |
| Wild type | +++ | Polar/focal | +++ | Polar/focal | +++ | +++ |
| II-3 | None | NAb | + | Focal | − | − |
| P30ΔI | +++ | Focal | ++ | Focal | − | − |
| P30ΔIA | ++ | Polar/focal | ++ | Polar/focal | − | + |
| P30ΔII | − | NDc | + | NT | − | − |
| II-7 | +++ | Polar/focal | + | Polar/focal | + | + |
| P30ΔIIIA | + | Polar/focal | + | Polar/focal | − | + |
| P30ΔIIIB | +/− | ND | + | Polar/focal | − | +/− |
| P30ΔIIIC | − | ND | + | Polar/focal | − | − |
| P30Sub7374 | − | NTd | + | NT | − | − |
| P30Sub7374SB | +++ | Polar/focal | +++ | NT | +++ | +++ |
| P30C81A and P30C81S | +++ | Polar/focal | +++ | Polar/focal | +++ | +++ |
+++, like wild-type level; ++, slightly lower than wild-type level; +, substantially lower than wild-type level; +/−, just detectable; −, not detected.
NA, not applicable.
ND, not detectable.
NT, not tested.
We also targeted the predicted TM domain and cytoplasmic domain I for substitution and deletion, respectively, to assess their roles in P30 function. Deletion of the cytoplasmic domain (P30ΔI) had no affect on stability, but surprisingly this derivative migrated at a higher molecular mass than wild-type P30 (Fig. 3A), raising the possibility that the deletion had removed the signal peptide processing site and prompting us to engineer a smaller domain I deletion derivative (P30ΔIA) lacking residues 58 to 68 (Fig. 1B). This derivative migrated with the expected mobility and at slightly reduced steady-state levels (Fig. 3B).
We initially replaced the P30 TM domain with that of FtsH, which has a predicted topology similar to that of P30, but this derivative was unstable (data not shown), prompting us to employ A-scanning substitutions. Several residues, including W73, F74, I75, P76, T77, F82, F84, S85, and L91, are highly conserved in P30 homologs (Fig. 1C and 2). While not conserved, C81 is the only C in P30 and therefore might participate in interchain disulfide bonding. All P30 TM domain substitution derivatives except P30W73A and P30F74A were consistently stable at wild-type levels (Fig. 3C and data not shown). P30W73A and P30F74A were observed at greatly reduced but in some transformants unusually variable levels (data not shown); this inconsistency raised the possibility of secondary mutations, and hence these derivatives were not characterized further. Interestingly, replacement of both residues (P30Sub7374) rendered P30 consistently unstable, while restoration of both residues to the original sequence (P30Sub7374SB) yielded wild-type levels of P30 (Fig. 3C; Table 1). C81 substitution (P30C81A and P30C81S) had no effect on stability.
Protein P65 is present at greatly reduced levels in mutants II-3 and II-7 and yet localizes correctly to the terminal organelle in each and thus requires the P-rich region of P30 for stability but not trafficking (20). To explore this relationship further, we examined P65 levels in mutant II-3 transformants producing the various recombinant P30 derivatives. Only P30ΔI and P30ΔIA of the domain deletion derivatives restored P65 to wild-type levels (Fig. 3A and B, respectively), whereas all TM domain substitution derivatives except P30Sub7374 conferred P65 stabilization, likely a function of P30 stability (Fig. 3C; Table 1).
P30 and P65 localization.
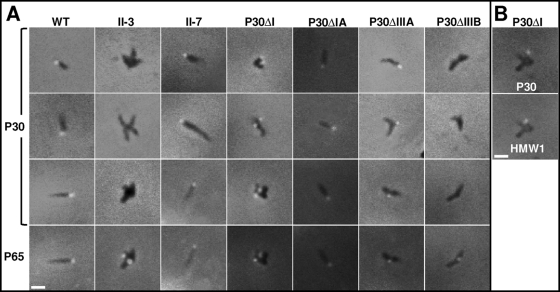
Like wild-type P30, both P30ΔIIIA and the P30 variant in mutant II-7 yielded a bright focus at the distal end of the terminal organelle by immunofluorescence microscopy (Fig. 4 A; Table 1), and thus the missing residues in each were not essential for proper trafficking. P30ΔI exhibited a polar fluorescent focus, but these cells were branched like those of mutant II-3 (34), making it difficult to determine if those foci corresponded to terminal organelles (Fig. 4A). HMW1 and TopJ (6, 39) were therefore used for spatial reference and localized just proximal to P30ΔI, like wild-type P30 (Fig. 4B and data not shown). P30ΔIA likewise yielded a polar fluorescent focus but conferred a more wild-type-like morphology (Fig. 4A). P30ΔIIIB, P30ΔIIIC, and P30ΔII were not detected (Fig. 4A and data not shown), in accordance with their instability, whereas all TM domain substitution derivatives except P30Sub7374 localized correctly, again, likely a function of P30 stability (data not shown). P65 formed distinct foci that colocalized with each P30 derivative, where present (Fig. 4A).
FIG. 4.
(A) Immunofluorescence localization of P30 and P65 in wild-type M. pneumoniae (WT), P30 mutants II-3 and II-7, and mutant II-3 containing the indicated domain deletion derivatives. The panels in rows 3 and 4 correspond to the same fields. (B) Immunofluorescence localization of P30 and HMW1 in the P30ΔI domain deletion mutant. Merged fluorescence and phase-contrast images of individual cells are shown. Antibody specificity was established previously (20, 34, 39). Bar, 1 μm.
P30 derivatives differ in ability to rescue cytadherence and motility.
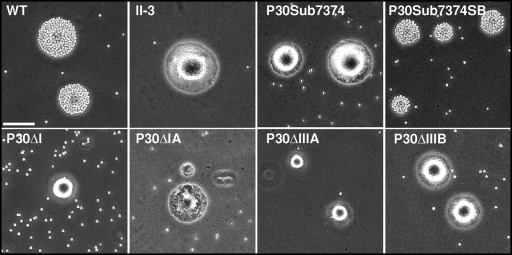
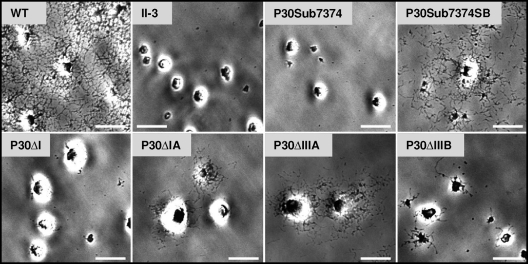
The wild-type P30 allele fully rescues HA and gliding motility in mutant II-3 (34). Somewhat surprisingly, no domain deletion derivative tested, including P30ΔI and P30ΔIA, which exhibit wild-type-like stability and localization, restored HA (Fig. 5; Table 1). P30ΔI likewise yielded no satellite growth, comparable to the nonmotile mutant II-3 (Fig. 6). P30ΔIA conferred only minimal satellite growth, much like that of P30ΔIIIA and indicative of very limited cell gliding, well below the wild-type level (Fig. 6). This was confirmed when gliding was assessed quantitatively, with P30ΔIA and P30ΔIIIA exhibiting mean corrected gliding velocities of only 2 to 3% of wild-type levels (Table 2), comparable to that of mutants II-7 and II-3R (16). P30ΔIA and P30ΔIIIA likewise exhibited reduced gliding frequencies, at 43 and 32% of wild-type levels, respectively. P30ΔIIIB conferred a satellite growth pattern indicative of very limited gliding, but we failed to quantify its velocity or frequency due to a paucity of single cells. All TM domain substitution derivatives except P30Sub7374 yielded wild-type HA and gliding (data not shown).
FIG. 5.
Qualitative hemadsorption analysis of wild-type M. pneumoniae (WT), P30 mutant II-3, and the indicated P30 domain deletion or amino acid substitution derivatives. Bar, 65 μm.
FIG. 6.
Satellite growth by wild-type M. pneumoniae (WT), P30 mutant II-3, and the indicated P30 domain deletion or amino acid substitution derivatives. Mycoplasmas were incubated in SP4 medium with 3% gelatin for 72 h. Bar, 15 μm.
TABLE 2.
Cell gliding velocities and frequencies for wild-type M. pneumoniae, P30ΔIA, and P30ΔIIIA
| Strain | Mean gliding velocity, μm s−1, ± SEM (% WT) | Mean corrected gliding velocitya, μm s−1, ± SEM (% WT) | % gliding frequency (% WT) |
|---|---|---|---|
| Wild type | 0.1478 ± 0.0097 (100) | 0.1775 ± 0.0091 (100) | 12.13 (100) |
| P30ΔIA | 0.0019 ± 0.0002 (1.28) | 0.0042 ± 0.0003 (2.36) | 5.21 (43) |
| P30ΔIIIA | 0.0032 ± 0.0008 (2.14) | 0.0056 ± 0.0008 (3.13) | 3.88 (32) |
The mean corrected gliding velocity is the total distance traveled by a cell divided by the total time of the field interval less time spent in resting periods(16).
DISCUSSION
M. pneumoniae protein P30 is fundamentally important in cytadherence, gliding motility, and cell division. Substitution within domain II or deletion within domain III of P30 confers reduced motility and cytadherence (16), consistent with a requirement for these extracellular regions of P30 in adherence and gliding function, a conclusion reinforced by the inhibition of cytadherence by antibodies targeting domain III (1, 36). More-specific roles for these and other P30 domains in adherence and gliding are not otherwise further defined, however.
P30 domain III is defined by 13 P-rich repeats followed by an additional 20 residues at the very C terminus. The importance of the P-rich repeats has been established (10, 34), but the significance of the region immediately following had not been explored previously. Our approach to engineering mutant P30ΔIIIA to target this region yielded two additional domain III deletion derivatives, which were included in this study. P30ΔIIIA was the only derivative of the three that was detectable at significant levels in Western immunoblots, but it conferred reduced cytadherence and motility phenotypes similar to those of mutant II-7, indicating that fully functional P30 requires both the P-rich repeats and the C-terminus region. It is not known if this involves correct folding, proper positioning of the C-terminal region by the P-rich repeats, or perhaps an alternative mechanism.
Domain II separates domain III from the membrane-spanning region, and residues 99 to 125 in domain II in particular are highly conserved in P30 homologs (Fig. 2) (19, 33). Deletion of residues 101 to 120 (P30ΔII) destabilized P30, much like C-terminal truncation. Interestingly, while wild-type M. pneumoniae exhibits a spontaneous loss of HMW2 (12, 22), this appeared to be more common specifically in mutant II-3 transformants with P30ΔII (data not shown). HMW2 and P30 are believed to incorporate early and late, respectively, in terminal organelle assembly (23), and in the absence of HMW2, no electron-dense core forms and P30 is unstable (4, 37, 44). However, the significance of domain II in this regard is not clear.
P30 domain I is believed to include a putative signal sequence and an intracytoplasmic region. Here we engineered and characterized P30 derivative P30ΔI, lacking residues 36 to 68 between the predicted signal processing site and the TM domain. P30ΔI was stable in mutant II-3, colocalized with other known terminal organelle proteins, and restored P65 to wild-type levels but failed to rescue HA or gliding motility. This finding is consistent with our previous observation that wild-type levels of P65 are not sufficient to confer gliding or cytadherence (16) and suggests that the putative cytoplasmic domain of P30 is essential for function. Significantly, the apparent size of P30ΔI raised the possibility that the domain I deletion eliminated the signal peptide processing site, which could cause improper folding and thus account for loss of function. P30ΔIA, lacking only residues 58 to 68 immediately preceding the predicted TM domain, migrated as expected for proper signal processing. Much like P30ΔI, P30ΔIA failed to rescue cytadherence and conferred only minimal gliding capacity, leading us to conclude that this predicted cytoplasmic domain is essential for P30 function, perhaps bridging the terminal organelle core with the surface-binding adhesive components of the P30 C-terminal domain. Electron cryotomography images of the distal end of the terminal organelle reveal tightly packed rows of proteins on both sides of the membrane, with those on the inner surface linked to the cytoskeletal core (18) and perhaps including the cytoplasmic domain of P30. Alternatively, or perhaps in addition, the P30 cytoplasmic domain might confer a conformational change to its extracellular domain, which is essential for function. In this regard, preliminary data indicate a difference in accessibility of P30ΔI, compared to that for wild-type P30 and P30ΔIIIA, to Streptomyces griseus protease type XIV (data not shown), consistent with the possibility that P30ΔI loss of function may involve altered conformation. Finally, these findings indicate that P30 processing likely occurs downstream of the site originally predicted from its hydropathy plot (9, 10), and based on the electrophoretic mobility of P30ΔI and P30ΔIA, we predict that this occurs between residues 38 and 58.
The P30 TM region separates domains I and II, and substitution here for W73 and F74 resulted in instability. Aromatic residues at the edge of α-helical integral membrane domains are thought to be important in helix positioning (5, 43). For example, W and F residues flanking the TM helix tend to push the helix into the lipid bilayer of microsomal membranes, with W favoring the lipid-water interface (5). Substitution of A for W flanking the second TM helix of the Tar chemoreceptor of Escherichia coli results in abnormal signaling (11), while replacing F in the helical TM domain of the major coat protein of bacteriophage M13 causes the entire TM domain to shift (27). Neither study reported altered protein stability, but we speculate here that substitution for W73 and F74 impacted protein positioning in the lipid bilayer, leading to accelerated turnover, perhaps a consequence of improper helix alignment for multimerization (16). In this regard, the P30 TM domain includes a G-xxx-G motif (G92 to G96) that reportedly contributes to TM helix-to-helix interactions (8, 35) and may likewise contribute to proper membrane topography for P30.
In conclusion, our findings (i) confirmed and extended the importance of the extracellular domain of P30 in mycoplasma adherence, gliding, and P65 stabilization, (ii) established that the TM domain is uniquely suited for P30 function and stability, and (iii) demonstrated that the putative cytoplasmic domain of P30 is required for normal gliding and adherence function but not for localization or stability. P30 is a novel protein with no structural homologs outside the mycoplasmas. A structure solution would likely provide valuable clues to the roles of P30 in adherence and gliding function, colonization, and virulence.
Supplementary Material
Acknowledgments
We thank Mitch Balish for his helpful comments and suggestions.
This work was supported by Public Health Service grants AI23362 and AI49194 from the National Institute of Allergy and Infectious Diseases to D.C.K.
Footnotes
Published ahead of print on 21 January 2011.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Baseman, J. B., et al. 1987. Identification of a 32-kilodalton protein of Mycoplasma pneumoniae associated with hemadsorption. Isr. J. Med. Sci. 23:474-479. [PubMed] [Google Scholar]
- 2.Biberfeld, G., and P. Biberfeld. 1970. Ultrastructural features of Mycoplasma pneumoniae. J. Bacteriol. 102:855-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biscardi, S., et al. 2004. Mycoplasma pneumoniae and asthma in children. Clin. Infect. Dis. 38:1341-1346. [DOI] [PubMed] [Google Scholar]
- 4.Bose, S. R., M. F. Balish, and D. C. Krause. 2009. Mycoplasma pneumoniae cytoskeletal protein HMW2 and the architecture of the terminal organelle. J. Bacteriol. 191:6741-6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, P., and G. von Heijne. 1999. The aromatic residues Trp and Phe have different effects on the positioning of a transmembrane helix in the microsomal membrane. Biochemistry 38:9778-9782. [DOI] [PubMed] [Google Scholar]
- 6.Cloward, J. M., and D. C. Krause. 2009. Mycoplasma pneumoniae J-domain protein required for terminal organelle function. Mol. Microbiol. 71:1296-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collier, A. M., W. A. Clyde, Jr., and F. W. Denny. 1971. Mycoplasma pneumoniae in hamster tracheal organ culture: immunofluorescent and electron microscopic studies. Proc. Soc. Exp. Biol. Med. 136:569-573. [DOI] [PubMed] [Google Scholar]
- 8.Curran, A. R., and D. M. Engelman. 2003. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr. Opin. Struct. Biol. 13:412-417. [DOI] [PubMed] [Google Scholar]
- 9.Dallo, S. F., A. Chavoya, and J. B. Baseman. 1990. Characterization of the gene for a 30-kilodalton adhesion-related protein of Mycoplasma pneumoniae. Infect. Immun. 58:4163-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dallo, S. F., A. L. Lazzell, A. Chavoya, S. P. Reddy, and J. B. Baseman. 1996. Biofunctional domains of the Mycoplasma pneumoniae P30 adhesin. Infect. Immun. 64:2595-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Draheim, R. R., A. F. Bormans, R. Z. Lai, and M. D. Manson. 2005. Tryptophan residues flanking the second transmembrane helix (TM2) set the signaling state of the Tar chemoreceptor. Biochemistry 44:1268-1277. [DOI] [PubMed] [Google Scholar]
- 12.Fisseha, M., H. W. Gohlmann, R. Herrmann, and D. C. Krause. 1999. Identification and complementation of frameshift mutations associated with loss of cytadherence in Mycoplasma pneumoniae. J. Bacteriol. 181:4404-4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn, T. W., M. J. Willby, and D. C. Krause. 1998. HMW1 is required for cytadhesin P1 trafficking to the attachment organelle in Mycoplasma pneumoniae. J. Bacteriol. 180:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasselbring, B. M., and D. C. Krause. 2007. Proteins P24 and P41 function in the regulation of terminal-organelle development and gliding motility in Mycoplasma pneumoniae. J. Bacteriol. 189:7442-7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasselbring, B. M., J. L. Jordan, R. W. Krause, and D. C. Krause. 2006. Terminal organelle development in the cell wall-less bacterium Mycoplasma pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 103:16478-16483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasselbring, B. M., J. L. Jordan, and D. C. Krause. 2005. Mutant analysis reveals a specific requirement for protein P30 in Mycoplasma pneumoniae gliding motility. J. Bacteriol. 187:6281-6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hegermann, J., R. Herrmann, and F. Mayer. 2002. Cytoskeletal elements in the bacterium Mycoplasma pneumoniae. Naturwissenschaften 89:453-458. [DOI] [PubMed] [Google Scholar]
- 18.Henderson, G. P., and G. J. Jensen. 2006. Three-dimensional structure of Mycoplasma pneumoniae's attachment organelle and a model for its role in gliding motility. Mol. Microbiol. 60:376-385. [DOI] [PubMed] [Google Scholar]
- 19.Hnatow, L. L., C. L. Keeler, Jr., L. L. Tessmer, K. Czymmek, and J. E. Dohms. 1998. Characterization of MGC2, a Mycoplasma gallisepticum cytadhesin with homology to the Mycoplasma pneumoniae 30-kilodalton protein P30 and Mycoplasma genitalium P32. Infect. Immun. 66:3436-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordan, J. L., K. M. Berry, M. F. Balish, and D. C. Krause. 2001. Stability and subcellular localization of cytadherence-associated protein P65 in Mycoplasma pneumoniae. J. Bacteriol. 183:7387-7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krause, D. C., D. K. Leith, and J. B. Baseman. 1983. Reacquisition of specific proteins confers virulence in Mycoplasma pneumoniae. Infect. Immun. 39:830-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krause, D. C., D. K. Leith, R. M. Wilson, and J. B. Baseman. 1982. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect. Immun. 35:809-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Layh-Schmitt, G., R. Himmelreich, and U. Leibfried. 1997. The adhesin related 30-kDa protein of Mycoplasma pneumoniae exhibits size and antigen variability. FEMS Microbiol. Lett. 152:101-108. [DOI] [PubMed] [Google Scholar]
- 26.Lipman, R. P., and W. A. Clyde, Jr. 1969. The interrelationship of virulence, cytadsorption, and peroxide formation in Mycoplasma pneumoniae. Proc. Soc. Exp. Biol. Med. 131:1163-1167. [DOI] [PubMed] [Google Scholar]
- 27.Meijer, A. B., R. B. Spruijt, C. J. Wolfs, and M. A. Hemminga. 2001. Membrane-anchoring interactions of M13 major coat protein. Biochemistry 40:8815-8820. [DOI] [PubMed] [Google Scholar]
- 28.Naot, Y., J. G. Tully, and H. Ginsburg. 1977. Lymphocyte activation by various Mycoplasma strains and species. Infect. Immun. 18:310-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, S. J., Y. C. Lee, Y. K. Rhee, and H. B. Lee. 2005. Seroprevalence of Mycoplasma pneumoniae and Chlamydia pneumoniae in stable asthma and chronic obstructive pulmonary disease. J. Korean Med. Sci. 20:225-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powell, D. A., P. C. Hu, M. Wilson, A. M. Collier, and J. B. Baseman. 1976. Attachment of Mycoplasma pneumoniae to respiratory epithelium. Infect. Immun. 13:959-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proft, T., H. Hilbert, G. Layh-Schmitt, and R. Herrmann. 1995. The proline-rich P65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insoluble fraction and exhibits size polymorphism in the strains M129 and FH. J. Bacteriol. 177:3370-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Razin, S., and E. Jacobs. 1992. Mycoplasma adhesion. J. Gen. Microbiol. 138:407-422. [DOI] [PubMed] [Google Scholar]
- 33.Reddy, S. P., W. G. Rasmussen, and J. B. Baseman. 1995. Molecular cloning and characterization of an adherence-related operon of Mycoplasma genitalium. J. Bacteriol. 177:5943-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romero-Arroyo, C. E., et al. 1999. Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J. Bacteriol. 181:1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russ, W. P., and D. M. Engelman. 2000. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 296:911-919. [DOI] [PubMed] [Google Scholar]
- 36.Schurwanz, N., E. Jacobs, and R. Dumke. 2009. Strategy to create chimeric proteins derived from functional adhesin regions of Mycoplasma pneumoniae for vaccine development. Infect. Immun. 77:5007-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seto, S., and M. Miyata. 2003. Attachment organelle formation represented by localization of cytadherence proteins and formation of the electron-dense core in wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol. 185:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobeslavsky, O., B. Prescott, and R. M. Chanock. 1968. Adsorption of Mycoplasma pneumoniae to neuraminic acid receptors of various cells and possible role in virulence. J. Bacteriol. 96:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevens, M. K., and D. C. Krause. 1991. Localization of the Mycoplasma pneumoniae cytadherence-accessory proteins HMW1 and HMW4 in the cytoskeletonlike Triton shell. J. Bacteriol. 173:1041-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waites, K. B., and D. F. Talkington. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17:697-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waites, K. B., M. F. Balish, and T. P. Atkinson. 2008. New insights into the pathogenesis and detection of Mycoplasma pneumoniae infections. Future Microbiol. 3:635-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallin, E., T. Tsukihara, S. Yoshikawa, G. von Heijne, and A. Elofsson. 1997. Architecture of helix bundle membrane proteins: an analysis of cytochrome c oxidase from bovine mitochondria. Protein Sci. 6:808-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willby, M. J., et al. 2004. HMW1 is required for stability and localization of HMW2 to the attachment organelle of Mycoplasma pneumoniae. J. Bacteriol. 186:8221-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.